Abstract
STUDY QUESTION
Can a targeted whole exome sequencing (WES) on a cohort of women showing a primary ovarian insufficiency (POI) phenotype at a young age, combined with a study of copy number variations, identify variants in candidate genes confirming their deleterious effect on ovarian function?
SUMMARY ANSWER
This integrated approach has proved effective in identifying novel candidate genes unveiling mechanisms involved in POI pathogenesis.
WHAT IS KNOWN ALREADY
POI, a condition occurring in 1% of women under 40 years of age, affects women’s fertility leading to a premature loss of ovarian reserve. The genetic causes of POI are highly heterogeneous and several determinants contributing to its prominent oligogenic inheritance pattern still need to be elucidated.
STUDY DESIGN, SIZE, DURATION
WES screening for pathogenic variants of 41 Italian women with non-syndromic primary and early secondary amenorrhoea occurring before age 25 was replicated on another 60 POI patients, including 35 French and 25 American women, to reveal statistically significant shared variants.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The Italian POI patients’ DNA were processed by targeted WES including 542 RefSeq genes expressed or functioning during distinct reproductive or ovarian processes (e.g. DNA repair, meiosis, oocyte maturation, folliculogenesis and menopause). Extremely rare variants were filtered and selected by means of a Fisher Exact test using several publicly available datasets. A case-control Burden test was applied to highlight the most significant genes using two ad-hoc control female cohorts. To support the obtained data, the identified genes were screened on a novel cohort of 60 Caucasian POI patients and the same case-control analysis was carried out. Comparative analysis of the human identified genes was performed on mouse and Drosophila melanogaster by analysing the orthologous genes in their ovarian phenotype, and two of the selected genes were fruit fly modelled to explore their role in fertility.
MAIN RESULTS AND THE ROLE OF CHANCE
The filtering steps applied to search for extremely rare pathogenic variants in the Italian cohort revealed 64 validated single-nucleotide variants/Indels in 59 genes in 30 out of 41 screened women. Burden test analysis highlighted 13 ovarian genes as being the most enriched and significant. To validate these findings, filtering steps and Burden analysis on the second cohort of Caucasian patients yielded 11 significantly enriched genes. Among them, AFP, DMRT3, MOV10, FYN and MYC were significant in both patient cohorts and hence were considered strong candidates for POI. Mouse and Drosophila comparative analysis evaluated a conserved role through the evolution of several candidates, and functional studies using a Drosophila model, when applicable, supported the conserved role of the MOV10 armitage and DMRT3 dmrt93B orthologues in female fertility.
LARGE SCALE DATA
The datasets for the Italian cohort generated during the current study are publicly available at ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/): accession numbers SCV001364312 to SCV001364375.
LIMITATIONS, REASONS FOR CAUTION
This is a targeted WES analysis hunting variants in candidate genes previously identified by different genomic approaches. For most of the investigated sporadic cases, we could not track the parental inheritance, due to unavailability of the parents’ DNA samples; in addition, we might have overlooked additional rare variants in novel candidate POI genes extracted from the exome data. On the contrary, we might have considered some inherited variants whose clinical significance is uncertain and might not be causative for the patients’ phenotype. Additionally, as regards the Drosophila model, it will be extremely important in the future to have more mutants or RNAi strains available for each candidate gene in order to validate their role in POI pathogenesis.
WIDER IMPLICATIONS OF THE FINDINGS
The genomic, statistical, comparative and functional approaches integrated in our study convincingly support the extremely heterogeneous oligogenic nature of POI, and confirm the maintenance across the evolution of some key genes safeguarding fertility and successful reproduction. Two principal classes of genes were identified: (i) genes primarily involved in meiosis, namely in synaptonemal complex formation, asymmetric division and oocyte maturation and (ii) genes safeguarding cell maintenance (piRNA and DNA repair pathways).
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by Italian Ministry of Health grants ‘Ricerca Corrente’ (08C621_2016 and 08C924_2019) provided to IRCCS Istituto Auxologico Italiano, and by ‘Piano Sostegno alla Ricerca’ (PSR2020_FINELLI_LINEA_B) provided by the University of Milan; M.P.B. was supported by Telethon-Italy (grant number GG14181). There are no conflicts of interest.
