Abstract
Light has been proposed to stimulate the translation of Chlamydomonas reinhardtii chloroplast psbA mRNA by activating a protein complex associated with the 5′ untranslated region of this mRNA. The protein complex contains a redox-active regulatory site responsive to thioredoxin. We identified RB60, a protein disulfide isomerase-like member of the protein complex, as carrying the redox-active regulatory site composed of vicinal dithiol. We assayed in parallel the redox state of RB60 and translation of psbA mRNA in intact chloroplasts. Light activated the specific oxidation of RB60, on the one hand, and reduced RB60, probably via the ferredoxin-thioredoxin system, on the other. Higher light intensities increased the pool of reduced RB60 and the rate of psbA mRNA translation, suggesting that a counterbalanced action of reducing and oxidizing activities modulates the translation of psbA mRNA in parallel with fluctuating light intensities. In the dark, chemical reduction of the vicinal dithiol site did not activate translation. These results suggest a mechanism by which light primes redox-regulated translation by an unknown mechanism and then the rate of translation is determined by the reduction-oxidation of a sensor protein located in a complex bound to the 5′ untranslated region of the chloroplast mRNA.
During photosynthesis, the light reactions produce deleterious by-products that lead to photoinactivation of the photosystem II reaction center and concurrent protein turnover (2, 37, 54). Thus, maintenance of photosynthetic capacity requires de novo synthesis and replacement of photodamaged proteins. The protein showing the highest rate of synthesis at high light intensity in higher plants and algal cells is D1 (a core protein of photosystem II encoded by psbA) (18, 37, 54). However, photoregulated protein synthesis is common to several additional photosynthetic proteins and is also observed during chloroplast biogenesis (5, 15, 30, 31, 36). Chloroplast gene expression is regulated by light during its biogenesis at transcriptional and multiple posttranscriptional events (reviewed in references 39, 40, 46, and 53). While in the mature chloroplast the demand for rapid response in gene expression, to accommodate an environment of fluctuating light intensities, probably selected translation as a major determinant of photoregulated gene expression (reviewed in references 11, 19, 39, and 53). Both initiation and elongation steps of translation have been implicated as light-regulated components of gene expression (5, 17, 27, 29, 56).
The 5′ untranslated region (5′UTR) of several chloroplast mRNAs is a major determinant of translation (9, 25, 38, 45, 48, 51, 52, 57, 58). In addition, genetic and biochemical evidence has suggested that nucleus-encoded factors are essential to the translation of chloroplast mRNAs in a gene specific manner (20, 26, 32, 51, 55, 57). The nucleus-encoded factors are thought to enter the chloroplast and mediate translational regulation via the 5′UTR of chloroplast mRNAs (38, 48, 51, 57, 58).
A set of mRNA-binding proteins which bind to the psbA 5′UTR with high affinity and specificity have been identified and purified from Chlamydomonas reinhardtii cells (14). psbA 5′UTR-binding proteins are composed of four proteins, RB38, RB47, RB55, and RB60. These form a complex (psbA 5′PC) which binds the mRNA through the RB47 protein. The level of binding of psbA 5′PC to the mRNA parallels the level of psbA mRNA translation and association with polyribosomes in light- and dark-grown wild-type C. reinhardtii and in several mutants lacking translation of psbA mRNA (14, 55, 56). This suggests that light regulates polyribosome association and translation of psbA mRNA by modulating the binding of psbA 5′PC to the 5′UTR.
Binding of psbA 5′PC to the mRNA is regulated by two light-responsive molecular mechanisms. ADP-dependent phosphorylation of RB60 inactivates psbA 5′PC at ADP levels attained in chloroplasts only in the dark (12). Modulation in vitro of psbA 5′PC-binding activity by a redox mechanism suggested that the complex contains a redox-responsive regulatory site. The reactivation of psbA 5′PC by dithiol reductants and not by monothiol reductants predicted that the regulatory site is composed of vicinal dithiols and is reduced in vivo by a thioredoxin-like protein (13). A reductive signal transduced by the ferredoxin-thioredoxin system of photosynthesis (6, 49) was proposed to reduce the regulatory vicinal dithiol site (VDS) within the psbA 5′PC, thereby activating translation of psbA mRNA (13).
Lately, three components of psbA 5′PC, RB38, RB47, and RB60, have been cloned. RB60 was shown to be a protein disulfide isomerase-like protein (28), and RB47 has high homology to poly(A)-binding proteins (55). RB38 shows no homology to proteins with known function (55). Based on sequence homology to thioredoxin-like proteins and redox regulation of recombinant RB47 in vitro, RB60 was proposed to function equivalently to chloroplast thioredoxins and transduce the redox signal directly from thioredoxin reductase to RB47 (28).
A “light” signal transduced by a thioredoxin-like protein has been proposed to activate psbA 5′PC by reduction of a regulatory VDS within the protein complex (13). This suggests that for perception of the reductive signal in vivo, the VDS has to be initially oxidized. Hence, we needed to establish, in parallel, the redox state in organello of the regulatory VDS and redox effects on D1 protein synthesis in intact chloroplasts. Here we show that the regulatory VDS is contained within RB60, the protein disulfide isomerase-like member of psbA 5′PC, identifying RB60 as the redox sensor protein of the 5′PC. We demonstrate that, similarly to activation of psbA 5′PC (13), photoregulated translation of D1 is stimulated by a dithiol but not by a monothiol reductant, further implicating VDS-containing proteins in redox-regulated translation of psbA mRNA. We found that illumination of chloroplasts triggers specific oxidation of RB60, which is counteracted dynamically by a reductive activity. Under increased light levels, chloroplasts exhibit a higher rate of psbA mRNA translation and contain a larger pool of reduced RB60, suggesting that a counterbalanced action of reductive and oxidizing activities modulates the translation of psbA mRNA in parallel with fluctuating light intensities.
MATERIALS AND METHODS
Alga growth conditions and chloroplast isolation.
