Abstract
Background
A large cluster of 59 cases were linked to a single flight with 146 passengers from New Delhi to Hong Kong in April 2021. This outbreak coincided with early reports of exponential pandemic growth in New Delhi, which reached a peak of > 400 000 newly confirmed cases on 7 May 2021.
Methods
Epidemiological information including date of symptom onset, date of positive-sample detection and travel and contact history for individual cases from this flight were collected. Whole genome sequencing was performed, and sequences were classified based on the dynamic Pango nomenclature system. Maximum-likelihood phylogenetic analysis compared sequences from this flight alongside other cases imported from India to Hong Kong on 26 flights between June 2020 and April 2021, as well as sequences from India or associated with India-related travel from February to April 2021 and 1217 reference sequences.
Results
Sequence analysis identified six lineages of SARS-CoV-2 belonging to two variants of concern (Alpha and Delta) and one variant of public health interest (Kappa) involved in this outbreak. Phylogenetic analysis confirmed at least three independent sub-lineages of Alpha with limited onward transmission, a superspreading event comprising 37 cases of Kappa and transmission of Delta to only one passenger. Additional analysis of another 26 flights from India to Hong Kong confirmed widespread circulation of all three variants in India since early March 2021.
Conclusions
The broad spectrum of disease severity and long incubation period of SARS-CoV-2 pose a challenge for surveillance and control. As illustrated by this particular outbreak, opportunistic infections of SARS-CoV-2 can occur irrespective of variant lineage, and requiring a nucleic acid test within 72 hours of departure may be insufficient to prevent importation or in-flight transmission.
Keywords: Genomic epidemiology, air travel-related outbreak, whole genome sequencing, SARS-CoV-2
Introduction
Since March 2020, increasingly stringent travel measures were implemented in Hong Kong to reduce the risk of SARS-CoV-2 importation,1,2 including pre-departure, on-arrival and post-arrival testing and quarantine of all passenger arrivals outside the home in designated quarantine facilities.3 On arrival, passengers undergo nucleic acid tests in the airport before transfer by shuttle bus to designated hotels for quarantine, with strict isolation of cases in a designated public hospital. Under penalties of fine and prosecution, hotel residents are not allowed to exit their rooms throughout quarantine, and no hotel staff may enter. Detection of cases on arrival or during quarantine initiates contact tracing and additional testing of contacts. In recognition of the emergence of SARS-CoV-2 variants of concern (VOC) and variants of interest (VOI) with public health significance,4 on-arrival quarantine was extended from 14 to 21 days on 25 December 2020.5 Travellers departing from high-risk countries are required to complete a health declaration and produce a negative nucleic acid test from an accredited laboratory within 72 hours of departure, and passenger flights from countries with widespread VOC circulation are classified as extremely high risk and temporarily banned.5
Multiple cases of SARS-CoV-2 were detected upon arrival of a direct flight from New Delhi to Hong Kong (DEL–HKG). The arrival of this flight in early April 2021 coincided with early reports of exponential growth of a catastrophic second wave of SARS-CoV-2 in India and immediately prior to widespread reports of healthcare infrastructure failure.5 Because India was at that time classified by the Hong Kong government as high risk, passengers were required to show documentation of a 21-day quarantine hotel reservation and proof of a negative nucleic acid test taken within 72 hours of departure. After six passengers were denied boarding, the Airbus A321neo was at 85% capacity with 146 passengers for a 6-hour flight. According to airline policy,6 World Health Organization (WHO) and local government sanitation guidelines were observed. The airline used thermal screening and social distancing during check-in and boarding, and airline staff members were equipped with gowns, gloves, masks and face shields. Upon arrival, passengers were again tested by RT-PCR, at which point five positive cases were detected. Cases among passengers continued to be detected throughout the duration of the 21-day quarantine period. For passengers sharing accommodations with positive cases, quarantine periods were extended to 30 days of post-arrival. Representing the largest known air travel-related outbreak of SARS-CoV-2,7 a total of 59 confirmed cases were detected from the 146 passengers onboard (an attack rate of 40%), though only 20% (12/59) of cases were reportedly symptomatic. Through whole genome sequencing and phylogenetic analysis, we aimed to infer the dynamics of SARS-CoV-2 transmission associated with this air travel-related outbreak.
