Abstract
Introduction:
Neuropeptides are signaling molecules originating in the neuroendocrine system that can act as neurotransmitters and hormones in many biochemical processes. Their exact function is difficult to characterize, however, due to dependence on concentration, post- translational modifications, and the presence of other comodulating neuropeptides. Mass spectrometry enables sensitive, accurate, and global peptidomic analyses that can profile neuropeptide expression changes to understand their roles in many biological problems, such as neurodegenerative disorders and metabolic function.
Areas Covered:
We provide a brief overview of the fundamentals of neuropeptidomic research, limitations of existing methods, and recent progress in the field. This review is focused on developments in mass spectrometry and encompasses labeling strategies, post-translational modification analysis, mass spectrometry imaging, and integrated multi-omic workflows, with discussion emphasizing quantitative advancements.
Expert Opinion:
Neuropeptidomics is critical for future clinical research with impacts in biomarker discovery, receptor identification, and drug design. While advancements are being made to improve sensitivity and accuracy, there is still room for improvement. Better quantitative strategies are required for clinical analyses, and these methods also need to be amenable to mass spectrometry imaging, post-translational modification analysis, and multi-omics to facilitate understanding and future treatment of many diseases.
Keywords: Imaging, Label-free, Mass Spectrometry, Multiplexing, Multi-omics, Neuropeptides, Peptidomics, PTMs, Quantitation, Stable Isotope Labeling
1. Introduction:
Regulation of the nervous system is a strictly controlled process influenced by a plethora of signaling peptides, neurotransmitters, hormones, and other modulating molecules [1,2]. These neuromodulators are critical to behavior [3–5], stress responses [6–11], maintaining homeostasis [12,13], and many other biological processes [14–16]. Neuropeptides, signaling peptides originating in the neuroendocrine system, are of particular interest as they have highly diverse function and structure, dynamic expression, and many sites of action [1]. Dysregulation of neuropeptides has been implicated in many diseases and biological states, including heavy metal toxicity [8], hypoxia [7], Alzheimer’s disease [17], depression [18], and others. As a result, comprehensive characterization of neuropeptides could have many benefits such as discovering biomarkers or elucidating important biological pathways involved. However, such global analysis is challenging not only due to many neuropeptides having low in vivo abundance and high structural diversity, but also because neuropeptide function is influenced by many factors, such as location, post-translational modifications, and the presence of other co-modulating neuropeptides [1].
This complexity inherent to all neuropeptidomic studies is exacerbated by the difficulties in determining possible neuropeptide sequences. Neuropeptides are produced by the select processing of precursor proteins (i.e., preprohormone) encoded within the genome [19]. These preprohormones contain a signaling sequence and the remaining prohormone. After cleavage of the signaling sequence, the prohormone is selectively and specifically cleaved by endopeptidases, such as various prohormone convertases, to produce several peptide sequences from a single precursor protein [20]. The peptides are then processed further and post-translationally modified to produce the bioactive neuropeptides [20]. The intricate pathways from genome to active neuropeptide, splice variants, and diversity of post-translational modifications lead to many possible peptide forms that are difficult to predict from genomics or even transcriptomics alone. This compounded with the fact that many model organisms do not have a fully sequenced genome to use as a reliable starting point for predicting a full neuropeptide database makes neuropeptide studies even more challenging.
Mass spectrometry (MS)-based approaches have led to enhanced workflows for detecting, characterizing, and quantifying neuropeptides in various samples. MS can provide detailed, high-accuracy information about the intact mass, sequence, modifications, and expression levels of a detected neuropeptide [1]. Furthermore, MS analyses can profile many analytes (neuropeptides) from a sample in a single experiment without requiring extensive a priori knowledge. This unbiased, untargeted method facilitates discovery-based neuropeptidomics. As neuropeptide function is influenced by the other neuropeptides present, methods to detect the suite of neuropeptides expressed in a sample are critical to understanding the underlying signaling mechanisms of neuropeptides.
Using various labeling and label-free strategies, MS can further provide useful quantitative information that can be used to determine neuropeptide expression differences between samples [21]. Alternative MS analysis methods, such as data-independent acquisition (DIA) can enable greater depth of coverage for the neuropeptidome [22]. These quantitative strategies can be applied not only to the detection and identification of neuropeptides, but also to identifying different forms of neuropeptides with post-translational modifications [23]. Additionally, there have been many developments in the field of MS imaging (MSI) to enable the detection of neuropeptides in specific locations of a tissue or tissue section [13,24,25]. This spatial distribution provides an additional level of information for quantitative neuropeptidomics. Furthermore, MS has applications in numerous -omics (e.g., proteomics, metabolomics), that can offer complementary information to the quantitative neuropeptidomic workflows. By combining the structure elucidation, quantitative information, spatial distribution, and analysis of correlated biomolecules, analysis by MS has greatly improved our understanding of neuropeptides and their roles in many biological processes. This review aims to highlight recent developments in the broader field of neuropeptidomics with an emphasis on quantitative strategies.
2. Quantitative Strategies:
As neuropeptide function has some dependence on concentration [26], being able to reliably identify and quantify neuropeptides is necessary for understanding their function, especially as it relates to different biological states. A recent review has been published detailing mass spectrometry strategies applied to functional neuropeptidomics [27]. These functional studies are challenging, however, due to the low concentrations (as low as femtomolar) in vivo [26] of neuropeptides, highlighting the continual need for improved quantitative methods. Although MS is not inherently quantitative, its widespread application and growth in the -omic fields has led to the development of various strategies for accurate and sensitive quantitation. The quantification of neuropeptides has been achieved with label-free methods that allow analysis of the neuropeptides without modification and minimal sample loss [28]; a variety of labeling strategies that facilitate more reliable quantitation and greater throughput analyses [29]; and more recently, DIA workflows to increase reproducibility and enable detection of more low abundance analytes and thus deeper profiling of the neuropeptidome [22]. These methods are of course not the only quantitative methods for neuropeptidomics and others have been reported and summarized in other reviews [30,31].
2.1. Label-free Quantitation:
In MS workflows, certain steps, such as desalting, are crucial, but researchers will often forgo extraneous steps to reduce sample loss for low abundance species like many neuropeptides. Label-free quantitation (LFQ) strategies may suffer from reduced throughput, but they often provide the least amount of sample processing (and fewer losses from it). In LFQ methods, samples are analyzed independently of each other, commonly using LC-MS or LC-MS/MS approaches [21,32]. Quantitation is typically performed by comparing signal intensities (e.g., chromatographic peak area). By examining the extracted ion chromatogram (XIC) of analytes, quantitation is achieved by comparing peak areas between sample runs. Controlling for run-to-run variability is critical for reliable quantitation as samples are run separately [28]. Software is typically used to align the XICs by retention time and filter data, such as ensuring the precursor/fragment ions and the charge states match between aligned peaks [33,34]. Additionally, normalization between analyses needs to be considered to facilitate more accurate comparisons between samples, as variations can arise from instrument calibration, sample preparation, and ambient temperature during analysis. Various methods and software packages for normalization exist and are systematically evaluated in a recent article by Välikangas et al [35]. Ye et al. demonstrated the efficacy of LFQ to quantify neuropeptide expression changes as a result of feeding in a rat model [36]. Anapindi, et al. similarly used XIC analysis to examine neuropeptide expression, but on a much larger scale; over 200 LC-MS runs were performed to identify and quantify over 1500 neuropeptides to examine their effect on chronic migraine and opioid-induced hyperalgesia [4]. Targeted approaches have also proven useful; Salem et al. performed relative quantitation of surrogate neuropeptides and their fragments to characterize the processing of pro-neuropeptides to mature neuropeptides [37]. Frequently, targeted approaches will employ parallel reaction monitoring (PRM) in which a targeted precursor ion and subsequent fragment ion are analyzed. Similarly, multiple reaction monitoring (MRM) analyses a single precursor ion and multiple fragment ions from that precursor ion. As these approaches minimize interference from other matrix components, sensitivity and throughput are greatly enhanced [38]. Although often used for measuring surrogate peptides from a targeted protein, MRM and PRM have seen use in peptidomics workflows, such as the analysis of orexin in mice cerebrospinal fluid [39] and the identification of endogenous signaling peptides in insects [40]. While MRM and PRM have high sensitivity, the simplicity of using XICs for quantitation allows it to be easily applied to novel and/or less developed MS workflows. Bianco et al., for example, used XICs to characterize differences between arginine and lysine vasopressin after analysis by Fourier transform ion cyclotron resonance (FTICR) MS with multiphoton dissociation [41].
LFQ workflows can also employ spectral counting for quantitation. Spectral counting assesses protein abundance by correlating concentration to the number of times a constituent peptide is identified, with the idea being that more abundant proteins will be identified more frequently in a single run [33]. The protein abundance index (PAI) is calculated from the normalized level of observed peptides per protein and its exponential modification (emPAI) is used to estimate protein concentrations [42]. In neuropeptidomic experiments, however, identifications are not made based on constituent peptides, as done in bottom-up proteomics, because the endogenous neuropeptide is generally not digested prior to LC-MS analysis. This greatly reduces the applicability of spectral counting in neuropeptidomic experiments, but it still has gained some use, typically with XIC information used for improved confidence. Southey et al. has shown that spectral counting and spectral indexing (based upon the cumulative intensity of product ions) provide more informative characterization of neuropeptides in the rat suprachiasmatic nucleus over XIC [28]. Other groups have published quantitative methods that utilize both XIC and spectral counting for characterizing endogenous peptides outside of the central nervous system. By comparing the peptidomes of patients with systemic juvenile idiopathic arthritis (SJIA) to healthy patients, LFQ methods enabled the detection of 17 potential biomarkers for SJIA in urine [43]. Similarly, Labas et al. used LFQ methods to characterize peptides in chicken semen to identify key peptides for phenotyping [44]. Although these applications do not address neuropeptides specifically, the peptidomics workflow is translatable and provides future directions for neuropeptide analyses. Even as other quantitative methods are developed, the scalability and ease of using LFQ will likely ensure its continued use in the future.
2.2. Label-based Strategies:
Although label-free quantitation is adaptable, requires fewer sample processing steps, and is amenable to analyses of many samples, it often requires a greater number of LC-MS runs and can suffer from run-to-run variability. Conversely, many labeling strategies exist that allow samples to be run simultaneously, enabling accurate multiplex quantitation with fewer control samples required, reduced instrument time, and decreased effects due to instrument variation [21]. For neuropeptide applications, most labeling reactions occur post-extraction and create a mass difference between channels via stable isotope incorporation to differentially label samples. Reductive dimethylation is frequently used to label neuropeptides as it targets primary amines (N-termini and lysine residues), common to most neuropeptides. Additionally, this reaction is low cost and easily accessible, requiring only isotopic formaldehyde and a reducing agent such as borane pyridine or cyanoborohydride [45]. Isotopic dimethylation has been used extensively in the Li lab to study neuropeptidomic changes in crustacean models [7,10,11,46,47]. Moreover, the group has shown the method is compatible with both electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) methods to provide complementary coverage of neuropeptides in a single sample [7]. Wilson et al. has developed an online dimethyl labeling system in which neuropeptides are derivatized on-column with either light or heavy reagents [48]. The Fricker lab has also published several articles using isotopic labeling for quantitative peptidomics. They have expanded the reductive dimethyl labeling scheme to an impressive five channels while improving accuracy with isotopic correction calculations [45]. Similarly, the lab has expanded the use of trimethylammoniumbutyryl (TMAB) chemical tags to a 4-plex [49,50]. TMAB tags add a permanent positive charge via a quaternary amine to N-termini and lysine residues of peptides using an amine-reactive NHS ester. These tags have been widely used in peptidomics to study numerous biological processes, including peptide degradation [51], prohormone processing [52], narcotic effects [53], and can be transferred to neuropeptidomic applications. These tags are effective, easily synthesized, and increase signal intensity by incorporating a permanent positive charge. This permanent charge, coming from a charged quaternary amine, does make the peptide more prone to dissociation, thus increasing instability and limiting application.
