Abstract
The TATA-binding protein (TBP)-associated factor TAFII250 is the largest component of the basal transcription factor IID (TFIID). A missense mutation that maps to the acetyltransferase domain of TAFII250 induces the temperature-sensitive (ts) mutant hamster cell lines ts13 and tsBN462 to arrest in late G1. At the nonpermissive temperature (39.5°C), transcription from only a subset of protein encoding genes, including the G1 cyclins, is dramatically reduced in the mutant cells. Here we demonstrate that the ability of the ts13 allele of TAFII250 to acetylate histones in vitro is temperature sensitive suggesting that this enzymatic activity is compromised at 39.5°C in the mutant cells. Mutagenesis of a putative acetyl coenzyme A binding site produced a TAFII250 protein that displayed significantly reduced histone acetyltransferase activity but retained TBP and TAFII150 binding. Expression of this mutant in ts13 cells was unable to complement the cell cycle arrest or transcriptional defect observed at 39.5°C. These data suggest that TAFII250 acetyltransferase activity is required for cell cycle progression and regulates the expression of essential proliferative control genes.
Precise regulation of gene transcription is intimately involved in many biological functions such as cell proliferation, differentiation, and development. Biochemical studies have demonstrated that one of the key players in the regulation of RNA polymerase II-dependent gene expression is the transcription factor IID (TFIID). TFIID is a multisubunit complex consisting of the TATA binding protein (TBP) and TBP-associated factors (TAFIIs) (7, 37). It is the only general factor that possesses sequence-specific DNA binding activity (15, 26), and the binding of TFIID to the promoter region nucleates the assembly of a functional transcription initiation complex (4, 38). The TAFII subunits of TFIID have been shown to interact with promoter-selective transcriptional regulators and play an essential role in transducing the activation signal from enhancer binding proteins to the basal machinery (40). Studies also have demonstrated that some TAFIIs are involved in directing core promoter function (39). Therefore, the TAFs play a crucial role in transcriptional regulation.
A functional link between gene transcription and cell cycle progression has been provided with the cloning of TAFII250 and the discovery that the gene encoding this largest subunit of TFIID is identical to CCG1 (cell cycle gene 1), a gene able to complement the late G1 arrest of the temperature-sensitive (ts) mutant hamster cell lines ts13 and tsBN462 (14, 28, 30, 32). Further characterization of ts13 and tsBN462 cells has revealed that both cell lines contain a single base pair change resulting in a glycine-to-aspartic acid amino acid substitution in the TAFII250/CCG1 protein (11). Furthermore, studies with the two systems have yielded comparable results and revealed that despite having a mutation in the general machinery, both cell lines do not exhibit a global defect in mRNA synthesis (21, 31, 36, 42). Instead, transcriptional regulation at only a subset of promoters appears to be altered at the nonpermissive temperature (39.5°C). Expression of the cell cycle regulators cyclin A, D1, and E is reduced dramatically at 39.5°C, while the levels of p21 and p27, cyclin-dependent kinase inhibitors, are upregulated (29, 31, 36, 42). By contrast, the expression of other growth-related genes such as c-fos and c-myc remains unaffected in the mutant cells. These findings strongly suggest that a TAFII250-dependent transcriptional defect is responsible for the ts13 mutant phenotype.
In Saccharomyces cerevisiae, mutant strains harboring conditional alleles of TAFII145, the yeast homologue of TAFII250, also arrest in late G1 under restrictive conditions (41). A genome-wide analysis of gene expression dependence on TAFII145 has been performed using high-density oligonucleotide arrays technology (16). Of the 5,441 genes that were scored, 16% displayed a dependence on TAFII145 similar to that observed for RNA polymerase II, suggesting that this subset of genes display a direct transcriptional requirement for TAFII145. Interestingly, many of the TAFII145-dependent genes are involved in either cell cycle progression, DNA repair, or DNA synthesis. Thus, these results further support our hypothesis that the cell cycle arrest observed in ts13 cells is due to the misregulated expression of cellular growth factors fundamental for progression through the G1 and S phases of the cell cycle.
