Abstract
Background
Oncostatin M (OSM) has been implicated in the pathogenesis of inflammatory bowel disease (IBD) and as a marker for nonresponsiveness to anti-tumor necrosis factor (TNF) therapy. We further unraveled the potential of OSM and related receptors as markers of diagnosis, prognosis, and therapy response in IBD.
Methods
We collected inflamed mucosal biopsies and serum from patients with Crohn disease (CD) and with ulcerative colitis: (1) newly diagnosed patients who were treatment-naïve, (2) patients initiating anti-TNF or (3) vedolizumab therapy, (4) postoperative patients with CD, and (5) multiple-affected families with IBD including unaffected first-degree relatives (FDRs). We measured the gene expression of mucosal OSM and its receptors OSMR/LIFR and co-receptor IL6ST, and the protein expression of serum OSM. Statistical significance was defined as P < 0.05.
Results
Newly diagnosed patients showed significantly increased mucosal OSM/OSMR compared with control patients, with the highest enrichment for OSM (fold change [FC] >17.9). Likewise, ileal OSM/OSMR were significantly upregulated in postoperative recurrent CD. Serum OSM was increased in newly diagnosed patients and postoperative patients with recurrent CD (FC ≥ 2.6). In families with IBD, higher serum levels were observed in FDRs than in control families (FC = 2.2). Furthermore, elevated colonic OSM/OSMR (but not serum OSM) were associated with the early need for biologic therapy (FC ≥ 1.9), and higher OSM was also predictive of primary nonresponse to both anti-TNF and vedolizumab therapy (FC ≥ 2.4). Immunohistochemistry highlighted mucosal OSM expression in macrophages.
Conclusions
We found that OSM is a diagnostic biomarker in the tissue and serum not only of newly diagnosed patients with IBD and postoperative patients with recurrent CD but also of their FDRs. Higher colonic OSM levels are furthermore associated with poor prognosis and with primary nonresponse to biologic therapies. Therefore, OSM could guide clinical decision-making.
Keywords: oncostatin M, biomarker, diagnostic, prognostic, therapy response, IBD
INTRODUCTION
Crohn disease (CD) and ulcerative colitis (UC) are chronic, relapsing inflammatory bowel diseases (IBDs) with increasing prevalence worldwide.1, 2 A single reference standard for IBD diagnosis does not exist.3 Often, the diagnosis is significantly delayed, leading to a postponed treatment initiation and subsequently affecting overall quality of life and disease progression.4 Together with the significant rate of primary and secondary resistance to current IBD treatments,5 novel therapeutic targets and biomarkers supporting these clinical needs are eagerly awaited.
Among potential targets and biomarkers, oncostatin M (OSM) has gained a lot of interest. In 2017, West, Hegazy, et al6 reported an increased expression of OSM and the OSM receptor-β (OSMR) in inflamed intestinal tissue from patients with IBD, where it drives intestinal stromal cell inflammation. The authors also showed that increased OSM predicts nonresponsiveness to anti-tumor necrosis factor (TNF) therapy.
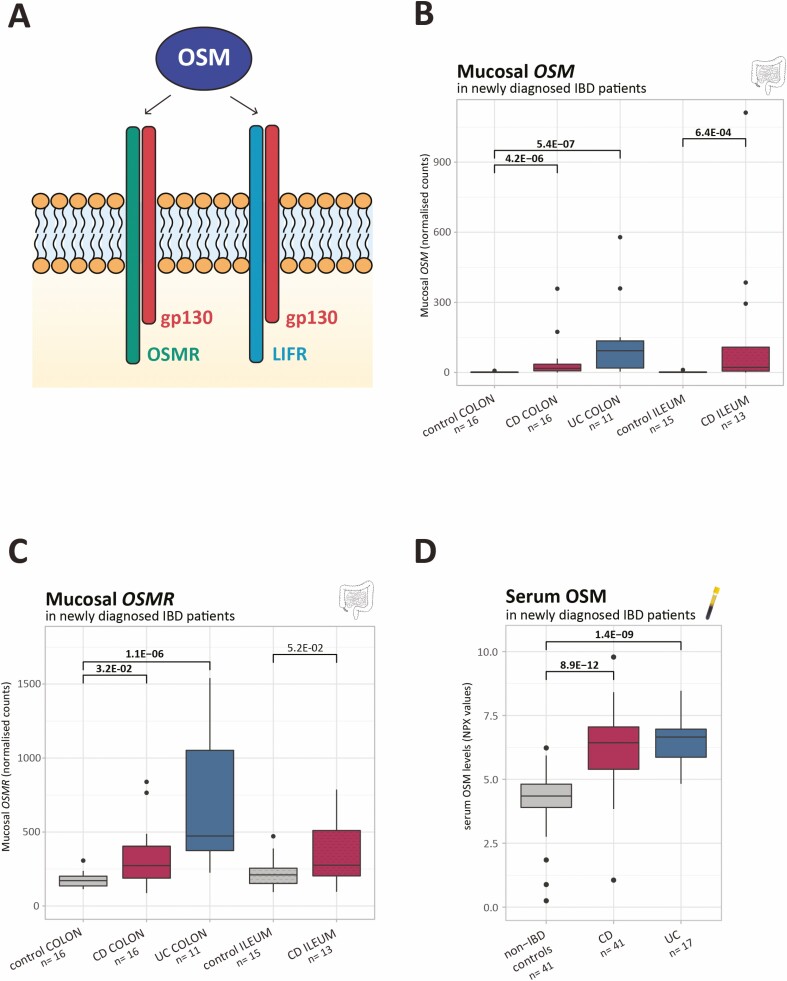
OSM is part of the interleukin (IL)-6 cytokine family and signals via a receptor complex composed of the gp130 co-receptor (similar for all IL-6 family members) and either OSMR or the leukemia inhibitory factor receptor-β (LIFR; Fig. 1A).7-9 In both cases (ie. OSMR or LIFR), formation of the heterodimeric complex can induce different signaling cascades (ie, JAK-STAT pathway, PI3K-Akt pathway), which depend on cell type and environmental conditions.10, 11
FIGURE 1.
Mucosal OSM and OSMR levels and serum OSM in newly diagnosed patients with IBD. A, OSM signals via a receptor complex composed of the gp130 co-receptor and either OSMR or LIFR. B-C, Boxplots of mucosal OSM and OSMR as measured by RNA sequencing (normalized counts). D, Boxplots of serum OSM as measured by the OLINK proximity extension technology (NPX values). Significant comparisons are highlighted in bold. NPX indicates normalized protein expression values.