One of the major limitations with isotopic labeling, as evidenced by Buchberger et al. [7] and others [45], is the added spectral complexity with higher multiplexing. This can lead to difficulties in accurately calculating relative abundances, even with isotopic corrections. Additionally, given the wide dynamic range of neuropeptides, it is possible (and perhaps likely) that peaks corresponding to downregulated neuropeptides could be too low in intensity to be quantified or even be detected. Even analytes of high enough intensity might still not be selected for fragmentation and tandem MS analysis by commonly used data-dependent acquisition (DDA) settings, simply due to the greater number of precursor peaks that come with an isotopic labeling workflow. Isobaric tagging–where neuropeptides are labeled with isotopically encoded tags that add the same mass but produce unique reporter ions upon tandem MS analysis–can offer advantages over conventional isotopic labeling. Figure 1 depicts the differences between isotopic (1A) and isobaric tagging (1B). These methods are capable of significantly higher order multiplexing, such as 8-plex with iTRAQ [54], 16-plex with TMTpro [55], or 21-plex with DiLeu [56], because only the MS2 spectra have added complexity due to the unique reporter ions. Moreover, the tags are strategically designed to create reporter ions in regions of the spectrum that do not contain other useful peaks. Isobaric tags have been used often for proteomics applications, but have had fewer applications with peptides, especially neuropeptides. Dimethylated leucine (DiLeu), for example, has been used to quantify neuropeptides in lobster brains as a function of growth cycle [15], and also as part of a multi-omic profiling of the mouse hypothalamus [57], but the low abundance of many neuropeptides still presents challenges. As quantitation occurs at the MS2 level, isobaric tagging requires analytes be selected for fragmentation and subsequent tandem MS analysis. Low abundance analytes, like many neuropeptides, are often omitted from tandem MS analysis using conventional DDA strategies. Recently, Sauer and Li have shown that optimization of the DDA parameters can facilitate a greater depth of coverage in the crustacean neuropeptidome. This enabled the characterization of many neuropeptides dysregulated in response to copper toxicity [8]. Similar optimization strategies have been used to improve proteomic coverage [58,59], highlighting a problem not unique to the field of neuropeptidomics. As instrumentation and analytical capabilities improve, it is likely that we will see an increased prevalence of isobaric and isotopic labeling methods reported for neuropeptides and other low abundance analytes.
Figure 1:
Comparison between isotopic (A) and isobaric (B) labeling. In isotopic labeling strategies, analytes are differentiated at the precursor mass level due to the incorporation of light and heavy tags. Isobaric workflows result in no differentiation at the precursor mass level, but upon fragmentation, unique reporter ions for each channel form, giving rise to quantitation based on relative intensities of reporter ions.
2.3. Data-Independent Acquisition:
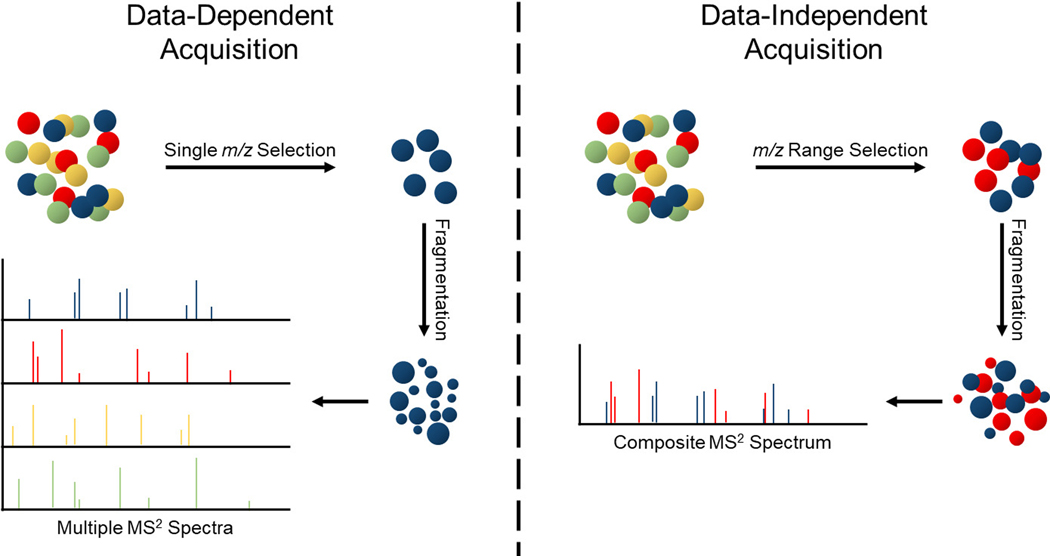
Conventional identification and quantitation of peptides and proteins by mass spectrometry typically involves DDA to trigger and obtain MS2 fragmentation spectra. Prioritizing fragmentation of the higher signal intensity ions, this acquisition method is biased towards higher abundance and more easily ionizable species. Fragmentation spectra are crucial to obtain confident identification of biomolecules. Neuropeptides are often of low abundance compared to other biological and neurological matrix components [60]; their analysis can benefit greatly by using a less biased fragmentation scheme. DIA methods were developed to overcome the limitations of DDA, by fragmenting all molecules within a desired m/z window, not only the highest signal intensity ions [61], as demonstrated in Figure 2. During DIA MS, all molecules within a m/z isolation window of user-defined width are fragmented, without a precursor selection to trigger fragmentation. Fragmentation ion spectra are collected from different isolation windows in a cyclic manner, from the beginning of the defined m/z survey range to the end before repeating. MS1 spectra are collected every cycle, or however often is desired. The use of DIA enables the identification, and therefore potential quantitation, of more molecules compared to DDA [61,63–65], including lower abundance molecules such as neuropeptides [22]. As a newer method, DIA MS does not have the breadth of application that DDA does and is not yet commonly applied to neuropeptide analyses. However, the field of neurobiology has seen great benefit from the adoption of quantitative DIA MS, as discussed in a recent review on its application to quantitatively analyze the brain proteome [66].
Figure 2:
Depiction of differences between DDA and DIA. In DDA, a single m/z is selected at a time for MS/MS. In DIA, several analytes are selected for simultaneous MS/MS across a wide m/z window and the composite mass spectrum is deconvoluted during data analysis to discern constituent analytes.
Although not all quantitative, a few MS analyses using DIA have recently been performed to better improve the characterization of neuropeptides. DIA MS was used in the untargeted quantification study of neuropeptide expression changes in response to feeding activity in crustacea [67]. The authors were able to detect and quantify 137 peptides directly from microdialysate with minimal sample preparation; no extraction or precipitation steps were required, demonstrating the utility of DIA for the quantification of limited samples [67]. While DIA MS is a powerful technique for untargeted quantification of neuropeptides, Saidi et al. demonstrated its limitations and higher variances during targeted quantification studies of neuropeptides found in animal spinal cord tissues [38]. Delaney and Li also evaluated the utility of DIA MS for the quantification of neuropeptides from crustacean neural tissue. While improving the technical and biological reproducibility of analysis and number of overall neuropeptides identified compared to DDA, the DIA method showed poor quantitative accuracy using a label free approach [22]. To improve LFQ of peptide hormones using DIA MS, a method was developed using an internal standard peptide, enabling accurate quantitation [68]. As the field of DIA MS quantification further advances, new strategies to improve its abilities to characterize neuropeptides will need to be developed.
The utility of DIA searches using a spectral library-free approach and database searches generated from FASTA sequences has been demonstrated for analysis and quantitation. Model spectra generated from these methods are not always as reliable for neuropeptides as the algorithms are often developed for tryptic peptides. PTMs and structural diversity of endogenous peptides complicate these matters, making it difficult to obtain good results without high-quality spectral libraries. Although identification through spectral library searches have been shown to increase identification and quantitation reproducibility [69], spectral library generation, requiring high amounts of starting material, may not always be feasible due to the limited sample concentration of neuropeptides. Database searching and spectral-free methods [70–74] will likely be more beneficial to the future study of neuropeptides by DIA MS.
Overall, DIA has been shown to improve sensitivity of analysis for both identification and LFQ [75], enabling consistent detection with up to a 10-fold increase in sensitivity compared to DDA, and improving the quantitative dynamic range of analysis [61,76]. Notably, the reproducibility and quantitative performance of DIA MS methods was evaluated by 11 sites worldwide, and reproducible quantitation of proteins was observed [76]. Reproducibility is very important for the quantitation of samples; the same analytes must consistently be identified across all conditions for comparison. A large limitation to DIA analysis is the duty cycle for each scan and total time it takes to cycle through the entire desired m/z range. Careful consideration is required when choosing parameters such as maximum ion injection time, automatic gain control target, isolation window width, and full m/z range, among others, as this may cause analytes to not be fragmented during the appropriate window before fully eluting during the LC gradient [77]. This issue is further exacerbated during DIA MS for quantitation as accurate quantitation requires data to be collected at multiple points across a peak profile. Further complicating quantitation by DIA MS, multiplexed label-based methods increase spectral complexity, although such methods have recently been utilized in a few proteomics experiments [78–81]. Additional information about the utility and considerations of DIA MS for neuropeptide analysis and quantitation, including software resources can be found in a recent comprehensive review detailing advances in the MS analysis of neuropeptides [manuscript under review]. More general reviews for proteomics also exist [64,72,82–84].
DIA can also be beneficial for the analysis of heavily modified analytes where MS signal intensity is distributed across multiple proteoforms, as this increases the number of precursors needed to be selected for fragmentation using DDA. A limitation to this method is the deconvolution of data; multiple precursor ions are co-fragmented in MS2 spectra and PTMs can further complicate fragmentation spectra. Although it has been shown to benefit the analysis of glycosylated proteins [85–87], this analysis has not yet been applied to the field of neuropeptidomics. Even with the limitations of DIA, it has been shown to successfully identify more peptides and neuropeptides, with improved reproducibly. With a limit of detection (LOD) in the amol range [61] and improved quantitation capabilities [76], the field of neuropeptidomics would benefit greatly from the adoption of quantitative DIA MS workflows.
3. Post-Translational Modifications:
Neuropeptides and other bioactive peptides are formed after enzymatic cleavage of larger precursors by peptidases. Additional enzymes can alter these peptides with PTMs, altering their structure, function, and stability, among other effects, contributing to the vast diversity of neuropeptides. Neuropeptide modifications can include amidation, phosphorylation, acetylation, glycosylation, and sulfation, to name a few [88]. Identifying and differentiating between these forms is crucial to understand molecular mechanisms in neurobiology, and thus these modified neuropeptides are investigated using MS by a variety of labs [10,89–91], thoroughly summarized in a few recent reviews [92]. The quantification of post-translationally modified neuropeptides faces additional challenges. Labeling approaches often target specific residues and moieties, so post-translational modification of these residues often inhibits quantitation via labeling. For example, many tags target primary amines and are therefore ineffective or less effective for peptides with acetylated N-termini. Further challenging analysis of modified peptides is the decreased ion signal intensity due to poor ionization efficiency of the modified peptides or from distribution of already low abundance neuropeptides across the differentially modified forms. This leads to the need for targeted analyses and enrichment strategies to detect and quantify peptides with PTMs [93–95], especially for those modified by highly dynamic glycosylation [95–98]. As MS considerations are more prominent for peptides modified by glycosylation [94,99,100], we will focus on the discussion of glycosylated peptides.
Estimated to modify potentially 33% of all known human peptide hormones [44], changes in glycosylation have a large impact on the role and efficacy of neuropeptides and other bioactive peptides [101–104]. It is therefore of interest to improve quantification abilities for these lower abundance peptides with decreased ionization efficiency compared to their non-modified counterpart [97]. During a targeted analysis to characterize insulin and other signaling peptides in pancreatic islets, Yu et al. discovered insulin to be glycosylated and found this form to be differentially regulated in mouse models of diabetes [105]. This demonstrates the need for more attention to be given to the analysis and quantification of modified signaling molecules. To this goal, Hansen et al. investigated the potential presence of glycans on atrial natriuretic peptide (ANP), a peptide hormone with its proteolytic degradation and potency being regulated by glycosylation. They characterized and quantified glycosylated ANPs using a targeted MS approach and demonstrated glycosylation on ANP to impact its stability, circulation time, and receptor activation in rats [106].