A strong correlation has been shown between the level of histone acetylation and the transcriptional activity of a chromosomal domain. Hyperacetylated histones have been found to predominate in regions of the genome that are actively transcribing genes (12), while hypoacetylated histones prevail within transcriptionally silenced domains (2). The current model is that acetylation of lysine residues in the basic amino terminal tails of the core histones destabilizes the higher-ordered structure of the nucleosomes rendering the DNA more accessible to the transcriptional machinery (8, 19). This intimate relationship between histone acetylation and transcriptional control was further strengthened by the discovery that the yeast transcriptional regulator GCN5 and the related human protein, hGCN5, both possess histone acetyltransferase (HAT) activity (3). GCN5 functions as an adapter protein and facilitates the action of acidic activators in yeast (9, 23). Mutations in GCN5 do not have a global effect on gene transcription, similar to that observed for the ts13 allele of TAFII250. Thus, GCN5 also appears to be selective in the genes that it regulates.
The human TAFII250 subunit of TFIID and its homologues in Drosophila and yeast have been reported to be HATs (24). Deletion analysis of the Drosophila and yeast proteins and sequence homology indicate that the HAT domain of human TAFII250 maps between amino acids 517 and 976. The ts13 mutation, which corresponds to amino acid 716 in the human protein, resides in the HAT domain of TAFII250. This finding, taken together with the studies on GNC5, has led us to hypothesize that the ts13 mutation compromises the HAT activity of TAFII250, resulting in the gene-selective defect in mRNA synthesis and ultimately cell cycle arrest. Consistent with this model, we report here that the HAT activity of the mutant form of TAFII250 present in ts13 cells displays temperature sensitivity in vitro. In addition, we demonstrate that TAFII250 HAT activity is necessary for cell proliferation and efficient transcription from the cyclin A and D1 promoters. Therefore, the HAT activity of TAFII250, the largest subunit of TFIID, appears to play a crucial role in the expression of key cell cycle control genes and the regulation of cell proliferation.
MATERIALS AND METHODS
Cell lines and cell culture.
ts13 cells were cultured in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (HyClone), l-glutamine, and penicillin-streptomycin and grown at 33.5°C in a humidified incubator containing 10% CO2. Sf9 cells were propagated in supplemented Hink's TNM-FH insect medium (JHR Biosciences) and grown in spinner culture at 27°C in the absence of CO2.
Luciferase reporter constructs.
Cyclin A- and c-fos-luciferase promoter constructs have been previously described (43). The cyclin D1 reporter plasmid was constructed by cloning the EcoRI-to-PvuII fragment of pD1-GO651 (kindly provided by Yue Xiong) into the SmaI site of pGL2-basic (Promega).
Expression and purification of recombinant proteins.
Histidine-tagged human TBP (His-hTBP) was expressed in bacteria as follows. Bacteria containing the TBP expression plasmid were grown at 37°C to an optical density at 600 nm of 0.5. Cells were diluted 10-fold and again grown at 37°C to an optical density at 600 nm of 0.5. Expression of His-hTBP was induced with the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 1 to 3 h of induction at 30°C, cells were harvested by centrifugation and resuspended in HIG buffer (25 mM HEPES-KOH [pH 7.6], 5 mM imidazole, 10% glycerol) containing 0.1% NP-40 and 400 mM KCl. After the addition of lysozyme (0.5 mg/ml) and 30 min of incubation at 4°C, the cells were lysed by sonication.
Human wild-type (WT) TAFII250, ts TAFII250, and HAT mutant (HATmt) TAFII250 were expressed as hemagglutinin (HA)-tagged proteins in Sf9 cells, using the baculovirus expression system. TAFII150 was expressed as a FLAG-tagged fusion protein in Sf9 cells. For protein production, ∼7.5 × 105 Sf9 cells were seeded per 10-cm dish and infected at a multiplicity of infection of 1 to 2. Approximately 48 h postinfection, the cells were lysed in 0.4 M HEMG buffer (25 mM HEPES [pH 7.6], 400 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5% NP-40) by sonication. Affinity purification was performed as follows. A 100-μl aliquot of cell extract was incubated with 1 to 2 μl of anti-HA ascites fluid for 1.5 h at 4°C, followed by addition of 25 μl of 50% slurry of protein A-Sepharose (PAS). After an additional 1 h at 4°C, the Sepharose beads were washed three times with 1 ml of wash buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.5% [vol/vol] NP-40, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, leupeptin [1 μg/ml], pepstatin [1 μg/ml]). The immobilized proteins were used immediately for HAT assays.
In vitro HAT assays.