In this study, we aimed to further unravel the potential of OSM and related receptors as markers of diagnosis, prognosis, and therapy response in IBD, both in the mucosa and in serum. We therefore investigated 5 different clinical scenarios: (1) newly diagnosed patients with IBD, (2) patients initiating anti-TNF or (3) vedolizumab therapy, (4) postoperative patients with CD 6 months after surgery, and (5) unaffected first-degree relatives (FDRs).
MATERIALS AND METHODS
Patient Recruitment and Sample Collection
We cross-sectionally studied mucosal biopsies and serum from patients with CD and with UC: (1) newly diagnosed patients, (2) patients initiating anti-TNF or (3) vedolizumab therapy, (4) postoperative patients with CD 6 months after ileocolonic resection with ileocolonic anastomosis, and (5) multiple-affected families with IBD, including unaffected FDRs. For each group, matched samples from non-IBD control patients were included as comparison. Baseline characteristics for each study cohort are listed in Supplementary Tables S1–5. An overview of the studied samples and groups can be found in Table 1. Ileal expression data generated in our study center were only available in the context of early IBD, ie, newly diagnosed and postoperative patients with recurrent CD.12
TABLE 1.
Overview of the Studied Samples and Corresponding Applied Definitions and Technologies
| Study Cohort | Subgroups | Serum | Mucosa (RNAseq*/microarray**) |
|---|---|---|---|
| Treatment-naïve newly diagnosed patients with IBD (within 6 months after diagnosis, naïve for biologics/immunosuppressives, no IBD-related surgery) | CD | 41 | 16 colonic* 13 ileal* |
| UC | 17 | 11 colonic* | |
| Control patients without IBD | 41 | 16 colonic* 15 ileal* | |
| Poor prognosis (need for biologic therapy within 2 years after diagnosis) | 38 | 18 colonic* | |
| Good prognosis | 13 | 9 colonic* | |
| Anti-TNF cohort | Remission (complete absence of ulcerations at month 6 [CD] or a Mayo endoscopic subscore 0-1 at week 8/14 [UC]) No remission | 91 95 | 13 colonic* 12 colonic* |
| Vedolizumab cohort | Remission (complete absence of ulcerations at month 6 [CD] or a Mayo endoscopic subscore 0-1 at week 14 [UC]) | 99 | 23 colonic* |
| No remission | 75 | 24 colonic* | |
| Postoperative patients with CD undergoing ileocecal resection with ileocolonic anastomosis | No postoperative recurrence (Rutgeerts score i0/i1 at month 6) | 36 | 8 neoterminal ileal** |
| Postoperative recurrence (Rutgeerts score ≥ i2b at month 614) | 46 | 24 neoterminal ileal** | |
| Control patients without IBD | 43 | 12 ileal** | |
| Multiple-affected families with IBD (minimum 3 FDRs with IBD) | Patients with IBD | 48 | — |
| Unaffected FDRs | 33 | — | |
| Members of control families without IBD | 40 | — |
RNAseq indicates RNA sequencing. *RNA sequencing based; **Microarray based.
Newly diagnosed patients with IBD who were treatment-naïve were included 6 months after diagnosis, were naïve for biologics/immunosuppressives, and had not had IBD-related surgery. A poor prognosis at the time of diagnosis was defined as the need for biologic therapy within 2 years after diagnosis. Endoscopic remission after anti-TNF or vedolizumab therapy was defined as the complete absence of ulcerations at month 6 (CD) or a Mayo endoscopic subscore of 0 to 1 at week 8/14 (UC). Patients with CD who underwent ileocecal resection with ileocolonic anastomosis and had a Rutgeerts score ≥ i2b at month 6, were considered as postoperative recurrent patients.13,14 Multiple-affected families with IBD were defined as those with at least 3 FDRs with IBD.
All biopsies were taken during endoscopy and stored in RNALater buffer (Ambion, Austin, TX) at –80°C. Biopsies from patients with IBD were collected at the most affected site, at the edge of the ulcerative surface. Control biopsies were collected from individuals undergoing screening colonoscopies and who had a normal endoscopy.
To localize OSM, resected intestinal tissue from patients with IBD undergoing surgery and resected unaffected intestinal tissue from control patients who did not hae IBD but did have colorectal cancer were collected. Tissue from these control patients was collected adjacent to the tumor-free resection margin.
Mucosal OSM, OSMR, LIFR, and IL6ST Expression Levels
Mucosal expression levels of OSM, OSMR, LIFR, and IL6ST (encode for the gp130 co-receptor) were obtained using microarray technology or RNA sequencing (Table 1).
For microarray analysis (postoperative CD cohort only), total RNA was extracted from biopsies using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and was analyzed via GeneChip Human Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA). Quality control and data analysis (R) were performed as previously described.12 The robust multichip average method15 was applied on Affymetrix raw data (.cel files), resulting in log2 expression values at the probe level.
We performed RNA sequencing on mucosal biopsies from newly diagnosed patients and patients initiating anti-TNF or vedolizumab therapy. Total RNA was extracted from the samples using the AllPrep DNA/RNA Mini kit (Qiagen) after tissue lysis using the FastPrep Lysing Matrix D tubes (MP Biomedicals, Brussels, Belgium). The integrity and quantity of RNA were evaluated with a 2100 Bioanalyzer and a Nanodrop ND-1000 spectrophotometer, respectively. After library preparation using the TruSeq Stranded mRNA protocol (Illumina, San Diego, CA), single-end RNA sequencing was performed on 500 ng total RNA (Illumina HiSeq 4000NGS). Raw sequencing data were then aligned to the reference genome (Hisat2 v.2.1.016), and absolute counts were generated using HTSeq.17 Counts were then normalized for library size using the R\DESeq2 package.18 Using the cellular xCell deconvolution method,19 macrophage enrichment scores from bulk RNA data were calculated. The effect of anti-TNF therapy on genes of interest in the colonic mucosa was investigated using publicly available microarray datasets (GEO GSE12251, GSE14580, GSE16879).20, 21
Serum OSM Levels
Relative serum OSM protein levels were quantified using proximity extension technology with an inflammation panel (Olink Proteomics AB, Uppsala, Sweden). Quality control and normalization of the data were performed using an internal extension control and an interplate control to adjust for intra- and interrun variation. Validation data can be found on the manufacturer’s website (https://www.olink.com). The final readout was expressed in normalized protein expression values, an arbitrary unit on a log2 scale.
Fecal Calprotectin
Fecal calprotectin was extracted using the Smart Prep extraction device (Roche Diagnostics, Mannheim, Germany), and concentrations were measured using the fCAL enzyme-linked immunosorbent assay kit (Bühlmann Laboratories AG, Schönenbuch, Switzerland).
