In the accompanying comment1, Kim et al. conclude that somatic gene recombination (SGR) and amyloid precursor protein (APP) genomic complementary DNAs (gencDNAs) in the brain are contamination artefacts and do not naturally exist. We disagree, and here we address the three types of analyses used by Kim et al. to reach their conclusions: informatic contaminant identification, plasmid PCR, and single-cell sequencing. Additionally, Kim et al. requested “reads supporting novel APP insertion breakpoints,” and we now provide ten different examples that support APP gencDNA insertion within eight chromosomes beyond wild-type APP on chromosome 21 from patients with Alzheimer’s disease. If SGR exists, as experimentally supported here and previously2,3, contamination scenarios become moot.
Our informatic analyses of data generated by an independent laboratory (Park et al.)4 complement, and are entirely consistent with, what Lee et al.2 presented via nine distinct lines of evidence, in addition to three from a prior publication3. Plasmid contamination was identified in a single pull-down dataset after publication of Lee et al.2; however, subsequent analyses did not alter any of our conclusions, including those of our prior publications3,5, and plasmid contamination-free replication of this approach by ourselves and others supported the original conclusions. Novel retro-insertion sites, alterations of APP gencDNA number and form within cell types from the same brain, and pathogenic SNVs that occur only in samples from patients with AD, all support the existence of APP gencDNAs produced by SGR.
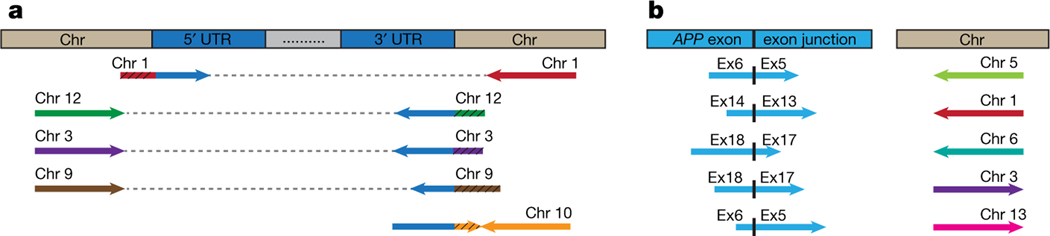
One predicted outcome of SGR is the generation of novel retro-insertion sites distinct from the wild-type locus, as we demonstrated using DNA in situ hybridization (DISH; Fig. 2n in Lee et al.). Analyses of independently published data sets4 produced by whole-exome pull-down of DNA from laser-captured human hippocampus or blood revealed ten different APP insertion sites within eight different chromosomes (Fig. 1, Supplementary Table 1). We identified clipped reads spanning APP untranslated regions (UTRs) and new genomic insertion sites on chromosomes 1, 3, 9, 10, and 12 (Fig. 1a; wild-type APP is located on chromosome 21). The corresponding paired-end reads mapped to the same inserted chromosome. We also identified reads spanning APP exon–exon junctions of gencDNAs that had mate-reads mapping to other genomic sites on chromosomes 1, 3, 5, 6, and 13 (Fig. 1b). We are unaware of contamination sources that could produce these results that are entirely consistent with our DISH data showing APP gencDNA locations distinct from wild-type APP. These new APP gencDNA insertion sites strongly support the natural occurrence of APP gencDNAs.
Fig. 1 |. Identification of novel APP insertion sites in the human genome.
a, Clipped reads spanning APP UTRs and novel chromosomal insertion sites were identified. The paired mate-reads of the clipped reads (black hatching) uniquely mapped to the same chromosomes. b, Discordant read-pairs were identified where one read spanned an APP exon–exon junction and the corresponding mate-read mapped to a novel chromosome. Each chromosome has a unique colour. Arrowhead direction represents the read orientation after mapping to the human reference genome. Arrows oriented in the same direction support sequence inversions. See detailed sequence and alignment information in Supplementary Table 1.
