Abstract
Yeast Rnt1 is a member of the double-stranded RNA (dsRNA)-specific RNase III family identified by conserved dsRNA binding (dsRBD) and nuclease domains. Comparative sequence analyses have revealed an additional N-terminal domain unique to the eukaryotic homologues of RNase III. The deletion of this domain from Rnt1 slowed growth and led to mild accumulation of unprocessed 25S pre-rRNA. In vitro, deletion of the N-terminal domain reduced the rate of RNA cleavage under physiological salt concentration. Size exclusion chromatography and cross-linking assays indicated that the N-terminal domain and the dsRBD self-interact to stabilize the Rnt1 homodimer. In addition, an interaction between the N-terminal domain and the dsRBD was identified by a two-hybrid assay. The results suggest that the eukaryotic N-terminal domain of Rnt1 ensures efficient dsRNA cleavage by mediating the assembly of optimum Rnt1-RNA ribonucleoprotein complex.
RNase III is a double-stranded-RNA (dsRNA)-specific endoribonuclease that introduces staggered cuts on each side of the RNA helix (28). In bacteria, RNase III is involved in processing pre-rRNA, tRNA, and phage polycistronic mRNA (7). Depletion of RNase III perturbs the expression level of about 10% of the bacterial proteins, suggesting a global role in gene regulation (10). Two eukaryotic homologues of RNase III were experimentally identified in Saccharomyces cerevisiae (Rnt1) (2) and Schizosaccharomyces pombe (Pac1) (14, 35, 41). In addition, database searches revealed homologues in the worm, mouse, and human (5, 35). Rnt1 was shown both in vivo and in vitro to process pre-rRNA (2, 18), three small nuclear RNAs (snRNAs) (1, 4, 40), and several small nucleolar RNAs (snoRNAs) (5, 6, 31). Similarly, Pac1 cleaves the 3′ end of U2 snRNA and the 3′ end of 25S rRNA (36, 37, 43). Also, it has been suggested that Pac1 plays a role in cell division, mating, and sporulation (14, 41). RNase III, Rnt1, and Pac1 cleave duplex RNAs longer than 20 nucleotides in vitro while their primary targets in vivo are intramolecular stem-loop structures (2, 33, 37). The basic features of the RNA cleavage mechanism appear to be similar for all three ribonucleases, but differences also exist that prevent free substrate exchange and genetic complementation (37).
Bacterial RNase III has two functionally and structurally separable subdomains: a C-terminal dsRNA-binding domain (dsRBD) and an N-terminal nuclease domain (8, 17). The dsRBD motif is located in the last 74 amino acids (aa) and adopts a tertiary fold consisting of two helices separated by three β-strands (17). This tertiary structure is conserved throughout the family of dsRNA binding proteins including the RNA-dependent kinase (PKR) (27) and the Drosophila staufen protein (3). The isolated dsRBD from Escherichia coli RNase III binds RNA to form a RNA-protein complex (17; A. Nicholson, personal communication). The solution structure of the bacterial RNase III dsRBD (17) and the protein-RNA cocrystal structure of frog dsRNA binding protein A (38) suggest multiple RNA-protein contacts involving the two α-helices and the loop between the first two β-strands of the dsRBD. The structure of the N-terminal nuclease domain of RNase III is not known, but many mutations have helped identify the main features required for RNA cleavage (8, 28). The nuclease domain contains two stretches of conserved acidic amino acid residues at positions 37 to 47 and positions 60 to 74 (7, 28). These amino acids play either a key role in catalysis or an essential structural role. Mutations in these two regions abolish RNA cleavage without affecting RNA binding (21, 28).
Yeast Rnt1 shares with bacterial RNase III the main structural features of the nuclease domain and dsRBD, suggesting that they have similar functions (2). However, unlike the bacterial enzyme, eukaryotic Rnt1 possesses an N-terminal domain. The N-terminal domain constitutes 36% of the total Rnt1 protein with no significant homology to other eukaryotic homologues of RNase III, and it has no known function. To determine the function of the N-terminal domain and verify the activities of dsRBD and the nuclease domain, we have constructed a series of deletions separating the different domains of Rnt1 and tested them for RNA binding and cleavage. Here we show that dsRBD is sufficient for RNA binding and that the nuclease domain is required for RNA cleavage. Direct analysis of the N-terminal deletion effects on RNA binding and cleavage reveals an auxiliary role ensuring efficient RNA cleavage. Deletion of the N-terminal domain reduces the processing of the 25S rRNA 3′ end by about 30% in vivo and slows growth by 35 to 40%. Biochemical and genetic assays suggest that the N-terminal domain influences Rnt1 function by mediating both inter- and intramolecular interactions.
MATERIALS AND METHODS
Strains and plasmids.
Yeast was grown and manipulated by standard procedures (11, 34). The ΔRNT1 cell is the haploid BMA64 strain carrying chromosomal disruption of RNT1 (6). Yeast PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4 gal80 (LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) was used for the yeast two-hybrid assays (15). λKH54, the E. coli strain AG1688 (MC1061 F′128 lacIq lacZ::Tn5), and plasmids pJH391, pFG157 and pKH101 (42) were used in the λ repressor system.
Plasmids used for protein expression were produced by cloning the PCR-amplified fragments of RNT1 in the bacterial expression vector pQE (Qiagen Inc., Mississauga, Ontario, Canada). pQE31/RNT1 was made by inserting a PCR fragment into the BamHI-SalI sites of pQE31 (primers 5′-CAAGCTTTTGGAT CCAATGGGCTC-3′ and 5′-CCATCATGGTCGACTAAAAGGAACG-3′). pQE31/ΔN-term was made by inserting a PCR fragment lacking the first 517 nucleotides of RNT1 into the BamHI-SalI sites of pQE31 (5′-GAAAATTTGGATCCAAGGAAGATG-3′ and 5′-CCATCATGGTCGACTAAAAGGAACG-3′). pQE31/ΔdsRBD was produced by inserting a DNA fragment containing a stop codon 1,038 nucleotides from the first AUG of RNT1. The resulting protein contains an extra 15 aa (FEASRCRKHSGRKGC) at the C terminus. pQE30/dsRBD was made by cloning the 403-nucleotide HindIII fragment in the HindIII site of pQE30. pQE31/N-term was made by deletion of the 3′-end EcoRV-HindIII fragment of pQE31/RNT1.