Immunohistochemistry
Immunohistochemical stainings were performed on 5 μm–thick glass-mounted sections prepared from formalin-fixed paraffin-embedded transmural bowel biopsies, using the BOND MAX autostainer (Leica Microsystems Ltd., Heerbrugg, Switzerland). Epitope retrieval was performed in citrate buffer (pH 6) at 98°C for 30 minutes. Rabbit polyclonal OSM antibody (ab198830, Abcam, Cambridge, UK) concentration was optimized, and a dilution of 1:200 was applied. The bound primary antibody was visualized using the BOND Polymer Refine Detection kit. All stains were evaluated by an IBD-experienced pathologist (G. De Hertogh). Microscopic images were generated using the Leica Application Suite V4.8.0. software, with a Leica DFC290 HD camera (Leica Microsystems Ltd.) mounted on a Leica DM2000 LED field microscope.
Statistical Analyses
Statistical analyses were conducted in R (R Development Core Team, Vienna, Austria). Comparisons were performed using Wilcoxon tests or 2 sample t tests (continuous variables) and χ 2 tests (categorical baseline characteristics), as appropriate. Continuous variables on plots were expressed as median with interquartile range. Correlations were evaluated using the Spearman r correlation coefficient. To assess the performance of univariate and multivariate logistic regression models, receiver operating characteristics (ROCs) and areas under the curve (AUCs) were analyzed, and the Delong test was performed to compare the AUCs (R\pROC package). Statistical significance was defined as P < 0.05.
Ethical Considerations
This study was carried out at the University Hospitals Leuven (Leuven, Belgium). The ethics committee of the University Hospitals Leuven approved the study (institutional review board approvals B322201213950/S53684, B322201110724/S52544, and B322201627472/S57662). All individuals gave written informed consent.
RESULTS
OSM and Related Receptors in Early IBD
Comparative analyses between early IBD models/unaffected FDRs and matched control patients without IBD
In newly diagnosed patients with CD and with UC, colonic mucosal OSM expression was upregulated as compared with control patients without IBD (fold change [FC] = 23.6, P = 4.2E-06; FC = 93.0, P = 5.4E-07, respectively; Fig. 1B). Although OSMR also showed increased expression in both CD and UC colons (FC = 1.6, P = 3.2E-02; FC = 2.7, P = 1.1E-06; Fig. 1C), LIFR did not (P = 1.6E-01; P = 2.9E-01; Supplementary Fig. 1A). Notably, IL6ST, which encodes for the gp130 co-receptor, was elevated in UC colons (FC = 1.5, P = 3.3E-03) but not in CD colons (P = 8.6E-02; Supplementary Fig. 1B).
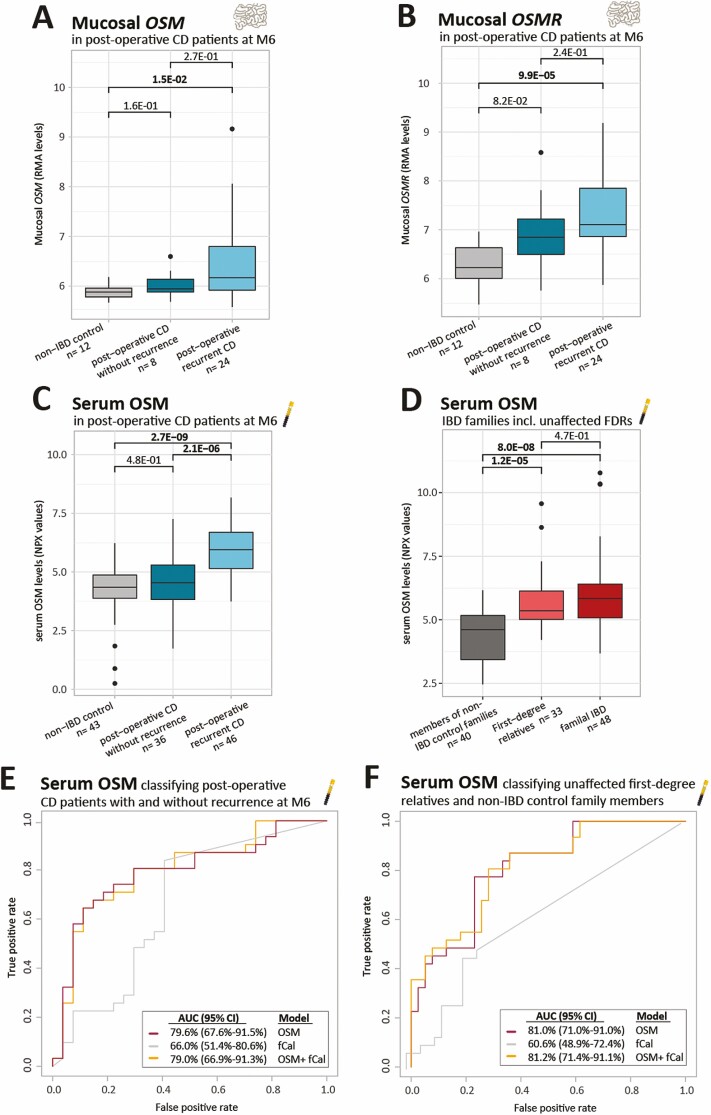
In the ileum of newly diagnosed patients with CD, the same observations were made with increased mucosal OSM (FC = 17.9, P = 6.4E-04) and borderline increased OSMR (FC = 1.4, P = 5.2E-02) as compared with the ileum of control patients without IBD (Figs. 1B, C). Limited or no differences were found for LIFR (P = 5.8E-02) and IL6ST (P = 1.7E-01), respectively (Supplementary Figs. 1A, B). Likewise, patients with CD with postoperative recurrent ileitis (Rutgeerts score ≥ i2b14), a good model to study the earliest mucosal CD lesions,12 had significantly higher OSM and OSMR levels in the ileal mucosa at month 6 than did control patients (FC = 2.1, P = 1.5E-02; FC = 2.2, P = 9.9E-05; Figs. 2A, B). No differences for LIFR and IL6ST were found (Supplementary Figs. 1C, D).
FIGURE 2.
Mucosal OSM and OSMR levels in postoperative patients with CD and serum OSM in postoperative patients with CD and multiple-affected families with IBD. A-B, Boxplots of mucosal OSM and OSMR as measured by microarray technologies (RMA values). C and D, Boxplots of serum OSM as measured by the OLINK proximity extension technology (NPX values). E and F, ROC analysis classifying (D) postoperative CD with and without recurrence or (F) unaffected FDRs of patients with IBD and members of control families without IBD based on serum OSM levels and fCal. Significant comparisons are highlighted in bold. fCal indicates fecal calprotectin; M6, six months after surgery; NPX, normalized protein expression values; RMA, robust multichip average.
