Abstract
In the Saccharomyces cerevisiae double-stranded RNA virus, programmed −1 ribosomal frameshifting is responsible for translation of the second open reading frame of the essential viral RNA. A typical slippery site and downstream pseudoknot are necessary for this frameshifting event, and previous work has demonstrated that ribosomes pause over the slippery site. The translational intermediate associated with a ribosome paused at this position is detected, and, using in vitro translation and quantitative heelprinting, the rates of synthesis, the ribosomal pause time, the proportion of ribosomes paused at the slippery site, and the fraction of paused ribosomes that frameshift are estimated. About 10% of ribosomes pause at the slippery site in vitro, and some 60% of these continue in the −1 frame. Ribosomes that continue in the −1 frame pause about 10 times longer than it takes to complete a peptide bond in vitro. Altering the rate of translational initiation alters the rate of frameshifting in vivo. Our in vitro and in vivo experiments can best be interpreted to mean that there are three methods by which ribosomes pass the frameshift site, only one of which results in frameshifting.
A 4.6-kb double-stranded RNA (dsRNA) virus, ScV-L-A (also known as ScV-L1), can be found in many laboratory strains of the yeast Saccharomyces cerevisiae. Like most fungal viruses, ScV-L-A is exclusively cytoplasmic, has no infectious cycle, and is known to be naturally transmitted only by budding or by cell fusion during mating (16). It is a member of the Totiviridae, which are dsRNA viruses with a single essential viral dsRNA (6).
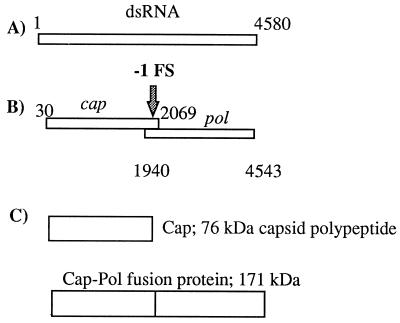
The viral genome has two open reading frames (ORFs) on its plus strand that overlap by 129 bases (Fig. 1). The first ORF is designated cap or gag (after the retrovirus nomenclature) and encodes the 76-kDa major capsid polypeptide. The second, pol, encodes the viral RNA-dependent RNA polymerase (12, 14, 21). Translation of pol as a 171-kDa Cap-Pol fusion protein is permitted by a programmed −1 ribosomal frameshift (12, 14, 21, 40) (Fig. 1). A second, separately encapsidated viral dsRNA (M1) encodes a secreted polypeptide toxin (k1 killer toxin) that provides a convenient phenotype for detecting synthesis of Cap and Cap-Pol (4, 34, 37).
FIG. 1.
(A) The 4.58-kb dsRNA genome of ScV-L-A. (B) The two ORFs, cap and pol, of the ScV-L-A plus strand, separated by the −1 frameshift within the overlapping region. (C) The ScV-L-A 76-kDa 0 frame protein product Cap and the 171-kDa fusion protein Cap-Pol.
Programmed, or directed, ribosomal frameshifting (PRF) is of particular value to viruses that package their plus strands, as it eliminates the need to splice their mRNAs and reduces the risk of packaging defective genomes. It also regulates the ratio of viral proteins synthesized. Examples of the phenomenon (both +1 and −1 shifts) have been found in a wide range of systems. In addition to the ScV systems, programmed translational frameshifting has been identified in retroviruses (17, 23, 30, 32); coronaviruses (5, 20); giardiaviruses, which are also members of the Totiviridae (41); two bacterial genes (3, 8); bacteriophage genes (7); astroviruses (28); the yeast EST3 gene (27); and the rat, mouse, Xenopus, and Drosophila ornithine decarboxylase antizymes (29) (reviewed in references 13 and 18). A significant number of cellular genes may utilize −1 PRF (19).
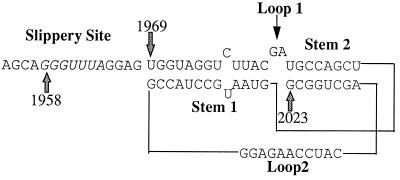
The ScV-L-A frameshifting site fits a fairly well described motif for promoting a −1 translational frameshift. There is a heptanucleotide slippery site (bases 1958 to 1964, GGGUUUA) and a region capable of forming a pseudoknot (bases 1969 to 2023) (14, 40) that together comprise the 71-nucleotide (nt) region that has been shown sufficient to support frameshifting (40) (Fig. 2). The existence of the stems of the pseudoknot and the requirement of the pseudoknot for frameshifting have been verified by mutagenesis (14, 40).
FIG. 2.
Possible structure of the ScV-L-A pseudoknot. The slippery site is in italics starting at base 1958. The second stem is shown with two more base pairs than in some formulations (14).
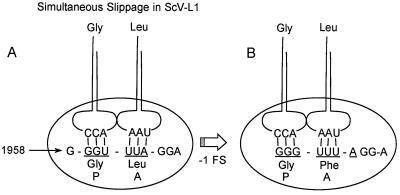
The proposed mechanism for the −1 translational frameshift, simultaneous slippage, was verified for Rous sarcoma virus by sequencing the frameshift product (22). In the case of L-A, the model predicts that the tRNAGly-tRNALeu in the P and A sites of the ribosome slips backward one base to pair with the Gly-Phe codons of the −1 pol reading frame (Fig. 3). Translation of the Cap-Pol fusion protein can then proceed in the −1 frame. This predicts a sequence of GLRS through the frameshift site, which has been verified by protein sequencing (T.-H. Tzeng and J. A. Bruenn, unpublished data). This frameshifting event has been shown to take place in vitro in rabbit reticulocyte lysates approximately 3.5% of the time a ribosome encounters the slippery site (40) and is important in regulating the stoichiometry of Cap and Cap-Pol in vivo (15). The −1 translational frameshifting event has been shown to require a ribosomal pause at the slippery site (38, 39), presumably because the ribosome is physically impeded by the intact pseudoknot just 3′ to the slippery site (39). The 5′ end of the ribosome paused at the slippery site has been previously mapped to bases 1946 and 1949 (39) by ribosomal heelprinting (42). Analysis of mutants demonstrates that ribosomal pausing is necessary but not sufficient for frameshifting (38, 39).
FIG. 3.
Simultaneous slippage model (22) shown here for ScV-L-A. A tRNAGly-tRNALeu pair in the 0 frame (underlined) P and A site of the ribosome slips back one nucleotide to pair with the −1 frame (pol) Gly-Phe codons, leaving mispaired nucleotides in the wobble position of each −1 frame codon.
