ABSTRACT
Laboratories submit all carbapenem-resistant Enterobacter, Escherichia coli, and Klebsiella species to the Alameda County Public Health Department (ACPHD). ACPHD evaluated 75 isolates submitted during 9 months for susceptibility to imipenem-relebactam (I-R) and, using whole-genome sequencing, identified β-lactamase genes. Of 60 (80%) isolates susceptible to I-R, 8 (13%) had detectable carbapenemase genes, including 4 KPC, two NDM, and two OXA-48-like; we described the relationship between the presence of β-lactamase resistance genes and susceptibility to I-R.
KEYWORDS: β-lactamase inhibitor, NDM, carbapenem resistance, imipenem-relebactam, whole-genome sequencing
INTRODUCTION
The U.S. Centers for Disease Control and Prevention (CDC) has designated carbapenem-resistant Enterobacterales (CRE), a group of organisms that cause infections with limited treatment options, as a top-tier public health threat that requires urgent and aggressive action, including development of new antibiotics (1). Although β-lactam combination agents such as ceftazidime-avibactam or meropenem-vaborbactam have become available, resistance to these agents has emerged; thus, evaluating resistance to other combination agents is a growing need (2–4).
While imipenem is an effective treatment for infections caused by class C cephalosporinases such as AmpC and extended-spectrum β-lactamase-(ESBL) enzymes, studies have shown that the combination of imipenem with relebactam (I-R) may be an effective treatment for patients with infections caused by bacteria with class A carbapenemases, such as the Klebsiella pneumoniae carbapenemase (KPC) (5–8). I-R has not been shown to be effective against bacteria with class B metallo-β-lactamase (MBL) genes, such as the New Delhi metallo-β-lactamase (NDM), and some studies have concluded that pathogens positive for class D oxacillinase carbapenemase genes (OXA) are also nonsusceptible (9–11).
Most studies examining I-R susceptibility have focused on isolates from single health care centers or large-scale sentinel surveillance programs (12, 13). Several publications utilized whole-genome sequencing (WGS) to identify genetic markers from isolates collected in multiple countries and academic centers (9, 12, 14).
This study is the first reported population-based assessment of CRE susceptibility to I-R and corresponding genetic characteristics in a U.S. health jurisdiction. Alameda County, population 1.67 million, is in the San Francisco Bay Area of California. Since June 2017, the Alameda County Public Health Department (ACPHD) mandates submittal of all isolates from E. coli, Klebsiella spp., and Enterobacter spp. identified by the clinical laboratory as resistant to one or more carbapenems to the ACPHD public health laboratory, where they undergo further genetic testing (15). Resistance genes are ascertained by WGS on the Illumina MiSeq platform. β-Lactamase genes are identified by a customized gene-calling workflow, built into Geneious software version 10.2, and confirmed in Resfinder version 3.2 (CGE webserver).
ACPHD assessed 75 CRE isolates submitted from July 2019 to April 2020 for antimicrobial resistance genetic markers using WGS and subsequently tested for susceptibility to I-R by broth microdilution per CLSI M100 guidelines using established MIC interpretative breakpoints for imipenem (13). We used the BD PhoenixSpec nephelometer, which is designed to measure the turbidity of microbial suspensions equivalent to standardized bacterial suspensions, and colonies were kept in a range between 0.49 and 0.51 for all samples. Isolates with detectable NDM or OXA carbapenemase genes were tested in triplicate to confirm susceptibility, and the median result was included in the analysis. Genomic data were deposited with links to accession number PRJNA788096 in the NCBI BioProject database.
To assess genetic relatedness, a phylogenetic analysis was performed using the k-mer distance method with the Complete algorithm for clustering, and the tree was drawn using Geneious version 10.2, created by Biomatters. Fisher’s exact test was used to assess statistically significant differences between proportions; all analyses were conducted in R version 4.0 (R Core Team).