Serum protein analysis showed increased OSM levels in newly diagnosed patients with CD and with UC vs control patients without IBD (FC = 4.3, P = 8.9E-12; FC = 5.0, P = 1.4E-09; Fig. 1D). Similarly, in postoperative patients with recurrent CD, elevated serum OSM levels were observed at month 6, in comparison to patients with CD without recurrence and to control patients (FC = 2.6, P = 2.1E-06; FC = 3.0, P = 2.7E-09; Fig. 2C). Remarkably, in multiple-affected families with IBD, higher OSM levels were found in unaffected FDRs as compared to matched control families (FC = 2.2, P = 1.2E-05), with levels in these FDRs similar to those of the affected relatives (P = 4.7E-01; Fig. 2D).
Diagnostic prediction model
We built univariate logistic regression models for mucosal OSM/OSMR and serum OSM levels and assessed their performance. The ROC analysis based on mucosal OSM showed high accuracy in discriminating between newly diagnosed patients with CD and with UC and control patients without IBD (AUC ≥ 86.2%; 95% confidence interval [CI] ≥ 69.1%; Table 2). Similar, serum OSM had a strong discriminative power, with an accuracy of 90.0% (95% CI, 83.0%-97.1%) and 94.5% (95% CI, 89.2%-99.9%) in patients with CD and with UC vs control patients (Table 2). Mucosal OSMR resulted in a significant AUC of ≥71.2% (95% CI ≥51.9%) to distinguish newly diagnosed patients with CD and with UC from control patients (Table 2).
TABLE 2.
ROC Curve Analyses for the OSM-Related Markers in All Study Cohorts
| Study Cohort | Outcome | Serum OSM Protein Levels,AUC (95% CI) | Mucosal Gene Expression Levels,AUC (95% CI) |
|---|---|---|---|
| Treatment-naïve newly diagnosed patients with IBD (within 6 months after diagnosis, naïve for biologics/immunosuppressives, no IBD-related surgery) | Patients with CD and control patients without IBD | 90.0% (83.0%-97.1%) | Colonic OSM: 92.3% (83.7%-100.0%) Colonic OSMR: 72.7% (52.8%-92.6%) Ileal OSM: 86.2% (69.1%-100.0%) Ileal OSMR: 71.2% (51.9%-91.6%) |
| Patients with UC and control patients without IBD | 94.5% (89.2%-99.9%) | Colonic OSM: 98.9% (96.1%-100.0%) Colonic OSMR: 98.3% (94.9%-100.0%)* | |
| Poor prognosis and good prognosis | 55.7% (35.3%-76.0%) | Colonic OSM: 77.8% (59.7%-95.9%) Colonic OSMR: 80.3% (62.8%-97.7%) | |
| Anti-TNF cohort | Remission and no remission | 52.2% (43.8%-60.6%) | Colonic OSM: 73.7% (53.7%-93.7%) Colonic OSMR: 76.3% (55.6%-96.9%) |
| Vedolizumab cohort | Remission and no remission | 51.0% (42.6%-59.4%) | Colonic OSM: 68.5% (52.9%-84.0%) Colonic OSMR: 62.0% (45.6%-59.4%) |
| Postoperative patients with CD undergoing ileocecal resection with ileocolonic anastomosis | Postoperative recurrence and control patients without IBD | 84.5% (76.5%-92.6%) | Ileal OSM: 75.0% (58.9%-91.1%) Ileal OSMR: 87.9% (76.2%-99.4%) |
| Postoperative recurrence and no postoperative recurrence | 79.6% (67.6%-91.5%) | — a | |
| Multiple-affected families with IBD (minimum 3 FDRs with IBD) | Unaffected FDRs and members of control families without IBD | 81.0% (71.0%-91.0%) | — |
*Significant accuracies. aSample size too small.
Early recurrence prediction model
Serum OSM 6 months after surgery was found as a marker for early postoperative recurrent CD, with an AUC of 79.6% (95% CI, 67.6%-91.5%) to distinguish postoperative patients with CD with recurrence from those without recurrence (Fig. 2E). This serum OSM model numerically outperformed fecal calprotectin concentrations, which had an accuracy of 66.0% (95% CI, 51.4%-80.6%; P = 1.8E-01; Fig. 2E).
Unaffected FDR prediction model
Serum OSM levels could also discriminate between unaffected and entirely asymptomatic FDRs and matched control families, with an accuracy of 81.0% (95% CI, 71.0%-91.0%; Fig. 2F). However, fecal calprotectin could not accurately classify unaffected FDRs and control families (AUC = 60.6%; 95% CI, 48.9%-72.4%). The discriminative power of serum OSM in unaffected FDRs thus outperformed fecal calprotectin (P = 9.1E-03; Fig. 2F).
OSM and Related Receptors Depending on Disease Prognosis
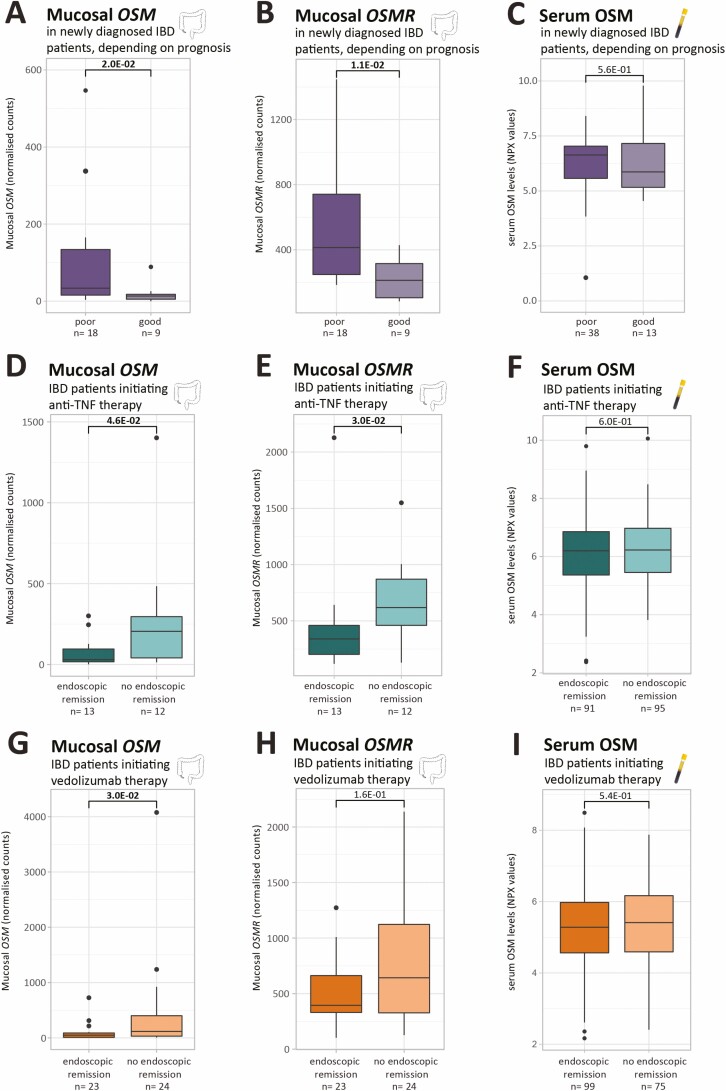
Elevated colonic expression of OSM, OSMR, and ILS6ST at diagnosis was associated with a worse prognosis as defined by the need for biologic therapy within 2 years after diagnosis (poor vs good prognosis: FC = 2.5, P = 2.0E-02; FC = 1.9; P = 1.1E-02; FC = 1.5, P = 1.7E-02, respectively; Figs. 3A, B, Supplementary Fig. 2B). Colonic LIFR and serum OSM did not differ depending on disease prognosis (P = 9.4E-01; P = 5.6E-01; Supplementary Fig. 2A, Fig. 3C).
