ABSTRACT
Antibiotic resistance among bacterial pathogens poses a major global health threat. Mycobacterium tuberculosis complex (MTBC) is estimated to have the highest resistance rates of any pathogen globally. Given the low growth rate and the need for a biosafety level 3 laboratory, the only realistic avenue to scale up drug susceptibility testing (DST) for this pathogen is to rely on genotypic techniques. This raises the fundamental question of whether a mutation is a reliable surrogate for phenotypic resistance or whether the presence of a second mutation can completely counteract its effect, resulting in major diagnostic errors (i.e., systematic false resistance results). To date, such epistatic interactions have only been reported for streptomycin that is now rarely used. By analyzing more than 31,000 MTBC genomes, we demonstrated that the eis C-14T promoter mutation, which is interrogated by several genotypic DST assays endorsed by the World Health Organization, cannot confer resistance to amikacin and kanamycin if it coincides with loss-of-function (LoF) mutations in the coding region of eis. To our knowledge, this represents the first definitive example of antibiotic reversion in MTBC. Moreover, we raise the possibility that mmpR (Rv0678) mutations are not valid markers of resistance to bedaquiline and clofazimine if these coincide with an LoF mutation in the efflux pump encoded by mmpS5 (Rv0677c) and mmpL5 (Rv0676c).
KEYWORDS: Mycobacterium tuberculosis, amikacin, antibiotic resistance, bedaquiline, clofazimine, drug susceptibility test, genomics, kanamycin
INTRODUCTION
Tuberculosis (TB) and its causative pathogen Mycobacterium tuberculosis complex (MTBC) are a major public health threat causing an estimated 10 million new cases of disease per year (1). Antibiotic resistance, in particular, poses a problem in controlling the TB epidemic (1). Owing to the inherently low growth rate of MTBC, genotypic drug susceptibility testing (DST) represents the only realistic option to inform the initial selection of the most appropriate treatment regimen (2). This raises the fundamental question of whether the effect and clinical interpretation of a marker for resistance depend on the presence of another mutation (i.e., epistasis) or whether the effect is universal.
Although it is known that the level of resistance conferred by resistance mutations in some genes can differ, the only well-understood epistatic mechanism that completely counteracts the effect of another mutation involves the whiB7 (Rv3197A) regulatory gene (3–5). Specifically, the overexpression of the whiB7 cannot confer streptomycin resistance in the vast majority of lineage 2 isolates because these have a frameshift in the tap (Rv1258c) efflux pump (6, 7). Yet, because the use of streptomycin has been downgraded in the most recent treatment guidelines by the World Health Organization (WHO), the clinical relevance of this example is limited (8).
Using whole-genome sequencing (WGS) data for 31,440 isolates, we set out to survey systematically whether other markers of resistance to more important antibiotics may be affected by epistasis if they involve the overexpression of a nonessential drug resistance gene. First, we analyzed the alkyl-hydroperoxidase ahpC (Rv2428), the function of which is not fully elucidated but may act as a compensatory mechanism for isoniazid (INH) resistance caused by katG mutations (i.e., the newly WHO-endorsed Cepheid Xpert MTB/XDR assay interrogates ahpC promoter mutations) (9, 10). Second, loss-of-function (LoF) mutations (i.e., frameshift indels, nonsense mutations, and mutations that abolish the start codon) in the transcriptional repressor mmpR (Rv0678), which is sequenced by several commercial targeted next-generation sequencing (tNGS) assays being evaluated by WHO, confer cross-resistance to bedaquiline (BDQ) and clofazimine (CFZ) via the overexpression of the nonessential efflux pump encoded by mmpS5 (Rv0677c) and mmpL5 (Rv0676c) (2, 11–13). Third, four promoter/upstream mutations for the eis (Rv2416c) acetyltransferase are responsible for kanamycin (KAN) resistance (14). Based on a recent review of allelic exchange results and data clinical from clinical isolates, WHO recognized one of those mutations (i.e., C-14T) as also conferring cross-resistance to amikacin (AMK), the only aminoglycoside (AG) now recommended for the treatment of TB (9, 14–16). In fact, the Xpert MTB/XDR already interprets this eis mutation accordingly, whereas the WHO-endorsed Hain GenoType MTBDRsl VER 2.0 (SL-LPA) assay will have to be updated accordingly (9). Finally, we included whiB7, as it also regulates eis and, therefore, could theoretically confer cross-resistance to both AGs rather than just to KAN (9, 14).
RESULTS
Summary characteristics of isolates.
Our sample consisted of 31,440 isolates that were geographically and genetically diverse. Based on the geographical metadata for 21,512 of these isolates, our analysis included 80 countries, of which 38 were represented by at least 50 isolates (Fig. S1 in the supplemental material). Fifty-four percent of the isolates belonged to lineage 4, but all other global MTBC lineages (i.e., lineages 1 to 3 and 5 to 6) were also covered. Lineage 7 was excluded, as only 35 genomes were publicly available (Fig. S1).
INH: ahpC upstream mutations in combination with ahpC LoF mutations.
We observed 57 unique single nucleotide polymorphisms (SNPs) in the upstream region of ahpC (Fig. 1A), of which 18 were homoplasic and occurred in at least 5 isolates, consistent with parallel evolution and known selection on this gene (Table 1; Table S1). We screened for frameshift indels, nonsense mutations, and mutations that abolish the start codon of ahpC given that these are the most likely types of mutations to confer an LoF phenotype. This yielded seven unique variants in eight isolates, of which just ahpC 323delC co-occurred with an upstream mutation in a single isolate (Table 2; Table S1). This particular upstream mutation (i.e., G-88A; Table 1; Table S1) is a marker for MTBC lineage 3 and correlates with only a 3-fold increase in the expression of ahpC, potentially by creating a new Pribnow box (7, 17). As a result, this SNP is not considered to be a marker of resistance (i.e., the Xpert MTB/XDR was designed not to detect it, unlike adjacent mutations), which means that this is not an example of epistasis (9). Indeed, this double mutant was phenotypically susceptible to INH at the critical concentration (CC) of 0.1 mg/liter using the Bactec Mycobacterial Growth Indicator Tube 960 system by Becton, Dickinson.
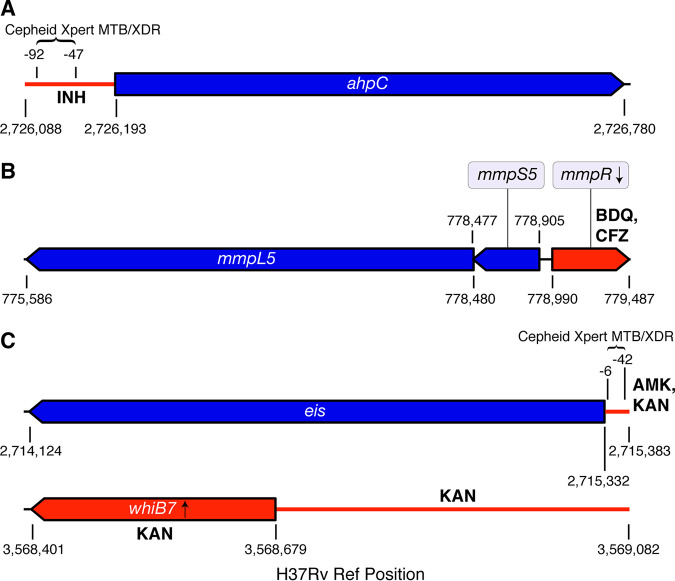
FIG 1.
Genomic regions interrogated. The nonessential genes conferring resistance or compensating for resistance are shown in blue, whereas the corresponding regulatory regions or nonessential regulators are shown in red along with the relevant antibiotic(s) (A, region associated with INH resistance; B, region associated with BDQ and CFZ resistance; C, regions associated with AMK and KAN resistance). For each of the four regions, we screened for any type of mutation in the upstream regions and likely LoF mutations in the coding regions (i.e., frameshift indels, nonsense mutations, and mutations that abolish the start codon, including synonymous mutations at the start codons of eis, mmpR, and whiB7, as these genes start with a valine). Unlike Cepheid, Hain has not disclosed the precise eis promoter region interrogated by its SL-LPA, which is why this information could not be included (67).
TABLE 1.