Keywords: whole-exome sequencing, primary ovarian insufficiency, case-control analysis, Drosophila comparative analysis and modelling, gene functional categorisation
Introduction
Primary ovarian insufficiency (POI) is a heterogeneous condition occurring in 1% of women under 40 years of age associated with both syndromic and non-syndromic forms. The POI disorder affects women’s fertility, leading to a premature loss of ovarian reserve, and is clinically characterised by primary or secondary amenorrhoea and hypergonadotropic hypogonadism. Genetic defects at the basis of POI, which cover at least half of POI aetiology, have been investigated and mainly detected in genes with a role in meiosis, gonadal development, DNA repair, hormonal signalling or transcriptional regulation, thus identifying POI as a complex genetically heterogeneous disorder resulting from defects in multiple complementary pathways (Bouilly et al., 2016). In recent years, array-comparative genomic hybridisation (array-CGH) or single-nucleotide polymorphism array (SNP-array) in cohorts of POI patients (Aboura et al., 2009; Dudding et al., 2010; Ledig et al., 2010; Quilter et al., 2010; Knauff et al., 2011; McGuire et al., 2011; Zhen et al., 2013; Norling et al., 2014; Jaillard et al., 2016; Tšuiko et al., 2016; Bestetti et al., 2019) and next-generation sequencing targeted to multigene panels or to the whole exome sequencing (WES) mainly performed on familial cases have shed light on new candidate POI genes (Caburet et al., 2014; de Vries et al., 2014; Qin et al., 2015; Bramble et al., 2016; Fauchereau et al., 2016; Carlosama et al., 2017; Moriwaki et al., 2017; He et al., 2019; Alvarez‐Mora et al., 2020). Furthermore, with the expanding application of gene panels and WES, data from several cohorts of patients have been processed in a search for pathogenic variants in genes known to be involved in POI pathogenesis or in ovarian function (Fonseca et al., 2015; Laissue, 2015; Bouilly et al., 2016; Desai et al., 2017; Tucker et al., 2018). However, only three of these studies have been performed on a large number of genes in cohorts of unrelated (Patiño et al., 2017; Jolly et al., 2019) and related POI patients (Yang et al., 2019).
In several cases, POI genetic defects have been identified, but many patients still lack a molecular diagnosis, suggesting that additional unknown genes are implicated in POI aetiology. Thus, the combination of the two types of genomic techniques might help to identify the molecular defects, as has been successfully proven (Tšuiko et al., 2016).
Diverse animal models have been used to investigate the role in the ovarian function of the genes identified by the above-mentioned approaches (França and Mendonca, 2019). Indeed, the use of animal models may be an important tool to gain insights into the POI condition as the basic mechanisms have not been completely elucidated. In particular, Drosophila melanogaster is considered a good model for the POI and many studies have documented the role of several genes in ovary development and function by studying the variability of ovariole number that has a direct role in the number of eggs produced (Lobell et al., 2017).
Here, we report on the first WES study on a large cohort (n = 41) of unrelated Italian POI patients (IPOI cohort) which enabled us to appoint potentially relevant variants in 542 selected genes, 48 of which were identified by our previous array-CGH study (Bestetti et al., 2019). Interestingly, a case-control study using publicly available datasets of controls and an ad-hoc cohort of Italian healthy females revealed a statistically significant correlation between POI and a number of variants and genes, and these data were further corroborated by screening 60 additional Caucasian POI patients.
Moreover, some of the identified genes were subject to comparative analysis in mouse and Drosophila and two of the most interesting orthologues were modelled in the fruit fly to test their effect on ovary structure and function. The armitage gene, the human MOV10 orthologue, coding for an RNA helicase and acting in germline and follicular cells, was identified by testing female fertility in very rare homozygous or trans-heterozygous armi mutants displaying complete sterility (Cook et al., 2004) and DMRT3 was recently identified as a candidate ovariole number gene (Lobell et al., 2017).
Finally, the combined genomic and functional approaches allowed us to confirm the maintenance across the evolution of some key genes safeguarding fertility and successful reproduction.
The overall study confirms that multiple genetic determinants contribute to POI. The extremely heterogeneous oligogenic basis of POI also emerges from the finding that a small number of patients from our Italian cohort were positive for both rare single-nucleotide variants (SNVs or Indels) and ovary-related copy number variants (CNVs; Bestetti et al., 2019). The identification of rare significant variants in patients negative for known CNVs points to the need for multiple genome-wide approaches to discover the molecular defects in POI patients and to enhance the definition of the molecular basis of POI.