Methods
Epidemiological information including date of symptom onset, date of positive-sample detection and travel and contact history for individual cases are summarized in Figure 2. Of the 59 RT-PCR confirmed SARS-CoV-2 cases, samples from 48 individuals contained adequate viral load for sequencing using previously described methods.8,9 Of those, 46 sequences were of sufficient coverage to be classified based on the dynamic Pango nomenclature system (https://pangolin.cog-uk.io/, accessed 10 May 2021)10; however, those with < 70% genome coverage were excluded from subsequent phylogenetic analysis, resulting in 43 complete or nearly complete SARS-CoV-2 genomes. We performed phylogenetic analysis to compare sequences from this outbreak alongside cases imported on 26 additional flights from India to Hong Kong between July 2020 and May 2021 (n = 51) (Table 1) as well as GISAID sequences from India or associated with India-related travel from February to April 2021 (n = 2660) and global reference sequences (n = 1217) including Wuhan-Hu-1 (MN908947.3) (accessed 20 May 2021) (Supplementary Table S1). After removing outliers based on a root-to-tip regression analysis in TempEst v.1.5.3,11 a maximum likelihood (ML) tree was generated for the final dataset (n = 3971) in IQ-TREE v.212,13 and dated using the least square dating (LSD2) method.14
Figure 2.
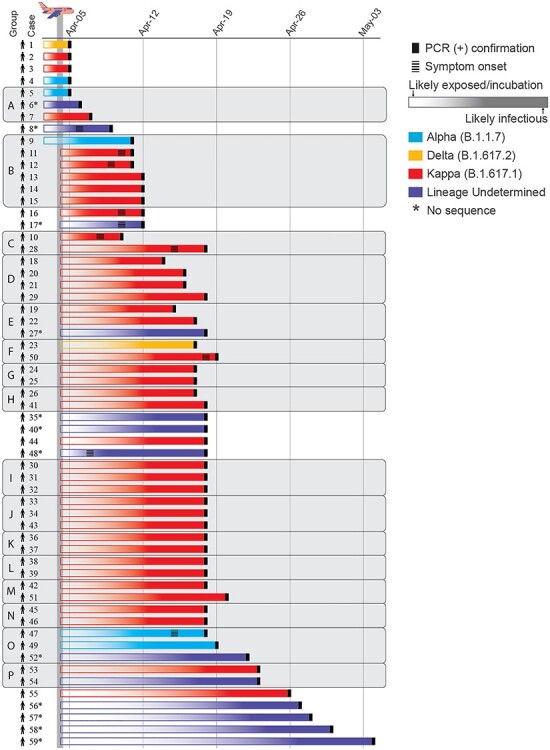
Timeline from exposure to detection for each of 59 SARS-CoV-2 cases associated with a flight from New Delhi to Hong Kong in April 2021. Colours represent WHO VOC/VOI and Pango lineage designations.10 Individuals travelling together are grouped and shaded in grey. Asterisk indicates cases from which samples were unavailable or unable to be sequenced.
Table 1.