Mutations detected in a global sample of MTBC clinical isolatesa
| Mutation position | Variant name | Gene position | Mutation type | Codon position | No. of isolates | No. of sublineages |
|---|---|---|---|---|---|---|
| 776210 | mmpL5 C2271A | 2271 | N | Y757* | 2 | 2 |
| 777499 | mmpL5 C982T | 982 | N | R328* | 2 | 2 |
| 777581 | mmpL5 C900A | 900 | N | Y300* | 293 | 2 |
| 778086 | mmpL5 395insC | 395 | Ins | 132 | 6 | 2 |
| 779127 | mmpR 138insG | 138 | Ins | 46 | 5 | 4 |
| 779181 | mmpR 192-198delG | 192 | Del | 64 | 20 | 4 |
| 779181 | mmpR 192-198insG | 192 | Ins | 64 | 86 | 2 |
| 779407 | mmpR 418insG | 418 | Ins | 140 | 6 | 1 |
| 2714753 | eis C580T | 580 | N | Q194* | 10 | 2 |
| 2715287 | eis 46insC | 46 | Ins | 16 | 3 | 2 |
| 2715305 | eis G28T | 28 | N | E10* | 6 | 2 |
| 2715330 | eis G3A | 3 | S | V1V | 6 | 2 |
| 2715342 | eis G-10A | −10 eis | I | 293 | 18 | |
| 2715344 | eis C-12T | −12 eis | I | 332 | 17 | |
| 2715346 | eis C-14T | −14 eis | I | 181 | 19 | |
| 2715369 | eis G-37T | −37 eis | I | 285 | 8 | |
| 2726105 | ahpC G-88A | −88 ahpC | I | 3,350 | 12 | |
| 2726141 | ahpC C-52A | −52 ahpC | I | 91 | 10 | |
| 2726141 | ahpC C-52T | −52 ahpC | I | 92 | 25 | |
| 2726145 | ahpC G-48A | −48 ahpC | I | 85 | 20 | |
| 3568487 | whiB7 193insG | 193 | Ins | 65 | 3 | 3 |
| 3568488 | whiB7 192delC | 192 | Del | 64 | 573 | 3 |
| 3568547 | whiB7 133delCA | 133 | Del | 45 | 2 | 2 |
| 3568626 | whiB7 54delA | 54 | Del | 18 | 61 | 2 |
| 3568779 | whiB7 T-100C | −100 whiB7 | I | 256 | 2 | |
| 3568857 | whiB7 C-178T | −178 whiB7 | I | 73 | 2 | |
| 3568921 | whiB7 G-242C | −242 whiB7 | I | 117 | 2 | |
| 3569029 | whiB7 A-350G | −350 whiB7 | I | 249 | 3 |
Mutations that occur in our sample of 31,440 clinical isolates within the mmpL5, mmpS5, mmpR, ahpC, eis, and whiB7 coding sequences and the oxyR-ahpC, eis-Rv2417c, whiB7-uvrD2 intergenic regions are shown (Fig. 1). Mutations in regulator regions (mmpR, oxyR-ahpC, eis-Rv2417c, and whiB7-uvrD2) reported in this table were among the four most commonly detected variants in each region. Mutations in regulated regions (mmpL5, mmpS5, ahpC, eis, and whiB7) reported in this table were present in at least two MTBC sublineages. The full set of mutations detected within these genomic regions is reported in Table S1 in the supplemental material. S, synonymous; N, nonsynonymous; I, intergenic.
TABLE 2.
Co-occurrence of regulator resistance mutations and regulon LoF mutationsa
| Mutation in regulator |
Mutation in regulated gene |
Isolate information with co-occurring mutations |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type of mutation | Variant name | Codon position | No. of isolates | Type of mutation | Variant name | Codon position | No. of isolates | No. of isolates with both mutations | Isolate sublineage(s) (no. of isolates) |
| Indel | mmpR 192-198delG | 64 | 20 | Indel | mmpL5 2028insA | 676 | 1 | 1 | 2.2.1.1.1 (1) |
| Indel | mmpR 192-198insG | 64 | 86 | Indel | mmpL5 606delC | 202 | 83 | 82 | 4.11 (82) |
| Indel | mmpR 207insA | 69 | 1 | Indel | mmpL5 1160insCGATG | 387 | 1 | 1 | 2.2.1 (1) |
| SNP | eis C-14T | 181 | Indel | eis 627insC | 209 | 1 | 1 | 2.2.1.1.1.i3 (1) | |
| SNP | eis C-14T | 181 | Indel | eis 486insCT | 162 | 2 | 2 | 4.1.i1.1.1.1 (2) | |
| SNP | eis C-14T | 181 | Indel | eis 473insT | 158 | 1 | 1 | 2.2.1.1.1.i3 (1) | |
| SNP | eis C-14T | 181 | Indel | eis 448delA | 150 | 7 | 7 | 2.2.1.1.1.i3 (7) | |
| SNP | eis C-14T | 181 | Indel | eis 400insG | 134 | 1 | 1 | 2.2.1.1.1 (1) | |
| SNP | eis C-14T | 181 | Indel | eis 279delCGGCGATGCGT | 93 | 1 | 1 | 2.2.1.1.1.i3 (1) | |
| SNP | eis C-14T | 181 | SNP | eis G39A | W13* | 1 | 1 | 2.2.1.1.1 (1) | |
| SNP | eis C-14T | 181 | SNP | eis G38A | W13* | 1 | 1 | 2.2.1.1.1 (1) | |
| SNP | eis C-14T | 181 | Indel | eis 16insC | 6 | 1 | 1 | 2.2.1.1.1 (1) | |
| SNP | eis C-14T | 181 | Indel | eis 15insC | 5 | 1 | 1 | 2.2.1.1.1 (1) | |
| SNP | eis C-14T | 181 | SNP | eis G3A | V1V | 6 | 6 | 1.1.1.1.1 (1), 2.2.1.1.1.i3 (5) | |
| SNP | ahpC G-88A | 3350 | Indel | ahpC 323delC | 108 | 1 | 1 | 3.1.1 (1) | |
| SNP | whiB7 T-147C | 1 | INDEL | whiB7 192delC | 64 | 573 | 1 | 1.2.1.1.1 (1) | |
| Indel | whiB7 −214delG | 9 | INDEL | whiB7 192delC | 64 | 573 | 1 | 1.2.1.1.1 (1) | |
| Indel | whiB7 −316insC | 2 | INDEL | whiB7 192delC | 64 | 573 | 1 | 1.2.1.1.1 (1) | |
A list of antibiotic resistance mutations in regulator regions (mmpR, oxyR-ahpC, eis-Rv2417c, and whiB7-uvrD2) that co-occur with LoF mutations in corresponding regulated regions (mmpL5, mmpS5, mmpR, ahpC, eis, and whiB7) within our sample of 31,440 clinical isolates is shown. More details can be found in Table S3 in the supplemental material.
BDQ/CFZ: mmpR LoF mutations in combination with mmpS5-mmpL5 LoF mutations.
We detected 91 fixed LoF variants in mmpL5-mmpS5-mmpR, of which 35 occurred in at least 2 isolates (Fig. 1B; Table S1). Frameshifts were most common (39/68) in mmpL5, followed by mmpR (21/68) and mmpS5 (8/68). Each gene harbored frameshifts in isolates from at least three MTBC major lineages, indicating parallel evolution (Table S1). The nonsense SNP mmpL5 Y300* was observed in 293 isolates and in 2 genetically distinct lineages, and the insC at nt395 of mmpL5 also occurred in 2 distant lineages (Table 1; Table S1). The mmpR delG in the homopolymer (HP) nt192-198 was observed in 20 isolates from three major lineages, whereas insG in the same HP was observed in 86 isolates from two major lineages (Table 1; Table S1). Noting the frequency of frameshifts in the homopolymer region of mmpR, we investigated nonfixed frameshift variants (i.e., that had within-sample allele frequencies of 10 to 75%) and recorded which isolates had >100× coverage of mmpR, mmpS5, and mmpL5. Frameshift variants at low to intermediate allele frequency were rare and occurred in a total of 6 isolates (3/7,435 in mmpR, 1/8,949 in mmpS5, and 2/6,217 in mmpL5). Two of these isolates had the frameshift insG in the aforementioned mmpR HP at 66% and 71% allele frequencies (Fig. 2; Table S2).
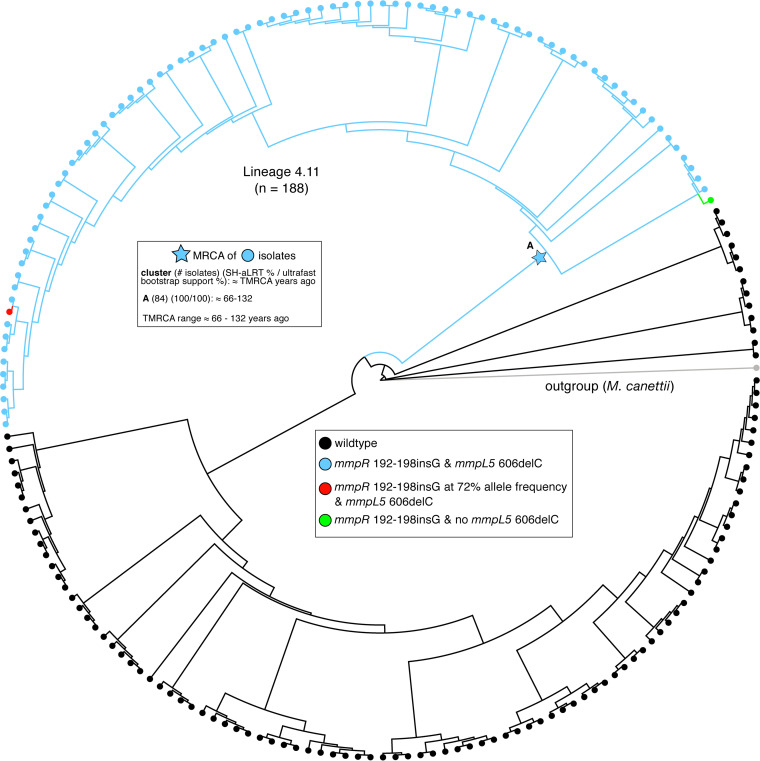
FIG 2.