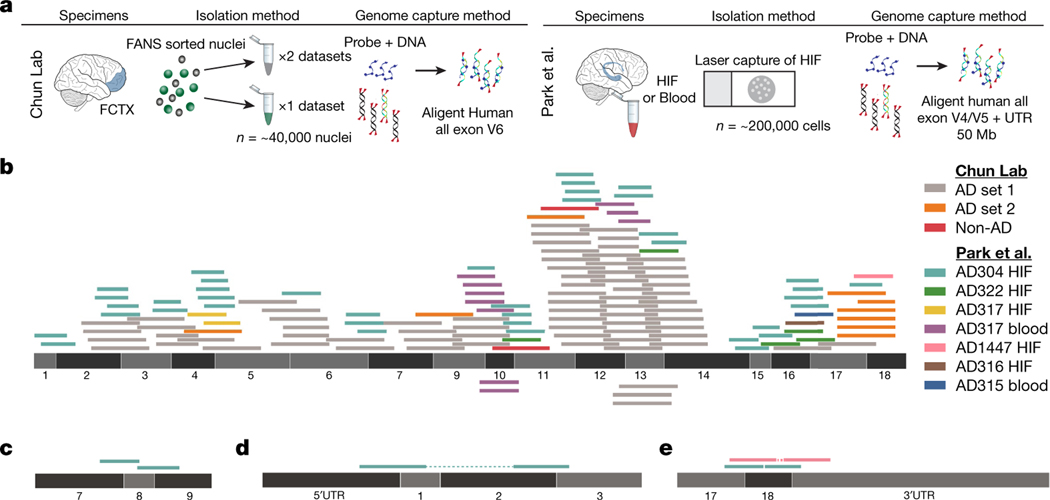
An APP plasmid contaminant (pGEM-T Easy APP) was found in our single pull-down dataset; however, we could not definitively determine which APP exon–exon reads resulted from gencDNAs as opposed to plasmid contamination, especially in view of the 11 other distinct and uncontaminated approaches that had independently supported and/or identified APP gencDNAs. Three other pull-down datasets from our laboratory were informatically analysed and found to contain APP gencDNA reads while being free from APP plasmid contamination by both VecScreen6 and subsequent use of the Vecuum script7 (Fig. 2a, b). Possible external source contamination noted by Kim et al. in two of three data sets could not definitively account for all APP exon–exon junctions.
Fig. 2 |. Identification of APP gencDNA sequences in ten new whole-exome pull-down datasets from two independent laboratories.
a, Method schematic depicting the standard protocol for whole-exome pull-downs and highlighted methodological differences between the independent laboratories (our lab and Park et al.4). b, APP-751 sequence with non-duplicate gencDNA reads from the ten new datasets; colour key indicates the source reads for all panels. c, Reads that map to junctions between APP exons 7, 8, and 9 that are absent from APP-751. d, e, Paired reads that represent a DNA fragment containing both an exon–exon junction and an APP 3′ or 5′ UTR.
The recent availability of independently generated datasets derived from patients with AD4 provided a test for the independent reproducibility of APP gencDNA identification. Five brain and two blood samples from individuals with sporadic AD (SAD) contained APP gencDNA sequences and were shown to be plasmid-free by Vecuum7 screening (Fig. 2a–e). In addition to exon–exon junction reads and novel insertion sites, we also identified APP UTR sequences paired with reads containing APP gencDNA exon–exon junctions (Fig. 2d, e). This may be explained by a key experimental design factor: the pull-down probes used by Park et al. contain sequences corresponding to the 5′ and 3′ UTRs of APP.