C. reinhardtii cw15 cells were grown in TAP medium (23), under a 12-h light/12-h dark regime at 25°C, to a density of approximately 107 cells/ml. Intact chloroplasts were collected from a 45%/70% interface of a discontinuous Percoll gradient by a previously described method (4, 22). The chlorophyll concentration was determined spectrophotometrically by Arnon's method (1). Isolated chloroplasts were kept in the dark for at least 30 min before being subjected to further treatments.
In organello translation and RNA isolation.
Protein synthesis in intact chloroplasts (200 μg of chlorophyll/ml) was performed as previously described (41) with slight modifications. All translation reaction mixtures included 10 mM MgATP. Dark-adapted chloroplasts were preincubated for 10 min in the dark or in the light in the presence or absence of 5 mM dithiothreitol (DTT), 10 mM β-mercaptoethanol (β-ME), or 10 mM diamide. Following preincubation, the chloroplasts were pulse-labeled for 10 min with [35S]methionine and then allowed to complete protein synthesis in the presence of 5 mM unlabelled methionine. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (34) and transferred onto nitrocellulose membranes. Radiolabeled proteins were detected by autoradiography. The location of proteins D1, D2, and the large subunit of ribulose bisphosphate carboxylase/oxygenase was determined by immunoblot assays using the corresponding antisera. Polysomal and total RNA was extracted from light- and dark-incubated chloroplasts as previously described (3). RNA extracted from equal amounts of chloroplasts (5 μg of chlorophyll) was fractionated, and RNA blots were hybridized with the 32P-labeled psbA cDNA probe.
Identification of VDS-containing proteins.
psbA 5′PC was isolated by RNA affinity purification as previously described (12). psbA RNA affinity-purified proteins were labeled with N-iodoacetyl-[125I]-3-iodotyrosine ([125I]IAIT) (21) in the presence of 0.5 mM DTT or with [14C]phenylarsine oxide ([14C]PAO) (33) for 20 min at room temperature. Labeled proteins were fractionated by SDS-PAGE (34). [125I]IAIT-labeled proteins were identified by autoradiography, and [14C]PAO-labeled proteins were blotted onto nitrocellulose membranes and visualized with a FujiX Bas 1000 PhosphorImager. Recombinant RB60 (rRB60; 140 ng), expressed from RB60 cDNA (accession no. AF036939), was incubated for 5 min with 1 mM DTT, oxidized with 5 mM dithionitrobenzoate, or modified with 5 mM n-ethylmaleimide for 5 min prior to labeling with [125I]IAIT (21) for 5 min at room temperature.
Cloning of RB60 cDNA and purification of recombinant RB60.
A lambda ZAPII cDNA expression library of C. reinhardtii was screened using antibodies raised against RB60 (12). Four clones were isolated from 105 plaques. In vivo excision of pBluescript from the lambda vector was performed as specified by the manufacturer (Stratagene, La Jolla, Calif.). Sequencing was performed with universal and sequence-specific primers, using the dideoxy-nucleotide terminator cycle sequencing method and an ABI model 373A sequencing system (Applied Biosystems, La Jolla, Calif.). Nucleotide sequences were analyzed using the Sequence Analysis Software Package (Genetics Computer Group, Madison, Wis.) (16). The open reading frames of all four clones were identical and encode the full-length sequence of RB60. A cDNA, containing the entire open reading frame, was subcloned into the pQE expression vector in frame with an amino-terminal His6 tag. Recombinant RB60 expression and purification was performed as specified by the manufacturer (Qiagen, Chatsworth, Calif.).
In organello labeling with [125I]IAIT.
Chloroplasts for labeling were treated in parallel with chloroplasts used for translation experiments. Following a 10-min incubation of chloroplasts in the dark or in the light, chloroplast proteins were labeled with [125I]IAIT (21) for 10 min at room temperature. RB60 was immunoprecipitated with a rabbit anti-rRB60 serum and protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc.) as recommended by the manufacturer. Total chloroplast proteins and immunoprecipitated RB60 were separated by SDS-PAGE (34) and blotted onto a nitrocellulose membrane. [125I]IAIT-labeled proteins were visualized by autoradiography. The amount of precipitated RB60 in each treatment was determined by an immunoblot assay using a mouse anti-RB60 serum. Total chloroplast proteins labeled with [125I]IAIT were precipitated with 10% trichloroacetic acid, and radioactivity was measured using a TRIATHLER multilabel tester (HIDEX, Turku, Finland).
RESULTS
Light-regulated translation in isolated intact chloroplasts.
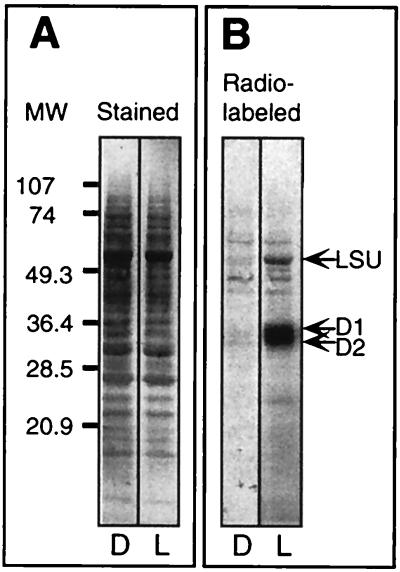
An isolated light-responsive intact-chloroplast translation system was established for C. reinhardtii to study redox signaling of photoregulated translation in chloroplasts. First, we tested whether fully developed chloroplasts contain the regulatory components controlling light-regulated translation. MgATP (10 mM) was included in all translation reaction mixtures to focus on photoregulatory events that are independent of the energy status of the chloroplast. A small set of proteins is clearly induced in chloroplasts incubated in the light, in contrast to those incubated in the dark (Fig. 1B). The D1, D2, and large subunit of ribulose bisphosphate carboxylase/oxygenase proteins, identified by the immunoblot assay, show the highest level of light induction in isolated intact C. reinhardtii chloroplasts. It is possible that the synthesis of additional proteins, which require a longer time for light induction or are unresolved in this electrophoresis system, is also light stimulated. Light did not stimulate the synthesis of several unknown proteins produced in the dark (Fig. 1B), indicating that light does not act as a general stimulator of protein synthesis. The same amounts of psbA mRNA (encoding the D1 protein) were detected in dark- and light-incubated chloroplasts, confirming that the amount of psbA mRNA present in the dark-incubated chloroplasts was not limiting D1 synthesis (Fig. 2, lanes Total). These results are similar to those found for whole cells (36) and indicate that translation of psbA mRNA in intact chloroplasts is regulated by light.