Materials and methods
Study design
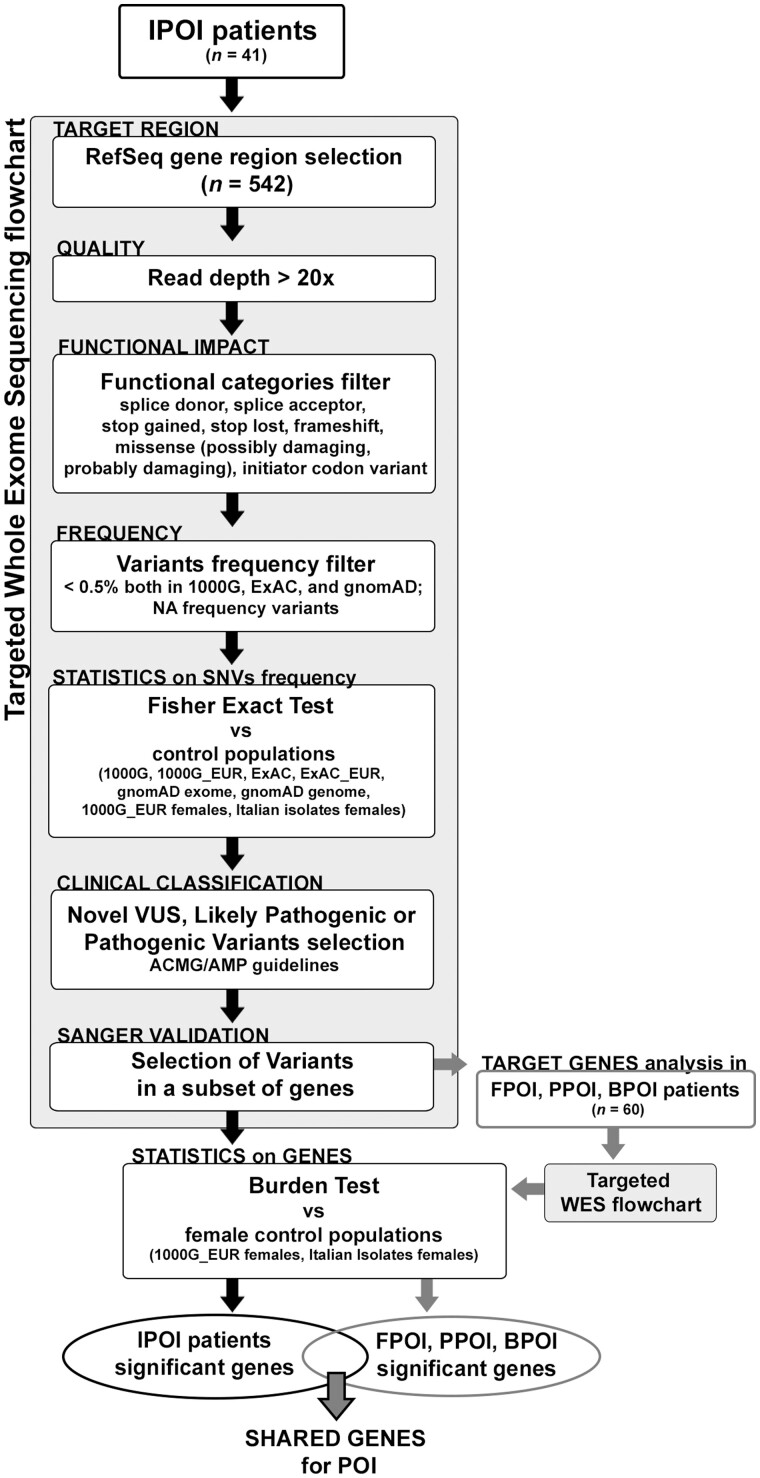
An Italian cohort of patients affected by early secondary amenorrhoea was collected and screened for pathogenic variants using a targeted-WES approach. As shown in Fig. 1, filtering steps were adopted to select the most promising variants. A Fisher Exact test was used to select the extremely rare variants comparing their frequency in several publicly available datasets. A Burden test was applied to highlight the most significant genes in two ad-hoc control female cohorts. To corroborate the obtained data, genes harbouring the validated and significant rare variants identified in the initial Italian cohort were sought in a novel cohort of POI patients to identify other pathogenic alterations. Genes identified as being significant by the Burden test in both patients’ cohorts were considered strong candidate genes for POI (Fig. 1), and their mouse and Drosophila orthologues were analysed in their ovarian phenotype. When applicable, a few were also functionally modelled in Drosophila to explore their role in fertility.
Figure 1.
Flowchart of the study design: filtering steps of the targeted-WES analysis. The genes with significant and Sanger validated SNVs/Indels identified in the Italian cohort (IPOI) were screened using the same approach in a second cohort of French American POI patients (FPOI, PPOI, BPOI). The genes appointed in both cohorts were subject to Burden test analysis and the shared genes were selected as candidates for POI. POI, primary ovarian insufficiency; SNV, single-nucleotide variant; WES, whole exome sequencing.
Ethical approval and consent to participate
Written informed consent to the research investigation, approved by the Ethical Clinical Research Committee of the San Raffaele Research Institute, was obtained from either the adult patients or the parents of underage patients.
Patients and controls
A total of 41 unrelated Italian Caucasian patients affected by non-syndromic POI were referred to the Division of Genetics and Cell Biology of the San Raffaele Research Institute in Milan and each gave their informed consent to the study. The patients (mean age 35 ± 8 years) presented with physiological primary (PA, n = 17) or early secondary (SA, n = 24) amenorrhoea occurring before age 25. The mean age at diagnosis was 20 ± 6 years and the hormone levels in the analysed patients were in the postmenopausal range (FSH >40 UI/l and/or estradiol <30 pg/ml). All patients were considered idiopathic because they did not have any POI-related conditions such as ovarian surgery or previous chemo- or radio-therapy. Of the 41 patients, 36 had 46,XX karyotype and/or were negative for FMR1 pre-mutation, while data were not available for 5 patients. The 17 primary amenorrhoea patients had previously been screened by array-CGH analysis (Bestetti et al., 2019). Clinical details of all patients are reported in Supplementary Table SI. WES data of an additional 60 POI patients, including 35 French women (FPOI; Yang et al., 2019) and 25 American women (19 in the PPOI and 6 in the BPOI cohorts), were collected at the Magee-Womens Research Institute of Pittsburgh, PA, USA and sent for targeted gene screening to have a replica of the Italian POI data.
