Supplemental Digital Content is available in the text.
Keywords: biomarkers, critical illness, genomics, innate immunity, proteomics, sepsis
OBJECTIVES:
Clinically deployable methods for the rapid and accurate prediction of sepsis severity that could elicit a meaningful change in clinical practice are currently lacking. We evaluated a whole-blood, multiplex host-messenger RNA expression metric, Inflammatix-Severity-2, for identifying septic, hospitalized patients’ likelihood of 30-day mortality, development of chronic critical illness, discharge disposition, and/or secondary infections.
DESIGN:
Retrospective, validation cohort analysis.
SETTING:
Single, academic health center ICU.
PATIENTS:
Three hundred thirty-five critically ill adult surgical patients with sepsis.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
Whole blood was collected in PAXgene Blood RNA collection tubes at 24 hours after sepsis diagnosis and analyzed using a custom 29-messenger RNA classifier (Inflammatix-Severity-2) in a Clinical Laboratory Improvement Amendments certified diagnostic laboratory using the NanoString FLEX platform. Among patients meeting Sepsis-3 criteria, the Inflammatix-Severity-2 severity score was significantly better (p < 0.05) at predicting secondary infections (area under the receiver operating curve 0.71) and adverse clinical outcomes (area under the receiver operating curve 0.75) than C-reactive protein, absolute lymphocyte counts, total WBC count, age, and Charlson comorbidity index (and better, albeit nonsignificantly, than interleukin-6 and Acute Physiology and Chronic Health Evaluation II). Using multivariate logistic regression analysis, only combining the Charlson comorbidity index (area under the receiver operating curve 0.80) or Acute Physiology and Chronic Health Evaluation II (area under the receiver operating curve 0.81) with Inflammatix-Severity-2 significantly improved prediction of adverse clinical outcomes, and combining with the Charlson comorbidity index for predicting 30-day mortality (area under the receiver operating curve 0.79).
CONCLUSIONS:
The Inflammatix-Severity-2 severity score was superior at predicting secondary infections and overall adverse clinical outcomes compared with other common metrics. Combining a rapidly measured transcriptomic metric with clinical or physiologic indices offers the potential to optimize risk-based resource utilization and patient management adjustments that may improve outcomes in surgical sepsis. Hospitalized patients who are septic and present with an elevated IMX-SEV2 severity score and preexisting comorbidities may be ideal candidates for clinical interventions aimed at reducing the risk of secondary infections and adverse clinical outcomes.
Sepsis afflicts over 1.7 million Americans annually and accounts for over 250,000 deaths in the United States alone (1). Sepsis remains the most common cause of death in the ICU (2, 3). Approximately 60–70% of all sepsis cases are diagnosed in the emergency department and 10–40% of total sepsis cases develop in currently hospitalized patients (2, 4, 5). Generally speaking, hospital-acquired sepsis and surgical sepsis, in particular, are associated with a higher frequency of septic shock and a two-fold increase in mortality compared with community-acquired sepsis (3, 4).
Identifying the severity of disease and likelihood of further deterioration of surgical sepsis patients with high accuracy and rapid turnaround remains a critical unmet need for hospitalized patients, since time to intervention and antimicrobial treatment is a critical determinant of outcome (6, 7). Current inhospital early warning systems for sepsis (modified early warning system) are used to alert healthcare providers to the possibility of sepsis (8). In many cases, the decision to initiate sepsis resuscitation bundles and more intensive management is an empiric decision made by the healthcare provider at the bedside, prior to documentary evidence of microbial infection, organ injury, or immunological dyscrasia. Existing clinical scores such as the Acute Physiology and Chronic Health Evaluation II (APACHE II) and Sequential Organ Failure Assessment (SOFA) score require 24 hours to properly calculate, making them impractical for immediate clinical decision-making. The quick SOFA assessment associated with the Sepsis-3 criteria has been recommended to identify patients at high risk of death, but its value is still controversial (9, 10). Other less common metrics such as procalcitonin, interleukin-6 (IL-6), and monocyte distribution width have all been promulgated as being predictive of sepsis severity (11, 12), although their predictive ability to diagnose either sepsis or its severity remains controversial (13). A rapid and clinically applicable diagnostic that can both diagnose infection and predict sepsis severity will be required to alter clinical management of the individual patient.
Here, we sought to validate the ability of a novel whole-blood host-immune transcriptomic metric for predicting outcome severity in a secondary use of a prospective cohort of critically ill surgical/trauma patients adjudicated as being septic. This metric of sepsis severity (IMX-SEV-2) is a component of a multibiomarker host-response approach to both diagnosing infections and predicting sepsis severity, the InSep test (Inflammatix, Burlingame, CA) (14). Intended as a point-of-care diagnostic with rapid turnaround times, InSep will provide the clinician with actionable information for upgrading management strategy and tailor ICU resources to individual patient needs.