RNT1 was expressed in vivo using the yeast expression vectors pCU425 (19), pGBDU series (15), and pACT2 (Clontech Laboratories, Inc., Palo Alto, Calif.). pCU425/ΔNT2 was generated by inserting a PCR fragment into the SmaI site of pCU425 (primers 5′-GAAAATTTCTCGAGAATGGAAGATG-3′ and 5′-AACAGCTATGACCATG-3′). BD/RNT1 was produced by inserting the RNT1 gene in the BamHI-SalI sites of pGBDU-C3. BD/ΔDS1 was generated by deleting a PstI fragment from BD/RNT1. BD/ΔCT was generated by a deletion of AvrII fragment from BD/RNT1. BD/NT1 was generated by deleting a PvuII-SalI fragment from BD/RNT1. BD/ΔNT1 was made by deleting a BamHI-PvuII fragment from BD/RNT1. BD/ΔNT2 was made by inserting a BD/RNT1 EcoRV fragment into the SmaI-EcoRV sites of pGBDU-C1. BD/ΔNT3 was generated by deleting an EcoRI fragment from BD/RNT1. BD-DS1 was generated by inserting BamHI-XhoI containing the last 383 nucleotides of RNT1 into pGBDU-C1. AD/RNT1 was made by inserting a BD/RNT1 fragment into the BamHI-BglII sites of pACT2. AD/NT2 was generated by inserting a blunt-ended BamHI-NheI fragment from pQE31/N-term in a blunt-ended BamHI site of pACT2. AD/ΔDS was generated by inserting a SmaI-HindIII fragment from BD/RNT1 into the pACT2 SmaI site. BD/NT2 was produced by inserting a SmaI-XhoI fragment from AD/NT2 into the SmaI-SalI sites of pGBDU-C3.
pJH/RNT1 used in the λ repressor assay was made by inserting a blunt-ended BglII-EcoRI fragment generated by partial digestion of BD/RNT1 into the blunt-ended SalI-BamHI sites of pJH391.
Protein purification.
All recombinant proteins in this work were produced either in E. coli BL21(DE3)pLysS (Promega Corp., Madison, Wis.), E. coli M15(pREP4) (Qiagen Inc.), or E. coli DH5αF′ (Life Technologies, Burlington, Ontario, Canada). Recombinant proteins were purified on a Ni-nitrilotriacetic acid agarose column (Pharmacia Biotech Inc., Baie d'Urfé, Québec, Canada) as described previously (16) with the following modifications. The first purification step was performed with Nickel buffer (25% glycerol, 1 M NaCl, 30 mM Tris [pH 8.0]). Protein fractions were pooled and passed through a second column with Nickel buffer without glycerol. Further purification of the N-terminal protein was performed on an HIC ISO column (Pharmacia Biotech Inc.) with a 0.05 to 1.5 M gradient of (NH4)2SO4 and 50 mM sodium phosphate buffer at pH 7.5. The pure protein was collected in the unbound fraction. All purifications were conducted using the AKTA explorer fast protein liquid chromatography system (Pharmacia Biotech Inc.). The protein fractions were dialyzed against dialysis buffer (50% glycerol, 0.5 M KCl, 30 mM Tris [pH 8.0], 0.1 mM dithiothreitol [DTT], 0.1 mM EDTA [pH 8.0]) and stored at −20 or −80°C for long-term storage. The identity of the two proteins produced by the plasmid pQE30/dsRBD (Fig. 1C) was confirmed by monitoring the expression patterns of the two proteins and using Western blot analysis. Tests for RNA binding and dimerization confirm that the two proteins have similar activities.
FIG. 1.
Purification and in vitro analysis of Rnt1 domains. (A) On top is shown a comparison of the functional domains of RNase III between bacteria, worms, and yeast. All members of the RNase III family possess a nuclease domain (NUCD) and a dsRNA binding domain (dsRBD). The N-terminal domain (NTD) is unique to the eukaryotic members of the RNase III family. Sequence alignment was performed using CLUSTALW (13). A schematic representation of Rnt1 fragments and the associated functional domains is shown at the bottom. The expressed segments of Rnt1 are indicated in amino acids on the left. (B) Illustration of the U5 3′-end model substrate. The cleavage sites are indicated by arrows. The numbers are relative to the U5 snRNA mature 3′ end. The dotted line represents the vector sequence (pRS315). (C) Purification of the N-terminal His6-tagged Rnt1 fragments. All proteins were purified on metal chelating affinity columns. The N-term protein was repurified using hydrophobic interaction chromatography. Aliquots from the last purification step of each protein were fractionated on SDS-PAGE and stained with Coomassie brilliant blue R. The upper band in the dsRBD lane corresponds to a readthrough of the natural stop codon of RNT1 to a bacterial stop codon downstream. The protein molecular weight markers are indicated on the left in thousands. (D) In vitro cleavage assay of Rnt1 derivatives, using the 3′ end of U5 as a model substrate. The 138-nucleotide substrate was incubated with no protein (lane 1), GST (lane 2), Rnt1 (lane 3), one of the four different Rnt1 deletions (lanes 4, 5, 7, and 8), or combinations of two different deletions (lanes 6 and 9). On the right, the position of the substrate and the different cleavage products are indicated as follows: S, full-length 138-nucleotide substrate; P1, 62-nucleotide 3′-end cleavage product; P2, 45-nucleotide 5′-end cleavage product; P3, middle 31-nucleotide cleavage product. The DNA molecular weight markers are indicated on the left. (E) Gel retardation assay of Rnt1 derivatives. The RNA was incubated with no protein (lane 1), Rnt1 (lane 2), one of the four different deletions (lanes 3, 4, 5, and 6), or GST (lane 7). The reaction was carried out in 25 mM KCl at 4°C, and the products were loaded on a 4% native gel. The positions of the shifted RNAs are indicated by solid arrowheads (Rnt, ΔNT, and DS) on the right. The positions of supershifted RNAs are indicated by open arrowheads (Rnt1S, ΔNTS, and DSS) on the left. The band indicated by the asterisk is a differently folded form of single-stranded RNA. The positions of the origin (ori) and unbound (Un) RNA are indicated on the right.
Enzymatic assays.
The radiolabeled RNA used as a substrate in the enzymatic assays was generated by T7 RNA polymerase in the presence of [α-32P]UTP. The RNA substrate was produced from a T7 promoter of plasmid pRS315/U5. To make this plasmid, a blunt-ended NheI fragment generated by PCR with primers 5′-CTTTTCTATTGCTAGCTTTCTAC-3′ and 5′-GCTAGCAAATGCTTCAATGAG-3′ was cloned in the blunt-ended XbaI site of pRS315. For the in vitro cleavage, 200 fmol of substrate was incubated for 10 min at 30°C in 10 μl of reaction buffer (30 mM Tris [pH 7.5], 5 mM spermidine, 10 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA [pH 7.5]). The general effects of salt, N-terminal deletion, or N-terminal addition were confirmed using a wide range of substrate and protein concentrations. The amount of KCl used is indicated in the description of each experiment. The reaction was stopped by addition of a stop buffer (20 mM EDTA [pH 7.5] and 0.1% bromophenol blue in formamide) and directly loaded on denaturing 8% polyacrylamide gel. The cleavage rate was calculated using the Molecular Analyst programs (Bio-Rad Industries, Hercules, Calif.).
Gel mobility shift assay and in-gel cleavage assay.
RNA binding reactions were performed using 2 fmol of radiolabeled RNA in 20 μl of binding buffer (20% glycerol, 30 mM Tris [pH 7.5], 5 mM spermidine, 0.1 mM DTT, 0.1 mM EDTA [pH 7.5]) for 10 min on ice. The amount of KCl and protein are indicated for each experiment. The reactions were fractionated on a 4% nondenaturing polyacrylamide gel at 0.5 V/cm2 and 4°C. The in-gel cleavage assay was performed by cutting the bands corresponding to different complexes formed in the gel mobility shift assay and incubating them in a cleavage buffer (30 mM Tris [pH 7.5], 5 mM spermidine, 0.1 mM DTT, 0.1 mM EDTA [pH 7.5], 20 mM MgCl2) at 30°C for 40 min. After the incubation period was complete, the gel pieces were removed and the RNA was extracted and loaded on 8% denaturing polyacrylamide gels.