Phylogeny of 188 sublineage 4.11 isolates. Isolates with both the mmpR 192-198insG and mmpL5 606delC variants are colored in blue (n = 82); isolates carrying neither variant are colored in black (n = 104). One isolate had the mmpR 192-198insG frameshift but not the mmpL5 606delC frameshift (green). Another isolate had the mmpL5 606delC frameshift and 72% of reads supporting the mmpR 192-198insG frameshift (red). Time to most recent common ancestor (TMRCA) estimates for the group of isolates with the mmpR 192-198insG and mmpL5 606delC variants are given in the upper left.
Three different LoF mutations in mmpR coincided with an LoF mutation in mmpL5 (Fig. 2). Of those, insG in the HP nt192-198, which had been repeatedly demonstrated to confer BDQ and CFZ resistance during in vitro selection experiments and patient treatment, occurred in 82 isolates, whereas the other two double mutations were observed in only a single isolate, respectively (18–23). All of the former 82 double-LoF mutants belonged to a monophyletic group within sublineage 4.11 that was mostly multidrug resistant (53 of 59 with known phenotypic data). Most double-LoF mutants were isolated in Lima, Peru, between 1997 and 2012 and represented 43% (82/188) of the isolates from the sublineage 4.11 in our data set (Fig. 2; Table 2; Tables S3 and S4). Among the 84 isolates with co-occurrence of mmpR and mmpL5 LoF, there were no SNPs in the other BDQ resistance locus, atpE.
We constructed a phylogeny of all 188 MTBC sublineage 4.11 isolates to study how the LoF mutations in mmpR and mmpS5-mmpL5 evolved (Fig. 2). The majority of isolates with the mmpR or mmpL5 frameshifts harbored both (82/84), but, based on the topology of the tree, we were unable to determine which of the two frameshifts arose first. Consequently, we could only date the common most recent common ancestor (MRCA). We approximated the age of the MRCA at 66 to 132 years prior to sampling (i.e., well before the use of BDQ or CFZ and likely before the introduction of thioacetazone, which is also exported by mmpS5-mmpL5) (24, 25).
KAN: eis upstream mutations in combination with eis LoF mutations.
We observed 23 unique LoF mutations upstream of eis (Fig. 1C), of which 10 were homoplasic and occurred in at least 5 isolates (Table S1). As expected, the classical G-37T, C-14T, C-12T, and G-10A mutations, which are known to confer KAN resistance based on allelic exchange and/or complementation experiments, were most frequent (Table 1) (14–16). Specifically, 881 isolates with either eis G-37T, C-12T, or G-10A and 179 isolates with eis C-14T did not have any of the other key AG resistance mutations in rrs (i.e., A1401G, C1402T, or G1484T) (see Table S5) (9).
We identified 30 unique LoF mutations in eis, of which 5 were homoplasic and occurred in at least 5 isolates (Table 1; Table S1). These LoF mutations never coincided with eis G-37T, C-12T, or G-10A, whereas this was the case for 21 eis C-14T mutants (i.e., 13 isolates with indels, 6 with a G3A-synonymous change that abolished the valine start codon, and 2 with nonsense mutations) (Table 2; Table S3). MIC data were available for five of these eis double mutants, which confirmed that they were susceptible to KAN, whereas seven eis C-14T control isolates with a wild-type eis-coding region were KAN resistant (Fig. 3; Table 3). The corresponding AMK MIC data mirrored the results for KAN.
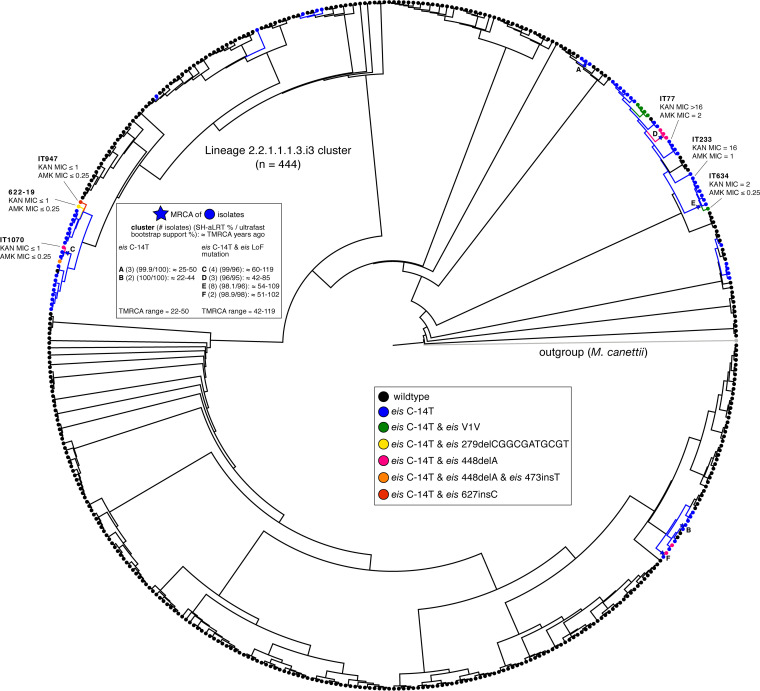
FIG 3.
Phylogeny of 444 sublineage 2.2.1.1.1.3.i3 isolates. Isolates with the eis C-14T promoter SNP and no-LoF variants in eis are colored in blue (n = 61), whereas isolates carrying both the eis C-14T promoter SNP and a LoF mutation in eis are colored according to the legend (n = 14). TMRCA estimates for groups of isolates with the eis C-14T promoter SNP are given in the top left. The MICs (mg/liter) for isolates from this sublineage are also included (MICs for isolates from other sublineages can be found in Table 3).
TABLE 3.
Overview of eis C-14T isolates with eis LoF mutations and corresponding MICs for KAN and AMKa
| Isolate ID | Present in 2.2.1.1.1.3.i3 cluster | rrs A1401G | rrs C1402T | rrs G1484T | eis C-14T | Eis LOF mutation | MIC (mg/liter) of: |
|
|---|---|---|---|---|---|---|---|---|
| KAN | AMK | |||||||
| IT184 | No | A | C | G | Yes | No | 8 | 0.5 |
| IT123 | No | A | C | G | Yes | No | 16 | 0.5 |
| 655-19 | No | A | C | G | Yes | No | 16 | 1 |
| IT233 | Yes | A | C | G | Yes | No | 16 | 1 |
| IT952 | No | A | C | G | Yes | No | 16 | 2 |
| IT524 | No | A | C | G | Yes | No | >16 | 2 |
| IT77 | Yes | A | C | G | Yes | No | >16 | 2 |
| 622-19 | Yes | A | C | G | Yes | Yes (279delCGGCGATGCGT) | ≤1 | ≤0.25 |
| IT1070 | Yes | A | C | G | Yes | Yes (448delA) | ≤1 | ≤0.25 |
| IT947 | Yes | A | C | G | Yes | Yes (627insC) | ≤1 | ≤0.25 |
| 168-19 | No | A | C | G | Yes | Yes (400insG) | ≤1 | ≤0.25 |
| IT634 | Yes | A | C | G | Yes | Yes (V1V) | 2 | ≤0.25 |
| SAMN02419559b | Yes | A | C | G | Yes | Yes (V1V) | ||
| SAMN02419535b | Yes | A | C | G | Yes | Yes (V1V) | ||
| SAMN02419543b | Yes | A | C | G | Yes | Yes (V1V) | ||
| SAMN02419586b | Yes | A | C | G | Yes | Yes (V1V) | ||
| SAMN07236283b | No | A | C | G | Yes | Yes (V1V) | ||
| SAMEA1016073 | Yes | A | C | G | Yes | Yes (448delA) | ||
| SAMEA1403685 | Yes | A | C | G | Yes | Yes (448delA) | ||
| SAMEA1403638 | Yes | A | C | G | Yes | Yes (448delA) | ||
| SAMN02584676b | Yes | A | C | G | Yes | Yes (448delA) | ||
| SAMN04633319b | Yes | A | C | G | Yes | Yes (448delA) | ||
| SAMN08376196b | No | A | C | G | Yes | Yes (W13*) | ||
| SAMN08709032b | No | A | C | G | Yes | Yes (W13*) | ||
| SAMN06210015b | No | A | C | G | Yes | Yes (16insC) | ||
| Peru2946 | No | A | C | G | Yes | Yes (486insCT) | ||
| Peru3354 | No | A | C | G | Yes | Yes (486insCT) | ||
| SAMN02584612b | No | A | C | G | Yes | Yes (15insC) | ||
| SAMN07956543b | Yes | A | C | G | Yes | Yes (448delA and 473insT) | ||
All 29 double mutants lacked the three classical AG resistance mutations in rrs (A, C, and G are the wild-type alleles for the A1401G, C1402T, and G1484T resistance mutations, respectively). Isolates that are part of the 2.2.1.1.1.3.i3 cluster are shown in Fig. 3. MICs were measured using either the UKMYC5 or UKMYC6 broth microdilution plates by Thermo Fisher Scientific (54). The provisional CCs for KAN and AMK for these plates are 4 and 1 mg/liter, respectively. Unlike for KAN, eis C-14T only has a modest effect on the MIC of AMK (i.e., the MIC distribution of this mutation spans the CC when the efflux pump is active, which is in line with data from other media) (9, 14–16, 68). Consequently, KAN is a more sensitive agent to detect an inactive efflux pump than AMK. More details can be found in Table S6 in the supplemental material.