MATERIALS AND METHODS
Study Design and Subject Enrollment
This is a secondary validation analysis in a previously reported cohort from a prospective, observational longitudinal study of hospitalized patients with surgical sepsis. We used the transcriptomic metric, IMX-SEV-2, to predict 30-day mortality when applied to samples obtained from a single-center prospective 1-year longitudinal cohort study (NCT02276417) of critically ill surgical patients on day 1 following diagnosis of sepsis. The parent cohort included 363 septic patients hospitalized in a 48-bed surgical ICU at a quaternary-care academic hospital between January 2015 and January 2020. Overall cohort inclusion criteria included: 1) age greater than or equal to 18 years, 2) clinical diagnosis of sepsis as defined by 2001 consensus guidelines, and 3) entrance into the electronic medical record (EMR)-based sepsis clinical management protocol. Exclusion criteria included any of the following: 1) refractory shock (death < 24 hr from sepsis protocol initiation) or inability to achieve source control (e.g., total bowel ischemic necrosis), 2) preadmission-expected lifespan less than 3 months, 3) patient/proxy not committed to aggressive management, 4) severe congestive heart failure (New York Heart Association Class IV), 5) Child-Pugh Class C liver disease or preliver transplant, 6) known HIV with CD4+ count < 200 cells/mm3, 7) patients receiving chronic corticosteroids or immunosuppressive agents, including organ transplant recipients, 8) pregnancy, 9) institutionalized patients, 10) inability to obtain informed consent within 96 hours of enrollment, 11) chemotherapy or radiotherapy within 30 days, 12) severe traumatic brain injury, and 13) spinal cord injury resulting in permanent sensory and/or motor deficits. Informed consent was obtained from each subject or their proxy. Detailed study cohort design and protocols used by the parent cohort have been published previously (15, 16). Detailed descriptions of the inclusion and exclusion criteria are contained in the Supplemental Methods and Results (http://links.lww.com/CCX/A813). Ethics approval was obtained from the University of Florida Institutional Review Board (201400611).
All enrolled subjects underwent prospective post hoc adjudication within a week of cohort enrollment by physician-investigators at to confirm sepsis diagnosis, severity, and source (17). Adjudicated nonseptic patients were excluded from this analysis. As the parent cohort was designed prior to the publication of Sepsis-3 consensus guidelines, patients were enrolled into the sampling cohort using 2001 sepsis consensus criteria definitions (18). Subsequently, patients were retrospectively readjudicated for sepsis and septic shock using the Sepsis-3 guidelines (19).
Hospital-acquired secondary infections were adjudicated by physician-investigators during primary data/chart review utilizing current United States Centers for Disease Control definitions and guidelines (20). Discharge disposition was prospectively classified based on known associations with long-term outcomes as either “good” (home with or without healthcare services or rehabilitation facility) or “poor” (long-term acute-care facility), skilled nursing facility, another acute care hospital, hospice, or inpatient death).
Individual clinical outcome variables included: 1) 30-day (all-cause) mortality, 2) development or absence of chronic critical illness (CCI), 3) discharge disposition, and 4) secondary infections. Inpatient clinical trajectory was defined as “early death,” “rapid recovery” (RAP), or “CCI.” Early death was defined as death within 14 days of sepsis onset. CCI was defined as an ICU length of stay (LOS) greater than or equal to 14 days with evidence of persistent organ dysfunction based on components of the SOFA score (21). Hospitalized patients who died after an ICU LOS greater than 14 days from the index hospitalization were classified as CCI (22). RAP patients were those discharged from the ICU within 14 days with resolution of organ dysfunction. Patients were defined as having an “adverse clinical outcome” if they experienced a secondary infection, CCI, poor discharge disposition, and/or mortality within the first 30 days.
Sample Collection and Analyses
Blood samples were collected in PAXgene tubes at 24 hours following initiation of EMR-based sepsis management protocols and were stored at –80°C for subsequent bulk analysis. RNA was extracted with the RNeasy Plus Micro Kit (QIAGEN, Germantown, MD). The IMX-SEV-2 classifier (Supplemental Table 1, http://links.lww.com/CCX/A813) was quantitated from 200 ng of RNA input using the 510(k)-cleared NanoString nCounter FLEX profiler (NanoString, Seattle, WA) according to a validated standard operating protocol in a Clinical Laboratory Improvement Amendments certified diagnostic laboratory.
Total leukocyte counts, absolute lymphocyte counts (ALCs), and C-reactive protein concentrations were determined at the University of Florida Health Clinical and Diagnostic Laboratories. Plasma IL-6 and glucagon-like peptide-1 (GLP-1) levels were determined using the Luminex MagPix platform (Austin, TX).
IMX Classifiers
We previously described the development of a 29 host-messenger RNA (mRNA) test, InSep (Inflammatix, Burlingame, CA) that uses machine learning algorithms IMX-SEV (severity metric trained on 30-d mortality), and IMX bacterial-viral-noninfected (BVN, for infection diagnosis) to produce three scores for the likelihood of bacterial infection, the likelihood of viral infection, and disease severity (14, 23). Here, we use the second-generation versions of the neural network–based classifiers, including IMX-BVN-2 and IMX-SEV-2. Each score is reported both as a continuous variable and stratified into preset “risk bands” to meet clinically actionable performance thresholds (14). For IMX-SEV-2, the score is broken into three categories: “likely,” “possible,” and “unlikely” of 30-day mortality, based on prior published work (see Supplemental Fig. 1, http://links.lww.com/CCX/A813) (14, 23).
Statistical Analyses
Descriptive data are presented as frequency and percentage, mean and sd, or median and 25th/75th percentiles. Fisher exact test and the Kruskal-Wallis test were used for comparison of categorical and continuous variables, respectively. Correlations among continuous and discrete variables were determined using Spearman test. Area under the receiver operating curve (AUROC) values and Hosmer-Lemeshow goodness-of-fit test were used to assess model discrimination and fit. The DeLong test was used to compare differences among receiver operating curves. False discovery correction for multiple comparisons was performed using the Benjamini-Hochberg procedure. The continuous Net Reclassification Index (cNRI) (24) was employed to measure the improvement in prediction performance gained by adding a clinical metric to the IMX-SEV-2 score. The cNRI was calculated using R (Version 4.0.5, https://www.r-project.org/). All remaining analyses were performed using SAS analytical software (Version 9.4, SAS Institute, Cary, NC). All significance tests were two-sided, with a p value less than or equal to 0.05 considered statistically significant.