In this report, the primary pause site is shown to be at base 1945 rather than 1946. The translational intermediate associated with a ribosome paused at this position is detected; using in vitro translation and quantitative heelprinting, the rates of synthesis, the ribosomal pause time, the proportion of ribosomes paused at the slippery site, and the fraction of paused ribosomes that frameshift are estimated. About 10% of ribosomes pause at the slippery site in vitro, and some 60% of these continue in the −1 frame. Ribosomes that continue in the −1 frame pause on the average about half a minute at the slippery site in vitro.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pGEM7ZCTU was constructed as previously described (40). Briefly, an L-A cDNA fragment of 396 bp (1783 to 2179) was inserted into a HindIII-Csp451 digested pGEM-7Zf(+) vector (Promega). The fragment contained the 71-nt minimal region necessary to support frameshifting. Plasmid 7zCTUSS was constructed by site-directed mutagenesis according to the method of Kunkel (26), with modifications as described elsewhere (40). In 7zCTUSS, base 1958 (G) is changed to an A to inhibit the −1 frameshift by preventing the simultaneous slippage of the tRNAGly-tRNALeu backward into the pol reading frame. Mutants 7ZCTUm3, a disruption of stem 1 of the pseudoknot, 7ZCTUm5, an inverted restoration of stem 1, and 7ZCTUd9, a deletion of the L-A bases beyond 2012, which truncates stem 2 of the pseudoknot, were constructed as described elsewhere (40).
In vitro transcription.
Transcripts for in vitro translations and heelprint analysis were made as described by Kreig and Melton (25) from the plasmids described above linearized with SspI (Bethesda Research Laboratories). Transcription for protected fragment analysis used [32P]UTP (specific activity, 3,000 Ci/mmol) at a final concentration of 1 μM and nonradioactive UTP at 0.1 mM. Transcripts for the pulse-chase translations and the heelprint showing artifacts (Fig. 5) were done with SspI-linearized pGEM constructs that yielded a 1,114-base message. Transcripts for the heelprint in Fig. 6, the translations in Fig. 8, and the protected fragment analysis were made with XbaI-linearized pGEM constructs that gave a 480-base transcript.
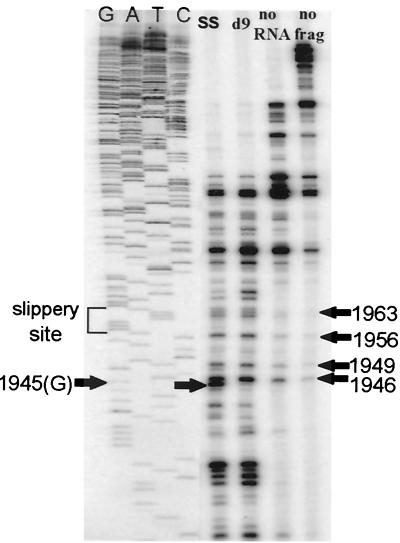
FIG. 5.
Heelprinting results with micrococcal nuclease. Lanes represent the SS and d9 mutants, control with mRNA left out of translation, and control with no reticulocyte or RNA used in the heelprinting reaction (no frag).
FIG. 6.
WT and mutant heelprints and a heelprint without added RNA (−).
FIG. 8.
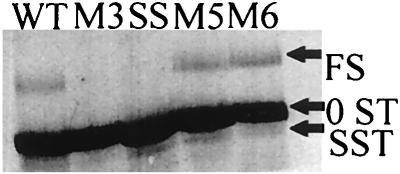
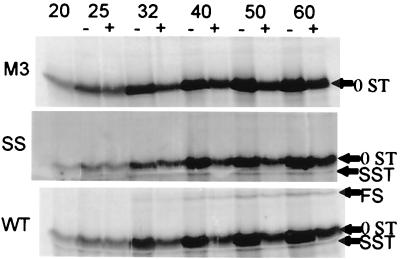
Standard translation of WT, SS, M3, M5, and M6 constructs. Positions of 0ST, SST, and FS products are indicated. These translations had no chase with nonradioactive methionine.
In vitro translation.
Translations were performed with rabbit reticulocyte lysates as previously described (39). A standard translation was at 26°C for 25 min without a chase with nonradioactive methionine. Translation products were expected to be 75 amino acids long (slipper site stop [SST] product), 110 amino acids 0 frame stop [0ST] product, and 234 amino acids (frameshift [FS] product) for SspI-linearized template transcripts of 1,114 bases. XbaI-linearized templates give a runoff translation FS product of 160 amino acids, while the SST and 0ST products are the same. For [35S]Met-labeled translations, the final volume of the reaction was 50 μl, of which 33 μl was lysate. Protected fragment translations were done in a 25-μl volume, of which 17.5 μl was lysate. In each translation, the FS and SST products have a single methionine (at the N terminus), while the 0ST product has three methionines.
Heelprinting.
Heelprinting (see Fig. 4) was performed as described elsewhere (39, 42), with additional modifications as follows in later heelprints. The final reaction volume was 25 μl, of which 17.5 μl was lysate. All translations and heelprints were done at room temperature and used about 3 μg of RNA. The Flexi (Promega) rabbit reticulocyte system was used. The nuclease digestion to degrade all RNA not protected by ribosomes was performed in 40 μl of 1 mM cycloheximide–3.5 mM magnesium acetate–3 mM CaCl2, first with micrococcal nuclease (final concentration, 10 U/μl; Boehringer Mannheim) at room temperature for 10 min, then with RNase V1 (Pharmacia) at a concentration of 17.5 U/ml for 10 min, and then with pancreatic RNase A (25 μg/ml) for an additional 5 min. The heelprinting reaction was completed as described previously (39) except that the nuclease-liberated monosomes with protected fragments were spun through the sucrose cushion at 70,000 rpm in a Beckman TLA 100.3 rotor for 30 min.
FIG. 4.
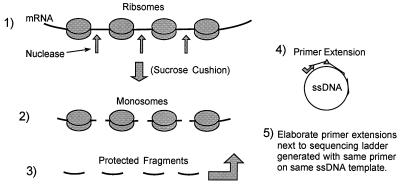
Heelprinting technique (42). In step 1, translating ribosomes are stopped on the template with cycloheximide and EDTA and unprotected mRNA is digested away, liberating monosomes. In step 2, the monosomes are collected through a sucrose cushion and the ribosomal proteins are stripped away, yielding protected RNA fragments. In step 4, the fragments are annealed to an ssDNA that has a region complementary to the message used in the translation. The heteroduplexes stop the extension of a primer at the 5′ ends of the RNA fragments that were protected from nuclease by the ribosome. In step 5, the primer extensions are elaborated next to a DNA sequencing ladder generated with the same primer on the same ssDNA template, thereby mapping the 5′ end of the protected RNA fragment.