RNase protection assay.
A probe complementary to the 3′ end of 25S rRNA and the 3′ external transcribed spacer (ETS) was produced by T7 transcription (2). Total RNA (10 μg) was incubated at 42°C for 12 h with 105 cpm of probe in 80% formamide hybridization buffer (25). The hybridization mix was digested with 2 μg of RNase T1 per ml for 1 h at 30°C, extracted with phenol-chloroform, ethanol precipitated, and loaded on a 6% polyacrylamide gel.
Gel filtration assay.
A Superdex 200 HR 10/30 column (Pharmacia Biotech Inc.) (10 by 300 to 310 mm) was equilibrated in gel filtration buffer (50 mM sodium phosphate [pH 7.5], 2 mM EDTA [pH 7.5], 0.5 M KCl) at 23°C and calibrated with low- and high-molecular-weight markers (Pharmacia Biotech Inc.). For sample application, 245 μg of each protein was applied to the column and 250-μl fractions were collected and analyzed on sodium dodecyl sulfate (SDS) gels (20).
Protein cross-linking.
Cross-linking experiments were performed as described previously (24). Purified proteins (0.3 μg) were incubated for 10 min at 30°C in 10 μl of gel filtration buffer with increasing concentration of freshly diluted glutaraldehyde (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada). The cross-linked proteins were analyzed by standard SDS-polyacrylamide gel electrophoresis (PAGE) and detected by silver staining.
Yeast two-hybrid assays.
Plasmids encoding the appropriate AD- and BD-RNT1 fusion were cotransformed in yeast PJ69-4A using a modified lithium acetate method (39). The cells harboring both plasmids were selected on SCD medium (11) lacking lysine, uracil, and leucine. The two-hybrid interactions were indicated by the ability of a pair of plasmids to activate the three test promoters in PJ69-4A (15). Three or four independent transformants for each plasmid pair were tested on medium lacking either adenine or histidine. The histidine-containing media were supplemented with 10 mM 3-aminotriazole to avoid basal expression of histidine (15). The activation of the third reporter gene was tested by β-galactosidase liquid assay as described earlier (32). Cells were harvested in mid-logarithmic phase, and their ability to hydrolyze o-nitrophenyl-β-d-galactopyranoside was measured as previously described (26).
λ repressor system.
The λ repressor assay was performed essentially as described previously (42). For the dot plaque assay, E. coli AG1688 transformed with either pJH/RNT1, pFG157, or pKH101 was grown to saturation in λ broth (1% tryptone, 0.25% NaCl, 0.2% maltose, 10 mM MgSO4, 50 μg of ampicillin per ml). A 300-μl volume of this bacterial culture was mixed in 3 ml of λ top agar (0.5% yeast extract and 0.7% agar in λ broth) and poured on a fresh plate of λ agar (λ broth, 1% agar), forming a bacterial lawn. Each lawn of bacteria was infected with a serial dilution of λKH54 phage lysate containing between 5 × 104 and 5 × 108 PFU. Infected lawns were incubated for 18 h at 30°C, and the sizes of the resulting plaques were measured.
Western blot analysis.
Yeast cells were grown to stationary phase in the appropriate SCD medium (11), and cellular proteins were extracted as previously described (39). Total proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (MSI, Westborough, Mass.). Western blot analysis was performed as described previously (12). Proteins were visualized using either monoclonal antibody against the Gal4 DNA binding domain or polyclonal antibody against the Gal4 activation domain (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The protein bands were visualized by enhanced chemiluminescence (ECL kit; Amersham, Arlington Heights, Ill.). The expression value of each fusion protein was estimated using the Molecular Analyst programs (Bio-Rad Industries).
RESULTS
The N-terminal domain of yeast RNase III is not essential for RNA binding and cleavage.
Analysis of the Rnt1 sequence reveals three distinct domains; a 127-aa C-terminal domain containing a 74-aa dsRBD motif, a 154-aa central domain containing the RNase III nuclease motif, and a 191-aa N-terminal domain lacking significant homology to known proteins (Fig. 1A). To determine the contribution of the various domains to Rnt1 function, we expressed them individually in bacteria and assayed their activity in vitro. Five different segments of Rnt1 were expressed as N-terminal His6-tagged proteins. The five proteins are full-length Rnt1 (Rnt1), Rnt1 lacking the C-terminal 150 aa including the 74-aa dsRBD motif (ΔdsRBD), the 191-aa protein representing the N-terminal domain (N-term), a protein lacking the first 171 aa of the N-terminal domain (ΔN-term), and a 127-aa protein containing the full dsRBD motif (dsRBD). Following expression in bacteria, these proteins were purified on two successive nickel affinity columns, with the exception of the N-term protein, which was repurified by hydrophobic interaction chromatography. All proteins were expressed in soluble form and were purified under native conditions to a purity of 85 to 95%, as judged by Coomassie blue-stained gels (Fig. 1C).
To determine the function of each expressed Rnt1 fragment, we have examined each derivative for RNA cleavage and RNA binding. The 3′ end of U5 snRNA (Fig. 1B) was used as a model substrate at low concentrations of monovalent salt to allow maximum cleavage (2, 22, 35). As shown in Fig. 1D, the full-length Rnt1 (lane 3) cleaved U5 at the expected in vivo sites (4), while no cleavage was seen when the RNA was incubated alone or with glutathione S-transferase (GST) (lanes 1 and 2, respectively). The ΔdsRBD, dsRBD, and N-term proteins did not cleave the RNA substrate (lanes 4, 5, and 8, respectively). Prolonged incubation or addition of different divalent metal ions did not enhance the activity of this set of proteins (data not shown). Mixing the dsRBD with the ΔdsRBD protein did not reconstitute enzyme function (lane 6), suggesting that the dsRBD and the nuclease domain are required in cis for RNA cleavage. Surprisingly, the ΔN-term protein, which lacks 36% of Rnt1 primary structure, cleaved U5 with an efficiency similar to that of the full enzyme (lane 7). Addition of the N-terminal domain to the ΔN-term protein had no noticeable effects (lane 9). We conclude that the N-terminal domain is not required for RNA cleavage under these conditions.
The ability of various Rnt1 domains to bind RNA was tested under conditions that allow RNA binding without cleavage (21). Radiolabeled U5 RNA transcripts were incubated with Rnt1 or derivatives in the absence of Mg2+ and fractionated on a polyacrylamide gel under native conditions. As expected, proteins containing the dsRBD motif including Rnt1, dsRBD, and ΔN-term bound to the RNA (Fig. 1E, lanes 2, 4, and 6, respectively) while proteins lacking the dsRBD motif did not (lanes 3 and 5). We conclude that the RNA binding activity of Rnt1 is restricted to the dsRBD, that RNA cleavage requires the nuclease domain, and that the N-terminal domain has no apparent effect on either binding or cleavage in vitro.