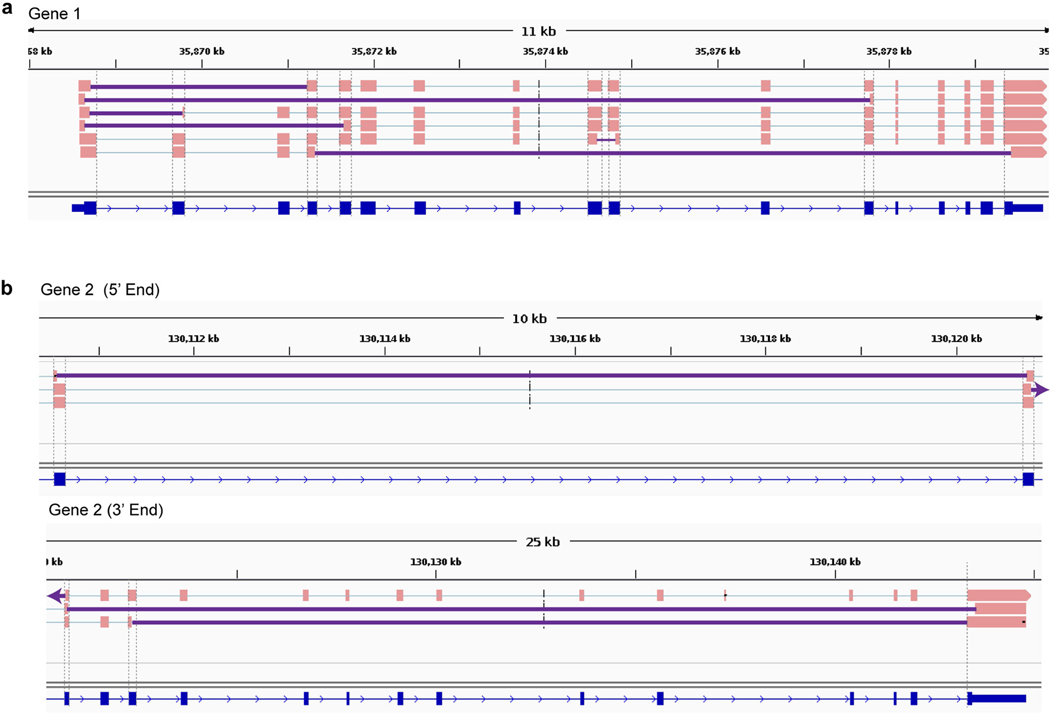
In addition to APP plasmid and amplicon contaminants, Kim et al. invoked genome-wide mouse and human mRNA contamination in the Park et al. data set. We cannot address conditions in the Park et al. laboratory but note that it is completely independent of our own. Kim et al. explain this by implicating the generation of DNA from mRNA, which requires reverse transcriptase activity. The Agilent SureSelect pull-down used by Park et al. and in our experiments do not use reverse transcriptase (Fig. 2a and Supplementary Methods), and we are unaware of any mechanism that would generate DNA from RNA in the absence of reverse transcriptase activity under the conditions used. An alternative explanation is the existence of gencDNAs that affect other genes, as we previously detected in non-APP intra-exonic junctions (IEJs) found in commercial cDNA Iso-Seq data sets (Extended Data Fig. 1). Additional validation would be required for new genes, but we note that an average of 450 Mb of extra DNA exists within cortical neurons from individuals with AD3 that could accommodate new gencDNA sequences. Kim et al. further invoked genome-wide mouse and human mRNA contamination in the Park et al. data set to account for APP gencDNAs, but this explanation conflicts with the available data. Mouse-specific single nucleotide polymorphisms (SNPs) in the Park et al. data set cannot account for all APP gencDNA-supporting reads: five of seven APP exon–exon junction sequences do not contain putative mouse-specific SNPs at the specific region reported by Kim et al. (Fig. 3; Kim et al. Fig. 2d). Most critically, the novel APP gencDNA insertion sites identified here cannot be explained by genome-wide mRNA contamination.
Fig. 3 |. Five APP gencDNA-supporting reads that span exon–exon junctions and do not contain mouse-specific SNPs.
APP gencDNA reads were identified that span the APP exon10–exon11 junction from the Park et al. datasets4. The reference sequences of human and mouse exons are indicated and the positions at which the nucleotides differ are highlighted. Five of the seven exon–exon junction-spanning reads do not contain mouse-specific SNPs.