HAT assays were performed essentially as described previously (24). Affinity-purified WT, ts mutant and HATmt TAFII250 proteins were washed once with 1 ml of assay buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride), and the immune complexes were resuspended in 300 μl of assay buffer; 30-μl aliquots of the immune complex mixture were preincubated at either 25 or 37°C for 1 h, followed by an additional 30-min incubation in the presence of 25 μg of core histones (Sigma type II-AS from calf thymus) and 0.25 μCi of [3H]acetyl coenzyme A (Amersham). The entire reaction was filtered through P81 paper and washed with 50 mM sodium carbonate (pH 9.2). Histone acetylation was measured by liquid scintillation. The amount of TAFII250 added to the reactions was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a bovine serum albumin standard and Coomassie blue staining.
TAFII250 protein interaction assays.
Immunopurified WT and HATmt TAFII250 proteins immobilized on anti-HA antibody bound to PAS beads were incubated with either crude Sf9 TAFII150- or bacterial TBP-containing cell extracts. After 2 h of incubation at 4°C, the retained proteins were washed extensively, dissociated from the beads, and separated by SDS-PAGE. Silver staining and Western blot analysis using anti-FLAG and anti-TBP antibodies were performed to visualize bound proteins.
Complementation of cell cycle arrest.
ts13 cells were seeded at ∼7.5 × 105 cells/10-cm-diameter dish and grown overnight at 33.5°C. Cells were transfected via calcium phosphate precipitation with 0.5 μg of CS2+MT, CS2+MT WT TAFII250, or CS2+MT HATmt TAFII250 expression vector. After 6 h at 33.5°C, the cells were washed with phosphate-buffered saline, fresh medium was added, and half of the plates were shifted to 39.5°C. The number of viable colonies at 39.5°C was determined after a 2-week period.
Transient transfection assays.
ts13 cells were maintained at 33.5°C or shifted to 39.5°C for 2 h before the introduction of DNA. Cells were transiently transfected as described above with 0.5 μg of the indicated CS2+MT construct, 0.5 μg of β-galactosidase expression vector, and 1 μg of luciferase reporter construct containing either the c-fos, cyclin A, or cyclin D1 promoter. The cells were washed after 6 h in the presence of DNA and harvested 24 h posttransfection in reporter lysis buffer (Promega). Luciferase activity was determined using standard protocols and normalized for transfection efficiency by β-galactosidase activity (13).
RESULTS
HAT activity of ts13 TAFII250 mutant protein is ts in vitro.
TAFII250, the largest subunit of TFIID, is a multifunctional protein that directly interacts with components of TFIID (44), as well as the cellular proteins Rb (33) and MDM2 (20). In addition, the TAFII250 protein possesses HAT (24) and kinase (6) activities. The functional role of these different properties of TAFII250 is not clearly understood.
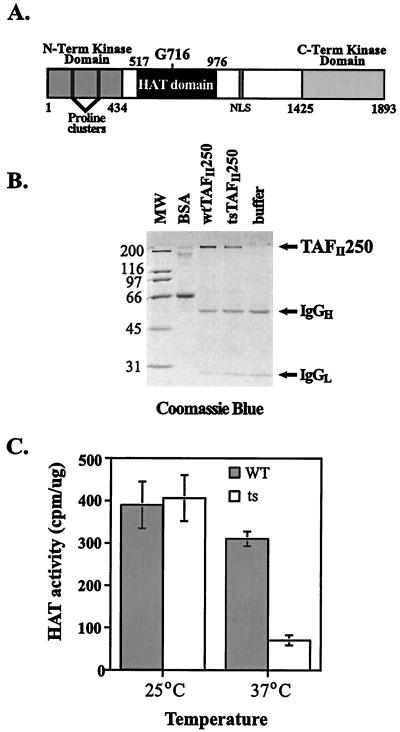
The ts mutant hamster cell line ts13 harbors a single missense mutation (glycine to aspartic acid) in the TAFII250 subunit of TFIID which induces the cells to arrest in the late G1 phase of the cell cycle at 39.5°C (11). The mutation responsible for the ts13 mutant phenotype maps to the HAT domain of the protein (Fig. 1A). To determine if this mutation disrupts TAFII250 HAT activity, we examined the ability of the WT and ts TAFII250 proteins to acetylate histones in vitro. Sf9 insect cells were infected with recombinant baculoviruses expressing an HA-tagged version of WT or ts TAFII250. The HA-tagged proteins were immunoaffinity purified from crude Sf9 cell lysates, using an anti-HA ascites fluid. The purified proteins, as shown in Fig. 1B, subsequently were added to reaction mixtures that contained the core histones and [3H]acetyl coenzyme A. It has been reported that activation of transcription in ts13 nuclear extracts is detected when the reactions are carried out at 25°C. However, the ability of the mutant cell extracts to support transcriptional activation is abolished at 37°C, suggesting that the ts TAFII250 present in the ts13 nuclear extract is functionally defective at the higher temperature. In addition, ts13 cells grow at a much lower rate and appear sickly at 37°C. For these reasons, we have elected to use 25 and 37°C as our permissive and nonpermissive temperatures, respectively, in vitro. When the reactions were incubated at 25°C, WT and ts TAFII250 displayed comparable levels of HAT activity (Fig. 1C). By contrast, when the reaction temperature was increased to 37°C, the ability of the ts mutant protein to acetylate histones was lower than that of its WT counterpart (Fig. 1C). Therefore, the HAT activity of TAFII250 is ts in vitro, suggesting that this function of TAFII250 is compromised in vivo in ts13 cells at 39.5°C.