FIG. 1.
Light-regulated translation in mature chloroplasts isolated from C. reinhardtii cells. Intact C. reinhardtii cw15 chloroplasts were collected from a 45%/70% interface of discontinuous Percoll gradient by published methods (4, 22). Intact chloroplasts were incubated in the dark for 30 min, and an aliquot was transferred for 10 min into the light (150 μmol m−2 s−1). The translational activities of the dark-incubated (D) and light-incubated (L) chloroplasts were then assayed by labeling newly synthesized proteins for 10 min with [35S]methionine. The nascent polypeptides were allowed to complete synthesis by incubation for an additional 5 min in the presence of excess nonradioactive methionine. Chloroplasts were lysed, and extracted proteins were fractionated by SDS-PAGE and blotted onto nitrocellulose membranes. (A) Amido Black staining of chloroplast proteins. (B) Autoradiograph of the same blot as in panel A showing the 35S-labeled proteins. LSU, large subunit of ribulose bisphosphate carboxylase/oxygenase.
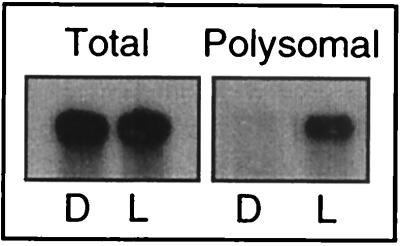
FIG. 2.
RNA blot showing light-regulated polysome association of psbA mRNA in mature chloroplasts isolated from C. reinhardtii cells. RNA was isolated from chloroplasts treated as for the experiment in Fig. 1. Equal amounts of total RNA and polysome-associated RNA were loaded on the gel. A 32P-labeled psbA cDNA was used to probe the blots.
To determine whether light controls the initiation step or the elongation step of translation, we assayed the association of psbA mRNA with ribosomes in dark- and light-incubated chloroplasts. The photoinduced recruitment of psbA mRNA to polysomes (Fig. 2, lanes Polysomal) shows that in mature chloroplasts, the initiation step of translation is light controlled. However, this does not exclude additional light-regulated steps, such as elongation. Identical treatment of chloroplasts prior to light incubation ensured that the dramatic activation of translation was due to the light treatment only and occurred within the short time span separating the two pools of light- and dark-treated chloroplasts. Taken together, these data suggest that a regulatory translational factor(s) controls the initiation of translation of psbA mRNA in response to light. These results are consistent with the proposed role of psbA 5′PC as a light-modulated trans-acting regulator of initiation of psbA mRNA translation (56).
Redox-regulated translation in isolated chloroplasts.
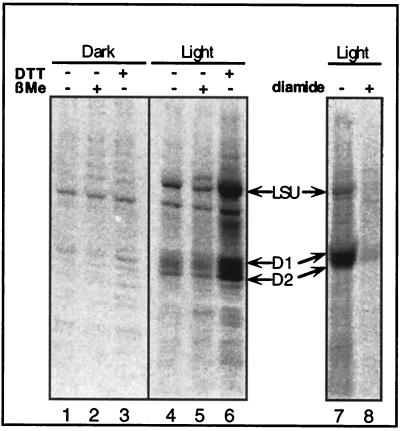
To test whether translation of psbA mRNA in intact chloroplasts is limited by an oxidized regulatory factor, we assayed whether translation of psbA mRNA could be enhanced by reducing agents (Fig. 3). The hallmark of a VDS is the insensitivity of its disulfide conformation to reduction by monothiols and its sensitivity to dithiol reductants. Thus, if translation of psbA mRNA is regulated by a VDS-containing factor, such as psbA 5′PC, it should be enhanced by DTT (a dithiol reagent) but not by β-ME (a monothiol reagent). Incubating illuminated chloroplasts with 5 mM DTT resulted in a clear increase in D1 synthesis (Fig. 3, lane 6) that was absent in illuminated chloroplasts treated with an equimolar thiol concentration of β-ME (lane 5). These data suggest that light-regulated translation of psbA mRNA in intact chloroplasts is controlled by a redox-responsive factor containing a regulatory VDS, such as psbA 5′PC, which upon reduction stimulates translation. Incubating illuminated chloroplasts with the thiol oxidant diamide diminished translation (lane 8). These properties are similar to those predicted by the in vitro studies of redox-regulated binding of psbA 5′PC (13), i.e., that thiol oxidation inhibits and dithiol reduction stimulates translation.
FIG. 3.
Light- and redox-regulated translation in mature chloroplasts. Shown is an autoradiograph of an SDS-PAGE blot, showing 35S-labeled proteins from chloroplasts treated as for the experiment in Fig. 1, except that a dithiol reductant (DTT), a monothiol reductant (β-ME), or a thiol oxidant (diamide) was added as indicated in the figure.
The stimulation of psbA mRNA translation by DTT also suggests that illuminating chloroplasts at 150 μmol m−2 s−1 was not sufficient to fully reduce the intrachloroplast pool of the redox-responsive factor(s) limiting translation under the experimental conditions used in this study. The DTT treatment enhanced the synthesis of other light-induced proteins (Fig. 3, compare lanes 6 and 4) but not of proteins synthesized in the dark (compare lanes 6 and 1). This implies that regulation by redox mechanisms via VDS-containing proteins may be common to light-activated translation of other transcripts. Interestingly, the responsiveness to DTT was lacking in chloroplasts incubated in the dark (lane 3). Also, treating dark-incubated chloroplasts with the thiol oxidant diamide did not activate the translation of psbA mRNA (data not shown). This indicates that, in contrast to illuminated chloroplasts (lane 6), translation is not limited by oxidation or reduction in the dark. These results also indicate that inhibition of translation in the dark is mediated by a nonredox component that is a prerequisite (a priming component) for redox regulation. Taken together, these results suggest that redox regulation is active only in illuminated chloroplasts in which oxidizing and reductive signals modulate the translation of light-induced proteins.