For the case-control analysis, available databases of 1000 Genomes Project phase 3 (1000GP3, IGSR: The International Genome Sample Resource. https://www.internationalgenome.org/home, accessed 14 December 2018), Exome Aggregation Consortium (ExAC, Cambridge, MA) and Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org, accessed 10 October 2019) were used (1000 Genomes Project Consortium, 2015; Lek et al., 2016; IGSR, 2018; Karczewski et al., 2020). Both the total and the European (or non-Finnish) population frequencies were consulted for statistical analysis. In addition, because males of unknown age are also included, sequencing data and frequencies of 263 European females were extracted from 1000GP3 and WES data (except for X chromosome) of 589 Italian healthy females from genetically isolated populations (Val Borbera, Friuli Venezia Giulia, Carlantino) and from Tuscany (1000GP3; Colonna et al., 2013) were also considered to have the best control cohorts for POI.
WES on Italian POI patients
Patients’ DNA was collected at the Division of Genetics and Cell Biology of the San Raffaele Research Institute. WES was performed on a HiSeq 2500 (Illumina, San Diego, CA, USA) at Magee Clinical Genomics Laboratory, Pittsburgh, PA, USA. Agilent SureSelect Human All Exon V5+UTRs Capture Kit (Agilent Technologies, Santa Clara, CA, USA) was used for exome capture. The average coverage of WES was 100× reads on the target regions of the capture kit (the quality scores of each sample are available upon request). Fastq data were aligned to the reference genome assembly GRCh37/hg19 using Burrows–Wheeler Aligner (Li and Durbin, 2009). For variant calling, GATK Haplotype Caller (Broad Institute, Cambridge, MA, USA) was used. VCF files were filtered based on 542 RefSeq genomic location of (i) genes known to be previously associated with female infertility, (ii) candidate genes identified by our previous array-CGH analysis on early-onset POI patients (Bestetti et al., 2019), (iii) known interactors of the strongest candidate array-CGH genes, (iv) genes with a role in ovary and (v) menopausal genes (Day et al., 2017; Supplementary Table SII). Variants in the target genomic regions were annotated using variant effect predictor (McLaren et al., 2016).
Variants filtering
A schematic view of the filtering step is shown in Fig. 1. In detail, variants with a coverage of total reads <20× were excluded. A filter for seven functional variant categories (missense, splice donor, splice acceptor, stop gained, stop lost, frameshift, initiator codon variant) considering an effect on the canonical gene transcripts was applied. Missense variants with a ‘possibly damaging’ or ‘probably damaging’ prediction based on PolyPhen-2 and SIFT were included. Searching for rare variants, a minor allele frequency (MAF) <0.5% in 1000GP3, ExAC and gnomAD was considered and variants never reported in these data sets were included. To select extremely rare variants, a case-control Fisher Exact test was performed using R.3.1.0 software. In detail, the frequency of the variants detected in the IPOI cohort was compared to the frequency of variants in the three publicly available databases of controls, i.e. 1000GP3, ExAC (both total and European/non-Finnish frequencies) and gnomAD (both exome and genome). In addition, the European females (n = 263, 1000GP3) and healthy females of Italian genetic isolates (n = 589) were also considered as control populations and their frequencies were determined according to the allele number of each population (i.e. 526 for 1000G European female, and 1178 for Italian females). The odds ratio (OR) with the 95% confidence interval and the adjusted P-value using Benjamini and Hochberg’s method were calculated for each variant. A P-value <0.05 assigned a statistically significant test. Only never reported and rare variants with a statistically significant frequency compared to each control population were considered.
Variant clinical classification and validation
SNVs/Indels not present in the 1000GP3, ExAC, gnomAD or the single-nucleotide polymorphism database (dbSNP; Sherry et al., 1999) were considered novel variants. Each gene variant was classified as benign, likely benign, of uncertain significance (VUS), likely pathogenic (LP) or pathogenic (P), according to the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG/AMP; Richards et al., 2015) and the related InterVar and Varsome software were consulted for variant classification (Li and Wang, 2017; Kopanos et al., 2019). Nonsense and frameshift variants in a gene where the loss of function is a known mechanism of disease (with caution for variants at the 3ʹ end of the gene) were recognised as very strong evidence for pathogenicity (PSV1). Very rare variants where the case-control study (Fisher Exact test) reached statistical significance, relative OR was >5.0 and the confidence interval around the OR did not include 1.0, were strongly recognised as evidence for pathogenicity (PS4) and the clinical classification was manually adjusted. Novel VUS, likely pathogenic or pathogenic variants in IPOI patients were selected for validation by Sanger sequencing. PCR reactions were performed using the AmpliTaq Gold® kit (Thermo Fisher Scientific) and the amplicons were purified and sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). Electropherograms were analysed using ChromasPro 1.5 software (Technelysium Pty Ltd., Tewantin, QLD, Australia). A full list of primers is available upon request.