BioSample accession numbers are shown.
The most common MTBC sublineage with the eis double mutants was 2.2.1.1.1.i3 (14/22 isolates). Of the 31,440 isolates, 444 belonged to this sublineage and clustered closely based on their pairwise SNP distance. The phylogeny of these 444 isolates showed that eis C-14T arose more than nine times independently (Fig. 3). We approximated the MRCA for the six groups of isolates that had high bootstrap support. For the two groups of isolates that only harbored the eis C-14T mutation, the MRCA was dated 22 to 50 years ago, in line with KAN’s introduction into clinical use in approximately 1958 (26). Of the 75 isolates with the eis C-14T promoter mutation, 14 also harbored an eis LoF mutation that arose 9 times independently (Fig. 3; Table 2). In each instance, the LoF variant emerged within a clade of eis C-14T mutants, suggesting that it appeared later in time. We compared the MRCA of the clades with double mutants to those with eis C-14T only. We found the MRCA of the double mutants to be older on average, suggesting that time and possibly fluctuating evolutionary pressures are needed for LoF to develop in an eis C-14T background.
KAN: whiB7 upstream mutations in combination with whiB7 or eis LoF mutations.
We found 116 unique SNPs upstream of whiB7 (Fig. 1C), of which 8 were homoplasic and occurred in at least 5 isolates (Table 1; Table S1). We identified 10 unique LoF mutations in whiB7, 2 of which (nt193insG and nt133delCA) evolved repeatedly, across 657 isolates. The most frequent mutation (nt192delC) occurred in 573 sublineage 1.2.1.1 isolates, which was in agreement with earlier findings (7). This was the only LoF mutation in whiB7 to coincide with an upstream mutation (i.e., in three isolates in total, each with a different upstream mutation) (Table 2; Table S3). Because none of these upstream mutations had been described in the literature, it was unclear whether these represented potential examples of epistasis (27, 28). Finally, no LoF mutations in eis were found in isolates harboring mutations upstream of whiB7.
DISCUSSION
Although our analysis did not yield any strong evidence for epistasis involving ahpC or whiB7, our finding that epistasis is possible due to LoF mutations in eis is not only relevant for AGs but has wider implications for the interpretation of sequencing data. First, with the exception of two synonymous mutations in aftA (Rv3792) and fabG1 (Rv1483) that confer ethambutol and ethionamide/INH resistance, respectively, by creating alternative promoters, synonymous mutations are typically excluded a priori from the analysis of WGS data (29, 30). We demonstrated that this assumption is not sound for start codons given that only one of the four triplets encoding valine can act as a start codon (i.e., GTC). Second, evidence for epistasis argues strongly that multivariate prediction approaches are needed for accurate resistance prediction from sequencing data.
It is notable that eis LoF mutations coincided only with eis C-14T mutants, even though isolates with eis G-37T, C-12T, and G-10A without any AG resistance mutations in rrs were almost five times more frequent in our data set (Table S5). We hypothesize that because eis C-14T leads to a greater upregulation of eis than the other three mutations, this comes at a fitness cost unless selective antibiotic pressure is maintained (14–16, 31). Indeed, molecular dating and the topology of the tree (Fig. 3) suggested that the LoF mutations arose independently on multiple occasions after the acquisition of eis C-14T. To our knowledge, this represents the strongest evidence to date for genotypic reversion from resistance to a susceptible phenotype for MTBC (32). We would like to stress, however, that even for AMK, this is a rare phenomenon given that of the 179 isolates that harbored eis C-14T without any AG resistance mutations in rrs, only 12% had concomitant eis LoF mutations (Table 2; Table S5). In other words, the cautious approach would be to still interpret eis C-14T as a marker for AMK resistance to construct a relevant regimen unless there is strong evidence that a particular isolate is affected by epistasis (e.g., unlike the SL-LPA, the tNGS assays by ABL and Deeplex actually interrogate part of the eis coding region) (2, 14). This initial treatment decision may then have to be adjusted based on the phenotypic DST result, depending on the laboratory capacity.
Because we did not have BDQ or CFZ MICs for any of the mmpR/mmpL5 double-LoF mutants, it remains to be determined whether these are examples of epistasis (in the case of the Peruvian cluster, this would be unrelated to antibiotic pressure, unlike for eis). We note, however, that indirect evidence exists that is in line with this prediction. Villellas et al. reported the BDQ MICs and mmpR sequence results for baseline isolates from the C208 phase 2b trial of BDQ, which featured five isolates with the same mmpR frameshift that we observed in the Peruvian cluster (Fig. 2) (33, 34). Three of the trial mutants were from South Africa and had 7H11 MICs of 0.25 to 1 mg/liter (i.e., ≥CC of 0.25 mg/liter and, thus, consistent with a functional efflux pump and resistant phenotype if an area of technical uncertainly is set at 0.25 mg/liter, as previously proposed) (34–36). In contrast, even the lowest BDQ concentration tested (i.e., 0.008 mg/liter) inhibited the growth of the remaining two trial mutants that were isolated in 2009 in Lima, Peru (N. Lounis, personal communication). Given that the Peruvian double-LoF cluster from this study was isolated in the same city during the same period, it is possible that the latter two trial isolates were from this cluster, although this remains to be confirmed using WGS data and, ideally, repeat MIC testing to exclude experimental error.
The possibility of epistasis underlines the need for comprehensive microbiological workup of the ongoing clinical trials of BDQ, particularly given that resistance to BDQ is now a criterion for the revised definition of extensively drug-resistant TB (37). mmpR, as well as mmpS5-mmpL5 and the corresponding intergenic region, has to be analyzed along with standardized MIC testing using an on-scale quality control strain for both BDQ and CFZ (35, 38–40). We recommend that discordances between genotypic and phenotypic DST results are confirmed by retesting and, where warranted, followed up with specialized testing (41). For example, the two aforementioned Peruvian results may actually be hypersusceptible to BDQ and CFZ (i.e., lower concentrations would have to be tested to determine the MIC endpoint) (7). If confirmed, this should also apply to mmpS5-mmpL5 LoF mutants with wild-type mmpR (e.g., just over half of the lineage 1.1.1.1 isolates in our data set had a nonsense mutation in mmpL5) and may have implications for the ongoing trials of TBAJ-876, TBAJ-587, TBI-166, and OPC-167832, as these agents are also exported by this pump (42–44).
MATERIALS AND METHODS
Sequencing data.
We limited this study to Illumina sequencing data, as it is the most widely used technology used in the TB field. Since we aimed to leverage public sequencing data for this study, we needed to implement a pipeline to quality control and process data of the same format (Illumina). This ensured only high-quality sequencing data ended up in our sample while only excluding a relatively small proportion of public samples sequenced with PacBio (Pacific Biosciences), Oxford Nanopore, or Ion Torrent sequencing technologies. We initially downloaded raw Illumina sequence data for 33,873 clinical isolates from NCBI (45). We identified the BioSample for each isolate and downloaded all of the associated Illumina sequencing runs. Isolates had to meet the following quality control measures for inclusion in our study: (i) at least 90% of the reads had to be taxonomically classified as belonging to MTBC after running the trimmed FASTQ files through Kraken (46), and (ii) at least 95% of bases had to have coverage of at least 10× after mapping the processed reads to the H37Rv reference genome (GenBank accession no. NC_000962.3).
Illumina sequencing FASTQ processing and mapping to H37Rv.
The raw sequence reads from all sequenced isolates were trimmed with version 0.20.4 Prinseq (settings, -min_qual_mean 20) (47) and then aligned to H37Rv with version 0.7.15 of the BWA-MEM algorithm using the -M settings (48). The resulting SAM files were then sorted (settings, SORT_ORDER = coordinate), converted to BAM format, and processed for duplicate removal with version 2.8.0 of Picard (http://broadinstitute.github.io/picard/) (settings, REMOVE_DUPLICATES = true, ASSUME_SORT_ORDER = coordinate). The processed BAM files were then indexed with SAMtools (49). We used Pilon (settings, –variant) on the resulting BAM files to generate VCF files that contained calls for all reference positions corresponding to H37Rv from pileup (50).