Sequenced SARS-CoV-2 genomes from cases imported to Hong Kong on 26 flights from July 2020 to May 2021
| Month of arrival | Location of origin | Pango lineage | Genome coverage (%) | Symptomatic/Asymptomatic |
|---|---|---|---|---|
| June 2020 | Mumbai | B.1 | 96.04 | Asymptomatic |
| B.1 | 99.3 | Symptomatic | ||
| B.1 | 99.3 | Symptomatic | ||
| July 2020 | Mumbai via Kuala Lumpur | B.1.210 | 99.3 | Symptomatic |
| B.1.210 | 99.3 | Symptomatic | ||
| July 2020 | New Delhi | B.1.36 | 98.49 | Asymptomatic |
| July 2020 | India (undetermined location) via Kuala Lumpur | B.1.1 | 99.3 | Symptomatic |
| July 2020 | Doha via India (undetermined location) | B.1.36.8 | 95.53 | Asymptomatic |
| August 2020 | New Delhi via Kolkata | B.1.36.36 | 99.3 | Asymptomatic |
| September 2020 | India (undetermined location) | B.1.36.18 | 99.3 | Asymptomatic |
| September 2020 | Delhi | B.1.369 | 99.3 | Asymptomatic |
| B.1.36 | 99.3 | Asymptomatic | ||
| September 2020 | Delhi | B.1.1 | 99.3 | Asymptomatic |
| October 2020 | Delhi | B.1.36.29 | 99.3 | Asymptomatic |
| B.1.36 | 99.29 | Asymptomatic | ||
| October 2020 | Chennai | B.1.1 | 97.18 | Asymptomatic |
| B.1.1 | 99.3 | Asymptomatic | ||
| October 2020 | Chandigarh via Mumbai | B.1.36 | 99.3 | Asymptomatic |
| B.1.36 | 99.3 | Asymptomatic | ||
| November 2020 | Mumbai | B.1.210 | 90.45 | Symptomatic |
| November 2020 | New Delhi | B.1.36 | 91.73 | Asymptomatic |
| December 2020 | Delhi | B.1.36 | 98.99 | Asymptomatic |
| B.1.36 | 97.86 | Symptomatic | ||
| B.1.36 | 97.85 | Asymptomatic | ||
| B.1.36 | 93.06 | Asymptomatic | ||
| December 2020 | Sri Lanka via Mumbai | B.1.1 | 83.21 | Asymptomatic |
| January 2021 | Mumbai | B.1 | 73.89 | Symptomatic |
| B.1.1.306 | 99.3 | Symptomatic | ||
| February 2021 | Dubai via Bangkok and India (undetermined location) | B.1.562 | 99.02 | Asymptomatic |
| March 2021 | Kolkata | B.1.1.7 | 99.25 | Asymptomatic |
| March 2021 | New Delhi | B.1.36.29 | 99.29 | Symptomatic |
| March 2021 | Kolkata | B.1.1.7 | 99.25 | Symptomatic |
| B.1.1.7 | 79.79 | Asymptomatic | ||
| B.1.1.7 | 91.63 | Asymptomatic | ||
| B.1.1.7 | 99.25 | Asymptomatic | ||
| March 2021 | Mumbai | B.1.617.2 | 99.28 | Symptomatic |
| B.1.617.1 | 99.3 | Symptomatic | ||
| April 2021 | New Delhi via Dubai | B.1.1.7 | 92.85 | Symptomatic |
| April 2021 | New Delhi via Kolkata | B.1.617.2 | 99.26 | Asymptomatic |
| B.1.617.2 | 99.26 | Symptomatic | ||
| B.1.617.2 | 99.26 | Asymptomatic | ||
| B.1.617.2 | 97.92 | Asymptomatic | ||
| B.1.1.7 | 99.2 | Symptomatic | ||
| B.1.1.7 | 99.26 | Symptomatic | ||
| B.1.617.2 | 99.21 | Symptomatic | ||
| B.1.617.2 | 99.26 | Asymptomatic | ||
| April 2021 | Doha via India (undetermined location) | B.1.617.2 | 99.26 | Asymptomatic |
| April 2021 | Mumbai | B.1.617.2 | 88.18 | Asymptomatic |