Statistical analysis with Burden test
Association between rare variants and POI phenotype was investigated by a gene-based Burden test using Rvtest (Morris and Zeggini, 2010; Zhan et al., 2016). The analysis confers a P-value to each gene tested by comparing the number and frequency of rare variants between cases and controls. In detail, only the genes with likely pathogenic, pathogenic or never reported VUS variants were considered for Burden tests. As control populations, the two ad-hoc female cohorts of 1000GP3 European and Italian isolates were selected for statistical testing. Each variant in the control populations was selected and filtered as previously performed for the IPOI patients sequencing data, searching for rare possibly pathogenic SNVs/Indels in the target genes. For Burden test analysis, a merged file between patients and control groups was built up. In the event that a variant was rare in a group but common (MAF >0.5%) in the other, both frequencies were included in the lists. The Bonferroni level of significance was applied.
Evaluation of IPOI identified genes on an independent Caucasian cohort of POI patients
The genes identified in IPOI patients with never reported VUS and likely pathogenic or pathogenic rare variants statistically significant for Fisher test were sought in the WES reads of 60 other POI patients to replicate our findings. VCF files were annotated and variants were filtered and classified as previously described. Rare variants present in more than four individuals were filtered out as probably being caused by polymorphic reads alignment. A Fisher exact test to select extremely rare variants and a gene-based Burden test were performed as mentioned above.
Comparative analysis between human, mouse and Drosophila orthologues
The presence of mouse or Drosophila orthologues among the 59 genes identified in the IPOI cohort was assessed using the MGI database (http://www.informatics.jax.org, accessed February 2021), FlyBase database (https://flybase.org, accessed June 2020) and literature data.
Drosophila strains and rearing conditions
Drosophila strains used in this work are listed below.
‘armi1/TM3’: the flies in this strain exhibit a mutation in the armitage gene on the third chromosome balanced with TM3, Sb chromosome. The stock number is BL8513 coming from Bloomington Center.
‘armiKG04664/TM3’: the flies of this strain exhibit a mutation in the armitage gene on the third chromosome balanced with TM3, Sb chromosome. The stock number is BL13979 coming from Bloomington Center.
‘dmrt93b-RNAi’: the flies of this strain express dsRNA for RNAi of dmrt93B (FBgn0038851) under UAS control. The RNAi construct is localised on the third chromosome and the stock number is BL27657.
‘C135-Gal4’: the flies in this strain express Gal4 protein (that binds UAS sequences) in specific follicle cells and in other tissues (larval brain, eye disc, gut, fat body, adult ovarian squamous, male access glands, seminal vesicles, ejaculatory duct, cyst cells and spermatocytes). The construct is localised on the third chromosome and the stock number is BL6978.
‘nanos-Gal4’: the flies in this strain express Gal4 in germ cells. The construct is localised on the third chromosome and the stock number is BL32563.
Flies were reared in plastic vials containing standard cornmeal food (100 g corn flour, 100 g sugar, 8 g agar, 100 g dried yeast powder, 3% Methyl para hydroxy benzoate in ethanol, for 1 l). All the fly stocks were maintained at 25°C.
Female fertility test
A total of 20 mutant females and controls were tested to control males. One individual female was mated with two males in each vial. After 4 days, the crosses were transferred to a fresh vial. The parental flies were removed from the last vial after an additional 4 days. The number of adult progeny from each vial was counted. A Student’s t-test was applied to evaluate differences between the two groups of data.
Drosophila ovary staining
Drosophila gonads were dissected in PBS 1X and fixed with 4% paraformaldehyde for 20 min, washed with PBT (1 × PBS with 0.5% Triton X-100), blocked with 5% Normal Goat Serum for 1 h and incubated with DAPI (100 ng/ml) for DNA labelling. Samples were examined at 40× magnification and captured using a laser-scanning confocal microscope (Zeiss LSM 700 on Axio imager M2).
Results
Targeted WES and statistical analyses on Italian POI patients
A targeted approach analysis of the WES data was adopted since no parental information was available to filter out inherited variants.