Empirical score for difficult-to-call regions.
We assessed the congruence in variant calls between short-read Illumina data and long-read PacBio data for a set of isolates that underwent sequencing with both technologies. Using 31 isolates for which both Illumina and a complete PacBio assembly were available, we evaluated the empirical base pair recall (EBR) of all base pair positions of the H37Rv reference genome. For each sample, the alignments of each high-confidence genome assembly to the H37Rv genome were used to infer the true nucleotide identity of each base pair position. To calculate the empirical base pair recall, we calculated what percentage of the time our Illumina-based variant calling pipeline, across 31 samples, confidently called the true nucleotide identity at a given genomic position. If Pilon variant calls did not produce a confident base call (pass) for the position, it did not count as a correct base call. This yields a metric ranging from 0.0 to 1.0 for the consistency by which each base pair is both confidently and correctly sequenced by our Illumina WGS-based variant calling pipeline for each position on the H37Rv reference genome. An H37Rv position with an EBR score of x% indicates that the base calls made from Illumina sequencing and mapping to H37Rv agreed with the base calls made from the PacBio de novo assemblies in x% of the Illumina-PacBio pairs. We masked difficult-to-call regions by dropping H37Rv positions with an EBR score below 0.9 as part of our variant calling procedure. Full details on the data and methodology can be found elsewhere (51).
Variant calling.
(i) SNP calling.
To prune out low-quality base calls that may have arisen due to sequencing or mapping error, we dropped any base calls that did not meet any of the following criteria: (i) the call was flagged as “pass” by Pilon, (ii) the mean base quality at the locus was >20, (iii) the mean mapping quality at the locus was >30, (iv) none of the reads aligning to the locus supported an insertion/deletion (indel), (v) there was a minimum coverage of 20 reads at the position, and (vi) at least 75% of the reads aligning to that position supported 1 allele (using the INFO.QP field, which gives the proportion of reads supporting each base weighted by the base and mapping quality of the reads, BQ and MQ, respectively, at the specific position). A base call that did not meet all filters (i to vi) was inferred to be low quality/missing.
(ii) Indel calling.
To prune out low-quality indel variant calls, we dropped any indel that did not meet any of the following criteria: (i) the call was flagged as “pass” by Pilon, (ii) the maximum length of the variant was 10 bp, (iii) the mean mapping quality at the locus was >30, (iv) there was a minimum coverage of 20 reads at the position, and (v) at least 75% of the reads aligning to that position supported the indel allele (determined by calculating the proportion of total reads [TD] aligning to the position that supported the insertion or deletion [IC and DC, respectively]). A variant call that met filters i, iii, and iv but not ii or v was inferred as a high-quality call that did not support the indel allele. Any variant call that did not meet all filters i, iii, and iv was inferred as low quality/missing.
(iii) Intermediate allele frequency indel calling.
To call indel variants in which the indel allele was detected at an intermediate frequency, we made the following modification to the “indel calling” filters outlined above. Filter v above was replaced with the following two filters: (vi) at least 10% but less than 75% of the reads aligning to that position supported the indel allele, and (vii) at least 10 reads support the indel allele. The mmpR analysis was restricted to isolates with 100× coverage across ≥99% of the gene.
SNP genotypes matrix.
We detected SNP sites at 899,035 H37Rv reference positions (of which 64,950 SNPs were not biallelic) among our global sample of 33,873 isolates. We constructed an 899,035 by 33,873 genotypes matrix (coded as 0:A, 1:C, 2:G, 3:T, 9:Missing) and filled in the matrix for the allele supported at each SNP site (row) for each isolate according to the “SNP calling” filters outlined above. If a base call at a specific reference position for an isolate did not meet the filter criteria, that allele was coded as “missing.” We excluded 20,360 SNP sites that had an EBR score of <0.90, another 9,137 SNP sites located within mobile genetic element regions (e.g., transposases, integrases, phages, or insertion sequences) (51, 52), and then 31,215 SNP sites with missing calls in >10% of isolates and 2,344 SNP sites located in overlapping genes (coding sequences). These filtering steps yielded a genotypes matrix with dimensions of 835,979 by 33,873. Next, we excluded 1,663 isolates with missing calls in >10% of SNP sites, yielding a genotypes matrix with dimensions 835,979 by 32,210. We used an expanded 96-SNP barcode to type the global lineage of each isolate in our sample (53). We further excluded 325 isolates that either did not get assigned a global lineage, were assigned to more than one global lineage, or were typed as lineage 7 (as only 35 isolates were typed as lineage 7, we reasoned that this sample and resulting phylogeny would be too small to add meaningful data to our convergent evolution analysis). We then excluded 41,760 SNP sites from the filtered genotypes matrix in which the minor allele count was 0, which resulted in a 794,219 by 31,885 matrix. To provide further MTBC lineage resolution on the lineage 4 isolates, we required an MTBC sublineage call for each lineage 4 isolate. We excluded 457 isolates typed as global lineage 4 but had no further sublineage calls and then again excluded 11,654 SNP sites from the filtered genotypes matrix in which the minor allele count was 0. The genotypes matrix used for downstream analysis had dimensions of 782,565 by 31,428, representing 782,565 SNP sites across 31,428 isolates. The global lineage (L) breakdown of the 31,428 isolates was 2,815 isolates in L1, 8,090 isolates in L2, 3,398 isolates in L3, 16,931 isolates in L4, 98 isolates in L5, and 96 isolates in L6.
Indel genotypes matrix.
We detected 53,167 unique indel variants within 50,576 H37Rv reference positions among our global sample of 33,873 isolates. We constructed a 53,167 by 33,873 genotypes matrix (coded as 1, high-quality call for the indel allele; 0, high-quality call not for the indel allele; and 9, missing) and filled in the matrix according to whether the indel allele was supported for each indel variant (row) for each isolate, according to the “Indel calling” filters outlined above. If a variant call at the reference position for an indel variant did not meet the filter criteria, that call was coded as missing. We excluded 2,006 indel variants that had an EBR score of <0.90, another 694 indel variants located within mobile genetic element regions, and then 207 indel variants located in overlapping genes (coding sequences). These filtering steps yielded a genotypes matrix with dimensions of 50,260 by 33,873. Next, we excluded any isolate that was dropped while constructing the SNP genotypes matrix to retain the same 31,428 isolates as described above. The genotypes matrix used for downstream analysis had dimensions of 50,260 by 31,428.
Mixed-allele frequency indel genotypes matrix.
After following the same filtering steps outlined above in “Indel genotypes matrix,” we detected 7,731 unique indel variants in our filtered sample of 31,428 isolates in which at least one isolate supported each indel variant at an intermediate allele frequency (10% ≤ intermediate allele frequency (AF) < 75%). We constructed a 7,731 by 31,428 genotypes matrix (coded as 0, high-quality call not for the indel allele, −9, missing; or 10 to 74, the percentage of reads supporting the indel allele) and filled the matrix according to whether the indel allele was supported at an intermediate allele frequency for each indel variant (row) for each isolate, according to the filters outlined above in “Intermediate allele frequency indel calling.” To determine the limit of detection for indels that might be present at lower allele frequencies, we calculated the number of isolates in our sample that have 100× coverage in ≥99% of the locus for mmpR (7,435), mmpS5 (8,949), and mmpL5 (6,217) (Table S2 in the supplemental material). We retained only frameshift indels yielding a genotypes matrix with dimensions of 5,925 by 31,428 and interrogated only the mmpR-mmpS5-mmpL5 chromosomal region for the presence of mixed indels (Table S2).
Inclusion and processing of 12 eis C-14T mutants with AG MICs.
We added 12 clinical eis C-14T mutants to the data set for which we had KAN and AMK MICs, and some of which had an LoF mutation in eis (ENA accession numbers available in Table S7). The MICs were determined using the UKMYC5 or UKMYC6 broth microdilution plates by Thermo Fisher Scientific, which are not WHO endorsed but have been used by WHO for its recent catalogue of resistance mutations (9, 54, 55). We processed the raw sequencing reads according to the methods described above to generate VCF files. We genotyped SNPs for these isolates at the 782,565 SNP sites and genotyped indels for the 50,260 indel variants previously identified using the same filters described above to construct 782,565 by 12 and 50,260 by 12 matrices, respectively.
During analysis, we observed that 3/12 isolates (IT947, 622-19, and 168-19) carried the eis C-14T promoter resistance mutation and no observed LoF mutation in eis but were phenotypically susceptible according to KAN MICs. Upon further inspection of the VCF files for these isolates, we found that all three isolates had an LoF mutation in eis that we originally did not detect per our variant calling methodology. We found that one isolate (622-19) had an 11-bp deletion in eis, which was not represented in the 50,260 indel variants since we restricted our analysis to indels ≤10 bp, and it consequently was excluded from our 50,260 by 12 matrix. Each of the other two strains, IT947 and 168-19, had a different 1-bp insertion in eis that was not identified in our original pool of 31,428 isolates, so it also was also not represented in the 50,260 by 12 matrix. We updated our variant call data by incorporating these newly identified variants (Table 2; Tables S1 and S3).