Out of 75 specimens, 27% had detectable carbapenemase genes, including 9 (60%) of 15 Klebsiella pneumoniae, 10 (42%) of 24 E. coli, and 1 (3%) of 29 Enterobacter cloacae complex isolates, representing 7 different carbapenemase alleles (Table 1). Out of 20 isolates with detectable carbapenemase genes, 14 (70%) also carried an ESBL gene, which was significantly greater than the proportion of isolates without carbapenemase genes that carried an ESBL gene (20%; P < 0.01) (see Table S1 in the supplemental material).
TABLE 1.
β-Lactamase genes and imipenem-relebactam susceptibility by genus and species
| Genus/species | No. of isolates (%) | I-R susceptible (%)a | No. of carbapenemase genes, n = 20 (27%)b |
No. of ESBL genes, n = 25, (33%)c |
No. of other genes, n = 60 (80%)d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC-2 | KPC-3 | NDM-1 | NDM-5 | NDM-7 | OXA-48 | OXA-181 | SHV | CTX-M | TEM-1 | AmpC | OXA | |||
| E. cloacae complex | 29 (39) | 28 (97) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 5 | 28 | 0 |
| E. coli | 24 (32) | 18 (75) | 0 | 0 | 1 | 6 | 1 | 1 | 1 | 0 | 10 | 9 | 8 | 2 |
| K. pneumoniae | 15 (20) | 7 (47) | 1 | 2 | 4 | 1 | 1 | 0 | 1 | 14 | 2 | 5 | 3 | 5 |
| K. aerogenes | 7 (9) | 7 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| Total | 75 | 60 (80%) | 2 | 2 | 5 | 7 | 2 | 1 | 2 | 15 | 12 | 19 | 43 | 7 |
Susceptible isolates were defined as having MIC less than 2 μg/ml; intermediate resistance was 2 μg/ml, and resistance was greater than 2 μg/ml.
Note that one K. pneumoniae isolate had detectable NDM-5 and OXA-181 genes. Columns are not mutually exclusive; a single isolate may have multiple ESBL or other genes present.
ESBL genes are defined as blaSHV or blaCTX-M.
Other classifications includes ACT group, blaTEM-1, AmpC, and noncarbapenemase, non-ESBL OXA genes (e.g., OXA-1, OXA-9).
A total of 60 (80%) isolates were susceptible to I-R, with an MIC of less than 2 μg/ml (MIC50, <0.5; MIC90, 8.0; range, <0.5 to >32). Among 75 isolates, 1 (1%) showed intermediate resistance (MIC, 2 μg/ml), and 14 (19%) were resistant to I-R (MIC, >2 μg/ml).
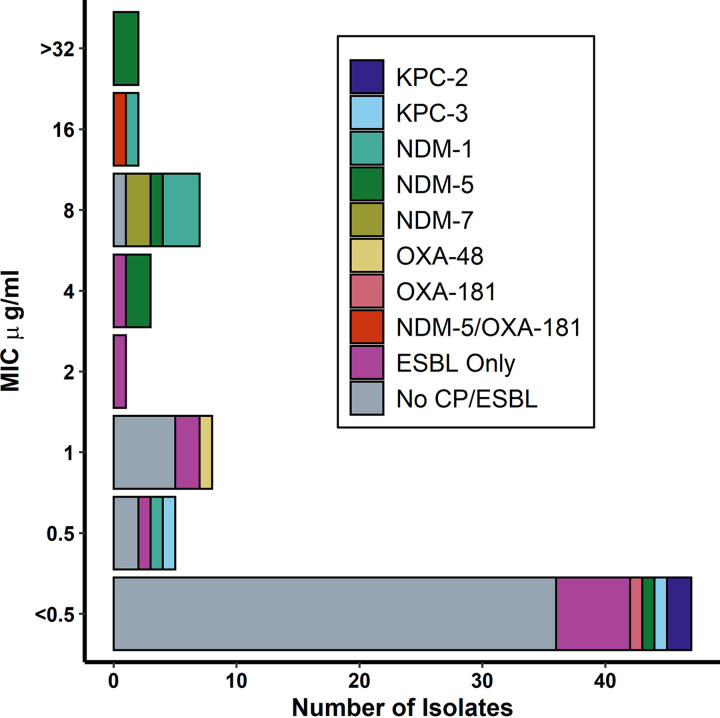
A significantly smaller proportion of isolates with detectable carbapenemase genes were susceptible to I-R (40%) compared to isolates without detectable genes (95%) (P < 0.01); of the 8 I-R-susceptible isolates with detectable carbapenemase genes, we identified 2 NDM, 2 OXA-48-like, and 4 KPC genes (Fig. 1). The single resistant isolate with OXA-48-like genes also contained an NDM-5 gene.