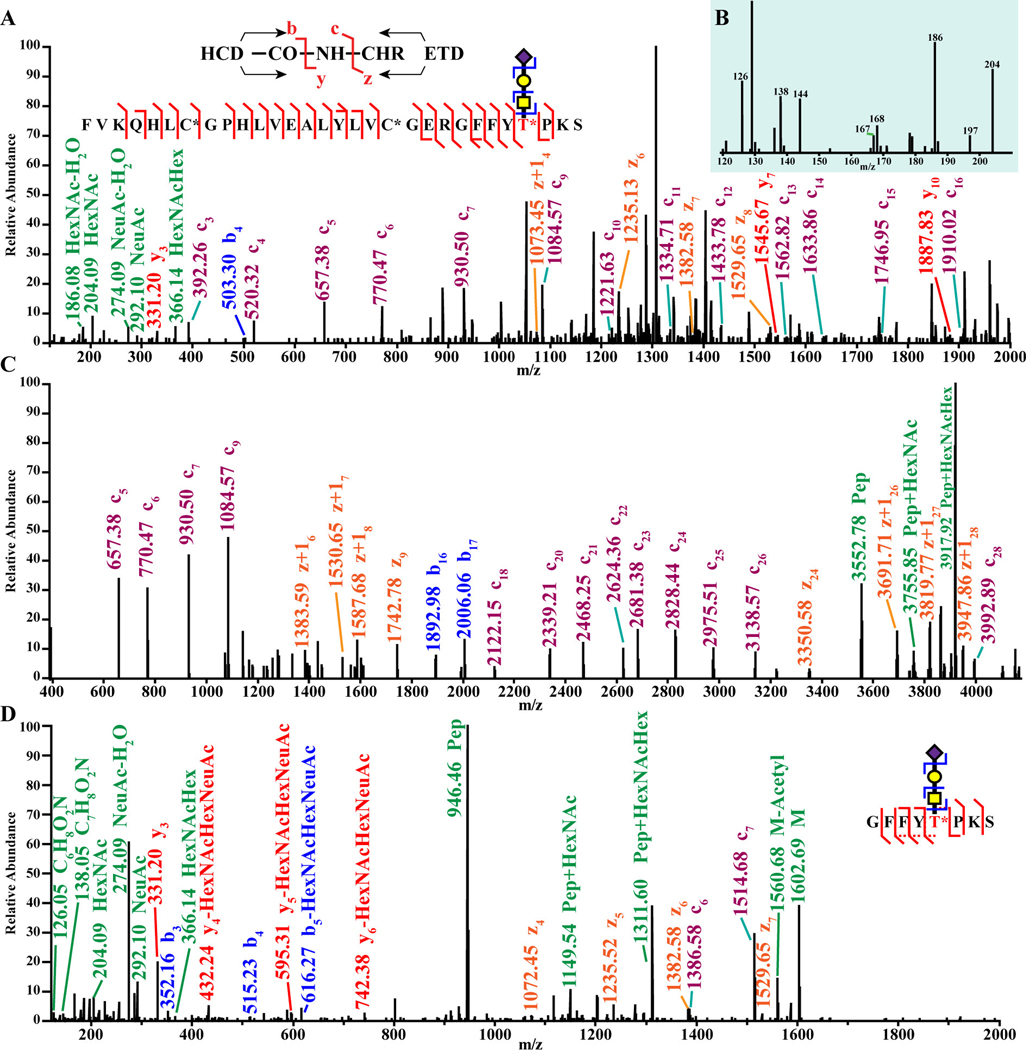
To improve the characterization of glycosylated neuropeptides, sensitive and accurate MS methods must be developed. Several dissociation methods have been investigated to better identify glycopeptides, namely collision-based [107–110] and electron-based [107,109,110] dissociation methods. Hybrid methods have also been developed to improve glycopeptide analysis such as electron-transfer/higher-energy collision dissociation (EThcD) [110], higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation [111], and HCD product ion-triggered EThcD [105]. EThcD has also been shown to be valuable for quantitative proteomics of phosphorylated biomolecules as well [112]. The type of fragmentation scheme used is important to consider during glycopeptide analysis. Riley et al. demonstrated the need for optimizing dissociation methods, finding peptides modified by N-glycans and O-glycans require different dissociation methods for optimal fragmentation [113]. The information-rich fragment ion spectra generated from EThcD, shown in Figure 3, is vital for the confident localization of O-glycans. To characterize glycosylated neuropeptides in crustaceans, Cao et al. utilized HCD triggered EThcD [23]. This study demonstrates the method’s utility for sensitive glyconeuropeptide analysis. Though not applied to endogenous peptides, Zhu et al. describe a three-part workflow for the in-depth investigation of proteins and glycoproteins in the central nervous system [114]. More broad strategies for the quantification of glycosylated proteins and digested peptides have been discussed thoroughly in a recent review [115]. Although glycoproteomics and its quantification is of great interest to the scientific community, we can see that there has been a disproportional amount of investigation into glycosylated neuropeptides, and even less with quantitative approaches applied. This is an underdeveloped field but as glycoproteomic strategies improve in the future, we expect to see them applied to the field of neurobiology towards quantifying endogenous peptides more readily.
Figure 3:
Sample spectra from mouse insulin. Using EThcD (A) and charge deconvolution (C) resulted in high sequence coverage due to production of both b/y- and c/z-ions. The spectrum in D highlights the glycan localization, and the low mass region is shown enlarged in (B). Figure reprinted with permission from Yu, et al. [105].
4. Mass Spectrometry Imaging:
As various modes of MS are being applied to neuropeptide discovery, there has been an increased emphasis in determining the biological relevance through localizing neuropeptides. MSI has been preferentially implemented in these studies due to the unique advantage of permitting targeted and untargeted detection of analytes within a tissue or cell while still providing spatial information [116,117]. To achieve this, many researchers often use MALDI, where the surface area of a tissue is portioned into data-attainable units (pixels) and imaged through several laser ablations. Quantitative MSI of neuropeptides can be achieved in different ways including absolute quantification using labels to create a calibration curve and semiquantitative spiking with an internal standard [116]. Here we discuss recent advances in MSI that enable quantitative and semiquantitative analysis of neuropeptides.
Within the field of neuropeptidomics, relative quantitation is widely practiced due to easier sample preparation and reduced costs. Information obtained by these relative quantitation methods can be increased through biological means such as immunohistology assays and Nissl staining [118,119]. These techniques are applied to tissue after MSI to normalize neuropeptide quantities with respect to number of nerves and free amines present. The relative abundance and spatial information from MSI is valuable, but still comes with challenges such as localized ion suppression, availability of software, duration of data collection, and having a sufficient MSI analyzer [120–123]. These issues are exacerbated by the low concentration of neuropeptides in tissue. There have, however, been advances in desorption electrospray ionization (DESI) and liquid extraction surface analysis (LESA) MSI in lipidomics that may help mitigate these issues [124–126]. Zemaitis et al. demonstrates an increased signal intensity in DESI-MSI compared to MALDI-MSI, leading to higher resolving power for lipids [124]. This can likely be attributed to a lack of matrix clusters, which in MALDI, contribute to a pattern of low ionization efficiency [124,125]. Some imaging techniques seek to circumvent these issues by performing DESI and MALDI on the same tissue, permitting both proteomic/peptidomic and metabolomic analyses [126]. The use of LESA shows increased signal intensity due to larger sampling size, making this method faster and more sensitive at the sacrifice of spatial resolution [126]. Both LESA and DESI come at the expense of spatial resolution; where MALDI can often offer a few micron spatial resolution, DESI is limited to 50–100 micron and LESA images often have pixel sizes of 1000 microns. These techniques can be coupled with subsequent LC-MS/MS to verify trends of relative abundances seen in imaging experiments or XIC for LFQ [117,127].
Practices are being explored in drug MSI for better absolute quantitation using more sensitive mass analyzers, such as FTICR, coupled with an increased number of internal standards as well as adjusting calibration curves to pixel deviation [123,128]. Techniques under development work to decrease the LOD such as those discussed in metabolomics works implementing surface-assisted laser desorption/ionization (SALDI) [129] and other matrix-free MSI methods where nanoparticles are used to coat the tissue to enhance ionization efficiency as well as spatial resolutions (Figure 4) [130,131]. These methods incorporate metals and have been found to retain spatial resolution of MALDI (~2–5 microns) while increasing ionization efficiency for small molecules. They are, however, presently limited to small molecules like drugs and metabolites. Such relative abundance techniques, combined with LFQ via LC-MS/MS back-correlation and software development, could greatly improve the accuracy and performance of quantitative MSI studies. The many forms of MSI complement the diversity of biological problems being studied, and advancements in MSI of neuropeptides will provide insights into the anatomy and physiology of neurological function and regulation. This in turn will help clinicians and other researchers identify and treat various health conditions.
Figure 4:
MSI enhancements from application of gold nanoparticles. Comparison of images from extracted neurotransmitters (b-d) to the optical image (a) shows differential expression based on grey/white matter regions of the brain. Image reprinted with permission from McLaughlin et al. [131]
5. Multi-omics:
The exact roles of neuropeptides are often difficult to discern when strictly observing only neuropeptides. Their receptors are typically proteins, such as G protein-coupled receptors (GPCRs) [132]; they regulate biochemical pathways with downstream metabolite products [133]; and are often co-expressed and co-released with small molecule neurotransmitters [134]. Multi-omic workflows have emerged in which elements of proteomic, peptidomic, metabolomic, etc. experiments are integrated to provide more comprehensive information [135–137]. As a result, multi-omic experiments have seen increased popularity in neuropeptide experiments in recent years.
5.1. Small Molecule Studies:
Characterizing neuropeptides and relevant small molecules in a single experiment is difficult due to stark differences in extraction efficiencies, solubilities, ionization efficiencies, fragmentation patterns, and subsequent data analysis. Specific applications have further constraints, such as matrix compatibilities for MSI. Nevertheless, researchers have made recent advances in combining neuropeptidomics with small molecule analyses (e.g., lipidomics and metabolomics), typically by processing the two separately, then co-analyzing the data. Keller et al. addressed the extraction issues using both acidified methanol and methanol/water/chloroform extractions to efficiently recover proteins, peptides, and metabolites. Two molecular weight cut-off (MWCO) filters were used to separate metabolites (<3 kDa), peptides (3–30 kDa), and proteins (>30 kDa) [137]. Although not specifically studying neuropeptides, the reported extraction methods (i.e., acidified methanol), have been used previously for neuropeptides [7,8,22,46]. Gutierrez et al. reported a similar strategy in which an acetone/chloroform precipitation was used to separate proteins and metabolites [136]. This method, dubbed SPOT (sample preparation for multi-omic technologies), is not described for the study of endogenous peptides, but could be adapted by adding a MWCO step similar to Keller et al. For some approaches, neuropeptides do not necessarily need to be isolated from metabolites prior to LC-MS analysis. Chen et al. used atmospheric pressure (AP) MALDI to study neuropeptides, lipids, and other biomolecules from the same tissue section, but incorporated different ionization methods [138]. This niche strategy eliminates issues with extraction and ionization efficiency differences by collecting data directly from the tissue section. The lack of tandem MS data, however, does require researchers to rely on accurate mass matching, possibly losing confidence in data interpretation
Often co-expressed and released with neuropeptides, neurotransmitters are another target for multi-omics. Wojnicz et al. report a method that combines short neuropeptides (4 residues) and metabolite analyses to study bovine cells without the need for separate extractions. Using synthetic standards, they created a calibration curve that allowed absolute quantitation of both neuropeptides and neurotransmitters [133]. These methods would likely have limited applicability to larger neuropeptides but are important in establishing multi-omic strategies. Similarly, zwitterion exchange has been used for online separation prior to LC-MS to quantify neurotransmitters and select neuropeptides (oxytocin and vasopressin) simultaneously from blood [139]. Alternative separation and sampling methods, like microdialysis coupled to LC-MS, have offered sensitive assays for quantifying both neuropeptides and neurotransmitters, summarized in a review by Zestos and Kennedy [140].