RESULTS
The parent study was enrolled a total of 363 sepsis patients that met 2001 consensus sepsis criteria and provided informed consent. Of these 363 patients, remaining RNA samples were available for this analysis in 333 subjects at 24 hours (± 6 hr), and two samples were obtained from additional subjects at less than 12 hours from the initiation of the sepsis management protocols. Of the 335 patients in this “overall” cohort, 316 subsequently met the criteria for sepsis or septic shock per Sepsis-3 criteria (19) (Supplemental Table 2, http://links.lww.com/CCX/A813).
Both cohorts represented an older population (median, 62 yr) with a significant burden of preexisting comorbidities (median Charlson comorbidity index, 3). Approximately 25% of Sepsis-3 patients met shock criteria; nearly 60% presented with or developed acute kidney injury, and nearly half progressed to multiple organ failure. More than half of the patients had either intra-abdominal or surgical site infection, whereas approximately 28% had either pneumonia or urinary tract infections. Secondary infections were also common in this Sepsis-3 cohort, with an overall rate of 2.1 infections per 100 person hospital days (Supplemental Table 3, http://links.lww.com/CCX/A813). Thirty patients (9.5%) died, and 194 patients (61.4%) had what was defined as an adverse clinical outcome; there was heavy overlap among the various measures of poor outcome (Supplemental Fig. 2, http://links.lww.com/CCX/A813).
Predicting Clinical Outcomes
The prognostic ability of the IMX-SEV-2 severity metric was analyzed both as a continuous variable and as likelihood distributions based on predetermined thresholds (Supplemental Fig. 1, http://links.lww.com/CCX/A813). For patients meeting Sepsis-3 criteria, the IMX-SEV-2 metric not only predicted 30-day mortality but also predicted development of CCI, discharge disposition, and frequency of secondary infections (Table 1). Table 2 presents the actual test characteristics of the IMX-SEV-2 metric when presented as likelihood distributions. Sensitivity and specificity were calculated by combining the “possible” group with the “likely” and “unlikely” groups, respectively.
TABLE 1.
Area Under the Receiver-Operator Curve for Outcome Variables
| Predictor | 30-d Mortality | Chronic Critical Illness Status | Discharge Disposition | Secondary Infection | Adverse Clinical Outcomea |
|---|---|---|---|---|---|
| Transcriptomic (Inflammatix-Severity-2) | |||||
| Severity metricb | 0.66 (0.54–0.78) | 0.68 (0.62–0.74) | 0.67 (0.61–0.73) | 0.71 (0.65–0.76) | 0.75 (0.69–0.80) |
| Severity bandsc | 0.68 (0.58–0.78) | 0.66 (0.60–0.71) | 0.64 (0.59–0.69) | 0.66 (0.61–0.71)d | 0.70 (0.65–0.74)d |
| Interleukin-6 | 0.68 (0.59–0.78) | 0.62 (0.56–0.68) | 0.59 (0.53–0.66)d | 0.68 (0.62–0.74) | 0.68 (0.62–0.75) |
| C-reactive protein | 0.60 (0.48–0.73) | 0.50 (0.42–0.58)d | 0.55 (0.47–0.62) | 0.54 (0.47–0.62)d | 0.51 (0.44–0.58)d |
| Absolute lymphocyte count | 0.54 (0.42–0.67)d | 0.58 (0.51–0.66)d | 0.62 (0.55–0.69) | 0.61 (0.54–0.68)d | 0.63 (0.56–0.70)d |
| Total WBC | 0.61 (0.50–0.73) | 0.58 (0.52–0.65)d | 0.55 (0.49–0.62)d | 0.53 (0.47–0.60)d | 0.58 (0.51–0.64)d |
| Glucagon-like peptide-1 | 0.62 (0.49–0.75) | 0.65 (0.58–0.72) | 0.64 (0.58–0.70) | 0.64 (0.57–0.70)d | 0.66 (0.60–0.72)d |
| Age | 0.71 (0.61–0.81) | 0.61 (0.54–0.67) | 0.70 (0.65–0.76) | 0.57 (0.50–0.63)d | 0.64 (0.58–0.71)d |
| Charlson comorbidity index | 0.73 (0.65–0.82) | 0.64 (0.57–0.70) | 0.69 (0.63–0.75) | 0.57 (0.50–0.64)d | 0.66 (0.60–0.72)d |
| Acute Physiology and Chronic Health Evaluation 2 score assessed at 24 hr after sepsis onset | 0.75 (0.66–0.84) | 0.75 (0.69–0.81) | 0.72 (0.66–0.78) | 0.63 (0.57–0.70) | 0.75 (0.69–0.80) |
aAdverse clinical outcome is the presence of either a secondary infection, chronic critical illness (CCI), a poor discharge diagnosis, and/or mortality within 30 d of sepsis adjudication (n = 197).
bArea under the receiver operating curve calculated based on transcriptomic metric predictor as a continuous variable.
cSee predetermined thresholds in Supplemental Figure 1 (http://links.lww.com/CCX/A813).
dBold values indicate statistical significance at p < 0.05 as different from severity metric.
Values were obtained in patients meeting Sepsis-3 criteria for 30-d mortality, development of CCI, discharge disposition (“good” vs “poor”), presence of secondary infection, or an adverse clinical outcome.