Biochemical identification of the regulatory VDS-containing protein of psbA 5′PC.
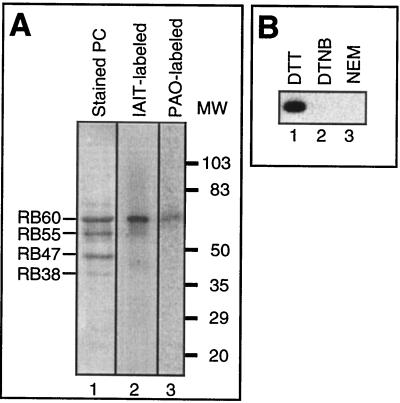
To characterize the redox state of the regulatory VDS of psbA 5′PC in photoregulated translation in chloroplasts, we first identified the protein containing it. A highly purified preparation of psbA 5′PC contains four major proteins, RB38, RB47, RB55, and RB60 (Fig. 4A, lane 1), each of which could potentially contain the regulatory VDS. To ensure the correct identification of the authentic VDS-containing protein(s), we used highly purified preparations of psbA RNA affinity-purified proteins (lane 1) in two independent experiments. (i) We first assayed for covalent labeling of highly reactive thiols of psbA 5′PC with [125I]IAIT under conditions that preferentially label VDS (21). Only one protein, RB60, was labeled by [125I]IAIT (Fig. 4A, lane 2). (ii) We used [14C]PAO, which specifically binds to VDS (33). [14C]PAO also bound only to the RB60 polypeptide (lane 3). These results indicate that RB60 is the only VDS-containing protein in psbA 5′PC and that it is the regulatory VDS-containing protein (13).
FIG. 4.
Identification of the regulatory VDS-containing protein. (A) Lanes: 1, Coomassie blue-stained SDS-PAGE of a highly purified preparation of psbA 5′PC (Stained PC) isolated from C. reinhardtii cells using psbA RNA-affinity chromatography; 2, PhosphorImage of SDS-PAGE of psbA 5′PC labeled with [125I]IAIT (IAIT-labeled); 3, autoradiograph of SDS-PAGE of psbA 5′PC labeled with [14C]PAO (PAO-labeled). (B) Autoradiograph of SDS-PAGE of recombinant RB60 labeled with [125I]IAIT. Lanes: 1, protein reduced with DTT; 2, protein oxidized with dithionitrobenzoate (DTNB); 3, protein modified with n-ethylmaleimide (NEM).
To use IAIT as a probe to characterize the redox state of RB60, we first verified that binding of [125I]IAIT to purified RB60 is dependent on the redox form of the protein. Equal amounts of purified recombinant RB60, expressed from RB60 cDNA, were incubated under different redox conditions with [125I]IAIT. As seen in Fig. 4B, [125I]IAIT bound only the reduced form of RB60 (Fig. 4B, lane 1). No binding was detected when [125I]IAIT was incubated with either oxidized RB60 with dithionitrobenzoate (lane 2) or RB60 modified with N-ethylmaleimide (lane 3). We concluded that [125I]IAIT can be used to probe the redox state of RB60.
Light modulates the redox state of RB60 and psbA mRNA translation in chloroplasts.
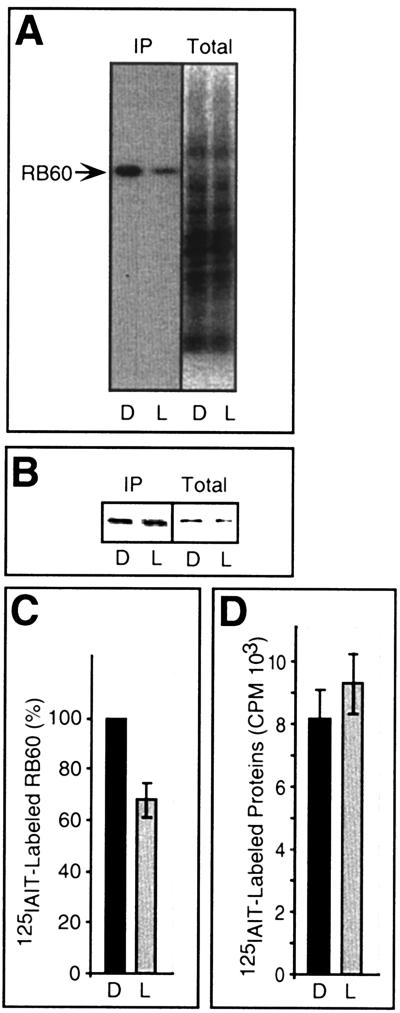
Previous results suggested that light regulates psbA mRNA translation by modulating the redox state of a VDS intrinsic to psbA 5′PC. Reduction of the VDS activates psbA 5′PC, and oxidation diminishes its activity (13). The enhancement of psbA mRNA translation by DTT in chloroplasts illuminated at 150 μmol m−2 s−1 (Fig. 3, lane 6) predicts that the pool of the VDS-containing factor controlling translation of psbA mRNA was partially oxidized. The insensitivity to DTT in dark incubated chloroplasts (lane 3) and the lack of response to oxidation by diamide imply that in contrast to illuminated chloroplasts, translation of psbA mRNA is not regulated by redox mechanisms in the dark. To determine the redox state of the pool of RB60 in the chloroplast under these two conditions, we isolated RB60 by immunoprecipitation from light- and dark-incubated chloroplasts that were labeled with [125I]IAIT (Fig. 5A, lanes IP). Labeling of RB60 was significantly lower in response to light, showing that the pool of RB60 is more oxidized in the light than in the dark (Fig. 5A, lanes IP, and Fig. 5C). These results are consistent with the stimulatory effect of DTT on psbA mRNA translation in the light and not in the dark and suggest that oxidation of RB60 is activated in the light.
FIG. 5.