| B.1.617.2 | 99.26 | Asymptomatic | ||
| B.1.617.1 | 92.9 | Asymptomatic | ||
| B.1.617.2 | 92.85 | Asymptomatic |
Results
Whole genome sequencing verified the presence of three distinct variants onboard the DEL–HKG flight, Alpha, Kappa and Delta,4 covering three Pango lineages10 B.1.1.7, B.1.617.1 and B.1.617.2, respectively. Kappa (B.1.617.1) was detected in the majority of passenger sequences (n = 39/46, 84.8%), followed by Alpha (B.1.1.7) in five passengers including a crew member who tested positive on arrival (n = 5/46, 10.9%) and Delta (B.1.617.2) in two passengers (n = 2/46, 4.3%). In an ML phylogeny, 37 of the Kappa sequences clustered together with zero or near-zero branch lengths suggesting a single transmission cluster in transit (inset in Figure 1A). However, one Kappa sequence was genetically distinct enough to infer pre-flight infection without further onboard transmission, despite the individual (Case 3) being symptomatic on arrival (Figures 1A and2). High genome coverage (>98%) of three of the five Alpha variant sequences are consistent with three separate introductions by (i) an infected crew member, (ii) an infected infant and (iii) an infected but asymptomatic adult in economy seating. Any of these three could represent the index case for onward transmission of the Alpha variant to two individuals from which only partial sequence could be obtained (identified as travel group O in Figure 2), testing positive on Days 14 and 15 post-arrival. The two Delta variant cases have nearly identical genomes (with >95% coverage), and the two individuals sat in close proximity but tested positive 12-day apart, suggestive of at least one onboard transmission of the Delta variant (Figures 1B and2). However, there were two additional nearby cases that could not be sequenced due to insufficient viral load, both travelling alone and testing positive on Days 14 and 30 post-arrival, and those cases may represent additional links in the inferred transmission chains.
Figure 1.
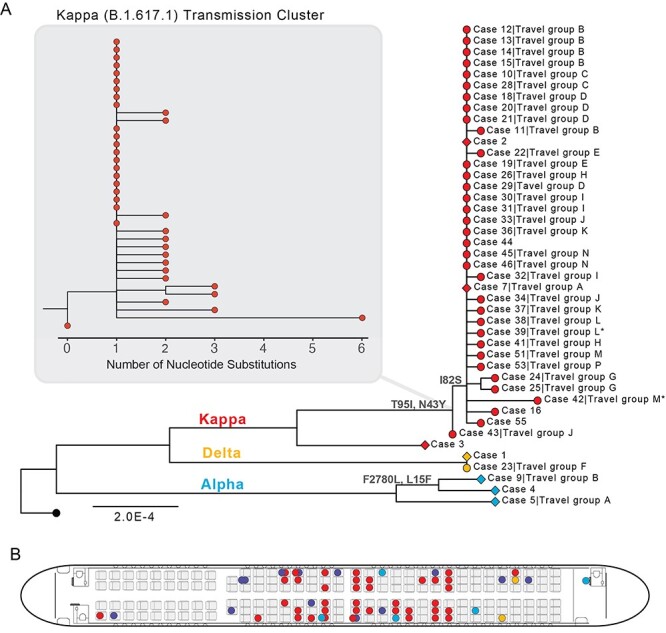
SARS-CoV-2 transmission associated with a flight from New Delhi to Hong Kong in April 2021. Colours represent WHO VOC/VOI and Pango lineage designations.10 (A) ML phylogeny of 43 genomes sequenced from 59 cases. Wuhan-Hu-1 (MN908947.3, black tip) was used to root the tree. Diamond tip shapes indicate index cases and circles represent secondary-transmission cases; tips are labelled with case numbers and travel groups. Asterisk denotes sequences with lower coverage (~73%). Nodes are labelled with observed amino acid mutations. The Kappa (B.1.617.1) transmission cluster is magnified to show nucleotide substitutions among samples. (B) Seat assignments of passengers testing positive. Seats with two positive cases indicate adults seated with children under the age of two.