Kim et al. used PCR of APP splice variant plasmids, which generated sequences containing IEJs. However, there are multiple discrepancies between this approach and our biological IEJs and gencDNAs. 1) The experimental conditions beyond our primer sequences were different: Kim et al. used twice the concentration of primers and more than one million times more template (250 pg APP plasmid is 4.6 × 107 copies versus about 40 gencDNA copies in our PCR of 20 nuclei; based on Lee et al.2 Fig. 5: DISH 16/17 averaged about 1.8 copies per SAD nucleus). 2) Both gencDNA and IEJ sequences can be detected with as few as 30 cycles of PCR, as we used in single molecule real-time sequencing (SMRT-seq) (Lee et al.2 Fig. 3) versus 40 cycles used by Kim et al. 3) The agarose gels in Kim et al. are uniformly and unambiguously dominated by a vastly over-amplified about 2-kb band (Kim et al. Fig. 1c and Extended Data Fig. 3a) that is never seen in human neurons despite our routine identification of myriad smaller bands (compare with Lee et al.2 Fig. 2b). We did observe an over-amplified about 2-kb band in our purposeful plasmid transfection experiments, which also used PCR; however, the formation of gencDNA and IEJs was comparatively limited, of sequences distinct from brain and critically, required both reverse transcriptase activity and DNA strand breakage (Lee et al.2, Fig. 4). 4) Finally, only 45 unique IEJs from the brains of individuals with AD and 20 from the brains of healthy controls were identified (Lee et al.2 Fig. 3 with some overlap, fewer than 65 total) compared to the 12,426 identified by Kim et al. (an approximately 200-fold increase over biological IEJs; Kim et al. Supplementary Table 1). We wish to note that microhomology regions within APP exons are intrinsic to the APP DNA sequence and that microhomology-mediated repair mechanisms involve DNA polymerases8,9. The PCR results of Kim et al. differ from our biological data but might inadvertently support the endogenous formation of at least some IEJs within DNA rather than requiring RNA.
Despite these differences between the non-biological plasmid PCR data generated by Kim et al. and our data, Kim et al. conclude that IEJs from our original study2 might have originated from contaminants. To eliminate this possibility, Lee et al.2 presented four lines of evidence for APP gencDNAs containing IEJs that are independent of APP PCR: two different commercially produced cDNA SMRT-seq libraries, DISH, and RNA in situ hybridization (RISH). The SMRT-seq libraries revealed IEJs within APP (Lee et al.2 Extended Data Fig. 1e) as well as other genes (Extended Data Fig. 1), which cannot be attributed to plasmid contamination or PCR amplification. The DISH and RISH results support the existence of APP gencDNAs and IEJs (see Supplementary Discussion and Lee et al.2 Fig. 2, Extended Data Figs. 1, 2) by using custom-designed and validated commercial probe technology (Advanced Cell Diagnostics, ACD), which was independently shown to detect exon–exon junctions10 and single-nucleotide mutations11. Thus, gencDNAs and IEJs can be detected in the absence of targeted PCR. Notably, the contamination proposed by Kim et al. cannot account for the marked change in the number and forms of APP gencDNAs that occurs with disease state. The change is also apparent when comparing cell types; signals are vastly more prevalent in neurons than in non-neuronal cells from the same brains of individuals with SAD when the samples are processed at the same time by DISH (Lee et al.2 Fig. 5). Independent peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) and dual-point-paint experiments from our previous work further support APP gencDNAs3 (Table 1). Critically, SMRT-seq identified 11 single-nucleotide variations that are considered pathogenic in familial AD and that were present only in our samples from individuals with SAD; none of them exist as plasmids in our laboratory.
Table 1 |.
Summary of targeted and non-targeted APP PCR methods and lines of evidence that support APP gencDNAs and IEJs
| Method | Targeted APP PCR | Support for the existence of IEJs and gencDNAs | Reference | |
|---|---|---|---|---|
| Approaches without targeted APP PCR | ||||
| 1 | RISH on IEJ 3/16 | None | IEJ 3/16 RNA signal is present in human SAD brain tissue | Lee et al.2 |
| 2 | Whole-transcriptome SMRT-seq | None | An independent commercial source identified IEJs in APP and other | Public dataseta, genes Lee et al.2this Reply |
| 3 | Targeted RNA SMRT-seq | None | RNA pull-down that identified APP IEJs | Public dataseta, Lee et al.2 |
| 4 | DISH of gencDNAs | None | IEJ 3/16 and exon–exon junction 16/17 showed increases in neurons compared to non-neurons from the same brain from an individual with SAD and to non-diseased neurons; J20 mice containing the APP transgene under a PDGF-β-promoter showed increased number and size of signal compared to non-neurons and wild-type mice | Lee et al.2 |
| 5 | Dual point-paint FISH | None | Identified APP CNVs of variable puncta size that were not always associated with Chr21 | Bushman et al.3 |