Protected fragment quantification.
Protected RNA fragments were generated by translation of gel-purified [32P]UTP-labeled RNA combined with cold transcripts generated from the same template. The transcription reactions containing the [32P]UTP were run on an 8 M urea–4.5% polyacrylamide gel. The positions of the full-length transcripts were visualized with a Molecular Dynamics PhosphorImager, after which they were cut from the gel and eluted. Approximately 106 cpm of the purified transcript was then combined with 3 μg of the corresponding cold transcript and used to program the rabbit reticulocyte translations. The reactions were then taken through the heelprinting procedure described above to the point where ribosome-protected RNA fragments were precipitated in ethanol. The fragments were then dissolved in 8 μl of diethyl pyrocarbonate-treated H2O and subjected to a modified version of the RNase reaction described by Myers et al. (31). In this procedure, 2 μl of protected fragments was added to each of three separate reactions containing 27 μl of a solution of 80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 0.4 M NaCl, and 1 mM EDTA; 2 μg of a 76-base oligonucleotide identical to the minus strand of L-A from bases 1985 to 1910 was added to one of the three reactions for each mutant. The SS hybridizations used an oligonucleotide identical to this with the single change of C to T at 1958. Each reaction was then brought to a final volume of 30 μl, heated to 90°C for 2 min, and placed at 45°C for 30 min to ensure hybridization of the RNA fragments to the oligonucleotide. The samples were spun briefly and then diluted to 60 μl with a solution containing 20 mM Tris-HCl (pH 7.5), 2 mM EDTA, 400 mM NaCl, and 200 mM LiCl. To one of the reactions with RNA fragments and no oligonucleotide and to the reaction with RNA fragments and oligonucleotide, 0.06 μg of pancreatic RNase (RNase A) was added. The reaction mixtures were incubated at room temperature for 20 min, after which 1 μl of 3′ RNAsin was added to each. The samples were phenol–chloroform-isoamylalcohol extracted twice, made to 0.3 M sodium acetate, diluted twofold with cold ethanol, incubated at −20°C for at least 1 h, spun for 20 min at 4°C, rinsed with 70% ethanol, and vacuum dried. The samples were then dissolved in 4 μl of 1× sequencing gel loading buffer (36), boiled for 2 min in a water bath, and resolved on an 8 M urea–8% polyacrylamide gel next to a DNA sequencing reaction as a size marker. The resulting bands for each mutant were quantified relative to that of the controls for that mutant, using a Molecular Dynamics PhosphorImager.
In vivo frameshifting assays.
Wild-type (WT) killer+ cells were transformed with a series of plasmid-based GCN2c-constitutively active alleles of the eIF-2c kinase, which inhibit translational initiation to various degrees from strong to weak (33) (kindly provided by A. Hinnebusch and T. Dever). PRF was measured with the tester pTI25 (0 frame control), pF8 or pJD104 (−1 or +1 ribosomal frameshift tester respectively), or pT124 (−1 frameshift suppression) (2, 14).
Statistics.
Standard errors in rates or ratios of rates were determined by calculating the limits of intercepts or slopes of straight lines with the statistical program StatView. Standard errors of individual values were determined from repeated measurements.
RESULTS
A number of parameters of pausing can be estimated in vitro. From the rate of in vitro translation and the time it takes all ribosomes to finish translating in the −1 frame after a chase with nonradioactive methionine, it is possible to estimate the maximum time a ribosome pauses at the slippery site. An independent estimate of the average pause time can be made from heelprinting. From the heelprinting data and from the ratio of the rates of synthesis of the SST and FS products, it is possible to derive independent estimates of the fraction of paused ribosomes that frameshift. Similarly, by altering the rates of initiation, it is possible to estimate the fraction of paused ribosomes that frameshift in vivo.
Heelprinting.
Heelprinting experiments with ribosomes translating the L-A message were repeated in order to develop conditions for making quantitative measurements of paused ribosomes. The heelprinting technique (outlined in Fig. 4) is designed to map the 5′ position of some factor (in this case a ribosome) that protects mRNA from nuclease digestion. The same primer and template are used to generate the sequencing ladder and the primer extension that becomes the heelprint. Where the primer extension is inhibited by a protected RNA fragment hybridized to the template, a band results, migrating on the gel to a position corresponding to the most 5′ nucleotide of the message protected by a ribosome from nuclease digestion. The position is identified by comparison with the adjacent sequencing ladder.
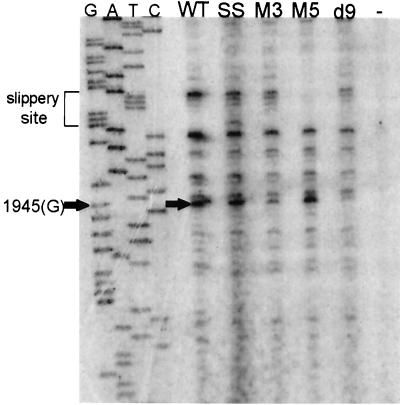
While this is primarily a mapping tool, the relative differences in intensities of bands at any particular position indicate qualitatively the number of ribosomes paused at the position in question within the population of messages being translated. The micrococcal nuclease heelprinting experiment shown in Fig. 5 includes the previously unpublished heelprint for the slippery site (SS) mutant which has base 1958 (G) altered to an A in order to prevent the −1 slippage of the ribosome into the pol reading frame. This mutant shows less than 1% of the frameshifting efficiency of the WT (14). The SS heelprint differs from the remaining lanes primarily at base 1945, misread as 1946 in reference 39, where a distinct band can be observed. This indicates that the 5′ end of the paused ribosome was positioned at base 1945. Since it is known that the rabbit reticulocyte ribosomes protect a region of mRNA of about 30 bases (39), this result is consistent with a ribosome being positioned with its P and A sites over the slippery site (1958) and verifies previous data showing that pausing is necessary but not sufficient for frameshifting (38, 39). Construct d9 truncates the pseudoknot at base 2012, thereby greatly diminishing the ribosome's propensity to pause at the slippery site (39). While the heelprints of the SS and d9 constructs have many bands in common, they clearly differ at position 1945, where the band present in the SS heelprint is entirely missing in the d9 heelprint.