5.2. Proteomics:
Although similar in structure to proteins, neuropeptides require different analytical workflow than proteins largely due to size differences. Most multi-omic workflows that combine proteomics with the study of endogenous peptides like neuropeptides analyze the two separately and interpret the combined data. For example, Liu et al. describes the use of label-free neuropeptidomics with multiplex DiLeu-labeled neuroproteomics, workflow highlighted in Figure 5, to profile changes in the mouse hypothalamus resulting from the gut microbiome [57]. This study demonstrated the impact of the gut microbiome on neurochemical processes [57]. Similarly, Chen et al. combined label-free and labeled data to interrogate proteomic, metabolomic, and peptidomic (translatable to neuropeptides) dysregulation in metabolic diseases [135]. Neuroproteomics has also revealed dysregulation of neuropeptide and neurotransmitter release with impacts in neuropsychiatric diseases, such as addiction, using LFQ MS [141]. Conversely, Hook et al. characterizes neuropeptide variants to inform neuroproteomics and precursor protein analysis to better understand proteolytic processing and how it relates to cell-cell signaling [88]. The study of endogenous peptides outside of the neuroendocrine system can also provide translatable methods for multi-omic neuropeptide studies. For example, Labas et al. do not specifically study neuropeptides, but report a LFQ assay to combine proteomic and peptidomic data to phenotype chicken semen in a multi-omic experiment using spectral counting and XICs from LC-MS data [44]. By omitting the digestion step typically used to study proteins (bottom-up proteomics), Li et al., describe the use of top-down MS methods to study microproteins and endogenous peptides in mouse brain tissue extracts [142]. Top-down MS methods like these are crucial for studying different protein and peptide forms but need adaptation to studying larger proteins and neuropeptides simultaneously.
Figure 5:
Multi-omic workflow for studying neuropeptides and proteins. Neuropeptides were extracted from the hypothalamus of germ-free (GF) and conventionally raised (ConvR) mice and analyzed using LFQ. Proteins were pelleted, digested, and differentially labeled with 10-plex DiLeu isobaric tags. The differentially labeled samples were pooled together and analyzed simultaneously to perform relative quantitation based on reporter ion intensities. Adapted from Liu et al. [53] with permission.
While several advances have been made to address MS concerns when studying classes of molecules with distinct chemical properties, such as sequential extractions, methods employing simultaneous co-analysis are far from being common. To fully understand the interplay between different types of biomolecules, improvements into interpreting large amounts of data is also required. As investigations into complicated multi-interaction diseases and biological functions are increasingly being pursued by the scientific community, we expect to see more extensive multi-omics experiments adopted into future workflows.
6. Conclusion:
The field of neuropeptidomics is constantly evolving and recent improvements have led to enhanced detection and quantitation of neuropeptides. Their low in vivo abundance, complex functions, and structural diversity make their analysis challenging, but MS analyses have mitigated many of these difficulties [143]. Isotopic labeling strategies, such as dimethyl labeling [45] and TMAB labeling [49], differentiate neuropeptides at the precursor ion level but are only used to compare a few experimental conditions. Enhanced multiplexing can be achieved with the incorporation of isobaric tagging because it does not significantly increase spectral complexity [8]. Labeling strategies do enable higher throughput and typically more reliable quantitation, but additional sample processing steps often cause sample loss which is detrimental for low abundance analytes like many neuropeptides. As a result LFQ methods are still common [141]. These methods are also more compatible with DIA methods, facilitating greater sensitivity and neuropeptidome coverage [22]. Quantitative strategies are also employed in imaging workflows to provide spatial distribution, utilizing a variety of normalization methods to ensure quantitation is accurate, summarized nicely in a review by Tobias and Hummon [116]. Analysis of PTMs, especially glycosylation, has greatly benefited from improvements in hybrid MS fragmentation method, such as the use of HCD-triggered EThcD to improve fragmentation of glycopeptides (and glyconeuropeptides) [23]. As neuropeptidomic workflows become more common and accessible, their findings can be incorporated into larger multi-omic workflows [57]. There is still much room for improvement in terms of technology and method development, but recent advances in neuropeptidomics have provided much insight into the complex signaling pathways involving neuropeptides, with impacts in the fields of biomarker discovery and drug development.
7. Expert Opinion:
The field of neuropeptidomics is instrumental to our understanding of neuromodulation and signaling. This in turn has applications in clinical settings in the areas of biomarker discovery, drug discovery, and drug action [132,134,144]. Neuropeptide dysregulation has been linked to many biological problems and diseases, such as Alzheimer’s disease [145], obesity [146], cancer [147], and depression [148]. The advances in neuropeptidomics have enabled researchers to better understand many of these conditions and the related signaling pathways. This can facilitate the discovery of new, more reliable biomarkers. Additionally, knowledge gained from these signaling pathways may provide new drug action sites or possibly even modifying neuropeptides to act as drugs themselves.
Adoption of neuropeptidomics in a clinical setting does still present challenges, however. As neurochemistry is incredibly complex, most studies are performed in organisms with simpler neuroendocrine systems, including some mammals like mice and rats [149], and many invertebrates, such as crustaceans [150] and nematodes [151]. Translation from these models to humans is difficult and, at the very least, will require more sophisticated, less invasive sampling methods (e.g., microdialysis). Clinical research involving humans also has the added complexity of genetic diversity. Biomarker discovery and genetic risk score assessments have historically been biased, and more and more research is demonstrating the importance of clinical research that accounts for sex, age, and racial diversity [152–159]. Additionally, MS analyses of neuropeptides, while sensitive, accurate, and quantitative, may not offer high enough throughput for clinical applications. Multiplexing techniques, such as the aforementioned DiLeu tags [8,56,160], can greatly improve throughput, but are not commercially available. Conversely, the tags that are commercially available (e.g., TMT) [55] are expensive and do not offer as high a degree of multiplexing. Furthermore, clinical applications could necessitate absolute quantitation, often requiring expensive isotopic peptide standards. The Li Lab has developed chemical tags that enable low cost, absolute quantitation. These isotopic DiLeu (iDiLeu) tags are used to create a calibration curve from a synthesized peptide and compare the target peptide to the calibration curve [161]. Methods like this still require synthetic peptides, albeit much cheaper as the isotopes are incorporated via chemical labeling, not during the synthesis of peptides. When the iDiLeu tags are combined with multiplex isobaric tagging reagents, the throughput of absolute quantification can be greatly enhanced [160], which could be highly beneficial for absolute quantification and biomarker validation with large cohort of clinical specimens.
Advances in quantitation are often aided by chemical labeling methods, but improvements in sample analysis are other avenues to consider. As mass spectrometers become more sophisticated, neuropeptidomic analyses are greatly enhanced. For example, increased scan times enable greater depth of coverage, especially for low level analytes like neuropeptides. Increased resolving power can help differentiate between neuropeptides with similar masses, and can facilitate greater multiplexing, as seen by the incorporation of mass defects in DiLeu 12- and 21-plex tags [56,160]. Newer instruments are also capable of performing alternative fragmentation methods, such as ETD and EThcD, to provide more detailed MS2 data. This is especially beneficial for analysis of PTMs like glycosylation [23]. DIA experiments have also offered many improvements, especially for low concentration analytes that normally are missed by DDA settings (e.g., neuropeptides) [22], despite being a relatively new method. These benefits are certain to increase as DIA sees more popularity and data analysis/deconvolution software improves. Advances in MALDI-MS have given rise to instruments with high acquisition rates and decreased laser size to generate MSI data with high spatial resolution without requiring extra time [161]. Further improvements in instrumentation will undoubtedly enable more robust neuropeptidomic experiments.
We speculate the field of neuropeptidomics to continue thrive on its current trajectory, but with increased prevalence, informing many biological studies. This is largely due to recent advances in quantitation and the many possibilities of its application for quantitative neuropeptidomics. As instrumentation enables faster analyses, either through increased scan times, or faster separation modes like capillary electrophoresis [162], LFQ could see increased use as throughput improves. Conversely, these advances could also lead to an increase in the use of labeling techniques as instruments are able to resolve miniscule mass differences to enable more accurate and sensitive analyses of neuropeptides. Ultimately these methods will both see continued use depending on application, but it will be interesting to see how they are incorporated into larger experiments. As neuropeptides have profound impacts on many biochemical and physiological processes, their study will undoubtedly be important in larger proteomic and metabolomic experiments, and we predict an increase in multi-omic studies, even at the single-cell level with continued improvements in instrumentation and microscale sample preparation. Advances in neuropeptide analyses are also not limited to quantitation. Spatial distribution information gained from MSI can be used to characterize neuropeptide function, receptors, etc. [118] Additionally, ion mobility MS (IM-MS) is routinely used to provide structural information that can be used to distinguish isobaric neuropeptides and better understand neuropeptide conformation and possibly functional roles dependent on its tertiary/quaternary structure [163,164]. As IM-MS and MSI methods are further developed, our understanding of neuropeptides will greatly improve. Combining the structural and quantitative aspects of mass spectrometry will provide richer characterization of neuropeptides, and thus has the power to lead to better biomarkers and drug design to help combat neurological disorders, obesity, and other common health concerns.
Article Highlights:
Developments in quantitative mass spectrometry have enabled greater sensitivity, higher throughput, and more comprehensive analyses of neuropeptidomics, improving understanding of the signaling pathways involved in many diseases.
Both isotopic and isobaric labeling strategies have seen increased usage, especially as instrument advancements enable greater multiplexing, and label-free neuropeptidomics remains common due to reduced sample loss and spectral complexity. Recent incorporation of data-independent acquisition strategy has benefits for both labeling and label-free methods.
Post-translational modification analysis remains challenging, but is in greater demand, especially with the discovery of glycosylated neuropeptides. These analyses have benefited from adapting glycoproteomics methods and improvements in instrumentation, such as the availability of ETD and EThcD for fragmentation.
Recent advances in normalization methods, matrix development, data analysis, etc., have enabled mass spectrometry imaging to not only be useful for localization, but also for quantitation of neuropeptides.
Neuropeptides play roles in diverse signaling pathways that involved a suite of co-modulating neuropeptides, proteins, neurotransmitters, and metabolites, highlighting the need for multi-omic workflows. These methods have seen increased use in recent years, facilitated by developments in analyte extraction and separations, differential labeling, and instrumentation.
Acknowledgments
Funding:
Preparation of this manuscript was funded in part by the National Science Foundation (CHE-1710140 and CHE-2108223) and National Institutes of Health through grants (R01DK071801, U01CA231081, RF1 AG052324, and P41GM108538). C.S.S. acknowledges a National Institute of Environmental Health Sciences fellowship as part of the National Ruth L. Kirschstein Research Service Award fellowship program (F31ES031859). A.P. was supported in part by the NIH Chemistry–Biology Interface Training Grant (T32 GM008505). O.L.R was supported by the UW-Madison/ACS Bridge to the Chemistry Doctorate Program. The UW/ACS Bridge Program is supported in part by the National Science Foundation Award NSF-1834545 to the American Chemical Society, supplemented by funds from the UW-Madison Graduate School, Office of Vice Chancellor for Research and Graduate Education and the Department of Chemistry.
L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Declaration of Interests:
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References:
Papers of special note have been denoted as (*) of interest, and (**) of considerable interest.