| 6 | PNA-FISH | None | APP exon copy number increases show variable signal size and shape with semiquantitative exonic probes | Bushman et al.3 |
| 7 | Agilent SureSelect targeted pull-down | None | Identified APP gencDNAs in brains from individuals with SAD; contains plasmid sequence contamination | Lee et al.2,this Reply |
| New #7 | Agilent all-exon pull-down | None | All-exon pull-downs, with no plasmid contamination by both Vecscreen and Vecuum, contain APP gencDNA sequences and evidence of gencDNA UTRs and novel insertion sites | Park et al.4, this Reply |
| Approaches with targeted APP PCR | ||||
| 8 | RT–PCR and Sanger sequencing | Oligo-dT primed and targeted APP primers | Novel APP RNA variants with IEJs; predominantly in neurons from individuals with SAD | Lee et al.2 |
| 9 | Genomic DNA PCR and Sanger sequencing | Yes | Identified APP gencDNAs with IEJs; predominantly in neurons from individuals with SAD | Lee et al.2 |
| 10 | Genomic DNA PCR and SMRT-seq | Yes | IEJ/gencDNAs were more prevalent in number and form in neurons from individuals with SAD compared to non-diseased neurons; identified 11 pathogenic SNVs that were present only in SAD samples | Lee et al.2 |
| 11 | APP-751 overexpression in CHO cells | Yes | IEJ and gencDNA formation required DNA strand breakage and reverse transcriptase | Lee et al.2 |
| 12 | Single-cell qPCR | Yes; individual exon | Intragenic exon 14 single-cell qPCR showed copy number increases in prefrontal cortical neurons over cerebellar neurons from the same brain of an individual with SAD | Bushman et al.3 |
CNV, copy number variation.
The Alzheimer brain Iso-Seq dataset was generated by Pacific Biosciences, Menlo Park, California. Additional sequencing information and analysis is provided at https://downloads.pacbcloud.com/public/dataset/Alzheimer_IsoSeq_2016/.
Kim et al. compared APP gencDNA copy number estimates from pull-down sequencing and DISH. However, a direct comparison is not possible since the two methodologies are fundamentally different. For example, pull-downs use solution hybridization on isolated DNA, whereas DISH uses solid-phase hybridization on fixed and sorted single nuclei. Moreover, the sequences targeted are not the same. Pull-down probes target wild-type sequences for endogenous and gencDNA loci, resulting in pull-down competition. By contrast, DISH probes target only gencDNA sequences to provide greater sensitivity. Competition by wild-type loci reduces the efficiency of capture, which is underscored by 32% to 40% of nuclei that do not contain gencDNAs and would contribute only wild-type sequences (Lee et al., Fig. 5c, f). Moreover, a majority of gencDNA positive nuclei (62% to 73%) showed two or fewer signals (Lee et al., Fig. 5c, f) which reduced the relative representation of gencDNA loci. As IEJs do not contain the full exon sequence, there is inefficient hybridization and a lack of sequence capture and detection. This limitation is overcome by SMRT-seq (Extended Data Fig. 1 and Lee et al., Extended Data Fig. 1e). Lastly, multiple other protocol variations exist, including tissue preparation, fixation, and hybridization conditions, which explain the hypothesized discrepancies.
Kim et al.’s third type of analysis yielded a negative result via interrogation of their own single-cell whole-genome sequencing (scWGS) data, which cannot disprove the existence of APP gencDNAs. An average of nine neurons from the brains of seven individuals with SAD were examined, raising immediate sampling issues required to detect mosaic APP gencDNAs. Kim et al. identified “uneven genome amplification”1,12–14 that resulted in about 20% of their single-cell genomes having less than 10× depth of coverage14 with potential amplification failure at one (~9% allelic dropout rate) or both alleles (~2.3% locus dropout rate)12,14. These limitations are compounded by potential amplification biases reflected by whole-genome amplification failure rates that may miss neuronal subtypes and/or disease states, which is especially relevant to single copies of APP gencDNAs that are as small as about 0.15 kb (but still detectable by DISH). Kim et al. state that the increased exonic read depth relative to introns reliably detects germline retrogene insertions in single cells from affected individuals (Kim et al., Fig. 3b); however, these data also demonstrate that increased exonic read depth is not observed in all cells—or even a majority in some cases—from the same individuals carrying the germline insertions of SKA3 (AD3 and AD4) and ZNF100 (AD2). These results demonstrate inherent technical limitations in the work by Kim et al. that prevent the accurate detection of even germline pseudogenes present in all cells, thus explaining an inability to detect the rarer mosaic gencDNAs produced by SGR. Kim et al.’s informatic analysis is also based on the unproven assumption that the structural features of gencDNA are shared with processed pseudogenes and LINE1 elements (Kim et al. Fig. 3a and Extended Data Fig. 1a), and possible differences could prevent straightforward detection under even ideal conditions as has been documented for LINE115. These issues could explain Kim et al.’s negative results.