Using the temporal window from arrival-to-detection as a proxy for SARS-CoV-2 incubation, we estimate that at least seven individual cases were likely infected prior to travel (Figures 1A and2). Forty-one cases were detected between Days 5 and 14 post-arrival, suggesting they were likely infected in transit. Of the cases infected in transit that were sequenced, 94.1% (n = 32/34) were related to a single Kappa variant transmission cluster, demonstrating potential for superspreading of SARS-CoV-2 on congested flights and reinforcing the importance of stringent travel-related testing. Another 11 cases were detected after the 14th day post-arrival, of which only five could be sequenced. At least two of these Kappa variant cases were likely infected by co-quarantined family members (Cases 51 and 53, detected on Days 16 and 19, respectively, Figures 1A and2). Case 59, detected 30 days of post-arrival, may be an example of an unusually long incubation period as this person travelled alone and was the only known case from this flight in their quarantine hotel (Figure 2). However, this was one of 13 cases not sequenced, and quarantine hotel information was missing for 15 cases in total (n = 15/59, 25.4%). Given that instances of SARS-CoV-2 transmission among epidemiologically unlinked cases during quarantine have been documented,15 we cannot rule out this possibility.
The age distribution of positive cases ranged from 4 months to 63 years (median age = 33 years), and notably, eight of the positive cases were detected in children under the age of two, who are generally exempt from masking requirements. Interestingly, members of at least two families were infected with independent transmission lineages, and sequencing revealed two instances of children infected with different variants than their family members (Travel groups A and B in Figures 1A and2). In a family of three individuals (Travel group A), one parent tested positive for Alpha, and the infant tested positive for Kappa (the other parent tested positive, but no sequence was recovered from this patient). As before, using arrival-to-detection as a proxy for incubation, it is likely that Travel group A was infected several days prior to departure. In a second family group (n = 6, Travel group B), one child was infected with Alpha, whereas the other five family members, two children and three adults, were infected with Kappa. The child (Case 9) who tested positive for the divergent Alpha variant at 5 days of post-arrival was likely infected prior to departure (Figures 1A and2), whereas the other five family members tested positive on Days 7–8 post-arrival, suggesting they were most likely infected in-transit.
As a major travel hub, Hong Kong’s practice of testing inbound passengers has proved a valuable source for sentinel SARS-CoV-2 surveillance and containment. Between June 2020 and April 2021, 51 cases of SARS-CoV-2 were detected on 26 other passenger flights from India to Hong Kong (Table 1).16 Among those flights, the second largest air travel-related SARS-CoV-2 cluster (n = 8 cases) was detected just 5 days after the DEL–HKG outbreak on another flight arriving from New Delhi (Table 1 and Supplementary Figure S1). All imported cases detected since mid-March 2021 belonged to Alpha (n = 8), Delta (n = 13) or Kappa (n = 2) lineages and originated from Kolkata, New Delhi and Mumbai, reflecting the predominant circulation of these variants across major Indian cities during the 2021 resurgence. Similar to the DEL–HKG outbreak, a majority (n = 33/51, 64.7%) of imported cases sequenced from the other 26 flights were asymptomatic (Table 1).
Discussion
The broad spectrum of disease severity and potentially long incubation period of SARS-CoV-2 pose a challenge for surveillance and control. Based on the mean incubation period of 5.7 days for SARS-CoV-217 and that infectiousness peaks prior to symptom onset,18 requiring a nucleic acid test within 72 hours of departure is clearly insufficient to eliminate the risk of SARS-CoV-2 importation or in-flight transmission. Other studies have described and modelled in-flight transmission of SARS-CoV-2,19 despite the use of precautionary measures such as pre-departure testing,20 symptom screening21 and health declarations.22 Thus, these results highlight the value of quarantine and testing on arrival to prevent virus introduction into the community. A recent review of in-flight transmission of SARS-CoV-2 shows secondary attack rates are highly variable,7 though admittedly so are adherence to and enforcement of mask mandates and distancing policies.23
Furthermore, as illustrated by this particular outbreak, opportunistic infections of SARS-CoV-2 can occur irrespective of variant lineage. Detection of multiple lineages within the same familial groups suggests that individuals in the same household likely exhibit similar behaviour and attitudes towards mask wearing, hand hygiene and other COVID-19 safety precautions. The high rate of transmission on this flight, particularly among children, suggests masking may not have been heavily enforced. It is also unclear whether individuals were required to stay seated during flight or allowed to unmask while eating and drinking. Further, it is difficult to control the hygienic behaviour of small children. Particularly on long flights, parents with small children may allow their children to move about the cabin or walk the aisles. Individuals travelling with children and infants may therefore pose a greater risk of transmission. This highlights the need to improve situational awareness by educating travellers on precautionary measures to minimize in-flight transmission.