TABLE 2.
Point Estimates of Specificity and Sensitivity for the Primary and Secondary Outcomes
| Severity Band | Dead at 30 d | Alive at 30 d | Sensitivity | Specificity | Likelihood Ratio |
|---|---|---|---|---|---|
| 30-d mortality | |||||
| Likely | 17 | 57 | 0.567 | 0.792 | 2.72 |
| Possible | 11 | 186 | |||
| Unlikely | 2 | 31 | 0.933 | 0.133 | 0.59 |
| Development of chronic critical illness | |||||
| Likely | N/A | N/A | 0.392 | 0.871 | 3.04 |
| Possible | N/A | N/A | |||
| Unlikely | N/A | N/A | 0.969 | 0.156 | 0.20 |
| Discharge status | |||||
| Likely | N/A | N/A | 0.355 | 0.857 | 2.48 |
| Possible | N/A | N/A | |||
| Unlikely | N/A | N/A | 0.965 | 0.160 | 0.22 |
| Secondary infection | |||||
| Likely | N/A | N/A | 0.382 | 0.840 | 2.38 |
| Possible | N/A | N/A | |||
| Unlikely | N/A | N/A | 1 | 0.160 | 0 |
| Adverse clinical outcomes | |||||
| Likely | N/A | N/A | 0.345 | 0.934 | 5.27 |
| Possible | N/A | N/A | |||
| Unlikely | N/A | N/A | 0.969 | 0.221 | 0.14 |
N/A = not applicable.
Values represent the number of subjects in each likelihood distribution category.
Sensitivity and specificity were calculated by collapsing the likely and possible, and possible and unlikely categories, respectively.
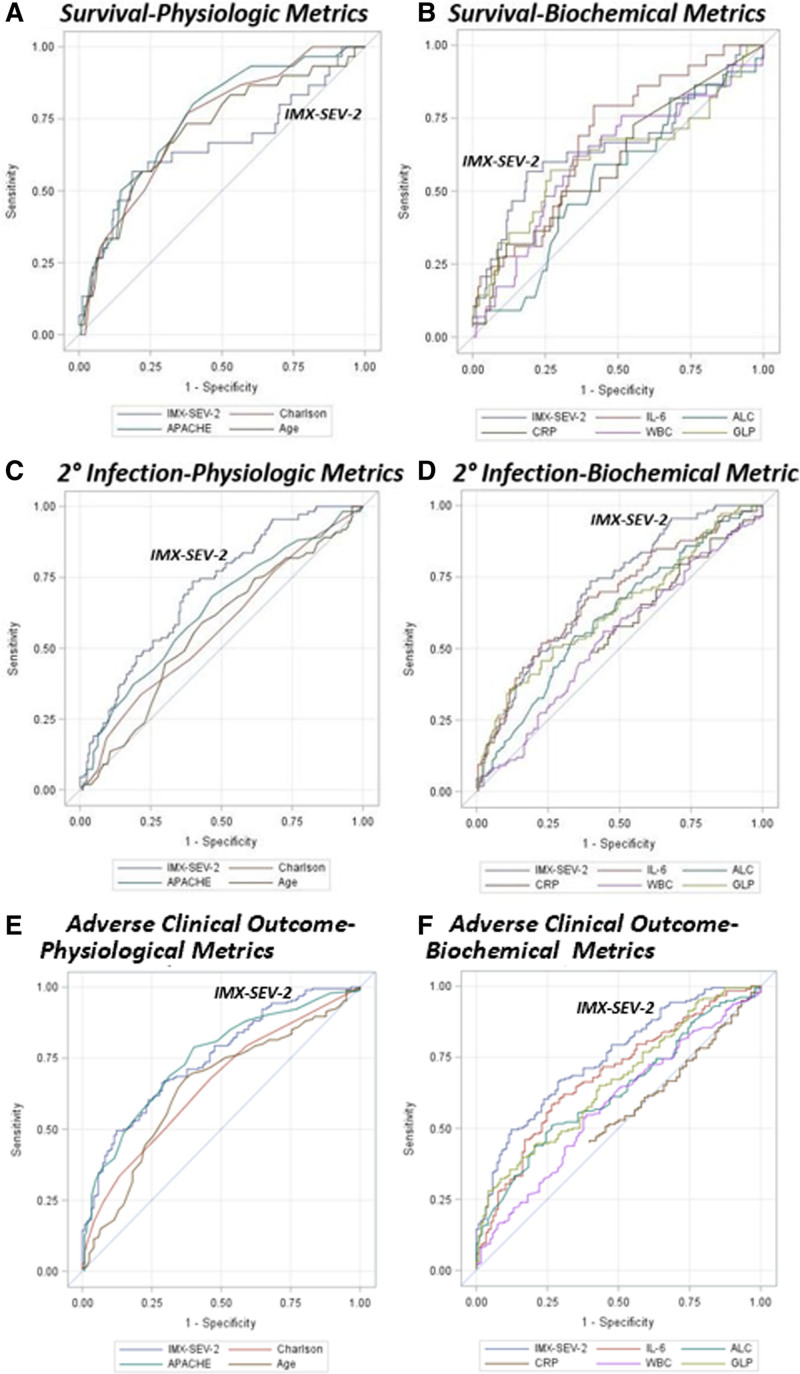
As shown in Table 1 and Figure 1, the continuous IMX-SEV-2 severity metric significantly better predicted the incidence of secondary infections than plasma C-reactive protein, plasma GLP-1, ALC, total WBC count, age, and Charlson comorbidity index (all p < 0.05). Additionally, the IMX-SEV-2 metric showed equivalency to predicting 30-day mortality and discharge disposition to other physiologic markers, and significantly improved (p < 0.05) predictive ability when compared with ALC for 30-day mortality and IL-6 for discharge disposition (Table 1). Finally, the IMX-SEV-2 severity metric better predicted (p < 0.05) the composite adverse clinical outcome than all of the other metrics other than IL-6 and APACHE II score.