- [1].Li L. and Sweedler JV (2008) Peptides in the Brain: Mass Spectrometry–Based Measurement Approaches and Challenges. Annual Review of Analytical Chemistry, 1, 451–83. 10.1146/annurev.anchem.1.031207.113053 [DOI] [PubMed] [Google Scholar]
- [2].Gäde G, Šimek P. and Marco HG (2021) Biochemically identified neuropeptides in a caddisfly (Trichoptera) and a pygmy mole cricket (Orthoptera: Caelifera: Tridactyloidea). Archives of Insect Biochemistry and Physiology, 106, 1–11. 10.1002/arch.21778 [DOI] [PubMed] [Google Scholar]
- [3].Habenstein J, Schmitt F, Liessem S, Ly A, Trede D, Wegener C. et al. (2021) Transcriptomic, peptidomic and mass spectrometry imaging analysis of the brain in the ant Cataglyphis nodus. Journal of Neurochemistry, 1–22. 10.1111/jnc.15346 [DOI] [PubMed] [Google Scholar]
- [4].Anapindi KDB, Yang N, Romanova EV, Rubakhin SS, Tipton A, Dripps I. et al. (2019) PACAP and other neuropeptide targets link chronic migraine and opioid-induced hyperalgesia in mouse models. Molecular and Cellular Proteomics, 18, 2447–58. 10.1074/mcp.RA119.001767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ueda D, Yonemochi N, Kamata T, Kamei J, Waddington JL and Ikeda H. (2021) Increase in neuropeptide Y activity impairs social behaviour in association with glutamatergic dysregulation in diabetic mice. British Pharmacological Society, 178, 726–40. 10.1111/bph.15326 [DOI] [PubMed] [Google Scholar]
- [6].Chen R, Xiao M, Buchberger A. and Li L. (2014) Quantitative neuropeptidomics study of the effects of temperature change in the crab cancer borealis. Journal of Proteome Research, 13, 5767–76. 10.1021/pr500742q [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Buchberger AR, Sauer CS, Vu NQ, DeLaney K. and Li L. (2020) A Temporal Study of the Perturbation of Crustacean Neuropeptides Due to Severe Hypoxia Using 4-Plex Reductive Dimethylation. Journal of Proteome Research, 19, 1548–55. 10.1021/acs.jproteome.9b00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Sauer CS and Li L. (2021) Mass Spectrometric Profiling of Neuropeptides in Response to Copper Toxicity via Isobaric Tagging. Chemical Research in Toxicology,. 10.1021/acs.chemrestox.0c00521 *Demonstrated the feasibility of using isobaric tags for multiplexed neuropeptidomics.
- [9].Buchberger AR, Vu NQ, Johnson J, Delaney K. and Li L. (2020) A Simple and E ff ective Sample Preparation Strategy for MALDI-MS Imaging of Neuropeptide Changes in the Crustacean Brain Due to Hypoxia and Hypercapnia Stress. Journal of the American Society for Mass Spectrometry, 31, 1058–65. 10.1021/jasms.9b00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu Y, Buchberger AR, Delaney K, Li Z. and Li L. (2019) Multifaceted Mass Spectrometric Investigation of Neuropeptide Changes in Atlantic Blue Crab, Callinectes sapidus, in Response to Low pH Stress. Journal of Proteome Research, 18, 2759–70. 10.1021/acs.jproteome.9b00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Y, Buchberger A, Muthuvel G. and Li L. (2015) Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics, 15, 3969–79. 10.1002/pmic.201500256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen R, Ma M, Hui L, Zhang J. and Li L. (2009) Measurement of Neuropeptides in Crustacean Hemolymph via MALDI Mass Spectrometry. Journal of the American Society for Mass Spectrometry, 20, 708–18. 10.1016/j.jasms.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].DeLaney K, Hu M, Hellenbrand T, Dickinson PS, Nusbaum MP and Li L. (2021) Mass spectrometry quantification, localization, and discovery of feeding-related neuropeptides in cancer borealis. ACS Chemical Neuroscience,. 10.1021/acschemneuro.1c00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu M, Helfenbein K, Buchberger AR, Delaney K, Liu Y. and Li L. (2021) Exploring the Sexual Dimorphism of Crustacean Neuropeptide Expression Using Callinectes sapidus as a Model Organism. Journal of Proteome Research, 20, 2739–50. 10.1021/acs.jproteome.1c00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jiang X, Xiang F, Jia C, Buchberger AR and Li L. (2018) Relative Quantitation of Neuropeptides at Multiple Developmental Stages of the American Lobster Using N, N -Dimethyl Leucine Isobaric Tandem Mass Tags. ACS Chemical Neuroscience, acschemneuro.7b00521. 10.1021/acschemneuro.7b00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mikołajczyk A. and Złotkowska D. (2019) Subclinical lipopolysaccharide from Salmonella Enteritidis induces neuropeptide dysregulation in the spinal cord and the dorsal root ganglia. BMC Neuroscience, BioMed Central. 1–13. 10.1186/s12868-019-0502-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ishii XM, Hiller AJ, Pham L, Mcguire MJ, Iadecola C. and Wang G. (2019) Amyloid-Beta Modulates Low-Threshold Activated Voltage- Gated L-Type Calcium Channels of Arcuate Neuropeptide Y Neurons Leading to Calcium Dysregulation and Hypothalamic Dysfunction. The Journal of Neuroscience, 39, 8816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Khairuddin S, Aquili L, Chin B, Lok T, Hoo C, Hui K. et al. (2020) Neuroscience and Biobehavioral Reviews Dysregulation of the orexinergic system : A potential neuropeptide target in depression. Neuroscience and Biobehavioral Reviews, Elsevier. 118, 384–96. 10.1016/j.neubiorev.2020.07.040 [DOI] [PubMed] [Google Scholar]
- [19].Ye H, Wang J, Zhang Z, Jia C, Schmerberg C, Catherman AD et al. (2015) Defining the neuropeptidome of the spiny lobster Panulirus interruptus brain using a multidimensional mass spectrometry-based platform. Journal of Proteome Research, 14, 4776–91. 10.1021/acs.jproteome.5b00627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fricker LD (2016) Proteolytic processing of Neuropepties. Neuromethods, 114, 209–20. [Google Scholar]
- [21].Protein LS, Anand S, Samuel M, Ang C, Keerthikumar S. and Mathivanan S. (2017) Chapter 4 Label-Based and Label-Free Strategies for Protein Quantitation. Proteome Bioinformatics, Methods in Molecular Biology, p. 31–43. 10.1007/978-1-4939-6740-7 [DOI] [PubMed] [Google Scholar]
- [22].Delaney K. and Li L. (2019) Data Independent Acquisition Mass Spectrometry Method for Improved Neuropeptidomic Coverage in Crustacean Neural Tissue Extracts. Analytical Chemistry, 5150–8. 10.1021/acs.analchem.8b05734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cao Q, Yu Q, Liu Y, Chen Z. and Li L. (2020) Signature-Ion-Triggered Mass Spectrometry Approach Enabled Discovery of N- And O-Linked Glycosylated Neuropeptides in the Crustacean Nervous System. Journal of Proteome Research, 19, 634–43. 10.1021/acs.jproteome.9b00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jiang S, Liang Z, Hao L. and Li L. (2016) Investigation of signaling molecules and metabolites found in crustacean hemolymph via in vivo microdialysis using a multifaceted mass spectrometric platform. Electrophoresis, 37, 1031–8. 10.1002/elps.201500497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vu NQ, Buchberger AR, Johnson J. and Li L. (2021) Complementary neuropeptide detection in crustacean brain by mass spectrometry imaging using formalin and alternative aqueous tissue washes. Analytical and Bioanalytical Chemistry, Analytical and Bioanalytical Chemistry. 413, 2665–73. 10.1007/s00216-020-03073-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maes K, Van Liefferinge J, Viaene J, Van Schoors J, Van Wanseele Y, Béchade G. et al. (2014) Improved sensitivity of the nano ultra-high performance liquid chromatography-tandem mass spectrometric analysis of low-concentrated neuropeptides by reducing aspecific adsorption and optimizing the injection solvent. Journal of Chromatography A, 1360, 217–28. 10.1016/j.chroma.2014.07.086 [DOI] [PubMed] [Google Scholar]
- [27].De Haes W, Sinay E.Van, Detienne G, Temmerman L, Schoofs L. and Boonen K. (2015) Functional neuropeptidomics in invertebrates. BBA - Proteins and Proteomics, Elsevier B.V. 1854, 812–26. 10.1016/j.bbapap.2014.12.011 [DOI] [PubMed] [Google Scholar]
- [28].Southey BR, Lee JE, Zamdborg L, Atkins N, Mitchell JW, Li M. et al. (2014) Comparing Label-Free Quantitative Peptidomics Approaches to Characterize Diurnal Variation of Peptides in the Rat Suprachiasmatic Nucleus. Analytical Chemistry, 86, 443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Boonen K, Haes W. De Houtven, Van J, Verdonck R, Baggerman G, Valkenborg D. et al. (2018) Chapter 9 Quantitative Peptidomics with Isotopic and Isobaric Tags. Peptidomics: Methods and Strategies, Methods in Molecular Biology, p. 141–59. [DOI] [PubMed] [Google Scholar]
- [30].Lee JE (2016) Neuropeptidomics: Mass Spectrometry-Based Identification and Quantitation of Neuropeptides. Genomics and Informatics, 14, 12–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Buchberger A, Yu Q. and Li L. (2015) Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annual Review of Analytical Chemistry, 8, 485–509. 10.1146/annurev-anchem-071114-040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang M, You J, Bemis KG and Fitzpatrick DPG (2008) Chapter 10 Label-Free Mass Spectrometry-Based Protein Quantification Technologies in Protein Biomarker Discovery. Methods in Pharmacology and Toxicology: Biomarker Methods in Drug Discovery and Development, p. 211–30. [Google Scholar]
- [33].Fricker L. (2018) Chapter 8 Quantitative Peptidomics : General Considerations. Peptidomics: Methods and Strategies, Methods in Molecular Biology, p. 121–40. [DOI] [PubMed] [Google Scholar]
- [34].Wu C, Monroe ME, Xu Z, Slysz GW, Payne SH, Rodland KD et al. (2015) An Optimized Informatics Pipeline for Mass Spectrometry-Based Peptidomics. Journal of the American Society for Mass Spectrometry, 26, 2002–8. 10.1007/s13361-015-1169-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Välikangas T, Suomi T. and Elo LL (2018) A systematic evaluation of normalization methods in quantitative label-free proteomics. Briefings in Bioinformatics, 19, 1–11. 10.1093/bib/bbw095 **Provides thorough evaluation and detailed discussion about most common normalization methods used in LFQ.
- [36].Ye H, Wang J, Tian Z, Ma F, Dowell J, Bremer Q. et al. (2017) Quantitative Mass Spectrometry Reveals Food Intake-Induced Neuropeptide Level Changes in Rat Brain : Functional Assessment of Selected Neuropeptides as Feeding Regulators * □. Molecular and Cellular Proteomics, © 2017 ASBMB. Currently published by Elsevier Inc; originally published by American Society for Biochemistry and Molecular Biology. 16, 1922–37. 10.1074/mcp.RA117.000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Salem J. Ben Nkambeu B., Arvanitis DN and Beaudry F. (2018) Deciphering the Role of EGL-3 for Neuropeptides Processing in Caenorhabditis elegans Using High-Resolution Quadrupole–Orbitrap Mass Spectrometry. Neurochemical Research, Springer US. 43, 2121–31. 10.1007/s11064-018-2636-2 [DOI] [PubMed] [Google Scholar]
- [38].Saidi M, Kamali S. and Beaudry F. (2019) Neuropeptidomics: Comparison of parallel reaction monitoring and data-independent acquisition for the analysis of neuropeptides using high-resolution mass spectrometry. Biomedical Chromatography, 33, 1–11. 10.1002/bmc.4523 [DOI] [PubMed] [Google Scholar]
- [39].Hopkins K, Mukherjee S, Ponce D, Mangum J, Jacobson LH and Hoyer D. (2021) Development of a LC-ESI-MRM method for the absolute quantification of orexin A in the CSF of individual mice. Medicine in Drug Discovery, The Authors. 11, 100102. 10.1016/j.medidd.2021.100102 [DOI] [Google Scholar]
- [40].Fleites LA, Johnson R, Kruse AR, Nachman RJ, Hall DG, Maccoss M. et al. (2020) Peptidomics Approaches for the Identification of Bioactive Molecules from Diaphorina citri. Journal of Proteome Research, 19, 1392–408. 10.1021/acs.jproteome.9b00509 [DOI] [PubMed] [Google Scholar]
- [41].Bianco G, Battista FG, Buchicchio A, Amarena CG, Schmitt-Kopplin P. and Guerrieri A. (2015) Structural characterization of arginine vasopressin and lysine vasopressin by fourier-transform ion cyclotron resonance mass spectrometry and infrared multiphoton dissociation. European Journal of Mass Spectrometry, 21, 211–9. 10.1255/ejms.1339 [DOI] [PubMed] [Google Scholar]
- [42].Shinoda K, Tomita M. and Ishihama Y. (2010) emPAI Calc — for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics, 26, 576–7. 10.1093/bioinformatics/btp700 [DOI] [PubMed] [Google Scholar]
- [43].Ling XB, Lau K, Deshpande C, Park JL, Milojevic D, Macaubas C. et al. (2010) Urine Peptidomic and Targeted Plasma Protein Analyses in the Diagnosis and Monitoring of Systemic Juvenile Idiopathic Arthritis. Clinical Proteomics, 6, 175–93. 10.1007/s12014-010-9058-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Labas V, Grasseau I, Cahier K. and Gargaros A. (2014) Qualitative and quantitative peptidomic and proteomic approaches to phenotyping chicken semen. Journal of Proteomics, Elsevier B.V. 112, 313–35. 10.1016/j.jprot.2014.07.024 [DOI] [PubMed] [Google Scholar]
- [45]. Tashima AK and Fricker LD (2018) Quantitative Peptidomics with Five-plex Reductive Methylation labels. Journal of the American Society for Mass Spectrometry, Journal of The American Society for Mass Spectrometry. 29, 866–78. 10.1007/s13361-017-1852-3 *By using isotopic corrections, they were able to achieve robust 5-plex quantitation of peptides.