To examine the function of the N-terminal domain in vivo, we cloned a set of RNT1 deletions in yeast expression vectors. The different deletion mutants were expressed in cells lacking the RNT1 gene, either directly using a copper-inducible promoter or as an N-terminal fusion with a nuclear localization signal from the GAL4 DNA binding domain (BD). As shown in Fig. 2A, constructs carrying the full BD-RNT1 fusion complemented the RNT1 knockout and enabled yeast cells to grow at both permissive (25°C) and restrictive (37°C) temperatures. In contrast, constructs carrying the GAL4 BD alone, a variety of deletions in the dsRBD, or the nuclease domain (BD-ΔDS1, BD-NT1, and BD-ΔCT) did not complement the knockout phenotype. Constructs carrying partial or complete deletions of the N-terminal domain (BD-ΔNT1 and BD-ΔNT2) allowed ΔRNT1 cells to grow at both the permissive and restrictive temperatures. However, cells expressing proteins with an N-terminal deletion grew more slowly than did cells expressing intact Rnt1 at both the permissive and restrictive temperatures (Fig. 2A). In rich liquid media, cells carrying RNT1 gene grew with a doubling time of 2.03 h while cells carrying partial (ΔNT1) or full (ΔNT2) deletion of the N-terminal domain grew 45 to 35% slower at 3.65 and 3.1 h, respectively; cells lacking the RNT1 gene grew with a doubling time of 13.16 h. These results show that a truncated version of Rnt1 lacking the N terminus is still active in vivo, albeit at a reduced level. Removal of the GAL4 nuclear localization signal or variations in the expression level of the ΔNT protein did not affect its ability to complement Rnt1 function (Fig. 2 and data not shown). This suggests that the effect of the N-terminal deletion on cell growth is not due to nuclear misslocalization. To ensure that the deletion of the N-terminal domain does not affect Rnt1 stability, we have compared the expression level of Rnt1 to that of its N-terminal deletion in vivo. As shown in Fig. 2D, the expression levels of plasmid-borne BD-ΔNT1 and BD-ΔNT2 fusion proteins are similar to that of BD-RNT1 in cells expressing a chromosomal copy of Rnt1 (lanes 5 to 7). In contrast, the expression level of the plasmid born ΔN-term fusion is much higher than Rnt1 fusion in cells lacking the chromosomal copy of Rnt1 (lanes 1 to 3). This suggests that the slow growth caused by the N-terminal deletion is not due to reduced expression level of Rnt1. Expression of both ΔN-term and N-term proteins in trans did not enhance cellular growth (data not shown). This result suggests that both domains are required in cis for optimum activity in vivo.
FIG. 2.
The N-terminal domain of Rnt1 is not essential for growth at 37°C. (A) Cells lacking the RNT1 gene were transformed with a series of RNT1 deletions fused with a nuclear localization signal from the GAL4 BD. The transformed yeast cells were streaked on minimal medium without uracil and incubated at 25 or 37°C. The position of each deletion is indicated on the right. The boxes illustrate deletions of the N-terminal domain (white), nuclease domain (light gray), and dsRBD (black). (B) Expression of the N-terminal deletion of Rnt1 from an inducible promoter without a nuclear localization signal. Segment 172 to 471 of Rnt1 was cloned under a copper-inducible promoter and transformed in cells lacking Rnt1. The cells were grown on minimal medium containing 100 μM Cu2+ at either 25 or 37°C. Boxes represent Rnt1 segments as described in panel A. (C) Mapping the 3′ end of 25S rRNA using an RNase protection assay. The RNA was extracted from cells lacking Rnt1 (lanes 4, 5, and 6), expressing Rnt1 (lane 3), or expressing N-terminal deletions (lanes 7, 8, and 9) with or without a nuclear localization signal. The RNA was hybridized to a probe complementary to the 3′ end of the 25S pre-rRNA and digested with RNase T1. The probe was also hybridized to E. coli tRNA as a control (lane 2). The positions of mature and extended 3′ ends are indicated on the right. The DNA molecular weight markers are indicated on the left. (D) Western blot analysis of Rnt1 and ΔN-term proteins. Proteins were extracted from RNT1 or ΔRNT1 cells expressing different deletions of Rnt1p fused to the Gal4 BD. Equal amounts of proteins were separated on an SDS-polyacrylamide gel and examined using monoclonal antibodies against the Gal4 BD. Protein extracts from untransformed cells (Un) were included as control. The upper protein band in each lane corresponds to the expected size of the fusion protein. The lower band corresponds to a smaller protein that may result from either a pre-mature stop or degradation at the protein C terminus.
To test the effect of the N-terminal deletion on the processing activity of Rnt1 in vivo, we monitored the level of mature 25S rRNA in cells expressing either Rnt1 or ΔN-term (Fig. 2C). An RNA protection assay was performed using a probe that spans the 3′ end of the 25S rRNA and includes sequences downstream (2). RNA from cells expressing Rnt1 protects the probe at one position corresponding to the mature 3′ end of 25S rRNA (Fig. 2C, lane 3). In contrast, RNA extracted from cells lacking Rnt1 protects the probe at multiple positions, corresponding to the 3′ end of unprocessed 25S pre-rRNA. RNA extracted from cells expressing ΔNT2 protects the probe at both mature and extended positions of the 25S rRNA (lanes 7 to 9). Quantification of the protected probe indicates that about 30% of the 3′ end of 25S rRNA is not processed in cells expressing the ΔN-terminal protein. Additional RNA protection assays indicated that the processing of U2 snRNA 3′ end is equally affected (data not shown), suggesting a general effect of the N-terminal deletion on Rnt1 processing activity. We conclude that the N-terminal domain is required for efficient RNA processing and normal cellular growth in vivo.
Deletion of the N-terminal domain impairs dsRNA cleavage at physiological salt concentrations in vitro.
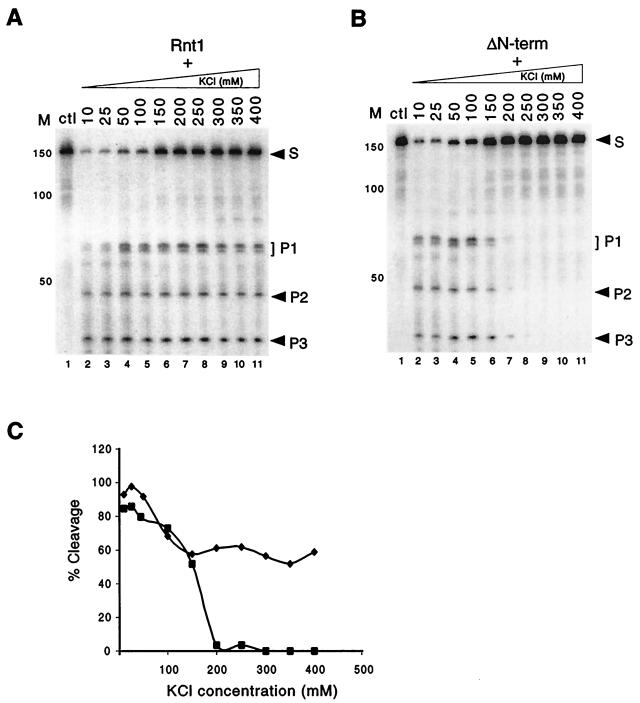
The apparent difference between the in vitro (Fig. 1) and in vivo (Fig. 2) activities of the ΔN-term protein may reflect differences in the reaction conditions. Physiological salt concentrations in yeast range between 150 and 200 mM (30), while in vitro cleavage tests were normally conducted at concentrations lower than 50 mM (Fig. 1) to allow maximal activity (1, 2). To examine this possibility, we assayed Rnt1 or ΔN-term cleavage of U5 over a range of KCl concentration from 10 to 400 mM. As shown in Fig. 3A, the Rnt1 cleavage rate diminished at KCl concentrations above 100 mM. From 150 to 400 mM KCl, the cleavage rate of Rnt1 remained more or less constant, with 60% of the substrate being cleaved. In contrast, ΔN-term cleaved the RNA substrate at a rate similar to that of Rnt1 at KCl concentrations below 50 mM and cleavage was suppressed completely at concentrations higher than 200 mM (Fig. 3B). At physiological salt concentrations (150 to 200 mM KCl), ΔN-term was about 30% to 40% less active than Rnt1 (Fig. 3C). This result is consistent with the reduced activity of ΔN-term observed in vivo (Fig. 2C). Thus, although we observed previously that the N-terminal domain did not affect Rnt1 activity at low salt concentration (Fig. 1), we conclude that the N-terminal domain is required for Rnt1 function in vitro at high concentrations of monovalent salt.