FIGURE 3.
Mucosal OSM and OSMR levels and serum OSM in newly diagnosed patients with IBD depending on disease prognosis and in patients with IBD initiating anti-TNF or vedolizumab therapy. A-B, D-E, G-H. Boxplots of mucosal OSM and OSMR as measured by RNA sequencing (normalized counts). C, F, I, Boxplots of serum OSM as measured by the OLINK proximity extension technology (NPX values). Significant comparisons are highlighted in bold. NPX indicates normalized protein expression values.
We then further assessed the prognostic accuracy of colonic OSM and OSMR gene expression levels and observed a significant AUC of 77.8% (95% CI, 59.7%-95.9%) and 80.3% (95% CI, 62.8%-97.7%), respectively (Table 2).
OSM and Related Receptors Depending on Therapy Response
Before anti-TNF therapy, colonic OSM and OSMR were upregulated in patients not achieving endoscopic remission (FC = 6.8, P = 4.6E-02; FC = 1.8, P = 3.0E-02, respectively; Figs. 3D, E). Baseline expression of LIFR and IL6ST was independent of response to anti-TNF (P = 1.00E+00; P = 2.7E-01; Supplementary Figs. 2C, D). In the vedolizumab cohort, increased colonic OSM levels were also observed in future nonresponders (FC = 2.4, P = 3.0E-02; Fig. 3G). No significant differences in OSMR, LIFR, and IL6ST expression between future vedolizumab responders and nonresponders could be observed (P = 1.6E-01; P = 7.9E-02; P = 6.4E-01; Fig. 3H, Supplementary Figs. 2E, F). At protein level, baseline serum OSM could not identify future anti-TNF or vedolizumab nonresponders (P = 6.0E-01; P = 5.4E-01; Figs. 3F, I). In the anti-TNF cohort, colonic OSM and OSMR differentiated responders from nonresponders with an AUC of 73.7% (95% CI, 53.7%-93.7%) and 76.3% (95% CI, 55.6%-96.9%), respectively (Table 2). The mucosal OSM signature predicted endoscopic response to vedolizumab with an accuracy of 68.5% (95% CI, 52.9%-84.0%) (Table 2).
Notably, paired transcriptomic data obtained before the first anti-TNF administration and 4 to 6 weeks after treatment initiation displayed a significant decrease in colonic OSM in endoscopic responders and responders (P = 6.1E-04; P = 3.3E-02, respectively), but levels were also significantly lower in responders after treatment than in nonresponders after treatment (P = 4.2E-07; Supplementary Fig. 3).
Lack of Correlation Between Mucosal OSM and Serum OSM
We then assessed the relationship between mucosal OSM and serum OSM levels within 86 patients with IBD (ie, 86.7% of all patients whose colonic data we studied). No significant correlations between tissue OSM and serum OSM were found within patients with IBD overall, patients with CD, or patients with UC (r = 0.09, P = 4.2E-01; r = 0.27, P = 1.0E-01; r = –0.04, P = 7.8E-01).
Immunohistochemical Localization of OSM in the Intestinal Wall
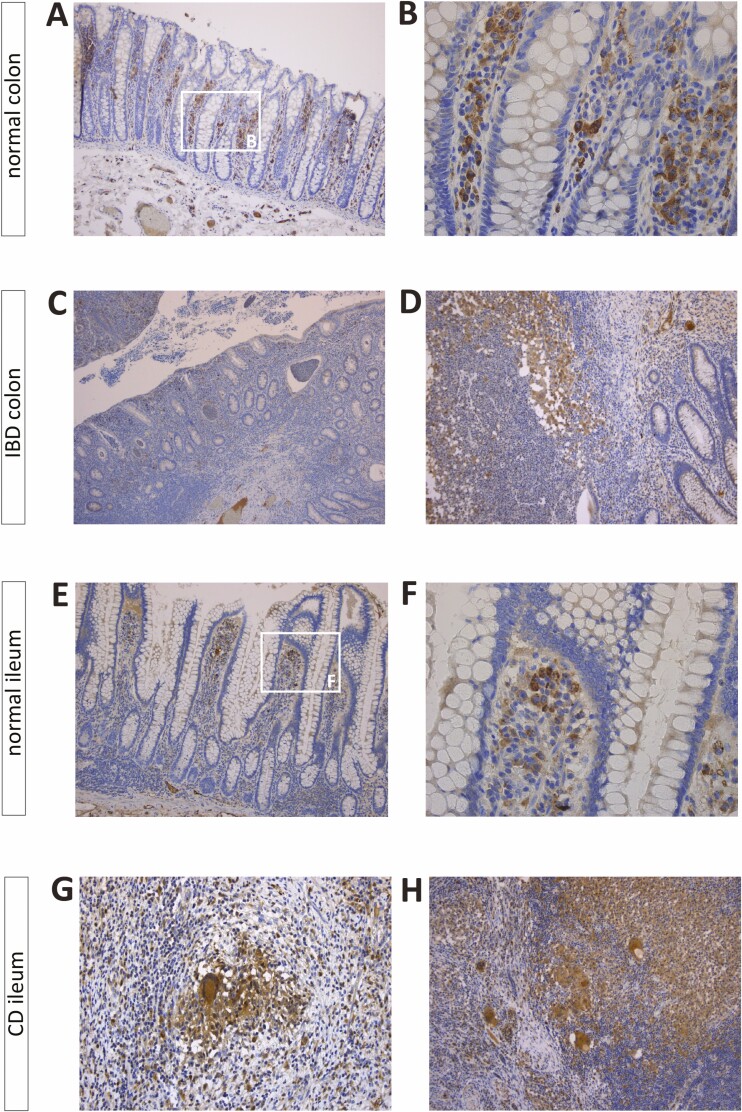
Immunohistochemical analysis of surgically resected intestinal tissue localized OSM in macrophages residing in the superficial lamina propria, irrespective of location (ileum, colon) and disease status (CD, UC, control; Figs. 4A-C4E, F). In CD, epithelioid granulomas and multinucleated giant cells also showed positive OSM staining (Figs. 4G, H). In penetrating CD, OSM expression was observed in macrophages lining the fissuring ulcer (Fig. 4D). These findings were further supported by a significant correlation between mucosal OSM mRNA levels and macrophage enrichment scores in CD and UC colons at the time of diagnosis (r = 0.90, P < 2.2E-16; r = 0.66, P = 3.1E-02, respectively; Supplementary Figs. 4A, B). This observation was mainly driven by macrophage M1 subtypes showing stronger correlations (CD: r = 0.84, P = 2.9E-05; UC: r = 0.60, P = 5.6E-02) than macrophage M2 subtypes (CD: r = 0.71, P = 2.7E-03; UC: r = 0.48, P = 1.4E-01; Supplementary Figs. 4C-F).