Targeted chromosomal regions.
We queried our SNP and indel matrices for the following types of mutations in the following regions of the H37Rv reference genome: (i) for mmpR-mmpS5-mmpL5, the coding sequences for mmpR (778990 to 779487), mmpS5 (778477 to 778905), and mmpL5 (775586 to 778480) for nonsense SNVs (single nucleotide variant), frameshift indels, missense SNVs that abolish the start codon and synonymous SNVs that abolish the start codon for mmpR which starts with a valine (we did not check for synonymous SNVs at the first codon for mmpS5 or mmpL5 because these coding sequences start with a methionine); (ii) for upstream ahpC-ahpC, the intergenic region oxyR-ahpC (2726088 to 2726192) for SNVs and indels and the coding sequence for ahpC (2726193 to 2726780) for nonsense SNVs, frameshift indels, and missense SNVs that abolish the start codon. We did not check for synonymous SNVs at the first codon for ahpC because the coding sequence starts with a methionine (and also serves as the initiation site); (iii) for upstream eis-eis, the intergenic region eis-Rv2417c (2715333 to 2715383) for SNVs and indels and the coding sequence for eis (2714124 to 2715332) for nonsense SNVs, frameshift indels, missense SNVs that abolish the start codon, and synonymous SNVs that abolish the start codon; and (iv) for whiB7 and whiB7, the intergenic region whiB7-uvrD2 (3568680 to 3569082) for SNVs and indels, and the coding sequence for whiB7 (3568401 to 3568679) for nonsense SNVs, missense SNVs that abolish the start codon, frameshift indels, and synonymous SNVs that abolish the start codon.
Antibiotic resistance mutations in rrs and atpE.
Resistance to aminoglycosides can occur as a result of mutations in the 1,400-bp region of the 16S rRNA (rrs), where rrs A1401G, C1402T, and G1484T mutations have all been implicated in aminoglycoside resistance (28, 56). To ensure that isolates were not aminoglycoside resistant directly from harboring one of these rrs mutations, we genotyped (with ≥75% allele frequency) the 1401, 1402, and 1484 nucleotide coordinates in rrs for the set of 12 added isolates with eis C-14T promoter resistance mutations and 17 other isolates (from our original set of 31,428 isolates) with coinciding eis C-14T promoter resistance mutation and eis LoF mutations (Fig. 3; Table 3; Table S3). None of these 29 isolates harbored any of the rrs A1401G, C1402T, or G1484T aminoglycoside resistance mutations (Table 3). Similarly, single nucleotide variants in the gene atpE, which encodes the BDQ target, have been associated with high-level BDQ resistance (11). We interrogated the genotypes for 29 SNP sites in atpE (SNPs that were present within our pool of 31,428 isolates) in the 84 isolates that harbored both a frameshift in mmpR and frameshift in mmpL5 (Fig. 1; Table 2) and found that none of the isolates carried a mutant allele at any of these SNP sites.
Phylogeny construction and assessment of convergent evolution.
To generate the trees, we first merged the VCF files of the isolates in the sample (188 lineage 4.11 isolates and 444 lineage 2.2.1.1.1.3.i3 isolates) with BCFtools (49). We then removed repetitive antibiotic resistance and low-coverage regions (53). We generated a multisequence FASTA alignment from the merged VCF file with vcf2phylip (version 1.5; https://zenodo.org/record/2540861#.YTfJ345KhPY). We constructed the phylogenetic tree with IQ-TREE (57). We used the mset option to restrict model selection to GTR models, implemented the automatic model selection with ModelFinder Plus (58), and computed the SH-aLRT test and bootstrap values with UFBoot (59) with 1,000 bootstrap replicates.
To quantify the number of independent mutational events (SNPs and indels) in the original sample of 31,428, we grouped isolates into 8 groups based on genetic similarity, 5 groups corresponding to global lineages 1, 2, 3, 5, and 6, and 3 groups for global lineage 4. We constructed eight phylogenies from these groups and then used the genotypes in conjunction with the phylogenies to assess the number of independent arisings for each mutation observed. We used an ancestral reconstruction approach to quantify the number of times each SNV arose independently in the phylogenies using SNPPar (60). This yielded a homoplasy score or an estimate for the number of independent arisings for each SNV (Table S1). To quantify the number of independent arisings for each indel, we developed a simple method to count the number of times each indel allele “breaks” the phylogenies. If a given mutant allele is observed in two separate parts of a phylogeny, then we can assume that this allele arose twice in the pool of isolates used to construct the tree. We calculated a homoplasy score by counting these topology disruptions for both SNVs and indels. The results for the SNVs were congruent with the homoplasy scores computed from the ancestral reconstructions, validating this approach for computing homoplasy scores for indels.
MRCA dating approximation.
To date the arising of a specific mutation within a group of isolates on a phylogeny, we looked for groups of isolates on the trees that carried the mutant allele of interest. We grouped isolates according to the following principles: (i) a group of isolates had to be a subtree of 2 or more monophyletic mutants, and (ii) we identified the MRCA of all mutants in that subtree assuming that reversion of mutations is impossible. For a given group, we checked that the MRCA of the isolates had an SH-aLRT of ≥80% and an ultrafast bootstrap support of ≥95%. If these conditions were satisfied, indicating high confidence in the branch, we then calculated the median branch length (SNPs per site) between the MRCA and the tips. We multiplied the median branch length (SNPs per site) by the number of sites in the SNP concatenate used to construct the tree to get the median branch length in SNPs per genome. Molecular clock estimates for MTBC range from 0.3 to 0.6 SNPs per genome per year, we divided the branch lengths in SNPs per genome by 0.3 SNPs per genome per year and 0.6 SNPs per genome per year to get upper and lower bound estimates for the MRCA age.
Data analysis and variant annotation.
Data analysis was performed using custom scripts run in Python and interfaced with iPython (61). Statistical tests were run with statsmodels (62), and figures were plotted using Matplotlib (63). NumPy (64), Biopython (65), and pandas (66) were all used extensively in data cleaning and manipulation. Functional annotation of SNPs was done in Biopython using the H37Rv reference genome and the corresponding genome annotation. For every SNP variant called, we used the H37Rv reference position provided by the Pilon (50)-generated VCF file to determine the nucleotide and codon positions if the SNP was located within a coding sequence in H37Rv. We extracted any overlapping coding DNA sequence (CDS) region and annotated SNPs accordingly; each overlapping CDS region was then translated into its corresponding peptide sequence with both the reference and alternate allele. SNPs in which the peptide sequences did not differ between alleles were labeled synonymous, SNPs in which the peptide sequences did differ were labeled nonsynonymous, and if there were no overlapping CDS regions for that reference position, then the SNP was labeled intergenic. Functional annotation of indels was also done in Biopython using the H37Rv reference genome and the corresponding genome annotation. For every indel variant called, we used the H37Rv reference position provided by the Pilon-generated VCF file to determine the nucleotide and codon positions if the indel was located within a coding sequence in H37Rv. An indel variant was classified as in-frame if the length of the indel allele was divisible by three; otherwise, it was classified as a frameshift.
Availability of genomes and scripts.
All MTBC genomes were collected from NCBI and are publicly available (Table S7). All packages and software used in this study have been noted in Materials and Methods. Custom scripts written in Python version 2.7.15 were used to conduct all analyses and interfaced via Jupyter Notebooks (https://github.com/farhat-lab/epistasis-KAN-BDQ-Mtbc-resistance).
ACKNOWLEDGMENTS
We thank Koné Kaniga and Nacer Lounis for sharing information about the C208 trial conducted by Janssen and Thomas Schön for helpful discussions regarding aminoglycoside resistance. We thank the members of the Farhat lab for helpful discussions and comments on the research project and manuscript.
R.V. was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE1745303. C.U.K. received an observership from the European Society of Clinical Microbiology and Infectious Diseases. M.R.F. was supported by NIH NIAID R01 AI55765. Portions of this research were conducted on the O2 High Performance Compute Cluster, supported by the Research Computing Group at Harvard Medical School.
C.U.K. is a consultant for Becton, Dickinson; the Foundation for Innovative New Diagnostics; and the TB Alliance. C.U.K.’s work for Becton, Dickinson involves a collaboration with Janssen and Thermo Fisher Scientific. C.U.K. worked as a consultant for QuantuMDx, the Stop TB Partnership, the WHO Global TB Program, and WHO Regional Office for Europe. C.U.K. gave a paid educational talk for Oxford Immunotec. Hain Lifescience covered C.U.K.’s travel and accommodation to present at a meeting. C.U.K. is an unpaid advisor to BioVersys and GenoScreen.
Footnotes
Supplemental material is available online only.