FIG. 1.
ts13 TAFII250 mutant protein displays ts HAT activity in vitro. (A) Schematic diagram of human TAFII250 protein. The positions of known functional domains are indicated. The homologous human amino acid residue mutated in ts13 cells (G716) maps to the putative HAT domain of TAFII250, suggesting that this enzymatic activity is compromised in ts13 cells. Term, terminal; NLS, nuclear localization signal. WT and mutant TAFII250 were purified from baculovirus-infected Sf9 extracts using immunoprecipitation techniques, run on SDS-polyacrylamide gels, and visualized by Coomassie blue staining to show purity. As a control, Sfa lysis buffer was subjected to immunoprecipitation. Positions of the immunoglobulin heavy (IgGH) and light (IgGL) chains are indicated. Positions of molecular weight standards (MW) are indicated in kilodaltons at the left. BSA, bovine serum albumin. (C) Purified TAFII250 proteins described for panel B were added to liquid HAT assays containing purified calf thymus histones and [3H]acetyl coenzyme A and incubated at 25°C (six experiments with 5 ≤ n ≤ 10) or 37°C (eight experiments with 5 ≤ n ≤ 10). The amount of acetylated histones was measured by liquid scintillation.
HAT-deficient TAFII250 protein is unable to complement the ts13 cell proliferative defect.
The reduction in TAFII250 HAT activity under conditions that cause growth arrest of ts13 cells suggests that this enzymatic activity is required for cell cycle progression, specifically through G1 into S phase. We propose that at 39.5°C, and not at the permissive temperature of 33.5°C, ts TAFII250 polypeptide adopts an altered conformation which disrupts HAT activity and potentially additional functions of the protein. The ability to detect the ts TAFII250 protein in extracts prepared from cells grown at 39.5°C suggests that the inactivation of TAFII250 is not a result of protein instability (31, 42). The protein kinase activity of the ts TAFII250 is unaffected by the ts13 mutation (R. Dikstein and R. Tjian, unpublished data). By conventional assays, we have not detected a significant difference between the interaction of WT and ts TAFII250 with other subunits of TFIID at 20°C and 37°C in vitro (E. H. Wang and R. Tjian, unpublished data). However, these findings do not rule out the possibility that the interaction of TAFII250 with other cellular proteins has been compromised and is responsible for the ts13 mutant phenotype.