Considering these points, we believe that our data and conclusions supporting SGR and APP gencDNAs remain intact and warrant their continued study in the normal and diseased brain.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data from Park et al. were deposited in the National Center for Biotechnology Information Sequence Read Archive database under accession number PRJNA532465. Data from the newly reported full exome pull-down data sets will be provided for the APP locus upon request.
Code availability
The source codes of the customized algorithms are available on GitHub at https://github.com/christine-liu/exonjunction.
Extended Data
Extended Data Fig. 1 |. IEJs identified from commercially available long-read transcriptome datasets in two genes other than APP.
Sequences containing IEJs were identified and shown for gene 1 (a) and gene 2 (b). Gene 2 is shown in two parts. Grey dashed lines show ends of RefSeq exons; solid purple lines denote IEJs. All splice isoforms were examined. The Alzheimer brain Iso-Seq dataset was generated by Pacific Biosciences, Menlo Park, CA, and additional information about the sequencing and analysis is available at https://downloads.pacbcloud.com/public/dataset/Alzheimer_IsoSeq_2016/.
Supplementary Material
Acknowledgements
We thank L. Wolszon and D. Jones for manuscript editing. Research reported in this publication was supported by the NIA of the National Institutes of Health under award numbers R56AG067489 and P50AG005131 (J.C.) and NINDS R01NS103940 (Y.K.). This work was supported by non-federal funds from The Shaffer Family Foundation, The Bruce Ford & Anne Smith Bundy Foundation, and Sanford Burnham Prebys Medical Discovery Institute funds (J.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests Sanford Burnham Prebys Medical Discovery Institute has filed the following patent applications on the subject matter of this publication: (1) PCT application number PCT/US2018/030520 entitled, ‘Methods of diagnosing and treating Alzheimer’s disease’ filed 1 May 2018, which claims priority to US provisional application 62/500,270 filed 2 May 2017; and (2) US provisional application number 62/687,428 entitled, ‘Anti-retroviral therapies and reverse transcriptase inhibitors for treatment of Alzheimer’s disease’ filed 20 June 2018. J.C. is a co-founder of Mosaic Pharmaceuticals.
Footnotes
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-020-2523-2.
References
- 1.Kim J et al. APP gene copy number changes reflect exogenous contamination. Nature 10.1038/s41586-020-2522-3 (2020). [DOI] [PMC free article] [PubMed]
- 2.Lee MH et al. Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature 563, 639–645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman DM et al. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. eLife 4, e05116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JS et al. Brain somatic mutations observed in Alzheimer’s disease associated with aging and dysregulation of tau phosphorylation. Nat. Commun 10, 3090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohrback S et al. Submegabase copy number variations arise during cerebral cortical neurogenesis as revealed by single-cell whole-genome sequencing. Proc. Natl Acad. Sci. USA 115, 10804–10809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummings JL, Morstorf T & Zhong K Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res. Ther 6, 37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J et al. Vecuum: identification and filtration of false somatic variants caused by recombinant vector contamination. Bioinformatics 32, 3072–3080 (2016). [DOI] [PubMed] [Google Scholar]
- 8.van Schendel R, van Heteren J, Welten R & Tijsterman M Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genet. 12, e1006368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sfeir A & Symington LS Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci 40, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splice variant case study: EGFRvIII detection in glioblastoma. https://acdbio.com/science/applications/research-areas/egfrviii (ACD, 2019).
- 11.Baker AM et al. Robust RNA-based in situ mutation detection delineates colorectal cancer subclonal evolution. Nat. Commun 8, 1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evrony GD et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 8, 1280–1289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evrony GD et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron 85, 49–59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohrback S, Siddoway B, Liu CS & Chun J Genomic mosaicism in the developing and adult brain. Dev. Neurobiol 78, 1026–1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from Park et al. were deposited in the National Center for Biotechnology Information Sequence Read Archive database under accession number PRJNA532465. Data from the newly reported full exome pull-down data sets will be provided for the APP locus upon request.