FIG. 3.
Deletions of the N-terminal domain of Rnt1 impairs RNA cleavage at physiological KCl concentrations. Cleavage of the U5 snRNA 3′ end by Rnt1 (A) or ΔN-term (B) in increasing salt concentrations is shown. In each panel, RNA incubated with GST under the same reaction conditions is included as a control (lane 1). The positions of the RNA substrate (S) and the cleavage products (P1, P2, and P3) are indicated on the right, and the DNA markers are indicated on the left. (C) Percent cleavage rate of Rnt1 (⧫) and ΔN-term (■) versus concentration of KCl. Autoradiographs of gels similar to these in panel A and B were scanned and quantified using the Bio-Rad gel analysis system. The data points shown are the average of two different experiments.
To determine the effect of monovalent salts on RNA binding, we carried out gel mobility shift assays of Rnt1, ΔN-term, and dsRBD under different salt concentrations. As shown in Fig. 4, Rnt1 formed one major complex with the RNA (Fig. 4A, lanes 6 to 8, and Fig. 4C, lanes 5 to 7) with a kd value of 195 nM. Surprisingly, ΔN-term bound the RNA more efficiently than Rnt1 did with a kd value of 73 nM in 5 mM KCl and 143 nM in 100 mM KCl (Fig. 4). ΔN-term formed two complexes with the RNA, an intermediate complex that formed at low protein concentration (Fig. 4A, lanes 9 to 13, and Fig. 4C, lanes 9 to 12) and a second complex that formed as the protein concentration was increased (Fig. 4A, lanes 13 to 16, and Fig. 4C, lanes 12 to 15). The intermediate ΔN-term complex and the Rnt1 complex appeared to have similar activities as judged by an in-gel cleavage assay (Fig. 5B and C, lanes 3 and 4, respectively). In contrast, the second complex formed by ΔN-term was less active and was sensitive to high concentration of monovalent salts (Fig. 5B and C, lanes 5). These results suggest that the deletion of the N-terminal domain influences the assembly of the RNA-protein complexes, favoring the formation of a less active protein-RNA complex. Further deletions removing the nuclease domain did not prevent the association of dsRBD with the RNA. As shown in Fig. 4, the dsRBD bound the RNA with a kd value of 145 nM in 5 mM KCl and 85 nM in 100 mM KCl. The dsRBD-RNA complexes showed a gradual and continuing shift as a function of the protein concentration (Fig. 4A and C). The heterogeneous complexes may represent binding of several proteins to a single RNA molecule or may be due to multiple protein-protein interactions. Salt concentrations ranging from 150 to 300 mM KCl, while reducing RNA cleavage (Fig. 3), did not have significant effects on the kinetic of binding for all three proteins (data not shown). We conclude that Rnt1 binding to RNA is mediated by the dsRBD and that deletion of the N-terminal domain does not decrease the binding efficiency.
FIG. 4.
Deletion of the N-terminal domain influences Rnt1 binding affinity. (A and C) Increasing concentrations of Rnt1, ΔN-term, or dsRBD were incubated with 2 fmol of U5 3′-end model substrate in 5 mM KCl (A) or 100 mM KCl (C). RNA incubated with GST under the same conditions is included as a control. The position of the gel origin (Ori) and unbound RNA (Un) are indicated on the left. (B and D) Quantitative analysis of RNA binding to Rnt1 (▴), ΔN-term (■), or dsRBD (●) were carried in either 5 mM KCl (B) or 100 mM KCl (D). The binding percentage was plotted versus the protein concentration. Each data point is the average of three experiments.
FIG. 5.
In-gel cleavage assay of Rnt1 and ΔN-term RNA-protein complexes. (A) A gel shift assay was conducted at 125 mM KCl as described in Materials and Methods. (B and C) The gel bands corresponding to each complex were cut and incubated in either 50 mM KCl (B) or 250 mM KCl (C) in presence of MgCl2 to allow RNA cleavage. At the end of the incubation period, the gel pieces were removed and the RNA was extracted and loaded on an 8% denaturing polyacrylamide gel (B and C). In panel A, the position of each Rnt1 complex is indicated on the left (bands 2 and 3) and the position of each ΔN-term complex is indicated on the right (bands 4 and 5). The band corresponding to the input RNA (band 1) was used as control. In panels B and C, the substrate and cleavage products are indicated on the right. The DNA molecular weight markers are indicated on the left.
Biochemical evidence for N-terminal domain- and dsRBD-mediated dimerization of Rnt1.
To examine the role of the N-terminal domain in the formation of active Rnt1 protein, we analyzed the conformation of Rnt1 and its derivatives in solution by size exclusion chromatography. Each protein was expressed in bacteria and purified as described in Fig. 1 before being loaded on a gel filtration column. Each column was calibrated with high- and low-molecular-weight markers prior to the sizing of each protein. Rnt1 eluted in three major peaks, the smallest corresponding to a dimer form and the other two corresponding to a tetrameric and a multimeric form (Fig. 6A). These protein complexes are not aggregates of denatured proteins, since all three forms were equally capable of cleaving the substrate RNA (data not shown). This result suggests that, similar to the bacterial RNase III (9, 22, 23), Rnt1 self-interacts to form a dimer in solution. Gel filtration of the ΔN-term resulted in only two peaks, one of which corresponded to the monomer form while the other corresponded to the dimer form (Fig. 6B). This result suggests that ΔN-term cannot self-interact as efficiently as Rnt1. For the dsRBD, only one peak corresponding to the dimer size was observed (Fig. 6C). This result suggests that a dimerization domain exists within the dsRBD, as observed with other dsRNA binding proteins (29). Notably, ΔdsRBD that lacks the dsRBD motif migrated as one large peak beyond the range of the column (data not shown). The N-term protein migrated on the column in the same fashion as the ΔdsRBD, forming only one peak of high molecular weight corresponding to a multiple protein complex (Fig. 6D). This result suggests that the N-terminal domain acts as a second dimerization signal for Rnt1.