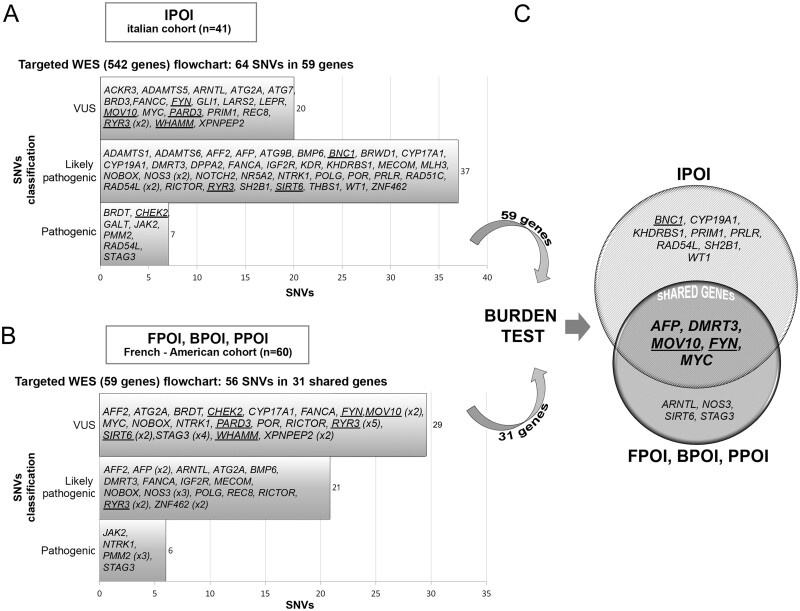
In the subset of 542 selected genes, 136 350 variants were identified in 41 POI Italian patients. Quality filtration revealed a total of 58 067 variants and 428 variations that satisfied the functional categories. Among them, 189 had either an MAF <0.5% or were never reported in 1000GP3, ExAC and gnomAD. Based on the Fisher Exact test and ACMG/AMP guidelines pathogenic, likely pathogenic and never reported variants were selected (n = 75) and 64 were validated by Sanger sequencing and submitted to the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar, accessed June 2020. Accession numbers SCV001364312 to SCV001364375; ClinVar database).
Out of the validated variants identified in 30 IPOI patients and involving 59 genes, 21 were novel and 43 were reported with a statistically significant Fisher test. There were 7 variants classified as pathogenic, 37 classified as likely pathogenic (1 novel in RAD54L), and 20 (all novel) classified as VUS (Fig. 2A). Regarding the functional impact on the protein, 1 frameshift, 2 stop-gain and 61 missense variants were identified. All SNVs/Indels were found in a heterozygous state except for a compound heterozygous state for REC8 (missense/deletion) and a homozygous condition for BMP6 in IPOI-02 and IPOI-34, respectively. Two SNVs in the NOS3 gene were identified whereas three variations were detected in RAD54L and RYR3. The remaining 53 genes presented only one rare variant each (Table I and Supplementary Table SIII).
Figure 2.
Flowchart of the results. (A) The Italian cohort (IPOI) of 41 individuals was analysed by means of a targeted (542 genes) WES. The upper left horizontal bars show the number of variants identified in IPOI according to the ACMG/AMP classification. The genes bearing the selected alterations (n = 59) are listed inside the bars. (B) The same approach was applied to a second cohort of French and American patients (FPOI, BPOI, PPOI) starting from the 59 genes identified in the Italian cohort. The bottom left histograms show the number of variants identified in FPOI, BPOI and PPOI according to the ACMG/AMP classification. The genes harbouring the selected alterations and shared with the Italian cohort (n = 31) are listed inside the bars. (C) Burden test analysis performed on the genes identified through targeted-WES in both cohorts of patients revealed significant genes in each cohort, which are shown in the Venn diagram. The shared significant genes are in bold and upper case letters inside the intersection. Genes identified by array-CGH analysis (Bestetti et al., 2019) and the genes encoding for interactors of the main array-CGH candidates, are shown underlined. ACMG/AMP, American College of Medical Genetics and Genomics; CGH comparative genomic hybridisation; POI, primary ovarian insufficiency; WES, whole exome sequencing.
Table I.
List of variants selected in the Italian POI cohort through targeted-WES.
| Gene | RefSeq | Coding cDNA | Protein | dbSNP | ACMG/AMP guidelines | ID IPOI |
|---|---|---|---|---|---|---|
| ACKR3 d | NM_020311 | c.T1003G | p.S335A | NA | VUS | IPOI28 |
| ADAMTS1 d | NM_006988 | c.C1304A | p.S435Y | rs375553171 | LP | IPOI37 |
| ADAMTS5 d | NM_007038 | c.G2176A | p.G726R | NA | VUS | IPOI03 |
| ADAMTS6 d | NM_197941 | c.G2840A | p.R947Q | rs1272612301 | LP | IPOI36 |
| AFF2 c | NM_001170628 | c.C2126G | p.S709W | rs188208167 | LP | IPOI24 |
| AFP d | NM_001134 | c.G1822A | p.G608R | rs146692547 | LP | IPOI22 |
| ARNTL d | NM_001030272 | c.G18C | p.M6I | NA | VUS | IPOI14 |
| ATG2A d | NM_015104 | c.C1954A | p.L652M | NA | VUS | IPOI40 |
| ATG7 d | NM_001144912 | c.C1144T | p.L382F | NA | VUS | IPOI07 |
| ATG9B d | NM_001317056.1 | c.1480G>T | p.E494X | rs996929151f | LP | IPOI04 |
| BMP6 d | NM_001718 | c.C409Af | p.L137M | rs747427445 | LP | IPOI34 |
| BNC1 a | NM_001301206 | c.C2252T | p.T751I | rs760137127 | LP | IPOI34 |