FIGURE 4.
Immunohistochemical localization of OSM in the intestinal mucosa. Immunohistochemical staining of resected normal colon from a control patient without IBD (A and B), resected colonic tissue from a patient with IBD (C: UC; D: CD), resected normal ileum from a control patient without IBD (E and F), and resected ileal tissue from a patient with CD (G and H) undergoing surgery. Original magnification ×50 (C), ×100 (A, D, E, H), ×200 (G), and ×400 (B and F).
Furthermore, OSM staining was also detected in endothelial cells (Supplementary Fig. 5G). Considering the enteric nerve system, in both control and IBD tissue there was a weak staining for OSM in ganglion cells of the submucosa and the myenteric plexus (Supplementary Figs. 5A-F, 5H-I).
DISCUSSION
The need for a personalized approach for patients with IBD is high.22, 23 Diagnostic delay results in postponed treatment initiation and may affect patient outcomes.4 Hence, we need to prioritize early diagnosis and effective treatment at the individual patient level. In this study, we investigated OSM and its receptors in tissue and serum from various IBD cohorts and unaffected FDRs and provided further evidence for its potential as a diagnostic, prognostic, and therapeutic biomarker.
A role for the OSM-OSMR axis in the pathogenesis of IBD was first discovered by a disease-susceptibility single-nucleotide polymorphism within the OSMR locus on chromosome 5.24 More recently, the landmark study by West, Hegazy, et al6 showed that OSM, out of 64 candidate cytokines, was the most highly and consistently expressed cytokine in the inflamed mucosa of patients with IBD. This upregulation of mucosal OSM was associated with an overexpression of OSMR, whereas LIFR and IL6ST (encoding for gp130) showed no or limited differences, respectively.6 In light of these results, those authors further investigated the functional role of the OSM-OSMR axis in IBD and suggested that OSM may act as an inflammatory amplifier and driver of disease chronicity. More specifically, they found that the binding of OSM to the OSMR-gp130 complex on intestinal stromal cells promotes chemokine, cytokine, and adhesion factor production.6
Similar to the findings by West, Hegazy, et al,6 we observed increased expression levels of OSM and OSMR in the colon and ileum of patients with IBD at an early disease stage: at diagnosis and also 6 months after surgery in recurrent CD ileitis. Postoperative recurrent CD is indeed a good model to study the earliest mucosal CD lesions.12 Likewise, at the serum level, we found increased OSM protein expression in newly diagnosed patients with IBD and postoperative patients with recurrent CD compared with control patients. Thus far, serum OSM has been measured in just 1 IBD study, in which OSM was indicated as UC-specific.25 However, as highlighted by those authors, their heterogeneous study cohorts were not ideal for the purpose of diagnostic biomarker identification. To further assess the utility of mucosal and serum OSM as diagnostic biomarkers for IBD, a comparison with OSM levels in other inflammatory gastrointestinal conditions is warranted.
Because increased OSM was observed at the time of diagnosis in the patients we studied (mucosal accuracy ≥86.2%; serological accuracy ≥90.0%), we asked whether studying OSM in the unaffected FDRs of patients with IBD might further support its potential as a diagnostic marker. In multiple-affected families with IBD, serum OSM levels were indeed elevated in FDRs compared with those in matched control families, and those levels could differentiate between the 2, with an accuracy of 81.0%, thereby outperforming fecal calprotectin (accuracy of 60.6%). Preliminary analysis of the GEM cohort showed that higher fecal calprotectin levels in FDRs are predictive for the development of CD.26, 27 Likewise, FDRs with serum OSM levels comparable to those observed in patients merit close follow-up to confirm whether these levels can predict disease onset. In the recent PREDICTS study, preclinical serum OSM was not selected as one of the features for a machine learning model predicting CD development.28 However, this finding does not necessarily imply that serum OSM at an individual level does not have the power to predict disease, so future work should investigate whether OSM is increased in those prediagnostic samples. Finally, we showed that serum OSM levels 6 months after surgery can predict early recurrent CD ileitis with an accuracy of 79.6%, as opposed to fecal calprotectin levels (66.0%). Because the majority of postoperative patients with recurrent CD are asymptomatic,29 their early mucosal lesions thus mimic what happens during the subclinical phase of CD before overt diagnosis is established.