Figure 1.
Area under the receiver operating curve performance curves for 30-d mortality (A and B) and incidence of secondary infection (C and D) using the continuous Inflammatix-Severity-2 (IMX-SEV-2) metric compared with other physiologic (A and C) and biochemical markers (B and D). ALC = absolute lymphocyte count, APACHE = Acute Physiology and Chronic Health Evaluation, CRP = C-reactive protein, GLP = glucagon-like peptide, IL-6 = interleukin-6.
Correlation Between Severity Index and Other Metrics
Spearman correlation coefficients were determined among the IMX-SEV-2 severity index and other biochemical and physiologic indices. The severity index was most closely correlated (in descending order) with plasma GLP-1 (ρ = 0.44), IL-6 (ρ = 0.43), and APACHE II (ρ = 0.34, all p < 0.001). In contrast, the IMX-SEV-2 metric was not correlated with either age or Charlson comorbidity index. Weak, but significant, associations were seen with total WBC (ρ = 0.31), ALC (ρ = –0.30; both p < 0.01) (Supplemental Table 4, http://links.lww.com/CCX/A813).
IMX-SEV-2 in Combination With Clinical Indices
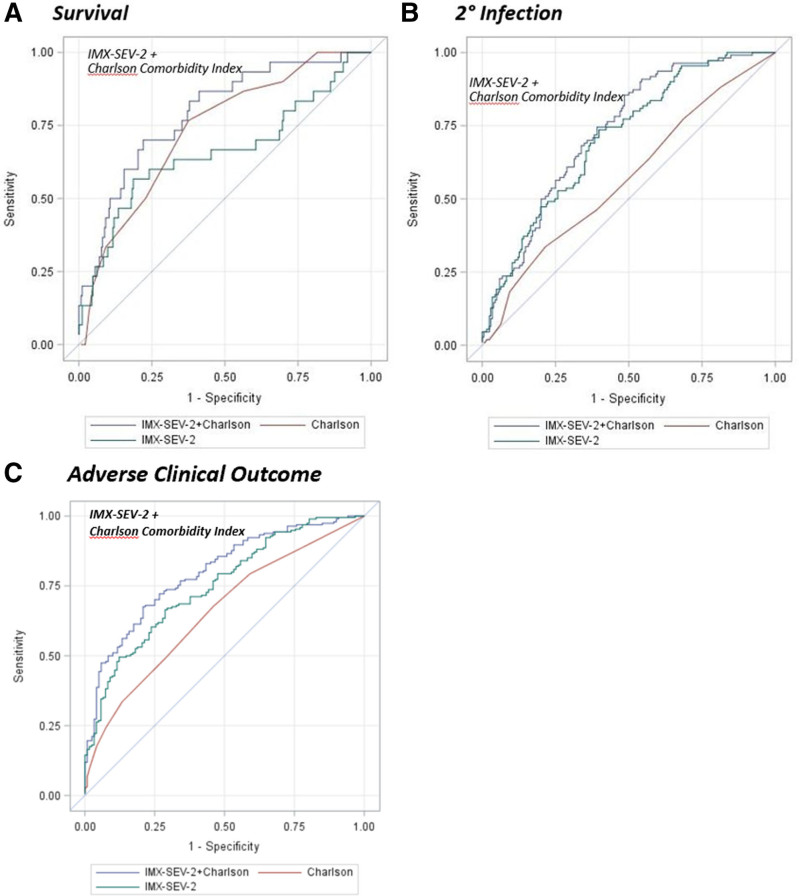
Because of the strong correlations between the IMX-SEV-2 severity metric with IL-6, GLP-1, and APACHE II, but not with the Charlson comorbidity score or age, an unbiased, multivariate logistic regression analysis was conducted with all of the biochemical and clinical metrics. Only the IMX-SEV-2 metric and the Charlson comorbidity index were selected as significant and independent predictors of 30-day mortality, with an enhanced composite AUROC of 0.79 (Table 3 and Fig. 2). Similarly, combining the IMX-SEV-2 severity metric with the APACHE II score or Charlson comorbidity index for predicting adverse clinical outcomes increased the AUROC to 0.81 (Table 3) and 0.80, respectively.
TABLE 3.
Combining Inflammatix-Severity-2 Metric With Clinical and Biochemical Indices to Predict 30-d Mortality and Adverse Clinical Outcomes
| Multivariate Model | OR (95% CI) | p |
|---|---|---|
| 30-d mortalitya | ||
| IMX-SEV-2 Severity metric (increase of 0.1) | 1.92 (1.40–2.64) | < 0.0001 |
| Charlson comorbidity index | 1.36 (1.17–1.57) | < 0.0001 |
| Adverse clinical outcomeb | ||
| IMX-SEV-2 Severity metric (increase of 0.1) | 4.42 (2.14–9.17) | < 0.0001 |
| Acute Physiology and Chronic Health Evaluation II | 1.11 (1.05–1.18) | 0.0003 |
IMX-SEV-2 = Inflammatix-Severity-2, OR= odds ratio.
aArea under the receiver operating curve (AUROC) (95% CI) = 0.79 (0.71–0.87).
bAUROC (95% CI) = 0.80 (0.74–0.87).