As an epidemiological investigation, this study has several limitations. Our transmission chain inferences are ultimately somewhat confounded by (i) cases that could not be sequenced, (ii) the highly variable incubation period of SARS-CoV-2 and (iii) the low-level genetic diversity between the samples. Furthermore, precise determination of the timing of exposure is confounded by a lack of detailed information on passenger movements during airport check-in, pre-flight boarding, the flight itself and during ground transportation to designated quarantine hotels. We cannot exclude the possibility that exposures may have happened before or after the flight. Conversely, it is also plausible that individuals testing positive in the 48 hours of post-arrival (Cases 6 and 7) could have been infected in flight. However, based on the clustering of cases in economy seating and insufficient spatial distancing between passengers onboard, it is highly likely that the majority of these transmission events occurred onboard.
The majority of air-travel associated SARS-CoV-2 outbreak investigations conducted early in the pandemic involved relatively few primary cases,24 often with traceable origins and discernible transmission patterns.25–27 Notably, there have also been reports of high-risk flights with no evidence of transmission,28 which suggest that the risk is highly variable and circumstantial. Just over a year into the COVID-19 pandemic, we describe transmission of three VOI/VOC lineages from at least seven index cases on a single flight. Other studies have applied similar methods (i.e. the comparison of viral sequences and epidemiological data) to infer probable transmission chains on long-haul flights,15,20 illustrating the potential risk for SARS-CoV-2 transmission during air travel. In agreement with our findings, studies using clinical and epidemiological data have similarly reported detection around 14 days of post-arrival in both symptomatic21 and asymptomatic secondary cases.29
Thus, as commercial flight volumes slowly rebound, travel-related surveillance remains essential. A spike in imported cases can serve as an indication of prevalence in places where testing is limited.16 The number of imported cases per flight increased considerably in April 2021 (Table 1), which reflects the high levels of community transmission in India at the time.
Following India’s strict initial lockdown,30,31 reported cases had steadily declined from September 2020 to February 2021, and control measures were gradually being relaxed; interstate travel had resumed and mass gatherings were allowed for elections, religious festivals and cricket matches.32 Under the belief they were flattening the curve, attitudes towards precautionary measures became lax at the individual and enforcement level. Then came a delayed and devastating second wave. Amid reports of under-testing and under-counting of cases,33 relatively few sequences were made available.34
Further attention to travel-related cases is warranted to elucidate risk factors and minimize air travel-related infection risk. Rapid tests could also be employed at departure airports as part of the check-in process to prevent individuals who are currently infectious from boarding an aircraft. This study highlights the value of physical distance and strict adherence to masking as well as the insufficiency of symptom-based surveillance to limit transmission of SARS-CoV-2.
Supplementary Material
Acknowledgements
We gratefully acknowledge the staff from the originating laboratories responsible for obtaining the specimens and from the submitting laboratories where the genome data were generated and shared via GISAID (Supplementary Table S1). We acknowledge the technical support provided by colleagues from the Centre for PanorOmic Sciences of the University of Hong Kong. We also acknowledge the Centre for Health Protection of the Department of Health for providing epidemiological data for the study.