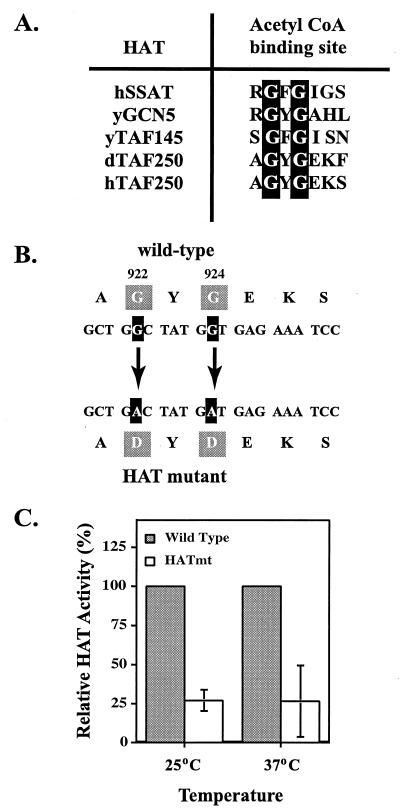
To test our hypothesis that only the disruption of TAFII250 HAT activity is required to induce cell cycle arrest, we specifically targeted the HAT domain of TAFII250. Our strategy involved identifying a potential acetyl coenzyme A binding site within the TAFII250 protein. Alignment of the consensus acetyl coenzyme A binding site in human spermine/spermidine acetyltransferase (hSSAT) with other known HATs such as GCN5, yeast TAFII145, Drosophila TAFII250, and human TAFII250 led to the identification of a potential binding site within the HAT domain of human TAFII250 (Fig. 2A). Studies on hSSAT have demonstrated that site-directed mutagenesis of the two glycine residues abolished acetyl coenzyme A binding and acetyltransferase activity (22). Based on these findings, we mutated the conserved glycine residues present in human TAFII250 (Fig. 2B). As expected, the HAT activity of the resulting mutant protein was less than that of WT TAFII250 (Fig. 2C). Furthermore, this reduction in HAT activity was observed at both 25 and 37°C in vitro.
FIG. 2.
Mutation of a putative acetyl coenzyme A (CoA) binding site compromises TAFII250 HAT activity. (A) A consensus sequence for an acetyl coenzyme A binding site has been identified in human (h), yeast (y), and Drosophila (d) HAT proteins including TAFII250 as shown. (B) Two conserved glycine residues present in human TAFII250 were mutated, as indicated, to aspartic acid (G922A and G924A) in order to ablate acetyl coenzyme A binding. (C) The TAFII250 HAT mutant described above and WT TAFII250 were expressed and immunoaffinity purified from recombinant baculovirus-infected Sf9 cells. The HAT activity of the purified WT and HAT mutant proteins was determined at 25°C (six experiments with 5 ≤ n ≤ 10) and 37°C (five experiments with 5 ≤ n ≤ 10) by liquid HAT assays. The relative HAT activity for the HAT mutant TAFII250 is expressed as a percentage of the activity detected with the WT protein (given a value of 100%) at each temperature.
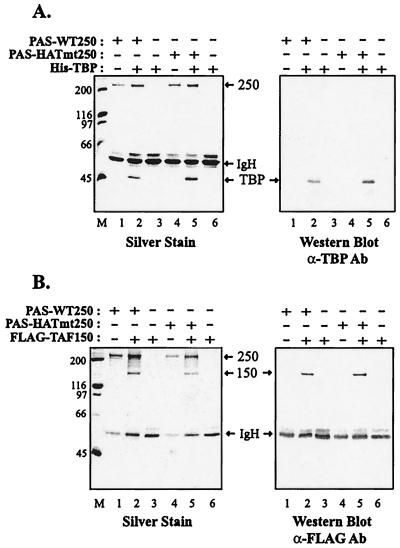
There exists the possibility that mutagenesis of the two glycine residues described above could alter additional functions of TAFII250. This outcome would hinder our ability to draw any conclusions about the function of TAFII250 HAT activity from studies with the mutant protein. Thus, we examined the ability of the TAFII250 HAT mutant to interact with TBP and TAFII150, two subunits of TFIID that directly contact TAFII250. In coimmunoprecipitation experiments, we observed that the HAT mutant immobilized on anti-HA PAS beads bound to TBP (Fig. 3A, lanes 2 and 5) and TAFII150 (Fig. 3B, lanes 2 and 5) as effectively as WT TAFII250.
FIG. 3.
TAFII250 HAT mutant retains TBP and TAFII150 binding. HA-tagged WT and HAT mutant TAFII250 proteins were immunopurified from recombinant baculovirus-infected Sf9 cell extracts and immobilized on anti-HA antibody (Ab) bound to PAS. The PAS-bound proteins, as indicated, were incubated with bacterial or Sf9 extracts overexpressing His-tagged TBP (A) or FLAG-tagged TAFII150 (B), respectively. The resulting dimers were dissociated from the PAS beads and analyzed by SDS-PAGE followed by silver staining and Western blot analysis using anti-TBP or anti-FLAG antibody as indicated. Positions of size markers are indicated in kilodaltons at the left.