FIG. 6.
Rnt1 dimerization is mediated by N-terminal and C-terminal signals. Size exclusion chromatography was carried out with Rnt1 (A), ΔN-term (B), dsRBD (C), and N-term (D) using a Superdex 200 HR 10/30 column. Each protein was loaded on columns precalibrated with 500 mM KCl. The protein content of each peak was verified by loading the corresponding fractions on SDS-polyacrylamide gels (shown below each chart). The size of each peak reflects the light absorbancy and not the amount of each protein. Peaks corresponding to the column end (end) are identified on top of each chart. The molecular weight marker is shown as a dotted line, and the size corresponding to each peak is indicated on top.
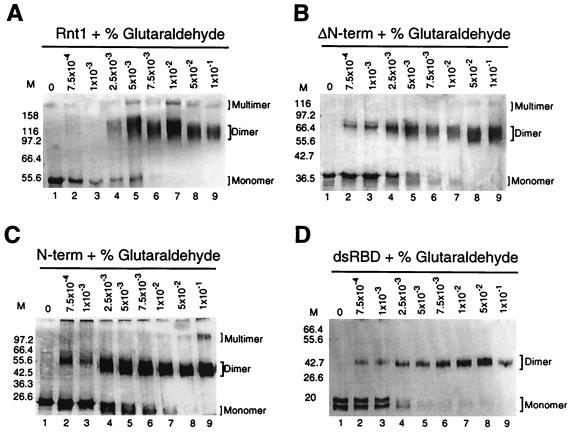
Protein cross-linking was used to further characterize the multimeric complexes of the purified Rnt1 derivatives. Each purified protein was incubated in 500 mM KCl with increasing glutaraldehyde concentrations (0 to 0.1%). The cross-linked proteins were analyzed by SDS-PAGE and visualized using silver stain. In the absence of glutaraldehyde, all proteins migrated as monomers with the expected molecular weights (Fig. 7). At increasing concentrations of glutaraldehyde, the monomeric bands were converted to bands corresponding to the dimer form for Rnt1 (Fig. 7A), ΔN-term (Fig. 7B), N-term (Fig. 7C), and dsRBD (Fig. 7D). For the ΔdsRBD, no dimerization was observed; instead, a band corresponding to a complex with high molecular weight was observed near the top of the gel (data not shown). The aggregation of ΔdsRBD may result from misfolding or denaturation of the protein. Bands corresponding to multimers can be seen in Rnt1 and N-term (Fig. 6A and C) and to a lesser extent in ΔN-term (Fig. 6B). Glutaraldehyde treatment of chymotrypsin or bovine serum albumin BSA (data not shown) under the same conditions did not change the migration of the monomeric forms. Addition of RNA or extensive treatments of the different proteins with RNase A, up to 50 mM DTT, or 25% glycerol did not affect the dimerization pattern (data not shown). However, increasing the protein concentration caused different degrees of protein multimerization (data not shown). These results indicate that Rnt1 dimerization is not RNA dependent and does not depend on disulfide bond formation. We conclude that Rnt1 can form a dimer through at least two dimerization signals, one in the N-terminal domain and the other in the dsRBD.
FIG. 7.
Glutaraldehyde cross-linking analysis of Rnt1 derivatives. Pure Rnt1 (A), ΔN-term (B), N-term (C), and dsRBD (D) were incubated in increasing concentrations of glutaraldehyde–500 mM KCl. The reaction mixtures were incubated for 10 min and then loaded on SDS-polyacrylamide gels and silver stained. The positions of monomer, dimer, and multimer are shown on the right. The molecular weight markers are indicated on the left in thousands.
In vivo evidence for N-terminal-mediated self-interactions of Rnt1.
To test Rnt1 dimerization in vivo and map its dimerization signals, we used the yeast two-hybrid assay. Two sets of plasmids carrying various segments of Rnt1 either fused to the Gal4 activation domain (AD) expressed from ADH1 promoter or fused to the Gal4 BD expressed from a truncated ADH1 promoter were used (Fig. 8A). The different plasmids were transformed in all pairwise combinations into yeast strain PJ69-4A (15) containing three different marker genes (HIS3, ADE2, and lacZ) under the control of three different test promoters (GAL1, GAL2, and GAL7, respectively). Real interactions can be scored using all three markers and may be quantified using a β-galactosidase liquid assay. The results shown in Fig. 8A indicate that the N-terminal domain fused to Gal4 AD (AD-NT2/1–191) can interact with itself (BD-NT2/1–191), the dsRBD (BD-DS1/344–471), and Rnt1 (BD-RNT/1–471). The N-term protein appears to interact directly with the dsRBD because they can be cross-linked in vitro in the absence of any other factors (data not shown). These results suggest that the enzyme may self-interact through interactions between the two N-terminal domains, the two dsRBDs, or the dsRBD and the N-terminal domain.
FIG. 8.
Yeast two-hybrid analysis of interactions between Rnt1 domains. (A) Summary of Rnt1 inter- and intramolecular interactions determined by two-hybrid analysis. Two sets of plasmids carrying the indicated segments of Rnt1 fused to either GAL4 BD (BD-fusion) or GAL4 AD (AD-fusion) were constructed. Different combinations of the two sets were introduced into yeast strain PJ69-4A carrying three reporter genes (ADE2, HIS, and lacZ) under the control of three different promoters. Interaction between any pair of BD and AD fusion proteins will lead to the activation of all three markers with different efficiencies depending on the promoter stringency. The expression level (PE) of each fusion protein was assayed by Western blot analysis of whole-cell extract with anti-BD or anti-AD antibodies. The expression level of each fusion is indicated as a percentage of the wild-type BD or AD expression level. The interaction level of each two plasmids is indicated as weak (+), moderate (++), and strong (+++). The strength of the interaction was also measured using the β-galactosidase liquid assay (LZ), and the average Miller units of three experiments are indicated. A schematic representation of the different constructs is shown on top and on the right of the table. White boxes indicate the N-terminal domain, light gray boxes indicate the nuclease domain, and black boxes indicate the dsRBD. (B) Comparison of the interaction strength between the different functional domains of Rnt1 using liquid β-galactosidase assays. An average of three experiments of each pair of plasmids was plotted using Miller points. (C) λ repressor assay of Rnt1 dimerization. Dot plaque assay of Rnt1 dimerization was conducted using E. coli AG1688 transformed with either λN or λN-Rnt1 fusion. The cells were poured as a lawn and infected with a 5-μl dilution of λKH54 lysates. The ability of Rnt1 to dimerize is measured by its ability to suppress λ infection and reduce bacterial lysis. The titer is indicated beside each spot.