FIG 1.
Carbapenemase and ESBL genes by imipenem-relebactam MIC (n = 75). Of 14 (19%) isolates that were resistant to I-R, 5 had detectable genes for NDM-5, 4 had NDM-1, 2 had NDM-7, and 1 was positive for both NDM-5 and OXA-18 genes. Two resistant isolates did not have detectable carbapenemase genes.
Twelve (86%) resistant isolates (n = 14) had detectable NDM genes. Interestingly, two (14%) of 14 NDM-positive isolates tested susceptible to I-R, indicating that the presence of an NDM gene does not always confer resistance against I-R (Table S1). A potential explanation for this finding is that genotype may not always lead to phenotypic expression of the carbapenemase; while it was beyond the scope of the current study, additional research should assess the expression of MBL genes using phenotypic and genotypic methods.
Conversely, genotypic factors such as porin mutations or undetected MBLs could play a role in explaining why 3 (20%) of 15 I-R-nonsusceptible isolates had no carbapenemase genes (16). We screened any carbapenemase-negative isolate with resistance to either imipenem and/or meropenem (n = 16) for the porin genes ompA, ompC, ompF, ompK35, and ompK36. The three nonsusceptible isolates without carbapenemase genes had at least one porin gene that was either absent or the open reading frame was interrupted by a stop codon, indicating the possible role of porins in I-R resistance (Table S1).
Although isolates were obtained through population-based surveillance of a single geographic area, we did not observe substantial relatedness among CRE isolates. The multiplicity of alleles and sequence types indicate a high level of genetic diversity, which is reflected by the overall lack of clustering in the phylogenetic tree (Fig. S1).
There are limitations to our study. Although our public health laboratory conducts routine genotypic surveillance of all CRE, we did not perform confirmatory antimicrobial susceptibility testing (AST) on isolates submitted by clinical laboratories; clinical lab AST results are listed for 71 (95%) isolates in Table S1. In addition, the number of isolates tested was not sufficient to make causal inferences about the association between antimicrobial resistance genes and I-R susceptibility, and our routine procedures for detection of antimicrobial resistance genes encoding β-lactamases did not exclude the possibility of efflux pumps, which also play a role in carbapenem resistance (17).
Despite these limitations, the genotypic and phenotypic assessment of a population-based sample of carbapenem-resistant Enterobacter spp., E. coli, and Klebsiella spp. demonstrated that certain genetic characteristics, such as the presence of NDM or OXA-48-like carbapenemases, may not be a sufficient cause of I-R resistance.
