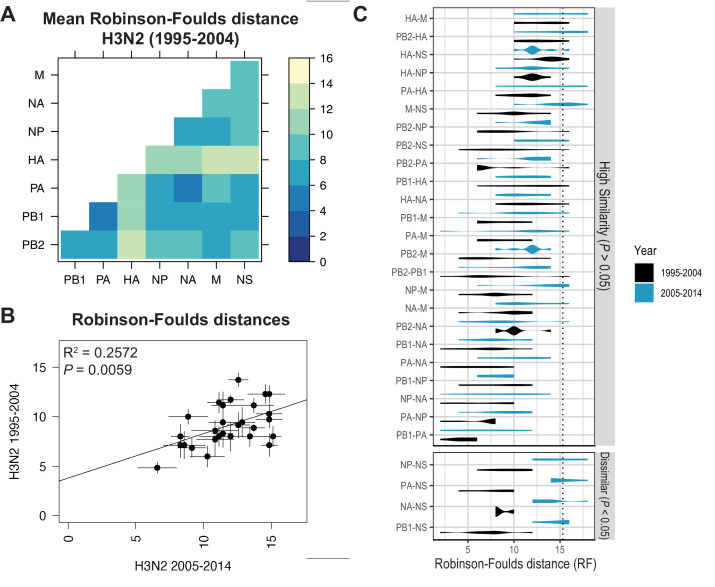
Figure 3. Parallel evolution of viral RNA (vRNA) segments from H3N2 viruses is conserved through antigenic drift.
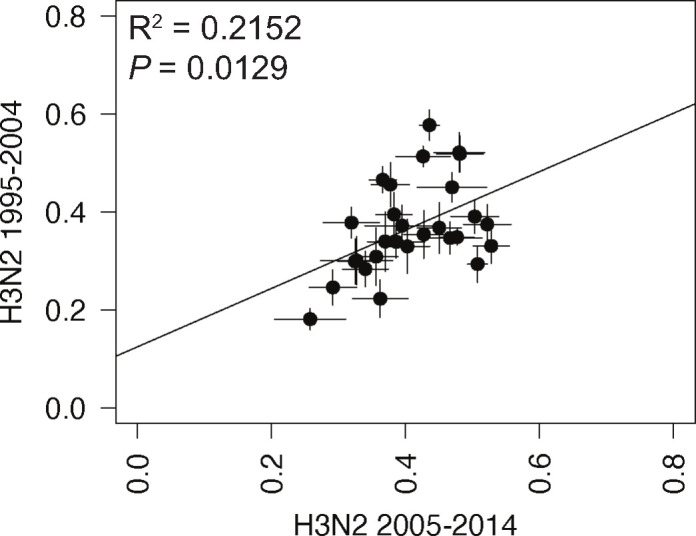
(A) Seven replicate maximum-likelihood trees were reconstructed for each vRNA gene segment from human H3N2 virus sequences (1995 to 2004) as described in Figure 1. Pairwise Robinson-Foulds distances (RF) were calculated between each pair of trees in each replicate. Mean tree distances were visualized in a heatmap. Refer to Figure 3—figure supplement 1 for the standard error of the mean (SEM) of each pair. (B) Mean RF of replicate trees from H3N2 viruses from 1995 to 2004 were plotted against those from 2005 to 2014. The line of best fit was determined by linear regression (solid line). The R2 and p-value are indicated. Error bars indicate the SEM of all replicates. (C) Replicate tree distances were plotted comparing H3N2 viruses from 1995 to 2004 (black) to H3N2 viruses from 2005 to 2014 (turquoise). ‘Dissimilar’ pairs are grouped where p < 0.05 (Mann-Whitney U test with Benjamini-Hochberg correction; exact p-values reported in Supplementary file 6). Dashed line, 95% confidence interval for tree similarity (determined by a null dataset; refer to Figure 2—figure supplement 3).
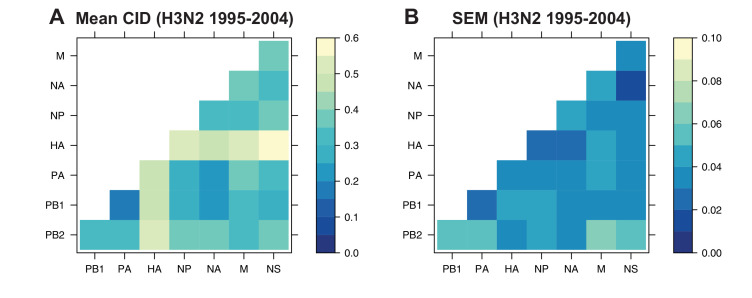
Figure 3—figure supplement 1. The standard error of the mean (SEM) of replicate Robinson-Foulds distances (RF).
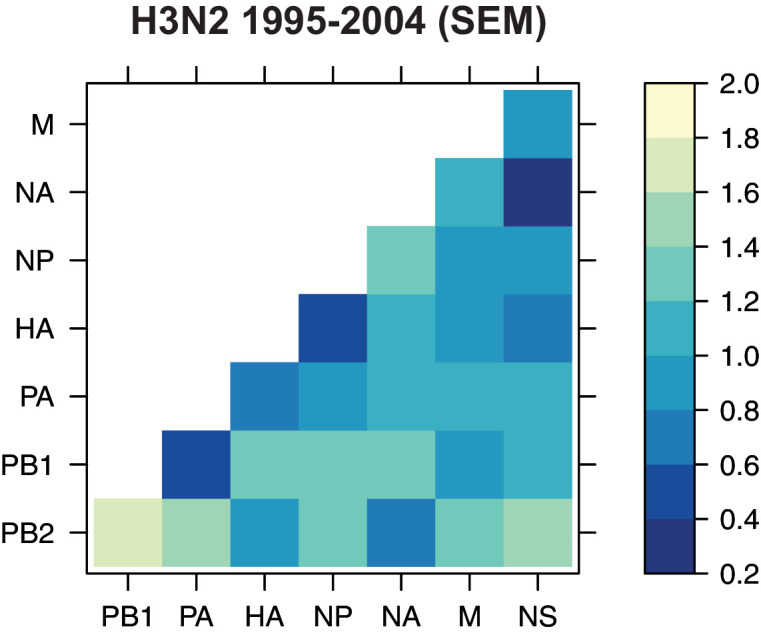
Figure 3—figure supplement 2. The mean clustering information distance (CID) of replicate viral RNA (vRNA) trees.
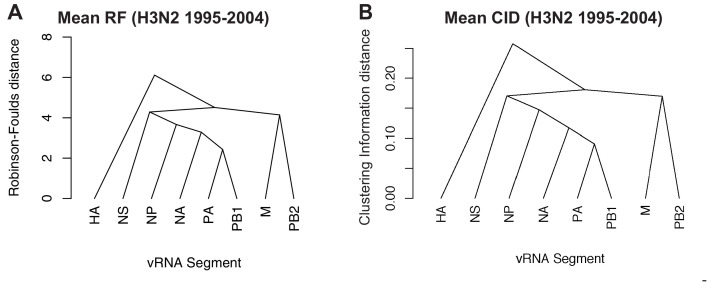
Figure 3—figure supplement 3. Networks determined from pairwise tree distances.
Figure 3—figure supplement 4. Linear regression of tree distances determined by clustering information distances (CID).