In vivo characterization of the redox state of RB60 and total proteins in dark- and light-incubated chloroplasts. (A) Autoradiograph of immunoprecipitation assays of RB60 (IP) conducted with extracts from isolated chloroplasts treated for 10 min in the dark (D) or light (L), followed by [125I]IAIT labeling. Total [125I]IAIT-labeled proteins (without immunoprecipitation) (Total) represent 5% of the amount used for immunoprecipitation assays. (B) The same blot as in panel A, probed with anti-RB60 sera showing equal loading of RB60 protein. (C) Quantification of [125I]IAIT-labeled RB60 (as determined by PhosphorImager analysis and normalized for equal amounts of precipitated RB60 protein) in 10-min light- and dark-treated chloroplasts. Values are means of six independent experiments, and the light value is expressed as percentage of the corresponding dark value (in each experiment designated as 100% of [125I]IAIT-labeled RB60). (D) Quantification of total chloroplast proteins labeled with [125I]IAIT, showing that light does not affect the global protein thiol state in the chloroplast. Each value is an average of three replications.
The addition of DTT to illuminated chloroplasts resulted in higher [125I]IAIT labeling of RB60 (data not shown), demonstrating that labeling of RB60 was limited by oxidation and not by some other unknown modification(s) of RB60. To determine whether light affects the redox state of the majority of chloroplast proteins in the same manner as it affects RB60, the amount of [125I]IAIT-labeling of total proteins in illuminated or dark-incubated chloroplasts was assayed. In contrast to RB60, the redox state of the majority of chloroplast proteins was not affected by illumination (Fig. 5A, lanes Total, and Fig. 5D). This is in accordance with previous results showing that the reduced/oxidized ratio of glutathione or ascorbate does not change in chloroplasts between light and dark conditions (24, 35). Thus, protein oxidation in the light is not a general chloroplast phenomenon but is specific to RB60 and, possibly, to other regulatory proteins.
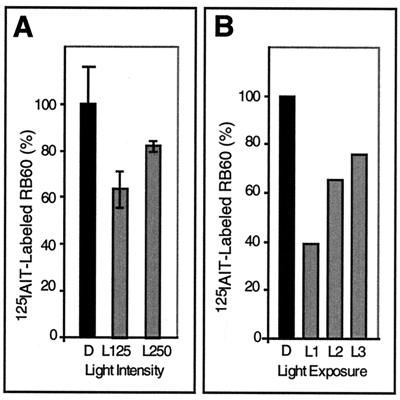
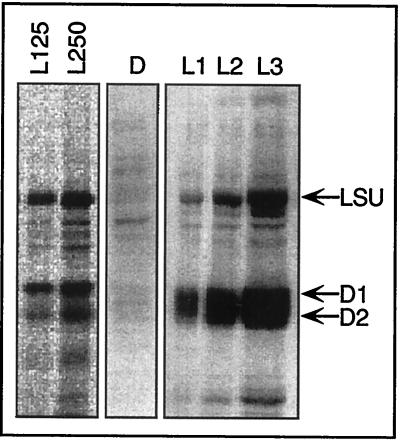
Perception of reductive signals in the intrachloroplast reducing environment requires a counteracting oxidizing activity. The partial oxidation of the pool of RB60 in the light compared to the dark (Fig. 5A, lanes IP, and Fig. 5C) suggests that RB60 oxidation is light activated and then is rendered receptive to the reductive photosynthesis-derived signal. This hypothesis predicts that the steady-state amount of reduced RB60 molecules and hence the rate of psbA mRNA translation in the light are consequences of antagonistic reducing and oxidizing activities. Thus, the pool of reduced RB60 should increase in response to increased light intensity. To test this, we characterized, in parallel, the redox state of RB60 (Fig. 6) and psbA mRNA translation (Fig. 7) under increased light regimes. We conducted two types of experiments: in the first, chloroplasts were exposed to increased light intensities (Fig. 6A and 7, lanes L125 and L250), while in the second, serial dilution of chloroplasts incubated at constant light intensity was used to obtain increased average chloroplast light exposure at lower chloroplast concentrations (Fig. 6B, and 7, lanes L1 to L3). The results of both experiments show that the pool of RB60 is reduced in response to increased light intensity (Fig. 6A) or higher average chloroplast light exposure (Fig. 6B) while the rate of psbA mRNA translation parallels the reduced state of RB60 (Fig. 7). These data are consistent with previous results obtained from in vitro biochemical analyses (13) and highlight an important component of redox regulation, i.e., the specific oxidation of RB60 in illuminated chloroplasts.
FIG. 6.
The redox state of the pool of RB60 in chloroplasts incubated in different light regimes. (A) The redox state of the pool of RB60 was determined as for the experiment in Fig. 5A, except that immunoprecipitation assays were performed using proteins of chloroplasts incubated in the dark (D) and under 125 (L125) or 250 (L250) μmol of light m−2 s−1. Quantification was performed as for the experiment in Fig. 5C, except that each value is an average of three replications (one replication of the dark treatment was designated as 100% of [125I]IAIT-labeled RB60). (B) The redox state of the pool of RB60 was determined as for the experiment in Fig. 5A, except that immunoprecipitation assays were performed with proteins of chloroplasts incubated in the dark or in the light (150 μmol m−2 s−1) at decreasing chloroplast concentrations, resulting in an increasing average light exposure of the chloroplasts. L1, 10 μg; L2, 5 μg; L3, 2.5 μg.
FIG. 7.
Effect of different light regimes on protein synthesis in mature isolated chloroplasts. Shown is an autoradiograph of protein labeling performed as for the experiment in Fig. 1, under the same light conditions as for the experiment in Fig. 6. LSU, large subunit of ribulose bisphosphate carboxylase/oxygenase.