Contributor Information
Vijaykrishna Dhanasekaran, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; HKU-Pasteur Research Pole, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Kimberly M Edwards, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; HKU-Pasteur Research Pole, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Ruopeng Xie, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; HKU-Pasteur Research Pole, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Haogao Gu, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Dillon C Adam, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Lydia D J Chang, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Sammi S Y Cheuk, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Shreya Gurung, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Pavithra Krishnan, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Daisy Y M Ng, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Gigi Y Z Liu, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Carrie K C Wan, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Samuel S M Cheng, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Dominic N C Tsang, Centre for Health Protection, Department of Health, The Government of Hong Kong Special Administrative Region, Hong Kong, China.
Benjamin J Cowling, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; Laboratory of Data Discovery for Health, Hong Kong Science and Technology Park, Hong Kong, China.
Malik Peiris, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; HKU-Pasteur Research Pole, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; Centre for Immunology and Infection, Hong Kong Science and Technology Park, Hong Kong, China.
Leo L M Poon, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; HKU-Pasteur Research Pole, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China; Centre for Immunology and Infection, Hong Kong Science and Technology Park, Hong Kong, China.
Author Contributions
V.D. and L.L.M.P. designed the study and were responsible for project supervision. K.M.E, R.X., H.G., D.C.A. and D.N.C.T. collected and analysed the data. L.D.J.C., S.S.Y.C., S.G., P.K., D.Y.M.N., G.Y.Z.L., C.K.C.W., and S.S.M.C performed genome sequencing. V.D., K.M.E and D.C.A. wrote the original draft, and all authors reviewed and edited the manuscript.
Funding
Health and Medical Research Fund, Food and Health Bureau of the Hong Kong SAR Government [grant number COVID190205]; National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [contract numbers U01AI151810, 75N93021C00016].
Conflict of interest: None declared.
References
- 1. Cowling BJ, Ali ST, Ng TWY et al. Impact assessment of non-pharmaceutical interventions against coronavirus disease 2019 and influenza in Hong Kong: an observational study. Lancet Public Health 2020; 5:e279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adam DC, Wu P, Wong JY et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat Med 2020; 26:1714–9. [DOI] [PubMed] [Google Scholar]
- 3. Leung GM, Cowling BJ, Wu JT. From a sprint to a marathon in Hong Kong. N Engl J Med 2020; 382:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krause PR, Fleming TR, Longini IM et al. SARS-CoV-2 variants and vaccines. N Engl J Med 2021; 385:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Government of the Hong Kong SAR Press Releases . Government tightens restrictions and compulsory quarantine requirements for persons arriving at Hong Kong who have stayed in places outside China. info.gov.hk. Hong Kong S.A.R, China: GovHK, 2020. [Google Scholar]
- 6. Vistara Airlines . Coronavirus update. ed. https://www.airvistara.com/th/en/coronavirus-update, 2021.
- 7. Freedman DO, Wilder-Smith A. In-flight transmission of SARS-CoV-2: a review of the attack rates and available data on the efficacy of face masks. J Travel Med 2020; 27:taaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sit THC, Brackman CJ, Ip SM et al. Infection of dogs with SARS-CoV-2. Nature 2020; 586:776–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gu H, Xie R, Adam DC et al. Sars-cov-2 under an elimination strategy in Hong Kong. medRxiv: the preprint server for health sciences, 2021, 2021.06.19.21259169.