The bands they do share can be identified as artifacts of two kinds: bands derived from adventitious hybridization of RNA fragments (probably from rRNA), and bands derived from preferred stops of the T4 DNA polymerase used for the primer extension. The latter variety of artifact is illustrated by the “no frag” lane: a heelprint performed on the single-stranded DNA (ssDNA) template without any reticulocyte or RNA used in the reaction. Note that in this negative control the T4 DNA polymerase used in the reaction has a tendency to stop at bases 1946, 1949, 1953, 1956, and 1963. Additionally, longer stop products can be seen in all lanes. The negative controls were repeated many times to ensure that the cause of the artifactual bands was not contaminated reagents. Artifactual stops seem to increase with the age of the polymerase (unpublished data). These appear to be the result of natural pauses by the T4 DNA polymerase on the template, and the major bands occur at pyrimidines just preceding A residues (bases 1949, 1953, 1956, and 1963). The authentic heelprint at base 1945 is a G preceding a pyrimidine. Additional bands, primarily of smaller size, are present in the “no RNA” lane (a heelprinting reaction carried out with an unprogrammed reticulocyte lysate). These are greatly reduced by more extensive RNase digestion of protected fragments (see below) and are therefore probably the result of adventitious hybridization of fragments of RNA from the reticulocyte lysate, presumably rRNAs.
Figure 6 shows the heelprint of the WT construct along with the SS mutant, M3 (disruption of stem 1 of the pseudoknot), M5 (pseudoknot restoration), d9 (pseudoknot truncation), and a negative control with no RNA protected fragments added to the heelprint. As in Fig. 5, the heelprint is at position 1945. The WT, SS, and M5 constructs all show a pronounced heelprint at base 1945. Disruptions of the pseudoknot (M3) and deletions of the pseudoknot (d9) yield much less signal at 1945, consistent with decreased ribosomal pausing in the absence of the pseudoknot. Many of the low-molecular-weight artifactual bands are missing from this set of heelprints, because their protected RNA fragments were produced by sequential digestion with micrococcal, V1, and pancreatic RNases. The artifactual bands resulting from T4 DNA polymerase hard stops are still visible at bases 1949, 1953, 1956, and 1963. It is clear from these data that the only position at which ribosomes are frequently paused on this message is over the slippery site, because the only prominent heelprint (after eliminating artifactual bands) is at base 1945.
Quantification of ribosome-protected fragments.
Using conditions such that the majority of ribosome-protected fragments are derived from ribosomes paused at the slippery site, it is possible to determine what proportion of ribosomes pause here.
We measured the protected RNA that could result from one or two ribosomes paused behind the pseudoknot i.e., from bases 1913 to 1989, a span which covers slightly more than the minimum 60 bases that two stacked ribosomes should cover. However, very few experiments show a heelprint in the vicinity of 1913, so under these conditions, few messages have more than one ribosome paused at the slippery site (not shown).
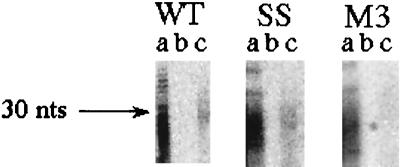
The molar fraction of RNA being translated which is in protected fragments from the region in which the heelprint occurs (f) is the fraction of ribosomes paused at the slippery site. The quantity f is estimated by PhosphorImager quantification from a gel in which the input fragments run through a mock hybridization and the input fragments hybridized to the 76-base region are elaborated on a sequencing gel (Fig. 7). f is the ratio of the amount of RNA in lane c divided by the amount of RNA in lane a. This is the fraction of ribosomes that are paused at the slippery site (see below) and turns out to be 0.132, or 13.2% ± 1.6%, in the WT, approximately the same in the SS mutant, and less in M3, as expected. If the amount of hybridization in the M3 control is taken as the background due to random isolation of fragments from this region, then the fraction of ribosomes paused at the slippery site is (13.2% ± 1.6%) − (6.7% ± 0.9%) = 6.5% ± 2.5%. This experiment was performed twice for each mutant and three times for the WT. The result agrees well with the proportion of paused ribosomes estimated from the relative rates of synthesis of the SST and FS products of 10.6% ± 2% (see below).
FIG. 7.
Quantitative protected fragment recovery. Protected RNA fragments from the WT, SS, and M3 constructs are shown after recovery from the hybridization procedure (see Materials and Methods) with no DNA oligonucleotide and no RNase (lanes a), no DNA with RNase (lanes b), or with DNA and with RNase (lanes c).
From the fraction f, the average pause time can be determined. In the heelprint experiments, the message is 480 bases long and contains 110 codons in the 0 frame. If ribosomes are randomly distributed along the message, this means that a ribosome should be located on any particular codon about 1/110 of the time. We are assuming that all ribosomes are in the 0 frame for this calculation; since 92 to 98% are in the 0 frame, little error should be introduced by this assumption. Since the rate of translation is 28.2 ± 7 amino acids/min (see below), this means that a ribosome should spend about 0.035 min/codon. However, about 6.5% of the ribosomes on the mRNA are located over the first slippery site codon. Hence ribosomes pause (0.065)/(1/110) = 7.2 times longer here than they should if dispersed randomly. Hence the average pause time is (0.035)(7.2) = 0.25 ± 0.07 min. If the ribosomes continuing in the 0 frame have a negligible pause and about 60% of the ribosomes continue in the −1 frame (see below), then the average pause is about 0.42 ± 0.11 min (0.25/0.6) for ribosomes in the −1 frame.
Identification of translation products.
Ribosomal pausing during −1 translational frameshifting has also been demonstrated by in vitro translation using the coronavirus infectious bronchitis virus pseudoknot (38). A transient translational intermediate of a size expected for a ribosome paused at the pseudoknot was identified. Ribosomal pausing (albeit at reduced levels) using a simple stem-loop with the same base pairs as the pseudoknot is also demonstrable. While ribosomes did pause in the latter construct, they did not frameshift, indicating that pausing may be required but is insufficient for frameshifting, as also demonstrated by heelprinting (39).
A similar translational analysis of L-A mRNAs from the WT, SS mutant pseudoknot disruptant mutant (M3), and pseudoknot restorations (M5 and M6) is shown in the standard translation of Fig. 8 (no chase with nonradioactive methionine). The 0ST product is the major product in each translation. The product resulting from a −1 frameshift (FS) is present only in the constructs capable of frameshifting (WT, M5, and M6). There is one additional product (SST) of the right size to be a peptide halted at the slippery site. The putative SST is present only where expected, in the SS, WT, M5, and M6 translations. It is not detectable in the M3 translation. The predicted and measured sizes of the translation products in the longer transcripts are as follows: 0ST (measured, 11.8 kDa; predicted, 12.0 kDa), FS (measured, 26.9 kDa; predicted, 27.8 kDa), and SST (measured, 8.4 kDa; predicted, 8.4 kDa). The identification of the FS product was confirmed by translating a shorter message (shown in Fig. 8), truncated at the XbaI site, producing an FS product with a predicted size of 18.4 kDa.