From a prognostic perspective, elevated colonic OSM and OSMR were associated with a worse disease prognosis, ie, the requirement of biologic therapy within 2 years after diagnosis. In contrast to mucosal OSM having a predictive power of 77.8%, serum OSM at diagnosis was not associated with a poor prognosis. The value of serum OSM as a prognostic biomarker needs further study because in patients with colorectal cancer, serum OSM levels increase in patients with lymphovascular involvement, higher tumor stage, and distant metastasis.30
The landmark study by West, Hegazy, et al6 also identified and validated mucosal OSM and OSMR expression as predictive biomarkers for anti-TNF responsiveness in IBD, with high pretreatment OSM being strongly associated with anti-TNF therapy failure. We confirmed the association between elevated mucosal OSM and OSMR and the absence of endoscopic response to anti-TNF, and those markers could predict response with an accuracy of 73.7% and 76.3%. Again, we could not translate the mucosal OSM signal into a serological OSM biomarker, nor a whole- blood OSM biomarker as previously reported,31 in contrast to other reported markers such as TREM1.31-33 A preliminary proteomic analysis of the large PANTS cohort confirmed that baseline serum OSM is independent of anti-TNF therapy response in CD.34 Although Bertani et al35 reported baseline serum OSM as a predictor for mucosal healing at week 54 in patients with CD treated with infliximab, the sample size of this study cohort was limited. Likewise, in a small cohort of pediatric patients with CD, an association was found between elevated plasma OSM levels and a poor biochemical response to infliximab.36
Because mucosal OSM closely correlates with histopathologic disease severity,6 we then questioned whether the mucosal OSM signal was predictive for the failure of anti-TNF therapy specifically. Indeed, colonic OSM was increased also in future vedolizumab nonresponders, with an accuracy of 68.5%. In addition, transcriptomic data from inflamed UC biopsies collected during the phase II TURANDOT study (anti-MAdCAM-1) showed that OSM expression was associated with the efficacy of the compound being investigated.37 The ROC analysis in this study resulted in a significant AUC of 83%.37 The differences in the results may be related to a more homogeneous and larger cohort that was studied in the trial. Overall, together with the observation of increased mucosal OSM in future nonresponders to corticosteroids,38 upregulated mucosal OSM can now be considered as a validated surrogate marker of a more refractory and difficult-to-treat disease.
Although OSM mainly displays a hematopoietic expression pattern, several studies have reported clear distinctions in the hematopoietic origin and profile of OSM expression in various disease states.39 Because previous reports on OSM protein expression in the intestinal mucosa are lacking, we localized OSM in resected tissues using immunohistochemistry and found a clear expression in macrophages residing in the superficial lamina propria and in epithelioid granulomas and multinucleated giant cells. Moreover, macrophage enrichment scores strongly correlated with mucosal OSM. Indeed, monocytes and macrophages have been shown to produce OSM,39 and West, Hegazy, et al6 reported the highest enrichment for mucosal OSM in antigen-presenting cells (B cells excluded). Their study also detected OSM gene expression—although to a lesser extent—in B and T cells,6 which we did not observe at the protein level in intestinal tissue. Furthermore, a recent high-resolution characterization of immune and stromal cells from ileal CD lesions showed that OSM is most highly expressed by inflammatory macrophages.40 Based on our results and previous findings, elevated mucosal OSM may be related to an increased accumulation of macrophages and especially proinflammatory M1 macrophages, typically seen in IBD lesions.41, 42
Overall, we are the first to study both mucosal and serological OSM and related receptors in different clinical settings related to IBD care. Moreover, postoperative patients with CD, unaffected FDRs, and patients stratified by early treatment escalation have never been studied in the context of OSM. Despite these strengths, a few limitations must be considered. First, although the Olink proximity extension technology and RNA sequencing are good screening tools, these only generate semi-quantitative data. Our findings thus not only require validation in larger, independent cohorts but also require the use of other methodologies to allow quantitative cutoffs for tissue and serum OSM and OSMR before translation into daily clinical practice. Second, the study cohort of newly diagnosed patients with IBD, especially at the mucosal site, was too limited to assess differences among patients based on disease location, behavior, extent, and prognosis at the ileal level. Likewise, the number of patients initiating biologic therapy from whom we obtained ileal expression data was too limited to study. Third, research focusing on OSM in other body fluids (eg, saliva or feces) should be considered. Finally, given the varying proportions of cell types within mucosal biopsies, we studied averaged expression patterns of all cells present in the biopsy. Although cell deconvolution provided us insight regarding the relative numbers of macrophage subtypes, single-cell transcriptomics would be more informative in this context.
CONCLUSIONS
In conclusion, OSM, a key player in the pathogenesis of IBD, is dysregulated in the serum and intestinal mucosa of newly diagnosed patients with IBD and early postoperative CD recurrence and in FDRs of patients with IBD. Hence, the diagnostic accuracy of OSM as a subclinical marker should be further examined. There is now consistent data that elevated colonic OSM points toward a more refractory phenotype. Future work should study whether early surgery may be preferred in these patients. Likewise, because anti-OSM antibodies have been developed (eg, NCT04151225; (https://clinicaltrials.gov/), it will be interesting to see whether patients with high OSM levels may benefit from anti-OSM therapy.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Vera Ballet and Eline Vandeput for an excellent job in maintaining the Biobank database; Helene Blevi, Tamara Coopmans, Nooshin Ardeshir Davani, Sophie Organe, and Willem-Jan Wollants for processing all patient samples and their technical support; and Eef Allegaert, Sarah Cumps, and Kathleen Van den Eynde for their technical assistance with the immunohistochemical stainings (Translational Cell and Tissue Research, KU Leuven).
Glossary
Abbreviations
- AUC
area under the curve
- CD
Crohn disease
- CI
confidence interval
- FC
fold change
- FDRs
unaffected first-degree relatives
- IBD
inflammatory bowel disease
- OSM
oncostatin M
- OSMR
oncostatin M receptor-β
- ROC
receiver operating characteristic
- TNF
tumor necrosis factor
- UC
ulcerative colitis
Author contributions: S. Verstockt contributed to the study concept and design, acquisition of data, analysis and interpretation of data, technical support, and drafting of the manuscript. B. Verstockt contributed to the study concept and design, acquisition of data, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. K. Machiels and M. Vancamelbeke contributed to the study concept and design, acquisition of data, and critical revision of the manuscript for important intellectual content. M. Ferrante contributed to material support and critical revision of the manuscript for important intellectual content. I. Cleynen contributed to the study concept and design, and critical revision of the manuscript for important intellectual content. G. De Hertogh contributed to the acquisition of data, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. S. Vermeire contributed to the study concept and design, acquisition of data, analysis and interpretation of data, material support, drafting of the manuscript, and study supervision.
Supported by: K. Machiels is a postdoctoral fellow and M. Ferrante and S. Vermeire are senior clinical investigators of the Research Foundation Flanders.
Conflicts of interest: B. Verstockt received financial support for research from Pfizer; lecture fees from AbbVie, Ferring Pharmaceuticals, Janssen, R-biopharm, and Takeda; and consultancy fees from Janssen and Sandoz. M. Ferrante received financial support for research from Janssen, Pfizer, and Takeda; consultancy fees from AbbVie, Boehringer Ingelheim, Celltrion, Ferring, Janssen, Lilly, Mitsubishi Tanabe, MSD, Pfizer, and Takeda; and speaker’s fees from AbbVie, Amgen, Biogen, Boehringer Ingelheim, Chiesi, Falk, Ferring, Janssen, Lamepro, Mitsubishi Tanabe, MSD, Pfizer, Takeda, Tramedico, Tillotts, and Zeria. G. De Hertogh’s company KU Leuven received fees for his activities as central pathology reviewer in clinical trials for Centocor, Takeda, Provention, Genentech, and CDX Diagnostics. S. Vermeire received financial support for research from MSD, AbbVie, Takeda, Janssen, and Pfizer; honoraria or consultation fees from AbbVie, MSD, Takeda, Ferring, Genentech/Roche, Shire, Pfizer Inc, Galapagos, Mundipharma, Hospira, Celgene, Second Genome, Progenity, GSK, Lilly, Arena, Gilead, and Janssen; and participated in company-sponsored speaker’s bureaus for AbbVie, MSD, Takeda, Ferring, Hospira, Pfizer, Janssen, and Tillots. For the remaining authors, no conflicts of interest were declared.