- 10. Rambaut A, Holmes EC, O'Toole A et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rambaut A, Lam TT, Carvalho LM, Pybus OG. Exploring the temporal structure of heterochronous sequences using tempest (formerly path-o-gen). Virus Evol 2016; 2:vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minh BQ, Schmidt HA, Chernomor O et al. Iq-tree 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020; 37:1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. To TH, Jung M, Lycett S, Gascuel O. Fast dating using least-squares criteria and algorithms. Syst Biol 2016; 65:82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eichler N, Thornley C, Swadi T et al. Transmission of severe acute respiratory syndrome coronavirus 2 during border quarantine and air travel, New Zealand (aotearoa). Emerg Infect Dis 2021; 27:1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu H, Chu DKW, Chang LDJ et al. Genetic diversity of SARS-CoV-2 in travelers arriving in Hong Kong. Emerg Infect Dis 2021; 27:2666–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xin H, Wong JY, Murphy C et al. The incubation period distribution of coronavirus disease 2019 (covid-19): a systematic review and meta-analysis. Clin Infect Dis 2021;12:ciab501. [DOI] [PubMed] [Google Scholar]
- 18. He X, Lau EHY, Wu P et al. Temporal dynamics in viral shedding and transmissibility of covid-19. Nat Med 2020; 26:672–5. [DOI] [PubMed] [Google Scholar]
- 19. Quilty BJ, Clifford S, CMMID nCoV working group2 et al. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-ncov). Euro Surveill 2020; 25:pi=2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swadi T, Geoghegan JL, Devine T et al. Genomic evidence of in-flight transmission of SARS-CoV-2 despite predeparture testing. Emerg Infect Dis 2021; 27:687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang N, Shen Y, Shi C et al. In-flight transmission cluster of covid-19: a retrospective case series. Infect Dis 2020; 52:891–901. [DOI] [PubMed] [Google Scholar]
- 22. European Centre for Disease Prevention and Control . Considerations relating to passenger locator data, entry and exit screening and health declarations in the context of covid-19 in the eu/eea and the uk. Stockholm: European Centre for Disease Prevention and Control, 2020. [Google Scholar]
- 23. Government of India Office of the Director General of Civil Aviation . Strict compliance of covid-19 protocols during air travel. New Delhi: No 4/1/2020-IR, 2020. [Google Scholar]
- 24. Thompson HA, Imai N, Dighe A et al. Sars-cov-2 infection prevalence on repatriation flights from Wuhan City, China. J Travel Med 2020; 27:taaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khanh NC, Thai PQ, Quach HL et al. Transmission of SARS-CoV 2 during long-haul flight. Emerg Infect Dis 2020; 26:2617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Speake H, Phillips A, Chong T et al. Flight-associated transmission of severe acute respiratory syndrome coronavirus 2 corroborated by whole-genome sequencing. Emerg Infect Dis 2020; 26:2872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karaca O, Hoehl S, Kohmer N et al. Assessment of SARS-CoV-2 transmission on an international flight and among a tourist group. JAMA Netw Open 2020; 3:e2018044–e2018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwartz KL, Murti M, Finkelstein M et al. Lack of covid-19 transmission on an international flight. Can Med Assoc J 2020; 192:E410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bae S, Shin H, Koo H et al. Asymptomatic transmission of SARS-CoV-2 on evacuation flight. Emerg Infect Dis 2020; 26:2705–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ministry of Home Affairs Government of India . Government of India issues orders prescribing lockdown for containment of covid-19 epidemic in the country, New Delhi: Press Information Bureau, Government of India 2020.
- 31. Ministry of Home Affairs Government of India . Guidelines on the measures to be taken by Ministries/Departments of Government of India, state/union territory governments and state/union territory authorities for containment of covid-19 epidemic in the country. Annexure to Ministry of Home Affairs Order No 40-3/2020-D, New Delhi: Ministry of Home Affairs, Government of India 2020.
- 32. Choudhary OP, Priyanka IS, Rodriguez-Morales AJ. Second wave of covid-19 in India: dissection of the causes and lessons learnt. Travel Med Infect Dis 2021; 43:102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh M. Incalculable tragedy: we may never know the truth about India’s covid-19 numbers. Asia Global Online, Asia Global Institute, The University of Hong Kong 19 May 2021. https://www.asiaglobalonline.hku.hk/incalculable-tragedy-we-may-never-know-truth-about-indias-covid-19-numbers [Google Scholar]
- 34. Mallapaty S. India’s massive covid surge puzzles scientists. Nature 2021; 592:667–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