- [46].Hu M, Helfenbein K, Buchberger AR, DeLaney K, Liu Y. and Li L. (2021) Exploring the Sexual Dimorphism of Crustacean Neuropeptide Expression Using Callinectes sapidus as a Model Organism. Journal of Proteome Research,. 10.1021/acs.jproteome.1c00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen R, Xiao M, Buchberger A. and Li L. (2014) Quantitative neuropeptidomics study of the effects of temperature change in the crab cancer borealis. Journal of Proteome Research, 13, 5767–76. 10.1021/pr500742q [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wilson RE, Jaquins-Gerstl A. and Weber SG (2018) On-Column Dimethylation with Capillary Liquid Chromatography-Tandem Mass Spectrometry for Online Determination of Neuropeptides in Rat Brain Microdialysate. Analytical Chemistry, 90, 4561–8. 10.1021/acs.analchem.7b04965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Berezniuk I, Sironi JJ, Wardman J, Pasek RC, Berbari NF, Yoder BK et al. (2013) Quantitative Peptidomics of Purkinje Cell Degeneration Mice. PLoS ONE, 8, 1–12. 10.1371/journal.pone.0060981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gelman JS, Wardman J, Bhat VB, Gozzo FC and Fricker LD (2012) Chapter 31 Quantitative Peptidomics to Measure Neuropeptide Levels in Animal Models Relevant to Psychiatric Disorders. Psychiatric Disorders: Methods and Protocols, Methods in Molecular Biology, p. 487–503. 10.1007/978-1-61779-458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wardman J. and Fricker LD (2011) Chapter 17 Quantitative Peptidomics of Mice Lacking Peptide-Processing Enzymes. Proprotein Covertases, Methods in Molecular Biology, p. 307–23. 10.1007/978-1-61779-204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhang Xin, Hui Pan, Peng Bonnie, Steiner Donald F., Pintar John E., Fricker LD (2010) Neuropeptidomic analysis establishes a major role for prohormone covertase-2 in neuropeptide biosynthesis. Journal of Neurochemistry, 112, 1168–79. 10.1111/j.1471-4159.2009.06530.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Berezniuk Iryna, Rodriguiz Ramona M., Zee Michael L., Marcus David J., Pintar John, Morgan Daniel J., Wetsel William C., Fricker LD (2017) ProSAAS-derived peptides are regulated by cocaine and are required for sensitization to the locomotor effects of cocaine. Journal of Neurochemistry, 143, 268–81. 10.1111/jnc.14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Moulder R, Bhosale SD, Goodlett DR and Lahesmaa R. (2018) Analysis of the plasma proteome using iTRAQ and TMT-based Isobaric labeling. Mass Spectrometry Reviews, 37, 583–606. 10.1002/mas.21550 [DOI] [PubMed] [Google Scholar]
- [55].Li J, Vranken JG Van Vaites L.P., Schweppe DK, Huttlin EL, Etienne C. et al. (2020) TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nature Methods, Springer US. 17. 10.1038/s41592-020-0781-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Frost DC, Feng Y. and Li L. (2020) 21-plex DiLeu Isobaric Tags for High-Throughput Quantitative Proteomics. Analytical Chemistry, 92, 8228–34. 10.1021/acs.analchem.0c00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Liu R, Wei P, Keller C, Salvatore N, Shi Y, Li Z. et al. (2020) Integrated Label-Free and 10-Plex DiLeu Isobaric Tag Quantitative Methods for Pro fi ling Changes in the Mouse Hypothalamic Neuropeptidome and Proteome: Assessment of the Impact of the Gut Microbiome. Analytical Chemistry, 92, 14021–30. 10.1021/acs.analchem.0c02939 **Used multiple quantitative methods to enhance functional studies in a multi-omic experiment.
- [58].Andrews GL, Dean RA, Hawkridge AM and Muddiman DC (2011) Improving proteome coverage on a LTQ-orbitrap using design of experiments. Journal of the American Society for Mass Spectrometry, 22, 773–83. 10.1007/s13361-011-0075-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hecht ES, McCord JP and Muddiman DC (2015) Definitive Screening Design Optimization of Mass Spectrometry Parameters for Sensitive Comparison of Filter and Solid Phase Extraction Purified, INLIGHT Plasma N-Glycans. Analytical Chemistry, 87, 7305–12. 10.1021/acs.analchem.5b01609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Svensson M, Skold K, Nilsson A, Falth M, Nydahl Katarina Svenningsson, P. and Andren PE (2007) Neuropeptidomics : MS Applied to the Discovery of Novel Peptides from the Brain. Analytical Chemistry, 15–21. [DOI] [PubMed] [Google Scholar]
- [61].Gillet LC, Navarro P, Tate S, Ro H, Selevsek N, Reiter L. et al. (2012) Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Molecular & Cellular Proteomics, 11, 1–17. 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chapman JD, Goodlett DR and Masselon CD (2014) Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrometry Reviews, 33, 452–70. 10.1002/mas.21400 [DOI] [PubMed] [Google Scholar]
- [63].Panchaud A, Scherl A, Shaffer SA, Haller PD Von Kulasekara, H.D. Miller S.I.,. et al. (2009) Precursor Acquisition Independent From Ion Count: How to Dive Deeper into the Proteomics Ocean. Analytical Chemistry, 81, 6481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Krasny L. and Huang PH (2021) Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Molecular Omics, Royal Society of Chemistry. 17, 29–42. 10.1039/d0mo00072h [DOI] [PubMed] [Google Scholar]
- [65].Panchaud A, Jung S, Sha SA, Aitchison JD and Goodlett DR (2011) Faster, Quantitative, and Accurate Precursor Acquisition Independent From Ion Count. Analytical Chemistry, 83, 2250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Li KW, Gonzalez-lozano MA, Koopmans F. and Smit AB (2020) Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome. Frontiers in Molecular Neuroscience, 13, 1–8. 10.3389/fnmol.2020.564446 *A recent review demonstrating the usefulness of DIA analyses in studying neurophysiology.
- [67].Schmerberg CM, Liang Z. and Li L. (2015) Data-Independent MS/MS Quantification of Neuropeptides for Determination of Putative Feeding-Related Neurohormones in Microdialysate. ACS Chemical Neuroscience, 6, 174–80. 10.1021/cn500253u [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Xiao P, Zhang F, Wang X, Song D. and Li H. (2021) Analysis of B-type natriuretic peptide impurities using label-free data-independent acquisition mass spectrometry technology. Clinical Chemistry Labratory Medicine, 59, 217–26. 10.1515/cclm-2020-0012 [DOI] [PubMed] [Google Scholar]
- [69].Fernandez-Costa C, Martinez-Bartolome S, McClatchy DB, Saviola AJ, Yu N-K and Yates JR (2020) Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. Journal of Proteome Research, 19, 3153–61. 10.1021/acs.jproteome.0c00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tsou C, Tsai C, Teo GC, Chen Y. and Nesvizhskii AI (2016) Untargeted, spectral library-free analysis of data-independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics, 16, 2257–71. 10.1002/pmic.201500526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen C, Hou J. and Tanner JJ (2020) Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. International Journal of Molecular Sciences, 21, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang F, Ge W, Ruan G, Cai X. and Guo T. (2020) Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools : A Glimpse in 2020. Proteomics, 20, 1–12. 10.1002/pmic.201900276 [DOI] [PubMed] [Google Scholar]
- [73].Pino LK, Just SC, Maccoss MJ, Searle BC, Pino LK, Just SC et al. (2020) Acquiring and Analyzing Data Independent Acquisition Proteomics Experiments without Authors Acquiring and Analyzing Data Independent Acquisition Proteomics Experiments without. Molecular and Cellular Proteomics, © 2020 Pino et al. 19, 1088–103. 10.1074/mcp.P119.001913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Searle BC, Pino LK, Egertson JD, Ting YS, Maccoss MJ, Maclean BX et al. (2018) Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nature Communications, 9, 1–12. 10.1038/s41467-018-07454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].He B, Shi J, Wang X, Jiang H. and Zhu H. (2019) Label-free absolute protein quantification with data-independent acquisition. Journal of Proteomics, Elsevier. 200, 51–9. 10.1016/j.jprot.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Collins BC, Hunter CL, Liu Y, Schilling B, Rosenberger G, Bader SL et al. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature Communications, Springer US. 8, 1–11. 10.1038/s41467-017-00249-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Venable JD, Dong M, Wohlschlegel J, Dillin A. and Iii JRY (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nature Methods, 1, 1–7. 10.1038/NMETH705 [DOI] [PubMed] [Google Scholar]
- [78].Zhong X, Frost DC, Yu Q, Li M, Gu TJ and Li L. (2020) Mass Defect-Based DiLeu Tagging for Multiplexed Data-Independent Acquisition. Analytical Chemistry, 92, 11119–26. 10.1021/acs.analchem.0c01136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Di Y, Zhang Y, Zhang L, Tao T. and Lu H. (2017) MdFDIA: A Mass Defect Based Four-Plex Data-Independent Acquisition Strategy for Proteome Quantification. Analytical Chemistry, 89, 10248–55. 10.1021/acs.analchem.7b01635 [DOI] [PubMed] [Google Scholar]
- [80].Zhang S, Di Y, Yao J, Wang Y, Shu H, Yan G. et al. (2021) Mass defect-based carbonyl activated tags (mdCATs) for multiplex data-independent acquisition proteome quantification. Chemical Communications, 57, 737–40. 10.1039/d0cc06493a [DOI] [PubMed] [Google Scholar]
- [81].Tian X, De Vries MP, Permentier HP and Bischoff R. (2020) A Versatile Isobaric Tag Enables Proteome Quantification in Data-Dependent and Data-Independent Acquisition Modes. Analytical Chemistry, 92, 16149–57. 10.1021/acs.analchem.0c03858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ludwig C, Aebersold R, Gillet L, Rosenberger G, Amon S. and Collins BC (2018) Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Molecular Systems Biology, 14, 1–23. 10.15252/msb.20178126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bilbao A, Varesio E, Luban J. and Strambio-de-castillia C. (2015) Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics, 15, 964–80. 10.1002/pmic.201400323 [DOI] [PubMed] [Google Scholar]
- [84].Huang X, Liu M, Nold MJ, Tian C, Fu K, Zheng J. et al. (2011) Software for quantitative proteomic analysis using stable isotope labeling and data independent acquisition. Analytical Chemistry, 83, 6971–9. 10.1021/ac201555m [DOI] [PubMed] [Google Scholar]
- [85].Ye Z, Mao Y, Clausen H. and Vakhrushev SY (2019) Glyco-DIA: a method for quantitative O-glycoproteomics with in silico-boosted glycopeptide libraries. Nature Methods, Springer US. 16, 902–10. 10.1038/s41592-019-0504-x [DOI] [PubMed] [Google Scholar]
- [86].Madsen JA, Farutin V, Lin YY and Smith S. (2018) Data-independent oxonium ion profiling of multi-glycosylated biotherapeutics. MAbs, Taylor & Francis. 10, 968–78. 10.1080/19420862.2018.1494106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lin C, Krisp C, Packer NH and Molloy MP (2018) Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. Journal of Proteomics, 172, 68–75. [DOI] [PubMed] [Google Scholar]
- [88]. Hook V, Lietz CB, Podvin S, Cajka T. and Fiehn O. (2018) Diversity of Neuropeptide Cell-Cell Signaling Molecules Generated by Proteolytic Processing Revealed by Neuropeptidomics Mass Spectrometry. Journal of the American Society for Mass Spectrometry, Journal of The American Society for Mass Spectrometry. 29, 807–16. 10.1007/s13361-018-1914-1 *Discerned processing steps to demonstrate how different mature neuropeptides can originate from the same preprohormone.