REFERENCES
- 1. Torres J, Mehandru S, Colombel JF, et al. Crohn’s disease. Lancet. 2017;389:1741–1755. [DOI] [PubMed] [Google Scholar]
- 2. Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. Lancet. 2017;389:1756–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maaser C, Sturm A, Vavricka SR, et al. ECCO-ESGAR guideline for diagnostic assessment in inflammatory bowel disease. J Crohns Colitis. 2019;13:144–164. [DOI] [PubMed] [Google Scholar]
- 4. Fiorino G, Danese S. Diagnostic delay in Crohn’s disease: time for red flags. Dig Dis Sci. 2016;61:3097–3098. [DOI] [PubMed] [Google Scholar]
- 5. Sabino J, Verstockt B, Vermeire S, et al. New biologics and small molecules in inflammatory bowel disease: an update. Therap Adv Gastroenterol. 2019;12:1756284819853208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators . Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gearing DP, Comeau MR, Friend DJ, et al. The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science. 1992;255:1434–1437. [DOI] [PubMed] [Google Scholar]
- 8. Mosley B, De Imus C, Friend D, et al. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J Biol Chem. 1996;271:32635–32643. [DOI] [PubMed] [Google Scholar]
- 9. West NR. Coordination of immune-stroma crosstalk by IL-6 family cytokines. Front Immunol. 2019;10:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hermanns HM. Oncostatin M and interleukin-31: cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev. 2015;26:545–558. [DOI] [PubMed] [Google Scholar]
- 11. Richards CD. The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm. 2013;2013:512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verstockt S, De Hertogh G, Van der Goten J, et al. Gene and mirna regulatory networks during different stages of Crohn’s disease. J Crohns Colitis. 2019;13:916–930. [DOI] [PubMed] [Google Scholar]
- 13. Rutgeerts P, Geboes K, Vantrappen G, et al. Predictability of the postoperative course of Crohn’s disease. Gastroenterology. 1990;99:956–963. [DOI] [PubMed] [Google Scholar]
- 14. Gecse K, Lowenberg M, Bossuyt P, et al. Sa1198 agreement among experts in the endoscopic evaluation of postoperative recurrence in Crohn’s disease using the Rutgeerts score. Gastroenterology. 2014;146:S– 227. [Google Scholar]
- 15. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 16. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis. 2010;16:2090–2098. [DOI] [PubMed] [Google Scholar]
- 21. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58:1612–1619. [DOI] [PubMed] [Google Scholar]
- 22. Hart AL, Lomer M, Verjee A, et al. What are the top 10 research questions in the treatment of inflammatory bowel disease? A priority setting partnership with the James Lind alliance. J Crohns Colitis. 2017;11:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noor NM, Verstockt B, Parkes M, et al. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol. 2020;5:80–92. [DOI] [PubMed] [Google Scholar]
- 24. Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2:224–234. [DOI] [PubMed] [Google Scholar]
- 25. Andersson E, Bergemalm D, Kruse R, et al. Subphenotypes of inflammatory bowel disease are characterized by specific serum protein profiles. Plos One. 2017;12:e0186142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kevans D, Silverberg MS, Borowski K, et al. ; GEM Project . IBD genetic risk profile in healthy first-degree relatives of Crohn’s disease patients. J Crohns Colitis. 2016;10:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S-H, Power N, Turpin W, et al. Sa1816; elevated fecal calprotectin in healthy first degree relatives of patients with Crohn’s disease is associated with future diagnosis of Crohn’s disease. Gastroenterology. 2019;156:S–413. [Google Scholar]
- 28. Torres J, Petralia F, Sato T, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. 2020;159:96–104. [DOI] [PubMed] [Google Scholar]
- 29. Rutgeerts P, Geboes K, Vantrappen G, et al. Natural history of recurrent Crohn’s disease at the ileocolonic anastomosis after curative surgery. Gut. 1984;25:665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gurluler E, Tumay LV, Guner OS, et al. Oncostatin-M as a novel biomarker in colon cancer patients and its association with clinicopathologic variables. Eur Rev Med Pharmacol Sci. 2014;18:2042–2047. [PubMed] [Google Scholar]
- 31. Verstockt B, Verstockt S, Dehairs J, et al. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. Ebiomedicine. 2019;40:733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verstockt B, Verstockt S, Blevi H, et al. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn’s disease patients? Gut. 2019;68:1531–1533. [DOI] [PubMed] [Google Scholar]
- 33. Gaujoux R, Starosvetsky E, Maimon N, et al. ; Israeli IBD Research Network (IIRN) . Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut. 2019;68:604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin S, Chanchlani N, Invergo BM, et al. Understanding the molecular mechanisms of anti-tnf treatment failure in patients with Crohn’s disease: a pilot serum proteomic analysis of the PANTS cohort. J Crohns Colitis. 2020;14:S067–S068. [Google Scholar]
- 35. Bertani L, Fornai M, Fornili M, et al. Serum oncostatin M at baseline predicts mucosal healing in Crohn’s disease patients treated with infliximab. Aliment Pharmacol Ther. 2020;52:284–291. [DOI] [PubMed] [Google Scholar]
- 36. Minar P, Lehn C, Tsai YT, et al. Elevated pretreatment plasma oncostatin M is associated with poor biochemical response to infliximab. Crohns Colitis 360. 2019;1:otz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou H, Xi L, Ziemek D, et al. Molecular profiling of ulcerative colitis subjects from the TURANDOT trial reveals novel pharmacodynamic/efficacy biomarkers. J Crohns Colitis. 2019;13:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haberman Y, Karns R, Dexheimer PJ, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Co mmun. 2019;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. West NR, Owens BMJ, Hegazy AN. The oncostatin M-stromal cell axis in health and disease. Scand J Immunol. 2018;88:e12694. [DOI] [PubMed] [Google Scholar]
- 40. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steinbach EC, Plevy SE. The role of macrophages and dendritic cells in the initiation of inflammation in IBD. Inflamm Bowel Dis. 2014;20:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Isidro RA, Appleyard CB. Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol. 2016;311:G59–G73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.