The strength of the interaction between each protein pair was measured using β-galactosidase assays and quantified using Miller units (26). As shown in Fig. 8B, the strongest interaction was detected between the two N-terminal domains (NT2/1–191) followed by the interaction between the N-terminal domain (BD-NT2/1–191) and ΔdsRBD (AD-ΔDS/1–321). The interaction between the N-terminal domain (AD-NT2/1–191) and the dsRBD (BD-DS1/344–471) was 10 times lower than the interaction between the N-terminal domains. Consistently, protein cross-linking assays showed that the N-term/N-term complex was more favored than the N-term/dsRBD complex (data not shown). Together, these results suggest that Rnt1 is capable of forming intermolecular interaction. Our results also suggest that Rnt1 has the ability to form an intramolecular complex. This conclusion is inferred from the ability of the N-term protein and dsRBD to interact.
Fusion of Rnt1 segments containing the nuclease domain to Gal4 AD activated the test promoter only when expressed with the N-terminal domain Gal4 BD fusion (Fig. 8A). Other fragments, including those proven to self-interact either biochemically (Fig. 6 and 7) or through the Gal4 BD fusion (Fig. 8A), failed to activate the test promoters when linked to the nuclease domain. Therefore, we could not directly test the intermolecular interaction of full-length Rnt1 by using the two-hybrid system. To confirm this interaction, we used a dimerization-dependent λ repressor fusion system in bacteria (42). Rnt1 was fused to the N-terminal DNA binding (DB) domain of λ phage (λN) and tested for dimerization. If dimerization occurs, the λ N-terminal domain will repress the transcription of genes required for the phage lytic growth and prevent λ superinfection. As seen in Fig. 8C, λ induced cell lysis is reduced when Rnt1 was fused to λ N confirming the self-dimerization of Rnt1. We conclude that Rnt1 function as a dimer with a dynamic conformation critically dependent on the protein interaction mediated by the N-terminal domains.
DISCUSSION
Yeast Rnt1 and the bacterial RNase III share the basic features required for dsRNA binding and cleavage (2). In addition to the nuclease domain and dsRBD, the yeast enzyme contains a 191-aa extension at the N terminus unique to the eukaryotic homologues of RNase III. We have found that the N-terminal domain favors the formation of stable or functional Rnt1 protein complexes. Deletion of the N-terminal domain reduces the processing activity of Rnt1 by 35 to 40% and makes it hypersensitive to monovalent salt (Fig. 2 and 3). Biochemical (Fig. 6 and 7) and genetic (Fig. 8) evidence indicates that the N-terminal domain can interact with itself and with the dsRBD. Together, our results suggest that the eukaryotic N-terminal domain enhances Rnt1 function by mediating the formation of optimum protein conformations.
Yeast RNase III has a novel functional domain.
The dsRBD and the nuclease domain of E. coli RNase III can be structurally and functionally separated (17; Nicholson, personal communication). The conserved C-terminal dsRBD is sufficient for dsRNA binding, and the N-terminal nuclease domain is sufficient for RNA cleavage. Yeast Rnt1 contains sequences homologous to both dsRBD and nuclease domain in addition to a unique 191-aa N-terminal domain. Here we provide evidence that yeast RNase III dsRBD is sufficient for dsRNA binding and that the nuclease domain is required for RNA cleavage. However, unlike the bacterial enzyme, deletion of the C-terminal dsRBD abolishes all RNA binding activity (Fig. 1) and the nuclease domain cannot cleave the substrate without the dsRBD, even with different divalent metal ions (Fig. 1 and data not shown). Deletion of the N-terminal domain did not affect the basic functions of Rnt1, suggesting that the protein structural and sequence elements required for RNA binding and cleavage are conserved among prokaryotes and eukaryotes. However, the efficiency of the RNA cleavage is diminished by the deletion of the N-terminal domain without significantly affecting the RNA binding efficiency (Fig. 3 and 4). Thus, deleting the N-terminal domain results in the formation of RNA-protein complexes in vitro that are either less productive or unstable under RNA cleavage conditions. These results suggest that the N-terminal domain is a functionally and structurally separate domain required for normal cell growth and efficient RNA cleavage. Database searches reveal the presence of two types of N-terminal domains among the eukaryotic homologues of RNase III (35). The first has homologies to the DEAD box ATPase-dependent helicase family and may be found in S. pombe (Pac 8), Caenorhabditis elegans, and Homo sapiens but not in the S. cerevisiae genome. The second type has no significant homology to known proteins and can be found in S. cerevisiae (Rnt1), S. pombe (Pac1), C. elegans, and H. sapiens genomes. Deletion of the Pac1 N-terminal domain appears to inhibit RNA cleavage, but the mechanism and extent of inhibition are not clear (14). The activity of the Pac1 N-terminal deletion was tested in crude bacterial extracts, preventing accurate measurements, and its ability to bind RNA was not examined. More tests with Pac1 and other eukaryotic homologues of Rnt1 are required to identify possible conserved functions of the N-terminal domain. Meanwhile, the results presented here suggest that in yeast the N-terminal domain of the eukaryotic RNase III is a distinct functional domain required for efficient RNA cleavage under physiological conditions.
Yeast RNase III self-interaction is mediated by N-terminal and C-terminal signals.
RNase III forms a dimer in solution and appears to function as a dimer (9, 20). Here we show that Rnt1 also forms an intermolecular complex mediated by signals located at the N-terminal and C-terminal domains. However, the mechanism of Rnt1 self-interaction and binding to dsRNA appears different from that of RNase III. Unlike RNase III, Rnt1 forms multiple protein complexes at high salt concentrations (Fig. 6 and 7). Multiple interactions may also occur after binding to RNA at high protein concentrations, suggesting that these protein interactions do not interfere with RNA association. In addition, the nature of Rnt1-RNA complexes appears to be different from those of RNase III, since their stability is not dependent upon divalent metal ions (19). These differences between RNase III and Rnt1 may be caused at least in part by the N-terminal domain of Rnt1. Analysis of Rnt1 derivatives suggests that the multiple protein complexes formed in solution are mediated in part by the N-terminal domain (Fig. 6 and 7). N-terminally deleted Rnt1 or a protein containing only the dsRBD multimerize less readily and form mainly dimers or remain as monomer in solution, similar to the bacterial RNase III (21). In contrast, proteins containing the N-terminal domain or lacking the dsRBD tend to form higher-molecular-weight complexes. These observations suggest that Rnt1 possesses two dimerization signals that may provide a dynamic switch between different complexes (see below).
Using yeast two-hybrid and λ repressor assays, we have confirmed that Rnt1 dimerizes and that the isolated dsRBD and N-terminal domain can self-interact. In addition, we have demonstrated an interaction between the dsRBD and the N-terminal domain. The interaction between these two Rnt1 domains suggests that Rnt1 can also form an intramolecular complex. Because the self-interaction of the N-terminal domain is much stronger in vivo (Fig. 8) and in vitro (Fig. 6 and 7) than the N-terminal domain/dsRBD interaction (Fig. 8 and data not shown), the kinetically most stable assembly should be a dimer involving self-interactions between the two N-terminal domains (Fig. 9A). Based on the observations that fragments lacking the N-terminal domain or containing the dsRBD by itself can form a dimer (Fig. 6 to 8), it is likely that the functional Rnt1 complex also includes an interaction between the two dsRBDs. Thus, the Rnt1 homodimer appears to be formed in parallel through an interaction between the two N-terminal domains and another between the two C-terminal domains. The formation of an Rnt1 dimer in a parallel configuration raises new questions about the mechanism of dsRNA cleavage. To explain the staggered cut introduced by RNase III at each side of the RNA helix, it was suggested that the bacterial enzyme dimerizes in an antiparallel configuration (head to tail) (27). Based on the evidence presented here, we propose that Rnt1 introduces the asymmetrical cuts by an alternative mechanism that allow asymmetrical positioning of the RNA helix with respect to the nuclease domains.