The successful generation of a temperature-independent TAFII250 HAT mutant (HATmt) enabled us to more directly test the requirement for TAFII250 HAT activity in cell proliferation. ts13 cells were transfected with a control, WT TAFII250, or HATmt TAFII250 expression vector and maintained at 33.5 or 39.5°C. While expression of WT TAFII250 resulted in cell proliferation (265 colonies/μg of DNA) at the nonpermissive temperature, no viable colonies were detected with the HATmt or control vector. The lack of complementation cannot be attributed to differences in protein expression, as comparable levels of the WT and HATmt TAFII250 proteins were detected in the transfected cell lysates by Western blot analysis (data not shown). These findings strongly indicate that the HAT activity of TAFII250 is necessary for cellular proliferation.
Efficient transcription from the cyclin A and D1 promoters requires TAFII250 HAT activity.
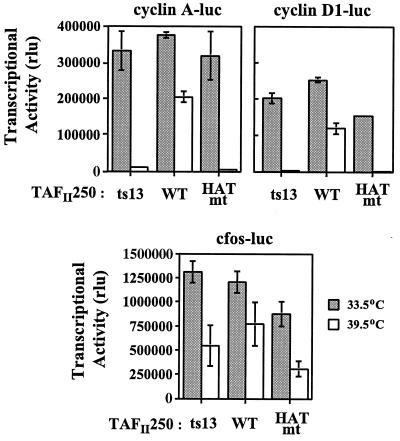
In addition to the inability to progress through the G1 phase of the cell cycle, ts13 cells exhibit a dramatic reduction in cyclin A and D1 gene transcription. This transcriptional defect is overcome by the exogenous expression of WT TAFII250 (29, 31, 36, 42). However, there is some question as to whether the decrease in cyclin A transcription is a direct consequence of the Gly690Asp mutation in TAFII250. On the other hand, there is general agreement that the cyclin D1 promoter is a direct target of TAFII250. Therefore, we tested whether expression of HATmt TAFII250 could overcome the cyclin A and D1 transcriptional defect observed at the nonpermissive temperature. ts13 cells were cotransfected with the control, WT, or HATmt TAFII250 expression vector along with luciferase reporter constructs containing either the ts cyclin A, ts D1, or resistant c-fos promoter. Approximately 24 h posttransfection, the transcriptional activity of each promoter construct was determined. In the presence of the control vector, transcription from the cyclin A and D1 promoters was markedly reduced at 39.5°C due to the presence of ts13 TAFII250 (Fig. 4). Expression of WT TAFII250 in the cells dramatically reduced the temperature sensitivity of the cyclin A and D1 promoters (Fig. 4). The HATmt TAFII250 was unable to restore transcriptional activity to cyclin A or cyclin D1 at 39.5°C (Fig. 4). By contrast, expression of the WT and HATmt proteins had no significant effect on c-fos promoter activity (Fig. 4). Therefore, the decrease in c-fos transcriptional activity observed at 39.5°C appears not to be TAFII250 dependent. We confirmed by Western blot analysis that comparable amounts of WT and HATmt TAFII250 were expressed at each temperature in the transfected ts13 cells (data not shown). Thus, the HAT activity of TAFII250 is required for transcription at only a subset of promoters.
FIG. 4.
Transcriptional requirement for TAFII250 HAT activity is promoter dependent. ts13 cells, maintained at permissive (33.5°C) or nonpermissive (39.5°C) temperature, were transiently transfected with the control (ts13) or indicated TAFII250 (WT or HATmt) expression vector and the cyclin A-, cyclin D1-, or c-fos-luciferase (luc) reporter construct. Approximately 24 h posttransfection, transcriptional activity of each promoter construct in relative light units (rlu) was determined and normalized for transfection efficiency. Note that the transcriptional activity of the c-fos promoter was greater than that of the cyclin promoters, as illustrated by the larger scale on the y axis. Presented are data from one representative experiment; similar results have been observed repeatedly.
DISCUSSION
Function for TAFII250 HAT activity in cell cycle progression.
Mammalian TAFII250 and the yeast homologue TAFII145 have been implicated in cell cycle progression, as mutations in these proteins lead to a defect in S-phase entry. TAFII250, the core subunit of the TFIID transcription initiation complex, is part of the basal transcriptional machinery. TFIID directly binds to core promoter DNA elements including the initiator sequence of select genes. Many of the TAF subunits of TFIID function as transcriptional coactivators, leading to an increase in mRNA synthesis in the presence of specific activators. The TAFII250 protein has been intensely characterized and possesses many activities, including protein binding, DNA binding, protein kinase, and HAT activities. However, the functions necessary for TAFII250's role in transcriptional coactivation and cell proliferation remain unclear.