DISCUSSION
The most compelling information on the way the redox signal is transduced by thioredoxins and regulates the activity of redox-responsive proteins comes from studies of chloroplasts. Reducing equivalents, generated from light by the concerted activities of photosystem II and photosystem I, are probably used via ferredoxin, ferredoxin-thioredoxin reductase, and thioredoxin to reduce regulatory redox-active sites of key proteins involved in CO2 assimilation, ATP synthesis, and translation of psbA mRNA (6, 13, 47, 49). An important characteristic shared by the regulated redox-responsive proteins is their preferential reduction by thioredoxins (mimicked by the dithiol reductant DTT and not by monothiol reductants, such as β-ME) (6). Here we identified RB60 as the subunit of psbA 5′PC containing a regulatory VDS (Fig. 4), previously implicated as prescribing preferential responsiveness of psbA 5′PC to thioredoxin and DTT (13). We show that photoregulated translation of D1 in intact chloroplasts is regulated at the level of initiation and is preferentially responsive to DTT (Fig. 2 and 3). These characteristics are consistent with the proposed role of psbA 5′PC as a light-modulated trans-acting regulator of initiation of psbA mRNA translation (56). Preferential responsiveness to DTT of photoregulated synthesis of the D2 protein and large subunit of ribulose bisphosphate carboxylase/oxygenase proteins was observed as well (Fig. 3), suggesting that the VDS-containing protein(s) may also regulate the translation of these photosynthetic proteins.
Similarly to the cytosol, the chloroplast contains an antioxidative system which under normal conditions maintains a reducing intrachloroplast environment (43). Regulation by redox mechanisms requires both reductive and oxidizing activities. Therefore, studies of redox signal transduction in intact chloroplasts, containing an active antioxidative system, are important to the elucidation of the molecular role of redox-active factors. Thioredoxin-regulated enzymes in chloroplasts incubated in the dark are activated by DTT, suggesting that they oxidize in the absence of the photosynthetic reducing potential in the dark (6). It was estimated that the fructose bisphosphatase/thioredoxin f system has a tendency for oxidation equivalent to disulfide bond formation in extracellular proteins (10). Here we show that treatment with the dithiol reductant DTT or the thiol oxidant diamide failed to stimulate the translation of psbA mRNA in dark-incubated chloroplasts (Fig. 3, lane 3). This suggests that translation of psbA mRNA is not limited by reduction or oxidation in the dark and that the inhibition of translation in the dark is mediated by a nonredox component. The inactivation of psbA 5′PC by ADP-dependent phosphorylation of RB60 at ADP levels attained in chloroplasts only in the dark (12) could potentially perform this function. The stimulation by DTT of psbA mRNA translation in illuminated chloroplasts (Fig. 3, lane 6) and the inactivation by diamide (lane 8) suggest that redox regulation is active in the light and that the pool of redox-regulatory protein controlling translation is partially oxidized in the light. Here we showed that the regulatory VDS, contained in RB60 (Fig. 4), is oxidized in illuminated chloroplasts in a specific manner (Fig. 5). The oxidation of RB60 in the presence of the antioxidative system in intact chloroplasts (Fig. 5) suggests that the redox state of RB60 is not functionally affected by of the global redox state of the chloroplast and is responsive only to reduction by thioredoxin-like proteins.
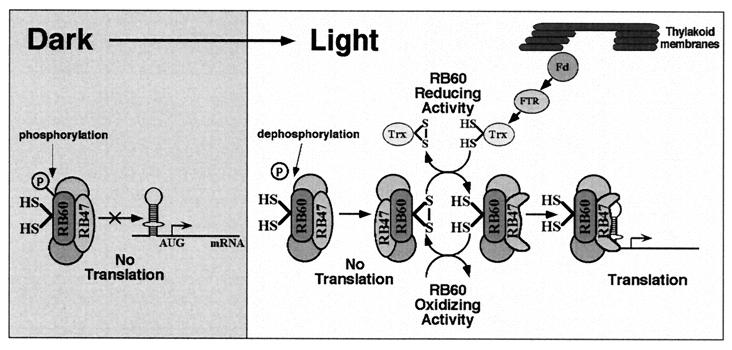
According to this hypothesis, the redox state of the pool of RB60 and, consequently, translation of psbA mRNA in the light is determined by a counterbalanced action of reductive and oxidizing activities (Fig. 8). The reductive activity is mediated by a thioredoxin-like protein, transducing photosynthetic reducing equivalents, and is proportional to the light intensity absorbed by photosynthesis. The nature of the oxidizing activity of RB60 is unknown and is being investigated by us. Light-triggered oxidation of RB60 could be mediated analogously to thioredoxin-regulated enzymes by autooxidation of RB60 induced by the priming pathway or alternatively by a specific light-activated oxidizing factor. Our hypothesis predicts that both translation of psbA mRNA and the pool of reduced of RB60 should increase in higher light intensity. To test this, we characterized in parallel the redox state of RB60 and psbA mRNA translation under increasing light intensity (Fig. 6 and 7). The results showed that the pool of RB60 becomes reduced in response to the increase in average chloroplast light exposure (Fig. 6) and that the rate of psbA mRNA translation parallels the reduced state of RB60 (Fig. 7). These results suggest a mechanism by which light primes redox-regulated translation by an unknown mechanism (Fig. 8). The inactivation of psbA 5′PC by ADP-dependent phosphorylation of RB60 (12) implicates protein phosphorylation and dephosphorylation as part of the priming event. Then, the rate of translation is determined, in parallel with changes in light intensity, by the reduction-oxidation of a sensor protein, RB60, located in a complex bound to the 5′UTR of psbA mRNA. A direct benefit of such a regulatory scheme is a dynamic capacity to stimulate translation under higher light intensities and to decrease translation as the light intensity diminishes. Such a dynamic regulation is required to maintain efficient energy conversion in an environment of fluctuating light levels.
FIG. 8.
A working model for redox signaling in light-regulated translation. The redox state of the regulatory VDS of RB60 controls the RNA-binding capacity of psbA 5′PC, which binds the RNA through the RB47 protein. Oxidation of the regulatory VDS inactivates and reduction activates binding of the PC to the 5′UTR of psbA mRNA and consequently psbA mRNA translation. Light regulates translation via a prerequisite (priming) redox-independent pathway and a redox-dependent pathway. The priming pathway probably involves dephosphorylation and specific oxidation of the pool of RB60 (either by autooxidation or by an RB60-specific oxidizing factor), rendering RB60 receptive to the reductive signal transduced by the redox-dependent pathway. In the redox-dependent pathway, light energy is captured by the thylakoid-associated photosynthetic complexes to produce reducing potential. Photosynthetic reducing equivalents are transduced by ferredoxin (Fd), ferredoxin-thioredoxin reductase (FTR), and thioredoxin (Trx) to reduce the regulatory VDS of RB60. Hence, in the light, a counterbalanced action of reducing and oxidizing activities modulates the redox state of the pool of RB60 and consequently the translation of psbA mRNA, in parallel with fluctuating light intensities. Transfer to the dark activates phosphorylation and inactivates the oxidation of RB60, resulting in an inactive form of psbA 5′PC that is not redox responsive. The encircled P denotes an inorganic phosphate.