| BRD3 d | NM_007371 | c.G1149A | p.M383I | NA | VUS | IPOI06 |
| BRDT d | NM_001242807 | c.1400delA | p.D467fs | rs1188709614 | P | IPOI01 |
| BRWD1 d | NM_018963 | c.G656A | p.R219H | rs372595428 | LP | IPOI08 |
| CHEK2 b , e | NM_007194 | c.A1169C | p.Y390S | rs200928781 | P | IPOI20 |
| CYP17A1 c | NM_000102 | c.T644G | p.V215G | rs1428700861 | LP | IPOI37 |
| CYP19A1 c | NM_001347256 | c.A383G | p.H128R | rs375975652 | LP | IPOI22 |
| DMRT3 d | NM_021240 | c.C1327T | p.P443S | rs764103256 | LP | IPOI40 |
| DPPA2 d | NM_138815 | c.T833C | p.L278S | rs1353278679f | LP | IPOI19 |
| FANCA d | NM_000135 | c.C3665T | p.P1222L | rs374537936 | LP | IPOI19 |
| FANCC d | NM_000136 | c.T1027G | p.Y343D | NA | VUS | IPOI10 |
| FYN e | NM_153047 | c.C206G | p.S69C | NA | VUS | IPOI36 |
| GALT d | NM_001258332 | c.C340T | p.R114C | rs111033750 | P | IPOI26 |
| GLI1 d | NM_001160045 | c.T1843C | p.Y615H | NA | VUS | IPOI19 |
| IGF2R d | NM_000876 | c.C451T | p.H151Y | rs756631085 | LP | IPOI36 |
| JAK2 d | NM_004972 | c.G436A | p.D146N | rs1299892808 | P | IPOI45 |
| KDR d | NM_002253 | c.C724T | p.L242F | rs587778428 | LP | IPOI25 |
| KHDRBS1 d | NM_001271878 | c.C1145T | p.P382L | rs750291697 | LP | IPOI02 |
| LARS2 d | NM_015340 | c.A326G | p.D109G | NA | VUS | IPOI36 |
| LEPR d | NM_001198687 | c.C709T | p.P237S | NA | VUS | IPOI28 |
| MECOM d | NM_001163999 | c.G2431A | p.G811S | rs767306816 | LP | IPOI37 |
| MLH3 d | NM_001040108 | c.A1387C | p.S463R | rs138974583 | LP | IPOI33 |
| MOV10 a | NM_001130079 | c.A2831G | p.Y944C | NA | VUS | IPOI09 |
| MYC d | NM_002467 | c.G478A | p.V160I | NA | VUS | IPOI04 |
| NOBOX c | NM_001080413 | c.G1440C | p.K480N | rs1006463439 | LP | IPOI37 |
| NOS3 d | NM_001160109 | c.G505A | p.E169K | rs765854160 | LP | IPOI25 |
| NOS3 d | NM_001160109 | c.C172T | p.P58S | rs752309888 | LP | IPOI26 |
| NOTCH2 d | NM_024408 | c.G5105A | p.R1702Q | rs999822357 | LP | IPOI33 |
| NR5A2 d | NM_001276464 | c.T465G | p.H155Q | rs749896579 | LP | IPOI10 |
| NTRK1 d | NM_001012331 | c.G2101A | p.E701K | rs747855434 | LP | IPOI47 |
| PARD3 a | NM_001184791 | c.C2854G | p.Q952E | NA | VUS | IPOI03 |
| PMM2 c d | NM_000303 | c.C323T | p.A108V | rs200503569 | P | IPOI35 |
| POLG b c | NM_001126131 | c.G1685A | p.R562Q | rs781168350 | LP | IPOI14 |
| POR d | NM_000941 | c.C1588T | p.P530S | rs375387233 | LP | IPOI03 |
| PRIM1 b | NM_000946 | c.G873T | p.W291C | NA | VUS | IPOI40 |
| PRLR d | NM_001204314 | c.T548G | p.L183W | rs748942718 | LP | IPOI21 |
| RAD51C d | NM_058216 | c.A1090G | p.S364G | rs587782565 | LP | IPOI26 |
| RAD54L b | NM_003579 | c.C1138T | p.R380W | rs150138364 | P | IPOI13 |
| RAD54L b | NM_003579 | c.A1883C | p.E628A | NA | LP | IPOI13 |
| RAD54L b | NM_003579 | c.C1900T | p.R634C | rs368491231 | LP | IPOI40 |
| REC8 d | NM_001048205 | c.A1057Cf | p.T353P | NA | VUS | IPOI02 |
| RICTOR d | NM_001285439 | c.A1325G | p.H442R | rs1329438711 | LP | IPOI30 |
| RYR3 a | NM_001036 | c.C2567G | p.T856S | NA | VUS | IPOI02 |
| RYR3 a | NM_001036 | c.A3092T | p.K1031M | rs753104655 | LP | IPOI03 |
| RYR3 a | NM_001036 | c.G6604A | p.A2202T | NA | VUS | IPOI21 |
| SH2B1 d | NM_015503 | c.T1846C | p.S616P | rs142515048 | LP | IPOI12 |
| SIRT6 a | NM_001321058 | c.G146A | p.R49H | rs771714154 | LP | IPOI44 |
| STAG3 c | NM_001282718 | c.C895T | p.R299X | rs774733445 | P | IPOI36 |
| THBS1 d | NM_003246 | c.C1060T | p.P354S | rs142519614 | LP | IPOI01 |
| WHAMM a | NM_001080435 | c.C734T | p.P245L | NA | VUS | IPOI01 |
| WT1 d | NM_000378 | c.A970G | p.S324G | rs121907908g | LP | IPOI36 |
| XPNPEP2 d | NM_003399 | c.G1081A | p.E361K | NA | VUS | IPOI03 |
| ZNF462 d | NM_021224 | c.C3515G | p.P1172R | rs1469441260 | LP | IPOI21 |
All variants reported had frequencies significant for Fisher Exact Test or were never reported in the population databases investigated (1000G, ExAC and gnomAD).