Contributor Information
Roger Vargas, Jr., Email: roger_vargas@g.harvard.edu.
Maha R. Farhat, Email: Maha_Farhat@hms.harvard.edu.
REFERENCES
- 1.World Health Organization. 2020. Global tuberculosis report. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/rest/bitstreams/1312164/retrieve. [Google Scholar]
- 2.Mohamed S, Köser CU, Salfinger M, Sougakoff W, Heysell SK. 2021. Targeted next-generation sequencing: a Swiss army knife for mycobacterial diagnostics? Eur Respir J 57:2004077. 10.1183/13993003.04077-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajileye A, Alvarez N, Merker M, Walker TM, Akter S, Brown K, Moradigaravand D, Schön T, Andres S, Schleusener V, Omar SV, Coll F, Huang H, Diel R, Ismail N, Parkhill J, de Jong BC, Peto TEA, Crook DW, Niemann S, Robledo J, Smith EG, Peacock SJ, Köser CU. 2017. Some synonymous and nonsynonymous gyrA mutations in Mycobacterium tuberculosis lead to systematic false-positive fluoroquinolone resistance results with the Hain GenoType MTBDRsl assays. Antimicrob Agents Chemother 61:e02169-16. 10.1128/AAC.02169-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro RA, Ross A, Kamwela L, Reinhard M, Loiseau C, Feldmann J, Borrell S, Trauner A, Gagneux S. 2020. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol Biol Evol 37:195–207. 10.1093/molbev/msz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagneux S. 2018. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 16:202–213. 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 6.Köser CU, Bryant JM, Parkhill J, Peacock SJ. 2013. Consequences of whiB7 (Rv3197A) mutations in Beijing genotype isolates of the Mycobacterium tuberculosis complex. Antimicrob Agents Chemother 57:3461–3461. 10.1128/AAC.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merker M, Kohl TA, Barilar I, Andres S, Fowler PW, Chryssanthou E, Ängeby K, Jureen P, Moradigaravand D, Parkhill J, Peacock SJ, Schön T, Maurer FP, Walker T, Köser C, Niemann S. 2020. Phylogenetically informative mutations in genes implicated in antibiotic resistance in Mycobacterium tuberculosis complex. Genome Med 12:27–28. 10.1186/s13073-020-00726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viney K, Linh NN, Gegia M, Zignol M, Glaziou P, Ismail N, Kasaeva T, Mirzayev F. 2021. New definitions of pre-extensively and extensively drug-resistant tuberculosis: update from the World Health Organization. European Respiratory J 57:2100361. 10.1183/13993003.00361-2021. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization. 2021. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/9789240028173. [Google Scholar]
- 10.World Health Organization. 2021. WHO operational handbook on tuberculosis. Module 3: diagnosis - rapid diagnostics for tuberculosis detection 2021 update. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/rest/bitstreams/1354706/retrieve. [Google Scholar]
- 11.Kadura S, King N, Nakhoul M, Zhu H, Theron G, Köser CU, Farhat M. 2020. Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother 75:2031–2043. 10.1093/jac/dkaa136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viljoen A, Dubois V, Girard‐Misguich F, Blaise M, Herrmann J, Kremer L. 2017. The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol Microbiol 104:889–904. 10.1111/mmi.13675. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto K, Nakata N, Mukai T, Kawagishi I, Ato M. 2021. Coexpression of MmpS5 and MmpL5 contributes to both efflux transporter MmpL5 trimerization and drug resistance in Mycobacterium tuberculosis. mSphere 6:e00518-20. 10.1128/mSphere.00518-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2018. Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/handle/10665/260470. [Google Scholar]
- 15.Zaunbrecher MA, Sikes RD, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 106:20004–20009. 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pholwat S, Stroup S, Heysell S, Ogarkov O, Zhdanova S, Ramakrishnan G, Houpt E. 2016. eis promoter C14G and C15G mutations do not confer kanamycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60:7522–7523. 10.1128/AAC.01775-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiner-Oms Á, Berney M, Boinett C, González-Candelas F, Young DB, Gagneux S, Jacobs WR, Parkhill J, Cortes T, Comas I. 2019. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nat Commun 10:3994. 10.1038/s41467-019-11948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andres S, Merker M, Heyckendorf J, Kalsdorf B, Rumetshofer R, Indra A, Hofmann-Thiel S, Hoffmann H, Lange C, Niemann S, Maurer FP. 2020. Bedaquiline-resistant tuberculosis: dark clouds on the horizon. Am J Respir Crit Care Med 201:1564–1568. 10.1164/rccm.201909-1819LE. [DOI] [PubMed] [Google Scholar]
- 19.de Vos M, Ley SD, Wiggins KB, Derendinger B, Dippenaar A, Grobbelaar M, Reuter A, Dolby T, Burns S, Schito M, Engelthaler DM, Metcalfe J, Theron G, van Rie A, Posey J, Warren R, Cox H. 2019. Bedaquiline microheteroresistance after cessation of tuberculosis treatment. N Engl J Med 380:2178–2180. 10.1056/NEJMc1815121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghodousi A, Rizvi AH, Baloch AQ, Ghafoor A, Khanzada FM, Qadir M, Borroni E, Trovato A, Tahseen S, Cirillo DM. 2019. Acquisition of cross-resistance to bedaquiline and clofazimine following treatment for tuberculosis in Pakistan. Antimicrob Agents Chemother 63:e00915-19. 10.1128/AAC.00915-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peretokina IV, Krylova LY, Antonova OV, Kholina MS, Kulagina EV, Nosova EY, Safonova SG, Borisov SE, Zimenkov DV. 2020. Reduced susceptibility and resistance to bedaquiline in clinical M. tuberculosis isolates. J Infect 80:527–535. 10.1016/j.jinf.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenkalb L, Carter J, Spitaleri A, Iqbal Z, Hunt M, Malone K, Utpatel C, Cirillo DM, Rodrigues C, Nilgiriwala KS. 2021. Deciphering bedaquiline and clofazimine resistance in tuberculosis: an evolutionary medicine approach. bioRxiv 10.1101/2021.03.19.436148. [DOI]
- 23.Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y. 2015. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70:2507–2510. 10.1093/jac/dkv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halloum I, Viljoen A, Khanna V, Craig D, Bouchier C, Brosch R, Coxon G, Kremer L. 2017. Resistance to thiacetazone derivatives active against Mycobacterium abscessus involves mutations in the MmpL5 transcriptional repressor MAB_4384. Antimicrob Agents Chemother 61:e02509-16. 10.1128/AAC.02509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Z, Lienhardt C, McIlleron H, Nunn AJ, Wang X. 2010. Global tuberculosis drug development pipeline: the need and the reality. Lancet 375:2100–2109. 10.1016/S0140-6736(10)60359-9. [DOI] [PubMed] [Google Scholar]
- 26.Ektefaie Y, Dixit A, Freschi L, Farhat MR. 2021. Globally diverse Mycobacterium tuberculosis resistance acquisition: a retrospective geographical and temporal analysis of whole genome sequences. Lancet Microbe 2:e96–e104. 10.1016/S2666-5247(20)30195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heyckendorf J, Andres S, Köser CU, Olaru ID, Schön T, Sturegård E, Beckert P, Schleusener V, Kohl TA, Hillemann D. 2018. What is resistance? Impact of phenotypic versus molecular drug resistance testing on therapy for multi-and extensively drug-resistant tuberculosis. Antimicrob Agents Chemother 62:e01550-17. 10.1128/AAC.01550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves AZ, Campbell PJ, Sultana R, Malik S, Murray M, Plikaytis BB, Shinnick TM, Posey JE. 2013. Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob Agents Chemother 57:1857–1865. 10.1128/AAC.02191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando H, Miyoshi‐Akiyama T, Watanabe S, Kirikae T. 2014. A silent mutation in mabA confers isoniazid resistance on Mycobacterium tuberculosis. Mol Microbiol 91:538–547. 10.1111/mmi.12476. [DOI] [PubMed] [Google Scholar]
- 30.Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN, Chatterjee D, Fleischmann R, Alland D. 2013. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet 45:1190–1197. 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanz-García F, Anoz-Carbonell E, Pérez-Herrán E, Martín C, Lucía A, Rodrigues L, Aínsa JA. 2019. Mycobacterial aminoglycoside acetyltransferases: a little of drug resistance, and a lot of other roles. Front Microbiol 10:46. 10.3389/fmicb.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson E, Lin S, Pinsky B, Desmond E, Banaei N. 2009. First documentation of isoniazid reversion in Mycobacterium tuberculosis. Int J Tuber Lung Dis 13:1347–1354. [PubMed] [Google Scholar]