What is the receptor(s) perceiving the light signal(s) activating translation of chloroplast mRNAs? Light regulates the expression of both nuclear and chloroplast genes. In the chloroplast, the light reactions of photosynthesis produce regulatory signals. Recent experiments have implicated the redox state of plastoquinone as a regulator of transcription of specific chloroplast genes and thylakoid protein phosphorylation (7, 44). A second reductive signal is probably transduced from photosystem I by the ferredoxin-thioredoxin system and regulates the activity of Calvin cycle enzymes, thylakoid-coupling factor I, translation of chloroplast mRNAs, and thylakoid protein phosphorylation (6, 7, 13, 50). Cytoplasmic and nuclear photoreceptors have been identified as regulators of plant development and adaptive responses to environmental changes (reviewed in references 8 and 42). At least two photoreceptors, PhyA and PhyB, affect chloroplast biogenesis (42).
It is possible that photoreceptors residing outside the chloroplast perceive the light signal regulating translation of chloroplast mRNA. Here we demonstrated that fully developed chloroplasts contain the components required for light signal perception and transduction and for regulation of psbA mRNA translation (Fig. 1 and 2). This suggests that the capacity to perceive and transduce the light signal activating translation is not dependent on light signals outside the chloroplast. Furthermore, we showed that the light signal has two components, a nonredox component and a redox component, and that the nonredox component is a prerequisite for the redox component (Fig. 3). The redox component is composed of a reductive signal, suggested to emanate from photosystem I (13), and a counteracting oxidizing component (Fig. 3, 5, and 6). Our data suggest that the light signals controlling both the oxidizing component and the priming nonredox component originate from within the chloroplast. The separation of chloroplast from cytoplasmic signaling pathways, resulting from studies of translation in isolated chloroplasts, may help elucidate the source of these light signals.
ACKNOWLEDGMENTS
We thank Z. Adam and E. Harel for help with chloroplast isolation procedure, and we thank G. Galili, M. Edelman, G. Schuster, and R. Fluhr for their critical reading of the manuscript. [14C]phenylarsine oxide was courtesy of T. Schäefer, and [125I]IAIT was a generous gift of C. Gitler.
A.D. holds The Judith and Martin Freedman Career Developmental Chair. This work was supported by a grant from Dorot Science Fellowships Foundation and a grant from the Minerva Foundation. T.T. is a recipient of a Feinberg Post-Doctoral Fellowship. A.L. is a recipient of a Feinberg Graduate School Fellowship.
REFERENCES
- 1.Arnon D. Copper enzymes in isolated chloroplasts. Polyphenol-oxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber J, Anderson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 3.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988;9:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belknap W R. Partial purification of intact chloroplasts from Chlamydomonas reinhardtii. Plant Physiol. 1983;72:1130–1132. doi: 10.1104/pp.72.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry J O, Breiding D E, Klessig D F. Light-mediated control of translational initiation of ribulose-1,5-bisphosphate carboxylase in amaranth cotyledons. Plant Cell. 1990;2:795–803. doi: 10.1105/tpc.2.8.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan B B. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 7.Carlberg I, Rintamaki E, Aro E M, Andersson B. Thylakoid protein phosphorylation and the thiol redox state. Biochemistry. 1999;38:3197–3204. doi: 10.1021/bi982506o. [DOI] [PubMed] [Google Scholar]
- 8.Cashmore A R, Jarillo J A, Wu Y J, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 9.Choquet Y, Stern D B, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman F A. Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA. 1998;95:4380–4385. doi: 10.1073/pnas.95.8.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clancey C J, Gilbert H F. Thiol/disulfide exchange in the thioredoxin-catalyzed reductive activation of spinach chloroplast fructose-1,6-bisphosphatase. Kinetics and thermodynamics. J Biol Chem. 1987;262:13545–13549. [PubMed] [Google Scholar]
- 11.Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danon A, Mayfield S P. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J. 1994;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danon A, Mayfield S P. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- 14.Danon A, Mayfield S P. Light-regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J. 1991;10:3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng X-W, Gruissem W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988;7:3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edhofer I, Muhlbauer S K, Eichacker L A. Light regulates the rate of translation elongation of chloroplast reaction center protein D1. Eur J Biochem. 1998;257:78–84. doi: 10.1046/j.1432-1327.1998.2570078.x. [DOI] [PubMed] [Google Scholar]
- 18.Fromm H, Devic M, Fluhr R, Edelman M. Control of psbA gene expression: in mature Spirodela chloroplasts light regulation of 32-kd protein synthesis is independent of transcript level. EMBO J. 1985;4:291–295. doi: 10.1002/j.1460-2075.1985.tb03628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillham N W, Boynton J E, Hauser C R. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- 20.Girard-Bascou J, Pierre Y, Drapier D. A nuclear mutation affects the synthesis of the chloroplast psbA gene production Chlamydomonas reinhardtii. Curr Genet. 1992;22:47–52. doi: 10.1007/BF00351741. [DOI] [PubMed] [Google Scholar]
- 21.Gitler C, Mogyoros M, Kalef E. Labeling of protein vicinal dithiols: role of protein-S2 to protein-(SH)2 conversion in metabolic regulation and oxidative stress. Methods Enzymol. 1994;233:403–415. doi: 10.1016/s0076-6879(94)33047-6. [DOI] [PubMed] [Google Scholar]
- 22.Goldschmidt-Clermont M, Malnoë P, Rochaix J-D. Preparation of Chlamydomonas chloroplasts for the in vitro import of polypeptide precursors. Plant Physiol. 1989;89:15–18. doi: 10.1104/pp.89.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorman D S, Levine R P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halliwell B, Foyer C. Properties and physiological function of a glutathione reductase purified from spinach leaves by affinity chromatography. Planta. 1978;139:9–17. doi: 10.1007/BF00390803. [DOI] [PubMed] [Google Scholar]