FIG. 9.
The N-terminal domain is a modulator of yeast RNase III activity. Hypothetical models of Rnt1 and ΔN-term function. (A) Yeast RNase III. The intramolecular interaction of the dsRBD and N-term stabilizes the inactive protein, while the intermolecular interaction mediated by N-term and dsRBD stabilizes the protein complex on the RNA. (B) ΔN-term. The N-terminal domain deletion from Rnt1 destabilizes the protein and weakens the protein-RNA complex, reducing the cleavage efficiency.
Is the N-terminal domain a regulator of Rnt1 function?
In addition to its role in Rnt1 dimerization, the N-terminal domain appears to influence RNA binding and cleavage. Accordingly, the physical interaction that we detected between the N-terminal domain and dsRBD could be related to a regulatory function. One interesting possibility illustrated in Fig. 9 is that the N-terminus-mediated protein interactions modulate Rnt1 function. The N-terminal domain may interact intramolecularly with the dsRBD. This interaction would be disrupted upon binding of the RNA substrate to trigger conformational changes leading to intermolecular interaction between the two N-terminal domains and RNA cleavage. This model would explain why the deletion of the N-terminal domain promotes dsRNA binding without increasing the cleavage rate (Fig. 3 and 4). We therefore suggest that the N-terminal domain has dual functions, as depicted in Fig. 9. The first function is to interact with the dsRBD to form a compact protein structure that would be stable in the absence of the RNA, and the second function is to self-interact upon RNA binding to stabilize the ribonucleoprotein complex leading to efficient RNA cleavage. However, other in vivo functions of the N-terminal domain such as the regulation of Rnt1 interaction with other cellular proteins remains a possibility.
ACKNOWLEDGMENTS
We thank Guillaume Chanfreau for the ΔRNT1 yeast strain and for suggesting the in-gel cleavage experiment, James Hu for the λ repressor kit, Dennis Thiele and Simon Labbé for the copper expression vectors, and Philip James for the two-hybrid plasmids and strains. We also thank Allen Nicholson for communicating unpublished results. We are indebted for Benoit Chabot, April Colosimo, Skip Fournier, Christine Gagnon, Michael Katze, Allen Nicholson, and Raymond Wellinger for critical reading of the manuscript.
This work was supported by grant MT-14305 from the Medical Research Council of Canada to S.A. The fast protein liquid chromatography apparatus used for protein purification was purchased by a grant from Canada Foundation for Innovation. S.A. is a Chercheur-Boursier Junior I of the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Abou Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 3.Bycroft M, Grunert S, Murzin A G, Proctor M, St Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. . (Erratum, 14:4385.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanfreau G, Elela S A, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 6.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Court D. RNA processing and degradation by RNase III. New York, N.Y: Academic Press, Inc.; 1993. [Google Scholar]
- 8.Dasgupta S, Fernandez L, Kameyama L, Inada T, Nakamura Y, Pappas A, Court D L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III—the effect of dsRNA binding on gene expression. Mol Microbiol. 1998;28:629–640. doi: 10.1046/j.1365-2958.1998.00828.x. . (Erratum, 30:679, 1998.) [DOI] [PubMed] [Google Scholar]
- 9.Dunn J J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fidelity of cleavage. J Biol Chem. 1976;251:3807–3814. [PubMed] [Google Scholar]
- 10.Gitelman D R, Apirion D. The synthesis of some proteins is affected in RNA processing mutants of Escherichia coli. Biochem Biophys Res Commun. 1980;96:1063–1070. doi: 10.1016/0006-291x(80)90060-1. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 14.Iino Y, Sugimoto A, Yamamoto M. S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J. 1991;10:221–226. doi: 10.1002/j.1460-2075.1991.tb07939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, de Martynoff G, Lou J, Hipskind R A, Nordheim A, Stunnenberg H G. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharrat A, Macias M J, Gibson T J, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbé S, Thiele D J. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Nicholson A W. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 22.Li H L, Chelladurai B S, Zhang K, Nicholson A W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993;21:1919–1925. doi: 10.1093/nar/21.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.March P E, Gonzalez M A. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res. 1990;18:3293–3298. doi: 10.1093/nar/18.11.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M E, Berk A J. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72:3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 27.Nanduri S, Carpick B W, Yang Y, Williams B R, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson A W. Structure, reactivity, and biology of double-stranded RNA. Prog Nucleic Acid Res Mol Biol. 1996;52:1–65. doi: 10.1016/s0079-6603(08)60963-0. [DOI] [PubMed] [Google Scholar]
- 29.Patel R C, Sen G C. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol Cell Biol. 1998;18:7009–7019. doi: 10.1128/mcb.18.12.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prista C, Almagro A, Loureiro-Dias M C, Ramos J. Physiological basis for the high salt tolerance of Debaryomyces hansenii. Appl Environ Microbiol. 1997;63:4005–4009. doi: 10.1128/aem.63.10.4005-4009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu L H, Henras A, Lu Y J, Zhou H, Zhou W X, Zhu Y Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds A, Lundblad V. Yeast vectors and assays for expression of cloned genes. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1998. [DOI] [PubMed] [Google Scholar]
- 33.Robertson H D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982;30:669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. New York, N.Y: Cold Spring Harbor; 1990. [Google Scholar]
- 35.Rotondo G, Frendewey D. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996;24:2377–2386. doi: 10.1093/nar/24.12.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotondo G, Gillespie M, Frendewey D. Rescue of the fission yeast snRNA synthesis mutant snm1 by overexpression of the double-strand-specific Pac1 ribonuclease. Mol Gen Genet. 1995;247:698–708. doi: 10.1007/BF00290401. [DOI] [PubMed] [Google Scholar]
- 37.Rotondo G, Huang J Y, Frendewey D. Substrate structure requirements of the Pac1 ribonuclease from Schizosaccharmyces pombe. RNA. 1997;3:1182–1193. [PMC free article] [PubMed] [Google Scholar]
- 38.Ryter J M, Schultz S C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsh E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Seipelt R L, Zheng B, Asuru A, Rymond B C. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu H P, Riggs M, Rodgers L, Wigler M. A gene from S. pombe with homology to E. coli RNAse III blocks conjugation and sporulation when overexpressed in wild type cells. Nucleic Acids Res. 1990;18:5304. doi: 10.1093/nar/18.17.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng X, Zhu H, Lashuel H A, Hu J C. Oligomerization properties of GCN4 leucine zipper e and g position mutants. Protein Sci. 1997;6:2218–2226. doi: 10.1002/pro.5560061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D, Frendewey D, Lobo Ruppert S M. Pac1p, an RNase III homolog, is required for formation of the 3′ end of U2 snRNA in Schizosaccharomyces pombe. RNA. 1999;5:1083–1098. doi: 10.1017/s1355838299990726. [DOI] [PMC free article] [PubMed] [Google Scholar]