Although a complete loss of TAFII250 expression is lethal (27), a single missense mutation, found in ts13 and tsBN462 cells, has the effect of altering transcription of select cell cycle regulators and leads to a proliferative block in late G1 (11, 31, 36, 42). Therefore, the ts13 cell line represents a good model system in which to identify TAFII250's role in transcriptional regulation and cell proliferation. Until now, no specific function of TAFII250 has been implicated in the ts13 mutant phenotype. Here, we present evidence that HAT activity of the ts13 TAFII250 mutant is ts in vitro. The importance of the hypothesized loss of TAFII250 HAT activity was examined further by targeting the acetyl coenzyme A binding pocket in TAFII250. The residues mutated are located within the TAFII250 HAT domain more than 100 amino acids away from the original Gly-to-Asp mutation and ideally should not affect the same microenvironment of the protein. Characterization of the acetyl coenzyme A binding mutant indicates that in vitro HAT activity is decreased at all temperatures compared to its WT counterpart. In complementation experiments, the TAFII250 HAT mutant was unable to overcome the ts13 transcriptional or proliferative defect, suggesting that TAFII250 HAT activity is necessary for the proper transcription of a select set of protein encoding genes essential for progression through the G1 phase of the cell cycle.
Transcriptional role for TAFII250 HAT activity.
The strong correlation between histone acetylation and transcriptional activity of a chromosomal domain has led to the current model that acetylation increases the accessibility of DNA to transcription factors. Many transcriptional regulatory proteins possess HAT activity, including p300/CREB binding protein (CBP), ACTR, SRC-1, P/CAF, GCN5, and TAFII250 (3, 5, 24, 25, 34, 45). The majority of these factors are considered coactivators that interact with a wide variety of nuclear receptors and enhancer binding proteins (35). Evidence for targeting HAT activity stems from studies on yeast GCN5, where mutagenesis of the HAT domain leads to a promoter-specific decrease in transcription (18). A number of different models can be proposed to account for this promoter-selective requirement for HAT activity. In the simplest case, HATs are recruited to only a subset of promoters via interactions with specific enhancer and/or upstream element binding proteins. However, this model cannot be applied to TAFII250, as it is a subunit of TFIID and thus an integral part of the general machinery present at all protein-encoding genes. If TAFII250 is found on all transcriptionally active genes, the requirement for TAFII250 acetyltransferase activity may be determined by the promoter context. The specific placement of nucleosomes away from transcription factor binding sites could render a promoter less dependent on histone acetylation. Alternatively, multiple HATs are recruited to some but not all promoters, and the mutation in ts13 cells uncovers promoters which recruit TAFII250 as the only HAT. Upon inactivation of TAFII250 HAT activity, transcription from these promoters would be severely compromised.
Transcription factors as potential substrates for TAFII250 HAT activity.
Recently p300/CBP, a well-characterized HAT, has been shown to acetylate the transcription factors p53 and GATA-1, stimulating their DNA binding activities (1, 10). These results have led to the hypothesis that acetylation of nonhistone proteins may have important regulatory functions. The specific activity of TAFII250's HAT activity is quite low compared to that of many other HATs. Therefore, it may be more accurate to classify TAFII250 as an acetyltransferase, as the true substrate for this activity may not be histones. Genetic analysis of the cyclin A promoter has led to the identification of a TAFII250-dependent upstream regulatory element that contains an activating transcription factor (ATF) binding site (43). One potential model is that certain forms of ATF or CREB are substrates for TAFII250 HAT activity. The acetylation of these proteins may enhance the ability of these regulatory proteins to bind DNA and activate gene transcription. In an examination of basal transcription factors, it has been reported that TFIIEβ (p34 subunit) can be acetylated by the TAFII250 protein in vitro, but the biological significance of this acetylation is unknown (17). Future studies may reveal other physiologically relevant substrates more indicative of TAFII250 function in vivo.
In conclusion, we have demonstrated a role for TAFII250 acetyltransferase activity in mediating efficient gene transcription. The acetylation event is likely to activate transcription from only a subset of genes, including those required for cell cycle progression. These results may represent a novel mechanism by which strict control of proliferative protein expression is maintained. Future studies will focus on the regulation of TAFII250 acetyltransferase activity and may unveil new and novel regulatory pathways involved in cell cycle control.