- 25.Hirose T, Sugiura M. Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J. 1996;15:1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen K H, Herrin D L, Plumley F G, Schmidt G W. Biogenesis of photosystem II complexes: transcriptional, translational, and posttranslational regulation. J Cell Biol. 1986;103:1315–1325. doi: 10.1083/jcb.103.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kettunen R, Pursiheimo S, Rintamaki E, Van Wijk K J, Aro E M. Transcriptional and translational adjustments of psbA gene expression in mature chloroplasts during photoinhibition and subsequent repair of photosystem II. Eur J Biochem. 1997;247:441–448. doi: 10.1111/j.1432-1033.1997.00441.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Mayfield S P. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Mullet J E. Ribosome-binding sites on chloroplast rbcL and psbA mRNAs and light-induced initiation of D1 translation. Plant Mol Biol. 1994;25:437–448. doi: 10.1007/BF00043872. [DOI] [PubMed] [Google Scholar]
- 30.Klein R R, Mullet J E. Control of gene expression during higher plant chloroplast biogenesis. Protein synthesis and transcript levels of psbA, psaA-psaB, and rbcL in dark-grown and illuminated barley seedlings. J Biol Chem. 1987;262:4341–4348. [PubMed] [Google Scholar]
- 31.Krupinska K, Apel K. Light-induced transformation of etioplasts to chloroplasts of barley without transcriptional control of plastid gene expression. Mol Gen Genet. 1989;219:467–473. [Google Scholar]
- 32.Kuchka M R, Mayfield S P, Rochaix J-D. Nuclear mutations specifically affect the synthesis and/or degradation of the chloroplast-encoded D2 polypeptide of photosystem II in Chlamydomonas reinhardtii. EMBO J. 1988;7:319–324. doi: 10.1002/j.1460-2075.1988.tb02815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kussmann M, Przybylski M. Tertiary structure-selective characterization of protein dithiol groups by phenylarsine oxide modification and mass spectrometric peptide mapping. Methods Enzymol. 1995;251:430–435. doi: 10.1016/0076-6879(95)51146-6. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Law M Y, Charles S A, Halliwell B. Glutathione and ascorbic acid in spinach (Spinacia oleracea) chloroplasts. The effect of hydrogen peroxide and of paraquat. Biochem J. 1983;210:899–903. doi: 10.1042/bj2100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malnoë P, Mayfield S P, Rochaix J-D. Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J Cell Biol. 1988;106:609–616. doi: 10.1083/jcb.106.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattoo A K, Pick U, Hoffman-Falk H, Edelman M. The rapidly metabolized 32,000-dalton polypeptide of the chloroplast is the “proteinaceous shield” regulating photosystem II electron transport and mediating diuron herbicide sensitivity. Proc Natl Acad Sci USA. 1981;78:1572–1576. doi: 10.1073/pnas.78.3.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayfield S P, Cohen A, Danon A, Yohn C B. Translation of the psbA mRNA of Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′ untranslated region. J Cell Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayfield S P, Yohn C B, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- 40.Mullet J E. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullet J E, Klein R R, Grossman A R. Optimization of protein synthesis in isolated higher plant chloroplasts. Identification of paused translation intermediates. Eur J Biochem. 1986;155:331–338. doi: 10.1111/j.1432-1033.1986.tb09495.x. [DOI] [PubMed] [Google Scholar]
- 42.Mustilli A C, Bowler C. Tuning in to the signals controlling photoregulated gene expression in plants. EMBO J. 1997;16:5801–5806. doi: 10.1038/sj.emboj.7590554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noctor G, Foyer C H. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 44.Pfannschmidt T, Nilsson A, Allen J. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- 45.Reinbothe S, Reinbothe C, Heintzen C, Seidenbecher C, Parthier B. A methyl jasmonate-induced shift in the length of the 5′ untranslated region impairs translation of the plastid rbcL transcript in barley. EMBO J. 1993;12:1505–1512. doi: 10.1002/j.1460-2075.1993.tb05794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 47.Ruelland E, Miginiac-Maslow M. Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci. 1999;4:136–141. doi: 10.1016/s1360-1385(99)01391-6. [DOI] [PubMed] [Google Scholar]
- 48.Sakamoto W, Chen X, Kindle K L, Stern D B. Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 1994;6:503–512. doi: 10.1046/j.1365-313x.1994.6040503.x. [DOI] [PubMed] [Google Scholar]
- 49.Schürmann P. Ferredoxin-thioredoxin system. Biothiols. 1995;252:274–283. doi: 10.1016/0076-6879(95)52030-9. [DOI] [PubMed] [Google Scholar]
- 50.Schwarz O, Schurmann P, Strotmann H. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem. 1997;272:16924. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- 51.Stampacchia O, Girard-Bascou J, Zanasco J L, Zerges W, Bennoun P, Rochaix J-D. A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staub J M, Maliga P. Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 1994;6:547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- 53.Stern D B, Higgs D C, Yang J J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 54.Wettern A K, Ohad I. Light-induced turnover of thylakoids polypeptides in Chlamydomonas reinhardtii. Isr J Bot. 1984;33:253–263. [Google Scholar]
- 55.Yohn C B, Cohen A, Danon A, Mayfield S. A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc Natl Acad Sci USA. 1998;95:2238–2243. doi: 10.1073/pnas.95.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yohn C B, Cohen A, Danon A, Mayfield S P. Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Mol Cell Biol. 1996;16:3560–3566. doi: 10.1128/mcb.16.7.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerges W, Girard-Bascou J, Rochaix J-D. Translation of the chloroplast psbC mRNA is controlled by interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol Cell Biol. 1997;17:3440–3448. doi: 10.1128/mcb.17.6.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zerges W, Rochaix J-D. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol Cell Biol. 1994;14:5268–5277. doi: 10.1128/mcb.14.8.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]