Kinetics of synthesis of translation products.
Translations of WT, SS, and M3 mRNAs terminated after 20 min are shown in the first lanes in Fig. 9. Immediately after the zero point was taken, the reaction was divided into two halves, one of which was brought to 40 μM cold methionine to chase transient products. The other half of the reaction was allowed to continue without dilution of the labeled methionine, and aliquots were taken from each half of the reaction as indicated. As in Fig. 8, the WT construct clearly shows the production of the FS, 0ST, and SST proteins, while the SS mutant shows the SST and 0ST products but not the FS product. The presence of the SST in both the WT and SS constructs but its absence in M3 is predicted by the heelprint shown in Fig. 6, which indicates that there is a ribosomal pause at the slippery site (heelprint at 1945) for the WT and SS constructs but not for M3. By PhosphorImager analysis, the accumulation of the translation products 0ST, SST, and FS shown in Fig. 9 can be quantified and the rate of synthesis can be estimated. The rate of synthesis of any in vitro translation product should be proportional to the number of ribosomes translating it. Hence, the frameshift efficiency can be estimated either from the relative rates of synthesis of the FS and 0ST products, corrected for the molarity of methionine in the products, or from the molar ratio of the total amounts of FS and 0ST that accumulate during synthesis. (The 0ST product has three methionines, while the SST and FS products each have only one, so the 0ST product is three times more intense per mole.) From the kinetics of synthesis of the FS and 0ST products (Fig. 10 and 11), the relative ratio of rates of synthesis, corrected for methionine content in the products, gives a frameshift efficiency of 6.1% ± 0.7%. From the accumulation of translation products after 60 min of synthesis (in the absence of a methionine chase), still in the linear portion of the curve, the molar proportion of FS product to total products is 5.2%, fairly close to measured in vivo rates of frameshifting (see below).
FIG. 9.
Pulse-chase in vitro translations. Reaction mixtures were incubated for 20 min and divided in half. One half was chased with cold methionine, and aliquots were taken from each half (with [+] or without [−] Met) at the indicated time points. Positions of the SST, 0ST, and FS products are indicated.
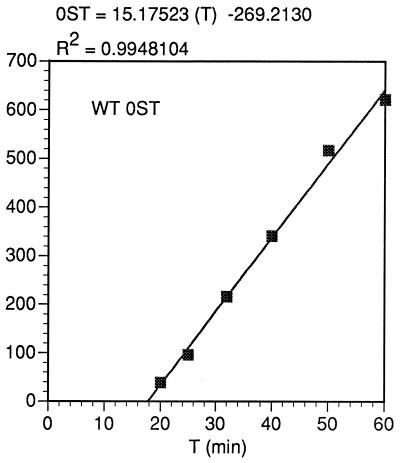
FIG. 10.
Kinetics of synthesis of the WT 0ST product from the data of Fig. 9. The ordinate is relative molar units, generated by the PhosphorImager volume report but divided by 3 to account for the difference in the number of methionines between the 0ST and the FS and SST products. The equation for the least squares line calculated is given.
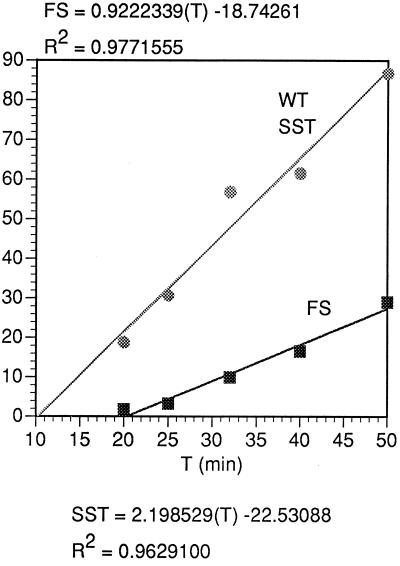
FIG. 11.
Initial kinetics of synthesis of the WT SST product and FS product, as in Fig. 10, from the same experiment as that of Fig. 10, in relative molar amounts.
After 20 min of synthesis with labeled methionine and 60 min of chase (a total of 80 min of synthesis) (from the data of Fig. 9), the molar ratio FS/(FS + 0ST + SST) approaches a value of 8.1%. This is probably by virtue of the delayed synthesis of already initiated FS product, while initiation of 0ST has decreased due to inactivation of ribosomes. This supposition is confirmed by examining the proportion of FS product synthesized after the chase compared to the total products synthesized after the chase, which is 9.1%.
Fraction of ribosomes that are paused from in vitro translations.
As above, the number of ribosomes translating an in vitro translation product should be proportional to its rate of synthesis. Hence the percentage of ribosomes that pause at the slippery site can be calculated from the relative initial molar rates of accumulation of the SST and 0ST products. From Fig. 10 and 11, this is 14.5% ± 2.2%. However, not all of this represents paused ribosomes. It is clear from Fig. 9 that not all of the SST product can be chased into FS or 0ST; some of it persists, presumably due to incorrect termination of ribosomes and release of SST (1, 24), which is not degraded as it would be in vivo.
The percentage of ribosomes which incorrectly terminate at the slippery site can be estimated from the amount of SST synthesized after the chase as a fraction of total synthesis. Any paused ribosomes that do continue synthesis result in product that appears in the FS and 0ST, and so only the incorrectly terminating ribosomes contribute to the amount of SST synthesized after the chase. The amount of SST synthesized after the chase is 3.9% of the total protein synthesized. Consequently, the rate of accumulation of SST by ribosomes that will continue past the slippery site either in the −1 frame or in the 0 frame is 14.5% ± 2.2 − 3.9% = 10.6% ± 2.2% of the rate of synthesis of total products, or the corrected fraction of ribosomes paused at the slippery site is 10.6% ± 2.2%. About a quarter of the SST synthesized is not a transient product but a termination product. This calculation of the fraction of ribosomes that pause assumes that synthesis of a terminal SST product generates an SST product but not a discernible pause. This value of 10.6% ± 2.2% compares well with the calculation from quantification of ribosome-protected RNA fragments (6.5% ± 2.5%; see above), supporting the assumption that the generation of terminal SST products does not create a discernible ribosomal pause.
Percentage of paused ribosomes that frameshift.