30-d Mortality: All biochemical and physiologic variables from patients meeting Sepsis-3 criteria were included in the multivariate logistic regression analysis. Only the InSep severity metric combined with the Charlson comorbidity index had independent predictive ability.
Adverse Clinical Outcome: The integrated outcomes included secondary infection, chronic critical illness, “poor” discharge disposition, and/or 30-d mortality. All biochemical and physiologic variables were included in the multivariate logistic regression analysis. The InSep severity metric combined with Acute Physiology and Chronic Health Evaluation II had independent predictive ability.
Figure 2.
Area under the receiver operating curve performance curves for 30-d mortality and incidence of secondary infection using the continuous Inflammatix-Severity-2 (IMX-SEV-2) metric combined with the Charlson comorbidity index compared with each alone.
A more clinically relevant measure of the usefulness of combining a clinical index with the IMX-SEV-2 metric employs the cNRI (24) although its use remains controversial (25). Using the cNRI to calculate the benefit of adding the IMX-SEV-2 metric to the Charlson comorbidity index improved the reclassification score (0.79; 95% bootstrap CI, 0.20–0.99; p < 0.05), suggesting a 79% improvement in the reclassification rate for 30-day mortality when the two metrics were combined. In addition, even though IMX-SEV-2 metric was strongly correlated with the APACHE II score, adding the predictive ability of the IMX-SEV-2 metric to the APACHE II score for 30-day mortality generated a cNRI metric (0.32; CI, 0.13–0.45; p < 0.05), indicative of a 32% reclassification rate.
Similar improvements in the reclassification score were seen for predicting adverse outcomes when adding the IMX-SEV-2 metric to the Charlson comorbidity index and APACHE II, respectively (cNRI, 0.71 and 0.69, both p < 0.05).
Infection Diagnosis With IMX-BVN-2
As the cohort contained only infected patients with clinically adjudicated bacterial sepsis who had received antibiotics in the past 24 hours, the likelihood ratios for the bacterial and viral infection metrics (IMX-BVN-2) were accurate but are less relevant and, therefore, have been relegated to Supplemental Tables 5 and 6 (http://links.lww.com/CCX/A813).
DISCUSSION
Surgical sepsis remains one of the most common and expensive hospital complications and has been a major target for healthcare quality improvements. Institution of early warning systems that can identify patients at risk of decompensation is now routinely used in academic health centers. The clinical challenge in sepsis with these early warning systems remains the timely diagnosis of microbial infection and the recognition of severity of the consequential inflammatory insult since both affect initial management and appropriate selection of intensity of monitoring and care.
The InSep acute infection and sepsis test interpret 29 host-immune mRNAs together with integrating classifiers to determine the likelihood of a bacterial or viral infection and the severity of the condition (14). Importantly, the IMX-SEV-2 severity neural-network classifier was trained on multiple transcriptomic databases from patients with both community- as well as hospital-acquired sepsis (23). Prospective validation of the IMX-SEV-2 metric for assessing the likelihood of 30-day mortality was recently obtained in a community-acquired sepsis cohort with AUROC scores exceeding 0.80 (26, 27).
When utilizing the predetermined IMX-SEV-2 severity band categories (Supplemental Fig. 1, http://links.lww.com/CCX/A813), the AUROC for predicting 30-day mortality in surgical sepsis was similar to other laboratory metrics and clinical indices. However, when considered as a continuous variable, the metric was significantly better at predicting both secondary infections (AUROC 0.71) and an overall adverse clinical outcome (AUROC 0.75) versus all the other metrics with the exception of IL-6 and APACHE II scores (Table 1). It is important to note that APACHE II is both a measure of physiologic derangement that is collinear to IMX-SEV-2 and is a composite score including age. Additionally, it is retrospectively calculated utilizing the most aberrant parameters measured within the first 24 hours of ICU admission. Thus, APACHE II is not a practically usable point-of-care assessment within the first hours after suspicion of sepsis diagnosis. The observation that the IMX-SEV-2 metric was at least equivalent and more often better than all other metrics at predicting a variety of clinical outcomes in this complicated hospitalized surgical sepsis population is remarkable.
It was not surprising that the IMX-SEV-2 severity metric was correlated with other measures of the inflammatory response (IL-6) or physiologic derangement (APACHE II) (Supplemental Table 4, http://links.lww.com/CCX/A813), since the IMX-SEV-2 severity metric incorporates transcriptomic changes that quantitate the magnitude of the host inflammatory response. Interestingly, the metric showed no significant correlation with the Charlson comorbidity score despite the fact that the 30-day mortality predictive properties were similar (Table 1). This suggests that the two metrics are independently predicting survival. This led us to examine whether combining the IMX-SEV-2 metric with clinical indices could improve the overall predictive power. Importantly, when the Charlson index was combined with the IMX-SEV-2 metric, the AUROC improved significantly to 0.79 and 0.81 for 30-day mortality and adverse clinical outcomes, respectively. The cNRI suggested that the improvement in 30-day mortality and adverse clinical outcome predictive ability were achieved by an increase in identifying true positives (sensitivity).