- [89].Mast DH, Checco JW and Sweedler JV (2020) Differential post-translational amino acid isomerization found among neuropeptides in aplysia californica. ACS Chemical Biology, 15, 272–81. 10.1021/acschembio.9b00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cao L, Qu Y, Zhang Z, Wang Z, Prykova I. and Wu S. (2017) Intact Glycopeptide Characterizatoin Using Mass Spectrometry. Physiology & Behavior, 176, 139–48. 10.1586/14789450.2016.1172965.Intact28363838 [DOI] [Google Scholar]
- [91].Han B, Fang Y, Feng M, Hu H, Qi Y, Huo X. et al. (2015) Quantitative Neuropeptidome Analysis Reveals Neuropeptides Are Correlated with Social Behavior Regulation of the Honeybee Workers. Journal of Proteome Research, 14, 4382–93. 10.1021/acs.jproteome.5b00632 [DOI] [PubMed] [Google Scholar]
- [92].DeLaney K, Phetsanthad A. and Li L. (2020) Advances in High-Resolution Maldi Mass Spectrometry for Neurobiology. Mass Spectrometry Reviews,. 10.1002/mas.21661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Fricker LD, Lim J, Pan H. and Che FY (2006) Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrometry Reviews, 25, 327–44. 10.1002/mas.20079 [DOI] [PubMed] [Google Scholar]
- [94].Hsu JL and Chen SH (2016) Stable isotope dimethyl labelling for quantitative proteomics and beyond. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 374. 10.1098/rsta.2015.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ren RJ, Dammer EB, Wang G, Seyfried NT and Levey AI (2014) Proteomics of protein post-translational modifications implicated in neurodegeneration. Translational Neurodegeneration, 3, 1–13. 10.1186/2047-9158-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Li G, Delafield DG and Li L. (2020) Improved structural elucidation of peptide isomers and their receptors using advanced ion mobility-mass spectrometry. TrAC - Trends in Analytical Chemistry, 124. 10.1016/j.trac.2019.05.048 [DOI] [Google Scholar]
- [97].Wohlgemuth J, Karas M, Eichhorn T, Hendriks R. and Andrecht S. (2009) Quantitative site-specific analysis of protein glycosylation by LC-MS using different glycopeptide-enrichment strategies. Analytical Biochemistry, Elsevier Inc. 395, 178–88. 10.1016/j.ab.2009.08.023 [DOI] [PubMed] [Google Scholar]
- [98].Chen CC, Su WC, Huang BY, Chen YJ, Tai HC and Obena RP (2014) Interaction modes and approaches to glycopeptide and glycoprotein enrichment. Analyst, 139, 688–704. 10.1039/c3an01813j [DOI] [PubMed] [Google Scholar]
- [99].Zhang H, Li X. jun, Martin DB and Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnology, 21, 660–6. 10.1038/nbt827 [DOI] [PubMed] [Google Scholar]
- [100].Dalpathado DS and Desaire H. (2008) Glycopeptide analysis by mass spectrometry. Analyst, 133, 731–8. 10.1039/b713816d [DOI] [PubMed] [Google Scholar]
- [101].Madsen TD, Hansen LH, Hintze J, Ye Z, Jebari S, Andersen DB et al. (2020) An atlas of O-linked glycosylation on peptide hormones reveals diverse biological roles. Nature Communications, Springer US. 11. 10.1038/s41467-020-17473-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Flintegaard TV, Thygesen P, Rahbek-Nielsen H, Levery SB, Kristensen C, Clausen H. et al. (2010) N-glycosylation increases the circulatory half-life of human growth hormone. Endocrinology, 151, 5326–36. 10.1210/en.2010-0574 [DOI] [PubMed] [Google Scholar]
- [103].Polt R, Dhanasekaran M. and Keyari CM (2005) Glycosylated neuropeptides: A new vista for neuropsychopharmacology? Medicinal Research Reviews, 25, 557–85. 10.1002/med.20039 [DOI] [PubMed] [Google Scholar]
- [104].Moradi SV, Hussein WM, Varamini P, Simerska P. and Toth I. (2016) Glycosylation, an effective synthetic strategy to improve the bioavailability of therapeutic peptides. Chemical Science, Royal Society of Chemistry. 7, 2492–500. 10.1039/c5sc04392a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yu Q, Canales A, Glover MS, Das R, Shi X, Liu Y. et al. (2017) Targeted Mass Spectrometry Approach Enabled Discovery of O-Glycosylated Insulin and Related Signaling Peptides in Mouse and Human Pancreatic Islets. Analytical Chemistry, 89, 9184–91. 10.1021/acs.analchem.7b01926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Hansen LH, Madsen TD, Goth CK, Clausen H, Chen Y, Dzhoyashvili N. et al. (2019) Discovery of O-glycans on atrial natriuretic peptide (ANP) that affect both its proteolytic degradation and potency at its cognate receptor. Journal of Biological Chemistry, 294, 12567–78. 10.1074/jbc.RA119.008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mechref Y. (2012) Use of CID/ETD mass spectrometry to analyze glycopeptides. Current Protocols in Protein Science, 1. 10.1002/0471140864.ps1211s68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Hart-Smith G. and Raftery MJ (2012) Detection and characterization of low abundance glycopeptides via higher-energy c-trap dissociation and orbitrap mass analysis. Journal of the American Society for Mass Spectrometry, 23, 124–40. 10.1007/s13361-011-0273-y [DOI] [PubMed] [Google Scholar]
- [109].Khatri K, Pu Y, Klein JA, Wei J, Costello CE, Lin C. et al. (2018) Comparison of Collisional and Electron-Based Dissociation Modes for Middle-Down Analysis of Multiply Glycosylated Peptides. Journal of the American Society for Mass Spectrometry, 29, 1075–85. 10.1007/s13361-018-1909-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yu Q, Wang B, Chen Z, Urabe G, Glover MS, Shi X. et al. (2018) Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)- enabled Intact Glycopeptide/Glycoproteome Characterization. Physiology & Behavior, 176, 139–48. 10.1007/s13361-017-1701-4.Electron-Transfer/Higher-Energy [DOI] [Google Scholar]
- [111].Singh C, Zampronio CG, Creese AJ and Cooper HJ (2012) Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. Journal of Proteome Research, 11, 4517–25. 10.1021/pr300257c [DOI] [PubMed] [Google Scholar]
- [112].Yu Q, Shi X, Feng Y, Kent KC and Li L. (2016) Improving Data Quality and Preserving HCD-Generated Reporter Ions with EThcD for Isobaric Tag-based Quantitative Proteomics and Proteome-wide PTM Studies. Physiology & Behavior, 176, 100–106. 10.1016/j.aca.2017.03.003.Improving [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113]. Riley NM, Malaker SA, Driessen MD and Bertozzi CR (2020) Optimal Dissociation Methods Differ for N- A nd O-Glycopeptides. Journal of Proteome Research, 19, 3286–301. 10.1021/acs.jproteome.0c00218 **Discusses the importance of fragmentation and dissociation methods for the analysis of glycopeptides and how these differ depending on the glycan/glycosylation site of interest.
- [114].Zhu R, Song E, Hussein A. and Kobeissy FH (2017) Chapter 9: Glycoproteins Enrichment and LC-MS/MS Glycoproteomics in Central Nervous System Applications. Neuroproteomics: Methods and Protocols, p. 213–27. 10.1007/978-1-4939-6952-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Delafield DG and Li L. (2021) Recent advances in analytical approaches for glycan and glycopeptide quantitation. Molecular and Cellular Proteomics, 20, 0–21. 10.1074/MCP.R120.002095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Tobias F. and Hummon AB (2020) Considerations for MALDI-Based Quantitative Mass Spectrometry Imaging Studies. Journal of Proteome Research, 19, 3620–30. 10.1021/acs.jproteome.0c00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Delaney K, Hu M, Hellenbrand T, Dickinson PS and Nusbaum MP (2021) Mass Spectrometry Quantification, Localization, and Discovery of Feeding-Related Neuropeptides in Cancer borealis. ACS Chemical Neuroscience, 12, 782–98. 10.1021/acschemneuro.1c00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hulme H, Fridjonsdottir E, Gunnarsdottir H, Vallianatou T, Zhang X, Wadensten H. et al. (2020) Simultaneous mass spectrometry imaging of multiple neuropeptides in the brain and alterations induced by experimental parkinsonism and L-DOPA therapy. Neurobiology of Disease, Elsevier. 137, 104738. 10.1016/j.nbd.2020.104738 [DOI] [PubMed] [Google Scholar]
- [119].Pratavieira M, Ribeiro A, Esteves FG, Sato KU, Malaspina O. and Palma MS (2018) MALDI Imaging Analysis of Neuropeptides in Africanized Honeybee (Apis mellifera) Brain: Effect of Aggressiveness. Journal of Proteome Research, 17, 2358–69. 10.1021/acs.jproteome.8b00098 [DOI] [PubMed] [Google Scholar]
- [120].Taylor AJ, Dexter A. and Bunch J. (2018) Exploring Ion Suppression in Mass Spectrometry Imaging of a Heterogeneous Tissue. Analytical Chemistry, 90, 5637–45. 10.1021/acs.analchem.7b05005 [DOI] [PubMed] [Google Scholar]
- [121].Cobice DF, Livingstone DEW, Mackay CL, Goodwin RJA, Smith LB, Walker BR et al. (2016) Spatial Localization and Quantitation of Androgens in Mouse Testis by Mass Spectrometry Imaging. Analytical Chemistry, 88, 10362–7. 10.1021/acs.analchem.6b02242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Källback P, Shariatgorji M, Nilsson A. and Andrén PE (2012) Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. Journal of Proteomics, Elsevier B.V. 75, 4941–51. 10.1016/j.jprot.2012.07.034 [DOI] [PubMed] [Google Scholar]
- [123].Kallback P, Vallianatou T, Nilsson A, Shariatgorji R, Schintu N, Pereira M. et al. (2020) Cross-validated Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging Quantitation Protocol for a Pharmaceutical Drug and Its Drug-Target Effects in the Brain Using Time-of-Flight and Fourier Transform Ion Cyclotron Resonance Analyzers. Analytical Chemistry, 92, 14676–84. 10.1021/acs.analchem.0c03203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Swales JG, Strittmatter N, Tucker JW, Clench MR, Webborn PJH and Goodwin RJA (2016) Spatial Quantitation of Drugs in tissues using Liquid Extraction Surface Analysis Mass Spectrometry Imaging. Scientific Reports, Nature Publishing Group. 6, 1–9. 10.1038/srep37648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhao C. and Cai Z. (2020) Three‐dimensional quantitative mass spectrometry imaging in complex system: From subcellular to whole organism. Mass Spectrometry Reviews, 1–19. 10.1002/mas.21674 [DOI] [PubMed] [Google Scholar]
- [126].Zemaitis KJ, Izydorczak AM, Thompson AC and Wood TD (2021) Streamlined Multimodal DESI and MALDI Mass Spectrometry Imaging on a Singular Dual-Source FT-ICR Mass Spectrometer. Metabolites, 11, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Theron L, Centeno D, Coudy-Gandilhon C, Pujos-Guillot E, Astruc T, Rémond D. et al. (2016) A Proof of Concept to Bridge the Gap between Mass Spectrometry Imaging, Protein Identification and Relative Quantitation: MSI~LC-MS/MS-LF. Proteomes, 4, 1–16. 10.3390/proteomes4040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Dewez F, Pauw E. De, Heeren RMA and Balluff B. (2021) Multilabel Per-Pixel Quantitation in Mass Spectrometry Imaging. Analytical Chemistryl, 93, 1393–400. 10.1021/acs.analchem.0c03186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Shrivas K, Hayasaka T, Sugiura Y. and Setou M. (2011) Method for Simultaneous Imaging of Endogenous Low Molecular Weight Metabolites in Mouse Brain Using TiO2 Nanoparticles in Nanoparticle-Assisted Laser Desorption/Ionization-Imaging Mass Spectrometry. Analytical Chemistry, 83, 7283–9. [DOI] [PubMed] [Google Scholar]