The relative numbers of paused ribosomes that pause and that go on in the −1 frame can be estimated from the ratio of rates of synthesis of FS and SST from Fig. 11. The percentage of paused ribosomes that frameshift at the slippery site is the ratio of FS rate to SST rate times 100 = 100 × (6.1 ± 0.7)/(10.6 ± 2.2) = 57.5% ± 23%. This assumes that all ribosomes translating in the −1 frame have undergone a detectable pause at the slippery site, an assumption justified by the demonstrable dependence of frameshifting on pausing (Fig. 5, 6, and 8).
Translation rate.
A number of calculations depend on knowing the actual rate of in vitro protein synthesis under our conditions. The average rate of translation can be estimated from the initial time of appearance of products of known size. The initial times of appearance of the control protein (luciferase, 552 amino acids) and the 0ST product (110 amino acids) are 33.5 ± 3 min (not shown) and 17.8 ± 0.5 min (Fig. 10), respectively. If both have the same initial delay before synthesis (mainly due to the time it takes to equilibrate the tRNAMet with labeled methionine), a rate of 28.2 ± 7 amino acids/min in the in vitro translation (considerably slower than in vivo) is calculated. A similar answer results from making the same assumption about the synthesis of luciferase and the SST. The initial delay is calculated to be 13.9 min, which agrees well with data obtained by chasing with nonradioactive methionine (see below).
Estimate of pause time from translation rates.
The minimum pause time in the −1 frame can be estimated from the intercepts of the SST and FS synthesis lines (Fig. 11). The SST is first visible at about 10 min (10.2 ± 1 min), and the FS appears at about 20 min (20.2 ± 2 min). Hence, it takes about 10 min (10.1 ± 3 min) minus the time it takes to translate the remaining 159 amino acids, or 10.1 ± 3 − (159/28.2 ± 7) = 4.5 ± 5 min, minimum, to resolve the paused ribosome at the slippery site in the −1 frame. The standard deviation here is very large, and the minimum pause time is not significantly different from zero.
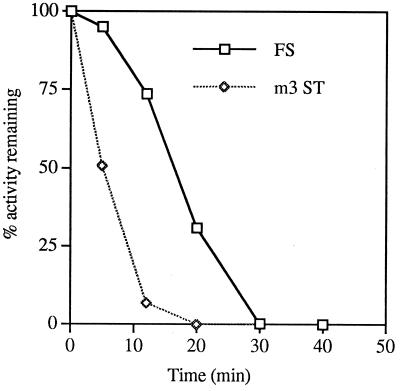
The maximum pause time is estimated from the difference between clearance times of ribosomes translating the 0ST and FS products by measuring translation activity remaining (Fig. 12) after chase for the WT and M3 mutant constructs. It takes 30 min to clear all ribosomes capable of making the FS product. It takes 20 min to clear all ribosomes capable of making the 0ST (in the M3 mutant, in which the stop is eliminated).
FIG. 12.
Translation activity remaining during the chase with nonradioactive methionine for the M3 0ST (m3 ST) and WT FS. The percent activity remaining (AR) is calculated from the plateau value of the PhosphorImager volume report for the FS or 0ST (P) and the value at 0 to 50 min after the chase of the volume report (V), as follows: AR = 100[(P − V)/P].
Note that most of this time is the time it takes for equilibration of nonradioactive methionine: the maximum time to clear the 0ST message of ribosomes (without a pause, as in M3) would be the time it takes to translate the entire 0ST message: [110/(28.2 ± 7)] = 3.9 ± 1 min. So 20 − 3.9 ± 1 = 16.1 ± 1 min (with standard error minimized [see below]) is the time for the methionine pool to reach equilibrium. This agrees well with the estimate of 13.9 min for tRNAMet pool equilibration calculated by assuming the same rates of synthesis of luciferase and the 0ST (see above).
The maximum time a ribosome pauses at the SST is 10 min minus the difference in time it takes to translate from the 0 frame termination codon to the −1 frame termination codon, or 10 − [(234 − 110)/(28.2 ± 7)] = 5.6 ± 1.5 min. Since the 10-min difference in clearance times has no easily calculated standard error, the standard error of 1.5 min is a minimum estimate.
Estimate of average pause time from translation chases.
The small percentage of ribosomes continuing on in the −1 frame takes much longer to clear, and the average pause time for these can also be calculated (Fig. 12) from the time it takes half of them to clear (16.5 min) minus the time it takes half the 0ST ribosomes to clear (6 min). Again, as above, subtracting the time it takes to translate from the 0 frame termination codon to the −1 frame termination codon gives 10.5 − (4.4 ± 1.5) = 6.1 ± 1.5 min for an average pause time. A meaningful standard error cannot be calculated for either the maximum or average pause times derived from Fig. 12, but the maximum pause time of 5.6 ± 1.5 min calculated above is not significantly different from the average pause time. The relatively long time necessary to equilibrate the tRNAMet pool makes this estimate of average pause time inherently inaccurate, but these translational measurements place an upper limit on the pause time.
Average pause times, estimated by two independent methods, for ribosomes continuing in the −1 frame were 6.1 ± 1.5 min (range, 4.5 ± 5 to 5.6 ± 1.5 min) calculated by in vitro translation and 0.42 ± 0.11 min calculated by heelprinting.
Ribosome spacing in vivo.
It is also possible to perturb −1 PRF in vivo in order to measure some of its characteristics. Since the RNA pseudoknot must denature to allow passage of the ribosome, the passage of subsequent ribosomes will be affected by (i) the relative rates of pseudoknot renaturation, (ii) ribosome transit speeds, and (iii) the temporal distance between elongating ribosomes. If pseudoknot renaturation rates and ribosome transit speeds are constant, then mutations or conditions affecting initiation rates could result in changes in the temporal spacing between elongating ribosomes. This would change the amount of time available for renaturation of the pseudoknot (which was denatured by a prior transiting ribosome) before it encountered the next transiting ribosome. Therefore, those conditions which decrease rates of translational initiation would result in elongating ribosomes being spaced further apart. Consequently, the pseudoknot would have more time to renature, leading to an increase in the percentage of ribosomes that encounter the renatured pseudoknot, increasing the percentage of paused ribosomes and thereby increasing the overall efficiency of −1 ribosomal frameshifting. Conversely, mutations or conditions which increase initiation rates would yield lower efficiencies of −1 ribosomal frameshifting. Further, since Ty1-directed +1 ribosomal frameshifting and frameshift suppression are independent of an RNA pseudoknot, changing the temporal spacing between elongating ribosomes should have no effect on these mechanisms.