ACKNOWLEDGMENTS
This study was supported in part by a grant from Merck & Co. (MISP; grant number 58488). The funders had no role in the design of the study or the analysis and interpretation of the results.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.U.S. Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. U.S. Department of Health and Human Services. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 2.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun D, Rubio-Aparicio D, Nelson K, Dudley MN, Lomovskaya O. 2017. Meropenem-vaborbactam resistance selection, resistance prevention, and molecular mechanisms in mutants of KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e01694-17. 10.1128/AAC.01694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dulyayangkul P, Douglas EJA, Lastovka F, Avison MB. 2020. Resistance to ceftazidime/avibactam plus meropenem/vaborbactam when both are used together is achieved in four steps in metallo-β-lactamase-negative Klebsiella pneumoniae. Antimicrob Agents Chemother 64:e00409-20. 10.1128/AAC.00409-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motsch J, Murta de Oliveira C, Stus V, Köksal I, Lyulko O, Boucher HW, Kaye KS, File TM, Brown ML, Khan I, Du J, Joeng H-K, Tipping RW, Aggrey A, Young K, Kartsonis NA, Butterton JR, Paschke A. 2020. RESTORE-IMI 1: a multicenter, randomized, double-blind trial comparing efficacy and safety of imipenem/relebactam vs colistin plus imipenem in patients with imipenem-nonsusceptible bacterial infections. Clin Infect Dis 70:1799–1808. 10.1093/cid/ciz530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livermore DM, Warner M, Mushtaq S. 2013. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 68:2286–2290. 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 7.Papp-Wallace KM, Barnes MD, Alsop J, Taracila MA, Bethel C, Becka S, van Duin D, Kreiswirth BN, Kaye KS, Bonomo RA. 2018. Relebactam is a potent inhibitor of the KPC-2 β-lactamase and restores imipenem susceptibility in KPC-producing Enterobacteriaceae. Antimicrob Agents Chemother 62:e00174-18. 10.1128/AAC.00174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. 2015. Activity of imipenem with relebactam against Gram-negative pathogens from New York City. Antimicrob Agents Chemother 59:5029–5031. 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haidar G, Clancy CJ, Chen L, Samanta P, Shields RK, Kreiswirth BN, Nguyen MH. 2017. Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00642-17. 10.1128/AAC.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush K. 2015. A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int J Antimicrob Agents 46:483–493. 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Lob SH, Hackel MA, Kazmierczak KM, Young K, Motyl MR, Karlowsky JA, Sahm DF. 2017. In vitro activity of imipenem-relebactam against Gram-negative ESKAPE pathogens isolated by clinical laboratories in the United States in 2015 (results from the SMART Global Surveillance Program). Antimicrob Agents Chemother 61:e02209-16. 10.1128/AAC.02209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Simmonds A, Stump S, Giddins MJ, Annavajhala MK, Uhlemann A-C. 2018. Clonal Background, resistance gene profile, and porin gene mutations modulate in vitro susceptibility to imipenem-relebactam in diverse Enterobacteriaceae. Antimicrob Agents Chemother 62:e00573-18. 10.1128/AAC.00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlowsky JA, Lob SH, Kazmierczak KM, Young K, Motyl MR, Sahm DF. 2018. In vitro activity of imipenem-relebactam against clinical isolates of Gram-negative bacilli isolated in hospital laboratories in the United States as part of the SMART 2016 program. Antimicrob Agents Chemother 62:e00169-18. 10.1128/AAC.00169-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt-Malan SM, Mishra AJ, Mushtaq A, Brinkman CL, Patel R. 2018. In vitro activity of imipenem-relebactam and ceftolozane-tazobactam against resistant Gram-negative bacilli. Antimicrob Agents Chemother 62:e00533-18. 10.1128/AAC.00533-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chea N, Bulens SN, Kongphet-Tran T, Lynfield R, Shaw KM, Vagnone PS, Kainer MA, Muleta DB, Wilson L, Vaeth E, Dumyati G, Concannon C, Phipps EC, Culbreath K, Janelle SJ, Bamberg WM, Guh AY, Limbago B, Kallen AJ. 2015. Improved phenotype-based definition for identifying carbapenemase producers among carbapenem-resistant Enterobacteriaceae. Emerg Infect Dis 21:1611–1616. 10.3201/eid2109.150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh-Moodley A, Perovic O. 2016. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis 16:536. 10.1186/s12879-016-1858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1 and Table S1. Download AAC.02288-20-s0001.pdf, PDF file, 0.6 MB (620.4KB, pdf)