- [130].Chu H, Unnikrishnan B, Anand A. and Mao J. (2018) Nanoparticle-based laser desorption/ionization mass spectrometric analysis of drugs and metabolites. Journal of Food and Drug Analysis, Elsevier Ltd. 26, 1215–28. 10.1016/j.jfda.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Mclaughlin N, Bielinski TM, Tressler CM, Barton E, Glunde K. and Stumpo KA (2020) Pneumatically Sprayed Gold Nanoparticles for Mass Spectrometry Imaging of Neurotransmitters. Journal of The American Society for Mass Spectrometry, 31, 2452–61. 10.1021/jasms.0c00156 [DOI] [PubMed] [Google Scholar]
- [132].Lolait SJ, Roper JA, Hazell GGJ, Li Y, Thomson FJ and Carroll AMO (2016) Chapter 10: Neuropeptide Receptors. Molecular Neuroendocrinology: From Genome to Physiology, p. 195–215. [Google Scholar]
- [133].Wojnicz A, Avendaño-Ortiz J, de Pascual R, Ruiz-Pascual L, García AG and Ruiz-Nuño A. (2016) Simultaneous monitoring of monoamines, amino acids, nucleotides and neuropeptides by liquid chromatography-tandem mass spectrometry and its application to neurosecretion in bovine chromaffin cells. Journal of Mass Spectrometry, 51, 651–64. 10.1002/jms.3794 [DOI] [PubMed] [Google Scholar]
- [134].Gantait AM, Bataineh YA, Surchi HS, Gantait A, Rani GT, Paul P. et al. (2020) Chapter 16: Neuropeptides and Neurotransmission. Frontiers in Pharmacology of Neurotransmitters, p. 553–77. [Google Scholar]
- [135].Chen D, Zhao X, Sui Z, Niu H, Chen L, Hu C. et al. (2020) A multi-omics investigation of the molecular characteristics and classification of six metabolic syndrome relevant diseases. Theranostics, 10, 2029–46. 10.7150/thno.41106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Gutierrez DB, Gant-Branum RL, Romer CE, Farrow MA, Allen JL, Dahal N. et al. (2018) An Integrated, High-Throughput Strategy for Multiomic Systems Level Analysis. Journal of Proteome Research, 17, 3396–408. 10.1021/acs.jproteome.8b00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Keller C, Wei P, Wancewicz B, Cross TWL, Rey FE and Li L. (2021) Extraction optimization for combined metabolomics, peptidomics, and proteomics analysis of gut microbiota samples. Journal of Mass Spectrometry, 56. 10.1002/jms.4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Chen B, OuYang C, Tian Z, Xu M. and Li L. (2018) A high resolution atmospheric pressure matrix-assisted laser desorption/ionization-quadrupole-orbitrap MS platform enables in situ analysis of biomolecules by multi-mode ionization and acquisition. Analytica Chimica Acta, Elsevier Ltd. 1007, 16–25. 10.1016/j.aca.2017.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Johnsen E, Leknes S, Wilson SR and Lundanes E. (2015) Liquid chromatography-mass spectrometry platform for both small neurotransmitters and neuropeptides in blood, with automatic and robust solid phase extraction. Scientific Reports, 5, 1–8. 10.1038/srep09308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zestos AG and Kennedy RT (2017) Microdialysis Coupled with LC-MS/MS for In Vivo Neurochemical Monitoring. AAPS Journal, The AAPS Journal. 19, 1284–93. 10.1208/s12248-017-0114-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Oliveros A, Starski P, Lindberg D, Choi S, Heppelmann CJ and Dasari S. (2017) Label-Free Neuroproteomics of the Hippocampal-Accumbal Circuit Reveals Deficits in Neurotransmitter and Neuropeptide Signaling in Mice Lacking Ethanol-Sensitive Adenosine Transporter. Journal of Proteome Research, 16, 1445–59. 10.1021/acs.jproteome.6b00830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Li W, Petruzziello F, Zhao N, Zhao H, Ye X, Zhang X. et al. (2017) Separation and identification of mouse brain tissue microproteins using top-down method with high resolution nanocapillary liquid chromatography mass spectrometry. Proteomics, 17, 1–5. 10.1002/pmic.201600419 [DOI] [PubMed] [Google Scholar]
- [143].Li L. and Sweedler JV (2008) Peptides in the Brain: Mass Spectrometry–Based Measurement Approaches and Challenges. Annual Review of Analytical Chemistry, 1, 451–83. 10.1146/annurev.anchem.1.031207.113053 [DOI] [PubMed] [Google Scholar]
- [144].Shetgaonkar GG and Kumar L. (2020) Chemistry of Neurochemicals : Psychopharmaceuticals and Neuropeptides. Principles of Neurochemistry, p. 41–70. [Google Scholar]
- [145].Li C, Wu X, Liu S, Zhao Y, Zhu J, Liu K. et al. (2019) Roles of Neuropeptide Y in Neurodegenerative and Neuroimmune Diseases. Frontiers in Neuroscience, 13, 1–11. 10.3389/fnins.2019.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Nillni EA (2018) Chapter 2: Neuropeptides Controlling Our Behavior. Energy Balance, Neuropeptide Hormones, and Neuroendocrine Function, p. 29–54. 10.1007/978-3-319-89506-2 [DOI] [Google Scholar]
- [147].Cheng Y, Tang X, Li Y, Zhao D. and Cao Q. (2019) Depression-Induced Neuropeptide Y Secretion Promotes Prostate Cancer Growth by Recruiting Myeloid Cells. Clinical Cancer Research, 25, 2621–32. 10.1158/1078-0432.CCR-18-2912 [DOI] [PubMed] [Google Scholar]
- [148].Urban JH (2020) Chapter 7: Neuropeptide Y and amygdala circuitry : Modulation of stress-related behavior [Internet]. 1st ed. Handb. Amygdala Struct. Funct. Elsevier B.V. 10.1016/B978-0-12-815134-1.00007-6 [DOI] [Google Scholar]
- [149].Salem J. Ben Nkambeu B. and Beaudry F. (2018) Characterization of neuropeptide K processing in rat spinal cord S9 fractions using high-resolution quadrupole–Orbitrap mass spectrometry. Biomedical Chromatography, 32, 1–9. 10.1002/bmc.4204 [DOI] [PubMed] [Google Scholar]
- [150].DeLaney KB and Amanda; Lingjun L. (2018) Chapter 17: Identification, Quantitation, and Imaging of the Crustacean Peptidome. Methods in Molecular Biology, p. 247–69. 10.1007/978-1-4939-7537-2_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Husson SJ, Mertens I, Janssen T, Lindemans M. and Schoofs L. (2007) Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Progress in Neurobiology, 82, 33–55. 10.1016/j.pneurobio.2007.01.006 [DOI] [PubMed] [Google Scholar]
- [152].Perry DJ, Wasserfall CH, Oram RA, Williams MD, Muir AB, Haller MJ et al. (2018) Application of a Genetic Risk Score to Racially Diverse Type 1 Diabetes Populations Demonstrates the Need for Diversity in Risk-Modeling. Scientific Reports, Springer US. 8, 1–13. 10.1038/s41598-018-22574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Guo W, Zhou M, Qiu J, Lin Y, Chen X, Huang S. et al. (2019) Association of LAG3 genetic variation with an increased risk of PD in Chinese female population. Journal of Neuroinflammation, Journal of Neuroinflammation. 16, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Philibert R. and Glatt SJ (2017) Optimizing the chances of success in the search for epigenetic biomarkers: Embracing genetic variation. American Journal of Medical Genetics Part B, 174B, 589–94. 10.1002/ajmg.b.32569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Devi R, Jie L, Yan T, Pei P, Hassan L, Ismail R. et al. (2021) Human tuberculosis and Mycobacterium tuberculosis complex: A review on genetic diversity, pathogenesis and omics approaches in host biomarkers discovery. Microbiological Research, Elsevier GmbH. 246. 10.1016/j.micres.2020.126674 [DOI] [PubMed] [Google Scholar]
- [156].Zhang F. and Finkelstein J. (2019) Inconsistency in race and ethnic classification in pharmacogenetics studies and its potential clinical implications. Pharmacogenomics and Personalized Medicine, 12, 107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Brown AF, Liang L, Vassar SD, Escarce JJ, Merkin SS, Cheng E. et al. (2018) Trends in Racial/Ethnic and Nativity Disparities in Cardiovascular Health Among Adults Without Prevalent Cardiovascular Disease in the United States, 1988 to 2014. Annals of Internal Medicine, 168, 541–9. 10.7326/M17-0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Babulal GM, Quiroz YT, Albensi BC, Arenaza-urquijo E, Astell AJ, Babiloni C. et al. (2019) Perspectives on ethnic and racial disparities in Alzheimer’s disease and related dementias: Update and areas of immediate need. Alzheimer’s & Dementia, 15, 292–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Landry BLG, Ali N, Williams DR, Rehm HL and Bonham VL (2018) Lack Of Diversity In Genomic Databases Is A Barrier To Translating Precision Medicine Research Into Practice. Health Affairs, 37, 780–5. [DOI] [PubMed] [Google Scholar]
- [160].Frost DC, Greer T. and Li L. (2015) High-resolution enabled 12-plex DiLeu isobaric tags for quantitative proteomics. Analytical Chemistry, 87, 1646–54. 10.1021/ac503276z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Greer T, Lietz CB, Xiang F. and Li L. (2014) Novel isotopic N,N-Dimethyl Leucine (iDiLeu) reagents enable absolute quantification of peptides and proteins using a standard curve approach. Journal of the American Society for Mass Spectrometry, 26, 107–19. 10.1007/s13361-014-1012-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Genangeli M, Heijens AMM, Rustichelli A, Schuit ND, Vittoria M, Di M. et al. (2019) MALDI-Mass Spectrometry Imaging to Investigate Lipid and Bile Acid Modifications Caused by Lentil Extract Used as a Potential Hypocholesterolemic Treatment. Journal of the American Society for Mass Spectrometry, Journal of The American Society for Mass Spectrometry. 30, 2041–50. 10.1007/s13361-019-02265-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Lombard-banek C, Yu Z, Swiercz AP, Marvar PJ and Nemes P. (2019) A microanalytical capillary electrophoresis mass spectrometry assay for quantifying angiotensin peptides in the brain. Analytical and Bioanalytical Chemistry, Analytical and Bioanalytical Chemistry. 411, 4661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Mast DH, Liao H-W, Romanova EV and Sweedler JV (2021) Analysis of Peptide Stereochemistry in Single Cells by Capillary Electrophoresis–Trapped Ion Mobility Spectrometry Mass Spectrometry. Analytical Chemistry,. 10.1021/acs.analchem.1c00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Li G, Delafield DG and Li L. (2020) Trends in Analytical Chemistry Improved structural elucidation of peptide isomers and their receptors using advanced ion mobility-mass spectrometry. Trends in Analytical Chemistry, 124. 10.1016/j.trac.2019.05.048 [DOI] [Google Scholar]