We tested the general validity of this hypothesis by transforming WT killer+ cells with a series of plasmid-based GCN2c constitutively active alleles of the eIF-2c kinase, which inhibit translational initiation to various degrees from strong to weak (33) (kindly provided by A. Hinnebusch and T. Dever). The results are summarized in Table 1. Examination of the data reveals that the ribosome spacing hypothesis is essentially true in that these mutations specifically affect programmed −1 but not +1 ribosomal frameshifting or frameshift suppression. Interestingly, the increases in −1 ribosomal frameshift efficiencies are only twofold in cells harboring the GCN2c alleles, regardless of the severity of the initiation defect. This result can be interpreted in at least two ways: (i) that in vivo the leading ribosome resolves the pseudoknot for immediately trailing ribosomes or (ii) that in vivo ribosome stacking at the pseudoknot prevents frameshifting (see Discussion).
TABLE 1.
Summary of experiments using different GCN2c alleles on URA3 CEN vectors to test the effects of initiation defects on ribosomal frameshiftinga
| Plasmid | GCN2c mutation | Relative growthb | Mean % PRFc ± SD (fold WT level)
|
% −1 frameshift suppressiond | |
|---|---|---|---|---|---|
| −1 | +1 | ||||
| p722 | None (WT) | ++++ | 3.4 ± 0.7 (1) | 5.9 ± 0.7 (1) | 0.044 |
| p1056 | E532K, E1522K | +++ | 6.3 ± 0.6 (1.8) | 5.8 ± 0.6 (1) | 0.043 |
| p1052 | M719V, E1537G | ++ | 6.4 ± 0.3 (1.8) | 6.2 ± 0.5 (1) | 0.044 |
| p1054 | E532K, E1537G | + | 6.4 ± 0.2 (1.8) | 5.9 ± 0.6 (1) | 0.046 |
All strains had strong killer phenotypes and stably maintained L-A and M1.
Corresponds to initiation competence.
As measured with pTI25 (0 frame control) or with pF8 or pJD104 (−1 or +1 ribosomal frameshift tester, respectively) (2, 14).
Suppression of a nonspecific frameshift mutation (pTI24/pTI25 × 100), in all cases approximately equal to the WT value.
The in vivo rates of frameshifting (3.5% ± 0.7% in the WT) are similar to those measured in vitro (6.1% ± 0.7%).
DISCUSSION
We have determined, by heelprinting and by analysis of in vitro translation products, that about 10% of ribosomes normally pause at the L-A slippery site and that many fewer pause when the downstream pseudoknot is disrupted. These measurements assume that ribosomes that incorrectly terminate at the pseudoknot do not detectably pause. By these same assumptions, about 60% of paused ribosomes frameshift. The extent of the pause at the slippery site can be calculated by several methods from in vitro translation data, all of which arrive at a pause of minutes, with some uncertainty. A calculation from the fraction of paused ribosomes, assuming a random distribution of ribosomes in the absence of pausing, gives a more exact pause time of about half a minute in vitro. All calculations of ribosomal pause time exceed by at least 10-fold the time it takes to make a single peptide bond.
Our in vitro experiments can best be interpreted to mean that ribosomes may suffer three fates at the frameshift site in L-A mRNA.
Fate 1.
About 10% of ribosomes pause at the pseudoknot. Another 4% of the ribosomes prematurely terminate. The remaining ribosomes pass the pseudoknot before it causes an obstruction. This could occur in two ways: the first ribosome translating the message could resolve the pseudoknot, and all subsequent ribosomes could encounter the unwound pseudoknot before it could re-form; or a subset of the Mof proteins or the translational surveillance complex (SC) (35) could recognize and resolve these RNA structures independent of elongating ribosomes.
Ribosomes that encounter the pseudoknot either pause at the pseudoknot or terminate there. Paused ribosomes may continue translation in two ways: either by a melting of the pseudoknot and a resumption of 0 frame translation or by a −1 translational frameshift (about 60% of the time in vitro).
Fate 2.
In the second mechanism, which permits a resumption of 0 frame translation after ribosomes are already paused at the pseudoknot, the MOF gene products seem to be involved, since altering their effectiveness alters the frequency with which the third method of passing the pseudoknot is chosen. Consequently, MOF gene products are probably involved in both the first and second mechanisms by which ribosomes pass the pseudoknot.
Fate 3.
The third form of resolution, which is of most interest, requires both the pseudoknot and the slippery site. The pseudoknot is necessary since a simple secondary structure of similar stability causes pausing but not frameshifting (38) and because a mutant with two to four bases of the second stem deleted (depending on how the pseudoknot is drawn) demonstrates ribosomal pausing but not frameshifting (39). The slippery site is necessary, as shown by the present results, since the SS mutant shows essentially normal pausing both by heelprint analysis and by in vitro translation yet reduces frameshifting to very low levels. Therefore, this third mechanism of resolution of the ribosome-pseudoknot complex must require a specific interaction between the pseudoknot and some component(s) of the translation machinery not yet defined. This resolution may be a lengthy process, since ribosomes pause (in vitro) for at least 10 times as long as it takes to make a single peptide bond.
Relationship between in vitro and in vivo −1 PRF.
The one experiment we have performed to change the kinetics of −1 PRF in vivo tells us that there are some differences between frameshifting in vivo and in vitro. Our preferred interpretation of the results is that decreasing the initiation rate results in an increase in frameshifting efficiency because the leading ribosome no longer clears the pseudoknot for trailing ribosomes; the pseudoknot now re-forms before a trailing ribosome arrives. The fact that there is a ceiling on frameshifting efficiency, twice the basal rate, regardless of how low the rate of initiation is most simply interpreted to mean that normally 50% of ribosomes pause at the pseudoknot, while with limited initiation, essentially all ribosomes pause. However, this would imply that only 7% of the paused ribosomes shift frame in vivo, while 60% do so in vitro.
The disparity between in vivo and in vitro results may be explained by the role of the Mof proteins. It is becoming clear that the process of remodeling nuclear to cytoplasmic ribonuclear particles is a key step in determining the posttranscriptional fate of mRNAs (11). There is evidence to support the notion that RNP remodeling by the first translating ribosome and its associated SC also plays a role in programmed −1 ribosomal frameshifting. For example, we have demonstrated that mutants of at least three factors present in the SC, Upf1p/Mof4p, Upf3p, and Sui1p/Mof2p, affect programmed −1 ribosomal frameshift efficiencies (9, 10, 35). We propose that one of the functions of the SC is to recognize and resolve complex tertiary mRNA structures. For example, one of the factors in the SC may specifically recognize the pseudoknot and recruit another (e.g., the Upf1p RNA helicase) to unwind this structure. Additionally, other, yet to be characterized Mof proteins, either components of the SC or integral ribosomal proteins that are part of the intrinsic ribosomal helicase, may play a role in recognizing and resolving the pseudoknot. By this model, resolution of the pseudoknot by the first ribosome or SC is part of the nuclear-to-cytoplasmic RNP remodeling process. Given that the nonsense-mediated mRNA decay machinery is not functional in translationally competent cell extracts (i.e., reticulocyte lysates), it is probable that the pseudoknot-resolving activity of the SC is also not functional in the in vitro system. This would explain why the frameshifting of paused ribosomes is so much higher in the reticulocyte system that in intact cells.