ACKNOWLEDGMENTS
We are indebted to K. White for preparation of His-hTBP bacterial cell extracts. We thank R. Tjian for providing the anti-HA ascites fluid, P. Verrijzer for the His-hTBP plasmid construct, R. Moon for the CS2+MT expression vector, and N. Davies for advice on in vitro HAT assays. We especially thank members of the Wang lab for valuable discussions and G. S. McKnight for critical reading of the manuscript.
E. L. Dunphy was supported in part by Public Health Service National Research Service Award T32 GM07270 from the National Institute of General Medical Sciences. This work was supported by research project grant RPG-98-201-CCG from the American Cancer Society and by startup funds from the Howard Hughes Medical Institute.
REFERENCES
- 1.Boyes J, Byfield P, Natakani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 2.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 3.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Lin R, Schiltz R, Chakravarti D, Nash A, Nagy L, Privalsky M, Nakatani Y. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 7.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;55:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 9.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 11.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 12.Hebbes T R, Clayton A L, Thorne A W, Crane R-C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbomel P, Bourachot B, Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984;39:653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- 14.Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder R G. The p250 subunit of native TATA-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993;362:179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- 15.Hoey T, Dynlacht B D, Peterson M G, Pugh B F, Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990;61:1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- 16.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 17.Imhof A, Yang X-J, Ogryzki V V, Nakatani Y, Wolfe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;9:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 18.Kuo M, Zhou J, Jambeck P, Churchill M, Allis C D. Histone acetyltransferase activity of yeast Gcn5 is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 20.Leveillard T, Wasylyk B. The MDM2 C-terminal region binds to TAF(II)250 and is required for MDM2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651–30661. doi: 10.1074/jbc.272.49.30651. [DOI] [PubMed] [Google Scholar]
- 21.Liu H T, Gibson C W, Hirschhorn R R, Rittling S, Baserga R, Mercer W E. Expression of thymidine kinase and dihydrofolate reductase genes in mammalian ts mutants of the cell cycle. J Biol Chem. 1985;260:3269–3274. [PubMed] [Google Scholar]
- 22.Lu L, Berkey K, Casero R. RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine acetyltransferase. J Biol Chem. 1996;271:18920–18924. doi: 10.1074/jbc.271.31.18920. [DOI] [PubMed] [Google Scholar]
- 23.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen H-T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 25.Ogryzko V, Schiltz R, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–965. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 26.Peterson M G, Tanese N, Pugh B F, Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 27.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 28.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 29.Rushton J J, Steinman R A, Robbins P A. Differential regulation of transcription of p21 and cyclin D1 conferred by TAFII250. Cell Growth Differ. 1997;8:1099–1104. [PubMed] [Google Scholar]
- 30.Sekiguchi T, Miyata T, Nishimoto T. Molecular cloning of the cDNA of human X chromosomal gene (CCG1) which complements the temperature-sensitive G1 mutants, tsBN462 and ts13, of the BHK cell line. EMBO J. 1988;7:1683–1687. doi: 10.1002/j.1460-2075.1988.tb02996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiguchi T, Noguchi E, Hayashida T, Nakashima T, Toyoshima H, Nishimoto T, Hunter T. D-type cyclin expression is decreased and p21 and p27 CDK inhibitor expression is increased when tsBN462 CCG1/TAFII250 mutant cells arrest in G1 at the restrictive temperature. Genes Cells. 1996;1:687–705. doi: 10.1046/j.1365-2443.1996.00259.x. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Z, Ruppert S, Robbins P D. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein associated factor TAFII250. Proc Natl Acad Sci USA. 1995;92:3115–3119. doi: 10.1073/pnas.92.8.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer T, Jenster G, Burcin M, Allis C D, Zhou J, Mizzen C, McKenna N, Onate S, Tsai S, Tsai M, O'Malley B. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 35.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanese N, Pugh B F, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 38.Van Dyke M W, Roeder R G, Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 39.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1128. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 40.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–341. [PubMed] [Google Scholar]
- 41.Walker S W, Reese J C, Apone L M, Green M R. Transcription activation in cell lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 42.Wang E H, Tjian R. Promoter selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250 in vitro. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 43.Wang E H, Zhou S, Tjian R. TAFII250 dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinzierl R, Dynlacht B D, Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–517. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenovirus oncoprotein E1A. Nature. 1993;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]