An important question is whether (and how) rapid and improved prognostication might make a difference in clinical practice. One potential argument is that IL-6 is available within hours in an increasing number clinical laboratories, whereas the SEV2 metric requires additional equipment and is currently for research use only. However, IMX-SEV-2 and IMX-BVN-2 are being developed for use at the point of care with a 30-minute turnaround measurement. The device is turnkey: a point-of-care blood sample tube is inserted directly into the device, and the results will be easily interpretable and actionable to the clinician with odds ratios and risk classifier bands presented within 30 minutes (14). The point-of-care availability of the technology allows more flexibility in deployment and implementation in austere settings, as it is not reliant on other existing and cumbersome equipment. As alluded to above, although metrics like AUROC and cNRI are not intuitive or useful to a bedside clinician, IMX-SEV-2 has preset interpretation bands tested here with high accuracy (low-severity group 97% sensitive and high-severity group 93% specific for combined poor outcomes). Critical care medicine is an art and a science, and one of the most common questions is whether to escalate organ support in a patient not clearly recovering. We posit that a patient with a “low-severity” result could be considered for a “stay-the-course” approach, whereas a “high-severity” result may benefit from earlier and more aggressive monitoring or organ support interventions. Additionally, the demonstrated high sensitivity of IMX-SEV-2 to rule out 30-day mortality or adverse clinical outcomes could make it valuable triage tool in critical care resource–limited environments or pandemic/disaster scenarios. Of course, any of these clinical hypotheses requires interventional testing before implementation.
Beyond standard clinical applications, improved prognostics have the potential to augment discussions with patients and caregivers seeking to understand the probability of functional recovery that aligns with the patient’s goals and values versus the probability of persistent illness and death. Clinicians’ responses to such queries often lack objectivity; a tool like IMX-SEV-2 could provide objective, data-driven, patient-specific probabilities of clinical outcomes that could be used to inform prognostic discussions with patients and caregivers. The impact of IMX-SEV-2 classifications on prognosis-based shared decision-making processes and subsequent resource use requires investigation.
Several caveats need to be considered. This is a single-center study that included only surgical patients with sepsis. Thus, these findings may not be applicable to other critically ill populations or settings. Additionally, a significant proportion of the patients (n = 123/316, 39%) were hospitalized for another medical condition, and sepsis was hospital-acquired and/or secondary to some injurious process. How that preexisting inflammatory process influenced the subsequent host response to the sepsis is unknown but is clearly a complicating factor. With that said, however, the findings emphasize the generalizability of the test and the power of using large datasets to generate transcriptomic signatures (28, 29).
Additionally, the metric was studied here in hospitalized patients already adjudicated to having sepsis. Blood samples were collected not at the time of sepsis suspicion but between 12 and 24 hours after the presumption of sepsis and onset of treatment bundles, including initiation of broad-spectrum antibiotics. The primary use of IMX-SEV-2 in hospitalized patients will be in individuals “suspected” of having sepsis based on their early warning system scores, prior to the initiation of sepsis-treatment bundles. A prospective validation of the metric in hospitalized patients suspected of sepsis is currently underway. Finally, given the varying incidence of composite outcome components, associations among predictions, classifications, and the composite outcome are subject to misinterpretation. We sought to minimize this potential source of bias by also reporting the individual components of the composite outcome with full transparency.
CONCLUSIONS
In this validation cohort of clinically adjudicated surgical sepsis patients who had received initial sepsis management, a novel host-response metric (IMX-SEV-2) measured at 24 hours accurately predicted secondary infections and adverse clinical outcomes. Furthermore, combining the IMX-SEV-2 severity metric at 24 hours after initiation of sepsis treatment with an assessment of comorbidities could predict adverse clinical outcomes with the greatest accuracy. Such information could alter management and impact survival in hospitalized patients with surgical sepsis. Prognostic enrichment would also be critical to potential subject selection for future precision medicine trials in sepsis and for shared decision-making among patients, caregivers, and clinicians to align resource use with patients’ goals and values.
ACKNOWLEDGMENTS
We thank for the efforts of the Sepsis and Critical Illness Research Center research nursing staff, Jennifer Lanz, Ruth Davis, Ashley McCray, and Jillianne Brakenridge, and research laboratory staff, Ricardo Ungaro, Marvin Dirain, Dina Nacionales, and Jaimar Rincon for the conduct of the parent clinical study. The authors also recognize the efforts of Mei Zhang from the Molecular Pathology Laboratory at Rocky Point for the Nanostring analyses for the Inflammatix-Severity-2 measurements.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Drs. Brakenridge, Liesenfeld, Sweeney, PL, and Moldawer conceived and designed the study. Drs. Brakenridge, Efron, Liesenfeld, Sweeney, and Moldawer wrote and edited the article. Dr. Starostik developed and validated the analytical methodologies for Inflammatix-Severity-2 (IMX-SEV-2), under CLIA-CAP accreditation. Drs. Ghita, Midic, and Wacker developed the statistical analysis plan and optimized the IMX-SEV-2 likelihood bands. Drs. Darden and Fenner edited the article, and composed the figures and tables.
Supported, in part, by U.S. Federal funds from the Department of Health and Human Services; Office of the Assistant Secretary for Preparedness and Response; Biomedical Advanced Research and Development Authority, Division of Research, Innovation, and Ventures program, under Contract 75A50119C00044. The original observational clinical study was funded under P50 GM-111152, awarded by the National Institute of General Medical Sciences.
Drs. Darden and Fenner were funded by a postgraduate training grant in burns, trauma, sepsis, and perioperative injury from the National Institute of General Medical Sciences (T32 GM-008721). Drs. Midic, Wacker, Liesenfeld, and Sweeney are employees of Inflammatix (Burlingame, CA). Drs. Brakenridge, Starostik, and Moldawer received research support for aspects of this study under Biomedical Advanced Research and Development Authority subcontract. Dr. Ghita has disclosed that she does not have any potential conflicts of interest.