A second interpretation of the increased frameshifting in vivo with decreased initiation rates is that when ribosomes stack up behind the pseudoknot, the first ribosome (on the slippery site) cannot move backward one base because of the ribosome behind it. The significance of the ceiling of twice the usual frameshifting efficiency would be that in yeast in vivo, under normal initiation conditions, 50% of the time ribosomes are stacked, but that a significant decrease in initiation rate results in no stacking at all. Again, the in vivo result is different from what one would expect in vitro, since there are very few ribosomes translating a message in vitro. In fact, in our system, we calculate fewer than two ribosomes per mRNA (results not shown). This would explain the absence of stacking as detected by heelprinting in vitro. This explanation makes no predictions about what fraction of ribosomes pause in vivo and so is not inconsistent with the in vitro data.
Pause time in vivo.
The time ribosomes spend paused at the pseudoknot is estimated by two independent methods, both of which arrive at a pause in the range of minutes. If the relationship between the pause time in vitro and the pause time in vivo is the same as the relative rates of translation, this would correspond to several seconds in vivo. However, since the slow rate of translation in vitro is probably due to the diffusion-limited accessibility of charged tRNAs and other components, which are probably closely associated with the translation complex in vivo, and since frameshifting probably requires only components closely associated with the ribosome both in vivo and in vitro, it is not clear that direct extrapolation of in vitro pause times to the in vivo situation is correct. It may be that pause times in vivo are more like those observed in vitro. At any rate, the pause time in vivo is a minimum of several seconds, or at least an order of magnitude longer than the time it takes to complete a single peptide bond.
ACKNOWLEDGMENTS
We thank A. Hinnebusch and T. Dever for plasmids.
We thank the NIH (grant GM58859 to J.D.D. and grant GM22200 to J.A.B.) and the NSF (grant MCB9727630 to J.A.B.) for support.
REFERENCES
- 1.Anderson R P, Menninger J R. Tests of the ribosome editor hypothesis. III. A mutant Escherichia coli with a defective ribosome editor. Mol Gen Genet. 1987;209:313–318. doi: 10.1007/BF00329659. [DOI] [PubMed] [Google Scholar]
- 2.Balasundaram D, Dinman J D, Wickner R B, Tabor C W, Tabor H. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blinkowa A L, Walker J R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostian K A, Elliott Q, Bussey H, Burn V, Smith A, Tipper D J. Sequence of the preprotoxin dsRNA gene of type 1 killer yeast: multiple processing events produce a two-component toxin. Cell. 1984;36:741–751. doi: 10.1016/0092-8674(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 5.Brierley I, Boursnell M E G, Binns M M, Bilimoria B, Blok V C, Brown T D, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruenn J A. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 1993;21:5667–5669. doi: 10.1093/nar/21.24.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condron B G, Gesteland R F, Atkins J F. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 1991;19:5607–5612. doi: 10.1093/nar/19.20.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigen W J, Caskey C T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Dinman J D, Kinzy T G, Peltz S W. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA trunover. Bioessays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Diamond M E, Dowhanick J J, Nemeroff M E, Pietras D F, Tu C-L, Bruenn J A. Overlapping genes in a yeast double-stranded RNA virus. J Virol. 1989;63:3983–3990. doi: 10.1128/jvi.63.9.3983-3990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3678. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Sherbeini M, Bostian K A. Viruses in fungi: infection of yeast with the k1 and k2 dsRNA killer viruses. Proc Natl Acad Sci USA. 1987;84:4293–4297. doi: 10.1073/pnas.84.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falk H, Mador N, Udi R, Panet A, Honigman A. Two cis-acting signal control ribosomal frameshift between human T-cell leukemia virus type II gag and pro genes. J Virol. 1993;67:6273–6277. doi: 10.1128/jvi.67.10.6273-6277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammell A B, Taylor R L, Peltz S W, Dinman J D. Identification of putative programmed −1 ribosomal frameshift signals in large DNA databases. Genome Res. 1999;9:417–427. [PMC free article] [PubMed] [Google Scholar]
- 20.Herold J, Siddell S G. An 'elaborated‘ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Icho T, Wickner R B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- 22.Jacks T, Madhani H D, Masiarz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 24.Janosi L, Mottagui-Tabar S, Isaksson L A, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–51. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieg P A, Melton D A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundblad V, Morris D K. Programmed translational frameshifting in a gene required for yeast teleomere replication. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 28.Marczinke B, Bloys A J, Brown D K, Willcocks M M, Carter M J, Brierley I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsufuji S, Matsufuji T, Miyazaka Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morikawa S, Bishop D H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers R M, Larin Z, Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985;230:1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- 32.Nam S K, Copeland T D, Hatanaka M, Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J Virol. 1993;67:196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez M, Wek R C, Vazquez de Aldana C R, Jackson B M, Freeman B, Hinnebusch A G. Mutations activating the yeast eIF-2a kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribas J C, Fujimura T, Wickner R B. Essential RNA binding and packaging domains of the Gag-Pol fusion protein of the L-A double-stranded RNA virus of Saccharomyces cerevisiae. J Biol Chem. 1994;269:28420–28428. [PubMed] [Google Scholar]
- 35.Ruiz-Echevarria M J, Yasenchak J M, Han X, Dinman J D, Peltz S W. The upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc Natl Acad Sci USA. 1995;95:8721–8726. doi: 10.1073/pnas.95.15.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Skipper N, Thomas D Y, Lau P C. Cloning and sequencing of the preprotoxin-coding region of the yeast M1 double-stranded RNA. EMBO J. 1984;3:107–111. doi: 10.1002/j.1460-2075.1984.tb01769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somogyi P, Jenner A J, Brierley I, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu C-L, Tzeng T-H, Bruenn J A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzeng T-H, Tu C-L, Bruenn J A. Ribosomal frameshifting requires a pseudoknot in the yeast double-stranded RNA virus. J Virol. 1992;66:999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang A L, Yang H-M, Shen K A, Wang C C. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translational frameshift. Proc Natl Acad Sci USA. 1993;90:8595–8599. doi: 10.1073/pnas.90.18.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolin S L, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]