- 33.Diacon AH, Pym A, Grobusch MP, de los Rios JM, Gotuzzo E, Vasilyeva I, Leimane V, Andries K, Bakare N, De Marez T, Haxaire-Theeuwes M, Lounis N, Meyvisch P, De Paepe E, van Heeswijk RPG, Dannemann B. 2014. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 371:723–732. 10.1056/NEJMoa1313865. [DOI] [PubMed] [Google Scholar]
- 34.Villellas C, Coeck N, Meehan CJ, Lounis N, de Jong B, Rigouts L, Andries K. 2017. Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 72:684–690. 10.1093/jac/dkw502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckert P, Sanchez-Padilla E, Merker M, Dreyer V, Kohl TA, Utpatel C, Köser CU, Barilar I, Ismail N, Omar SV, Klopper M, Warren RM, Hoffmann H, Maphalala G, Ardizzoni E, de Jong BC, Kerschberger B, Schramm B, Andres S, Kranzer K, Maurer FP, Bonnet M, Niemann S. 2020. MDR M. tuberculosis outbreak clone in Eswatini missed by Xpert has elevated bedaquiline resistance dated to the pre-treatment era. Genome Med 12:104. 10.1186/s13073-020-00793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmo C, Millard J, Brien K, Moodley S, van Dorp L, Lutchminarain K, Wolf A, Grant AD, Balloux F, Pym AS, Padayatchi N, O'Donnell M. 2020. Bedaquiline resistance in drug-resistant tuberculosis HIV co-infected patients. Eur Respir J 55:1902383. 10.1183/13993003.02383-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alagna R, Cabibbe AM, Miotto P, Saluzzo F, Köser CU, Niemann S, Gagneux S, Rodrigues C, Rancoita PVM, Cirillo DM. 2021. Is the new WHO definition of extensively drug-resistant tuberculosis easy to apply in practice? Eur Respir J 58:2100959. 10.1183/13993003.00959-2021. [DOI] [PubMed] [Google Scholar]
- 38.Kaniga K, Cirillo DM, Hoffner S, Ismail NA, Kaur D, Lounis N, Metchock B, Pfyffer GE, Venter A. 2016. A multilaboratory, multicountry study to determine bedaquiline MIC quality control ranges for phenotypic drug susceptibility testing. J Clin Microbiol 54:2956–2962. 10.1128/JCM.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schön T, Köser CU, Werngren J, Viveiros M, Georghiou S, Kahlmeter G, Giske C, Maurer F, Lina G, Turnidge J, van Ingen J, Jankovic M, Goletti D, Cirillo DM, Santin M, Cambau E, ESGMYC . 2020. What is the role of the EUCAST reference method for MIC testing of the Mycobacterium tuberculosis complex? Clin Microbiol Infect 26:1453–1455. 10.1016/j.cmi.2020.07.037. [DOI] [PubMed] [Google Scholar]
- 40.Schön T, Matuschek E, Mohamed S, Utukuri M, Heysell S, Alffenaar J-W, Shin S, Martinez E, Sintchenko V, Maurer FP, Keller PM, Kahlmeter G, Köser CU. 2019. Standards for MIC testing that apply to the majority of bacterial pathogens should also be enforced for Mycobacterium tuberculosis complex. Clin Microbiol Infect 25:403–405. 10.1016/j.cmi.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köser C, Robledo J, Shubladze N, Schön T, Dolinger D, Salfinger M. Guidance is needed to mitigate the consequences of analytic errors during antimicrobial susceptibility testing for TB. Int J Tuber Lung Dis, in press. [DOI] [PubMed] [Google Scholar]
- 42.Hariguchi N, Chen X, Hayashi Y, Kawano Y, Fujiwara M, Matsuba M, Shimizu H, Ohba Y, Nakamura I, Kitamoto R, Shinohara T, Uematsu Y, Ishikawa S, Itotani M, Haraguchi Y, Takemura I, Matsumoto M. 2020. OPC-167832, a novel carbostyril derivative with potent antituberculosis activity as a DprE1 inhibitor. Antimicrob Agents Chemother 64:e02020-19. 10.1128/AAC.02020-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Converse PJ, Upton AM, Mdluli K, Fotouhi N, Nuermberger EL. 2021. Comparative efficacy of the novel diarylquinoline TBAJ-587 and bedaquiline against a resistant Rv0678 mutant in a mouse model of tuberculosis. Antimicrob Agents Chemother 65:e02418-20. 10.1128/AAC.02418-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, Wang B, Fu L, Zhu H, Guo S, Huang H, Yin D, Zhang Y, Lu Y. 2019. In vitro and in vivo activities of the riminophenazine TBI-166 against Mycobacterium tuberculosis. Antimicrob Agents Chemother 63:e02155-18. 10.1128/AAC.02155-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. 2000. GenBank. Nucleic Acids Res 28:15–18. 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vargas R, Freschi L, Marin M, Epperson LE, Smith M, Oussenko I, Durbin D, Strong M, Salfinger M, Farhat MR. 2021. In-host population dynamics of Mycobacterium tuberculosis complex during active disease. Elife 10:e61805. 10.7554/eLife.61805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503. 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freschi L, Vargas R, Hussain A, Kamal SM, Skrahina A, Tahseen S, Ismail N, Barbova A, Niemann S, Cirillo DM. 2020. Population structure, biogeography and transmissibility of Mycobacterium tuberculosis. bioRxiv 10.1101/2020.09.29.293274. [DOI] [PMC free article] [PubMed]
- 54.Fowler PW, CRyPTIC Consortium . 2021. Epidemiological cutoff values for a 96-well broth microdilution plate for high throughput research antibiotic susceptibility testing of M. tuberculosis. medRxiv 10.1101/2021.02.24.21252386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rancoita PMV, Cugnata F, Gibertoni Cruz AL, Borroni E, Hoosdally SJ, Walker TM, Grazian C, Davies TJ, Peto TEA, Crook DW, Fowler PW, Cirillo DM, for the CRyPTIC Consortium . 2018. Validating a 14-drug microtiter plate containing bedaquiline and delamanid for large-scale research susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 62:e00344-18. 10.1128/AAC.00344-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kambli P, Ajbani K, Nikam C, Sadani M, Shetty A, Udwadia Z, Georghiou SB, Rodwell TC, Catanzaro A, Rodrigues C. 2016. Correlating rrs and eis promoter mutations in clinical isolates of Mycobacterium tuberculosis with phenotypic susceptibility levels to the second-line injectables. Int J Mycobacteriol 5:1–6. 10.1016/j.ijmyco.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edwards DJ, Duchêne S, Pope B, Holt KE. 2020. SNPPar: identifying convergent evolution and other homoplasies from microbial whole-genome alignments. bioRxiv 10.1101/2020.07.08.194480. [DOI] [PMC free article] [PubMed]
- 61.Pérez F, Granger BE. 2007. IPython: a system for interactive scientific computing. Comput Sci Eng 9:21–29. 10.1109/MCSE.2007.53. [DOI] [Google Scholar]
- 62.Seabold S, Perktold J. 2010. Statsmodels: econometric and statistical modeling with python. Proc 9th Python Sci Conf 57:92–96. 10.25080/Majora-92bf1922-011. [DOI] [Google Scholar]
- 63.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 64.Van Der Walt S, Colbert SC, Varoquaux G. 2011. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 13:22–30. 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- 65.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKinney W. 2010. Data structures for statistical computing in Python. Proc 9th Python Sci Conf 445:56–56. 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- 67.Hain Lifescience. 2017. GenoType MTBDRsl VER 2.0 - molecular genetic assay for identification of the M. tuberculosis complex and its resistance to fluoroquinolones and aminoglycosides/cyclic peptides from sputum specimens or cultivated samples package insert. IFU-317A-04. Hain Lifescience, Nehren, Germany. [Google Scholar]
- 68.Gygli SM, Keller PM, Ballif M, Blöchliger N, Hömke R, Reinhard M, Loiseau C, Ritter C, Sander P, Borrell S, Collantes Loo J, Avihingsanon A, Gnokoro J, Yotebieng M, Egger M, Gagneux S, Böttger EC. 2019. Whole-genome sequencing for drug resistance profile prediction in Mycobacterium tuberculosis. Antimicrob Agents Chemother 63:e02175-18. 10.1128/AAC.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information. Download AAC.01164-21-s0001.pdf, PDF file, 0.2 MB (235.7KB, pdf)
Table S1. Download AAC.01164-21-s0002.xlsx, XLSX file, 0.04 MB (36.8KB, xlsx)
Table S2. Download AAC.01164-21-s0003.xlsx, XLSX file, 0.01 MB (12KB, xlsx)
Table S3. Download AAC.01164-21-s0004.xlsx, XLSX file, 0.01 MB (15.2KB, xlsx)
Table S4. Download AAC.01164-21-s0005.xlsx, XLSX file, 0.01 MB (15KB, xlsx)
Table S5. Download AAC.01164-21-s0006.xlsx, XLSX file, 0.01 MB (9.3KB, xlsx)
Table S6. Download AAC.01164-21-s0007.xlsx, XLSX file, 0.01 MB (13.9KB, xlsx)
Table S7. Download AAC.01164-21-s0008.xlsx, XLSX file, 0.6 MB (724.5KB, xlsx)