The entire clinical data set used in the preparation of this report is freely accessible through the website https://www.ctsi.ufl.edu/research/laboratory-services/ctsi-biorepository-2/scirc-specimens-archive/ under management of the University of Florida Clinical and Translational Science Institute Biorepository.
REFERENCES
- 1.Gaieski DF, Edwards JM, Kallan MJ, et al. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med. 2013; 41:1167–1174 [DOI] [PubMed] [Google Scholar]
- 2.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014; 5:4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchman TG, Simpson SQ, Sciarretta KL, et al. Sepsis among Medicare beneficiaries: 1. The burdens of sepsis, 2012-2018. Crit Care Med. 2020; 48:276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee C, Wang R, Zhang Z, et al. ; CDC Prevention Epicenters Program. Epidemiology of hospital-onset versus community-onset sepsis in U.S. hospitals and association with mortality: A retrospective analysis using electronic clinical data. Crit Care Med. 2019; 47:1169–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markwart R, Saito H, Harder T, et al. Epidemiology and burden of sepsis acquired in hospitals and intensive care units: A systematic review and meta-analysis. Intensive Care Med. 2020; 46:1536–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu VX, Fielding-Singh V, Greene JD, et al. The timing of early antibiotics and hospital mortality in sepsis. Am J Respir Crit Care Med. 2017; 196:856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Wang J, Liu G, et al. Prompt admission to intensive care is associated with improved survival in patients with severe sepsis and/or septic shock. J Int Med Res. 2018; 46:4071–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton DJ, Graves KK, Kukhareva PV, et al. Modified early warning score-based clinical decision support: Cost impact and clinical outcomes in sepsis. JAMIA Open. 2020; 3:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruangsomboon O, Boonmee P, Limsuwat C, et al. The utility of the rapid emergency medicine score (REMS) compared with SIRS, qSOFA and NEWS for predicting in-hospital mortality among patients with suspicion of sepsis in an emergency department. BMC Emerg Med. 2021; 21:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellhammar L, Linder A, Tverring J, et al. NEWS2 is superior to qSOFA in detecting sepsis with organ dysfunction in the emergency department. J Clin Med. 2019; 8:E1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polilli E, Sozio F, Frattari A, et al. Comparison of monocyte distribution width (MDW) and procalcitonin for early recognition of sepsis. PLoS One. 2020; 15:e0227300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brakenridge SC, Moore FA, Mercier NR, et al. Persistently elevated glucagon-like peptide-1 levels among critically ill surgical patients after sepsis and development of chronic critical illness and dismal long-term outcomes. J Am Coll Surg. 2019; 229:58–67. e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunsolus IL, Sweeney TE, Liesenfeld O, et al. Diagnosing and managing sepsis by probing the host response to infection: Advances, opportunities, and challenges. J Clin Microbiol. 2019; 57:e00425–e00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducharme J, Self WH, Osborn TM, et al. A multi-mRNA host-response molecular blood test for the diagnosis and prognosis of acute infections and sepsis: Proceedings from a clinical advisory panel. J Pers Med. 2020; 10:E266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loftus TJ, Mira JC, Ozrazgat-Baslanti T, et al. Sepsis and critical illness research center investigators: Protocols and standard operating procedures for a prospective cohort study of sepsis in critically ill surgical patients. BMJ Open. 2017; 7:e015136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft CA, Moore FA, Efron PA, et al. Computer versus paper system for recognition and management of sepsis in surgical intensive care. J Trauma Acute Care Surg. 2014; 76:311–317 [DOI] [PubMed] [Google Scholar]
- 17.Stortz JA, Mira JC, Raymond SL, et al. Benchmarking clinical outcomes and the immunocatabolic phenotype of chronic critical illness after sepsis in surgical intensive care unit patients. J Trauma Acute Care Surg. 2018; 84:342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy MM, Fink MP, Marshall JC, et al. ; SCCM/ESICM/ACCP/ATS/SIS. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003; 31:1250–1256 [DOI] [PubMed] [Google Scholar]
- 19.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stortz JA, Cox MC, Hawkins RB, et al. Phenotypic heterogeneity by site of infection in surgical sepsis: A prospective longitudinal study. Crit Care. 2020; 24:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: For the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016; 315:762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brakenridge SC, Efron PA, Cox MC, et al. Current epidemiology of surgical sepsis: Discordance between inpatient mortality and 1-year outcomes. Ann Surg. 2019; 270:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayhew MB, Buturovic L, Luethy R, et al. A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections. Nat Commun. 2020; 11:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leening MJ, Vedder MM, Witteman JC, et al. Net reclassification improvement: Computation, interpretation, and controversies: A literature review and clinician’s guide. Ann Intern Med. 2014; 160:122–131 [DOI] [PubMed] [Google Scholar]
- 25.Kerr KF, Wang Z, Janes H, et al. Net reclassification indices for evaluating risk prediction instruments: A critical review. Epidemiology. 2014; 25:114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Safarika A, Wacker JW, Katsaros K, et al. : A 29-mRNA host response test from blood accurately distinguishes bacterial and viral infections among emergency department patients. Intensive Care Med Exp. 2021; 9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer W, Kappert K, Galtung N, et al. A novel 29-messenger RNA host-response assay from whole blood accurately identifies bacterial and viral infections in patients presenting to the emergency department with suspected infections: A prospective observational study. Crit Care Med. 2021; 49:1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney TE, Perumal TM, Henao R, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. 2018; 9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweeney TE, Azad TD, Donato M, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018; 46:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.