Genes identified through array-CGH analysis of primary amenorrhoea patients (Bestetti et al., 2019).
Menopausal genes (Colonna et al., 2013).
OMIM POI genes or genes previously associated to POI.
Other ovary-related genes(Colonna et al., 2013; Patiño et al., 2017).
Interactor of the most promising candidates from previous array-CGH analysis (Bestetti et al., 2019).
BMP6: homozygous variant; REC8: compound heterozygous variant.
Frequency reported in dbSNP with <0.00001 MAF.
dbSNP, single-nucleotide polymorphism database; LP, likley pathogenic; MAF, minor allele frequency; NA, not available; P, pathogenic; POI, primary ovarian insufficiency; VUS, variant of uncertain significance.
Out of 30 POI patients with candidate variants, 12 carried only 1 SNV, while the remaining 18 had multiple variants. No differences between PA and SA in the number of variants (one or >1), were observed. Seven patients were identified with predicted pathogenic variants: IPOI-1 had a frameshift in the ovary-related gene BRDT (rs1188709614); IPOI-13 and IPOI-20 had missense variants in the menopausal genes RAD54L (rs150138364) and CHEK2 (rs200928781), respectively; IPOI-26, IPOI-35 and IPOI-45 presented with non-synonymous SNVs in the ovary-related genes GALT (rs111033750), PMM2 (rs200503569) and JAK2 (rs1299892808); and IPOI-36 was a carrier of a stop gain in the POI-OMIM STAG3 gene (rs774733445; Fig. 2A).
In the present cohort, 17 primary amenorrhoea patients were previously screened by array-CGH (Bestetti et al., 2019). In this group, seven PA patients presented both rare selected SNVs/Indels (1–3) and ovary-related CNVs, whereas seven and three PA patients presented only rare selected SNVs/Indels and ovary-related CNVs, respectively (Supplementary Fig. S1).
The selected 64 variants identified in the Italian POI cohort were not present in the two investigated ad-hoc female control cohorts, supporting their actual rarity and consistent with the Fisher Exact test that showed a significantly rare frequency compared to the publicly accessed databases (Table I and Supplementary Table SIII).
To further validate the results obtained from the Italian POI cohort, a second cohort of 60 POI patients with early secondary amenorrhoea was screened to search for pathogenic variants in the selected 59 genes identified in IPOI. We used the same flowchart and revealed in 32 patients 56 variants: 31 were never reported and for 25 of them a significance for the Fisher test was obtained. The 56 variants were identified in 31 out of the selected 59 genes: 6 classified as pathogenic, 21 classified as likely pathogenic and 29 classified as VUS (Fig. 2B). With a single exception, variants were found in a heterozygous state and included 52 missense, 2 frameshift, 1 splicing and 1 stop-gain. None of the alterations were shared with the IPOI cohort (Supplementary Table SIII).
As rare variants can be also identified in control populations, to evaluate the frequency of the SNVs/Indels in the selected genes, and pick out the most constrained/candidate genes, we applied a Burden test for rare variants. The analysis performed using two ad-hoc selected female populations (1000GP3 and Italian Isolates), revealed 13 and 9 significant genes in the IPOI cohort and FPOI-BPOI-PPOI, respectively. Five genes (AFP, DMRT3, MOV10, FYN and MYC) were significant in both cohorts and were thus considered the most significant candidate genes (Fig. 2C and Supplementary Table SIV). AFP encodes alpha fetoprotein, a plasma protein mainly expressed during foetal life in the yolk sack and liver, and high levels of AFP are present in pregnant women to stop follicular growth (Zong et al., 2019). Concerning DMRT3, double sex and mab-3-related transcription factor 3, its involvement in sexual development is known, but there is scarce knowledge of its specific role in the ovary, whereas MOV10 gene, encoding an RNA helicase phosphorylated after DNA damage, exerts a role in RNA stability and regulation of the network of mammalian germ cells (Fu et al., 2019). Finally, FYN, a proto-oncogene belonging to the membrane-associated Src-like kinase family, encodes a VLDLR interactor (Bestetti et al., 2019) in the Reelin pathway, and the well-known MYC proto-oncogene encodes a transcription factor binding E-box elements involved in cellular proliferation.
Comparative analysis in Mus musculus and D. melanogaster of the human identified POI genes
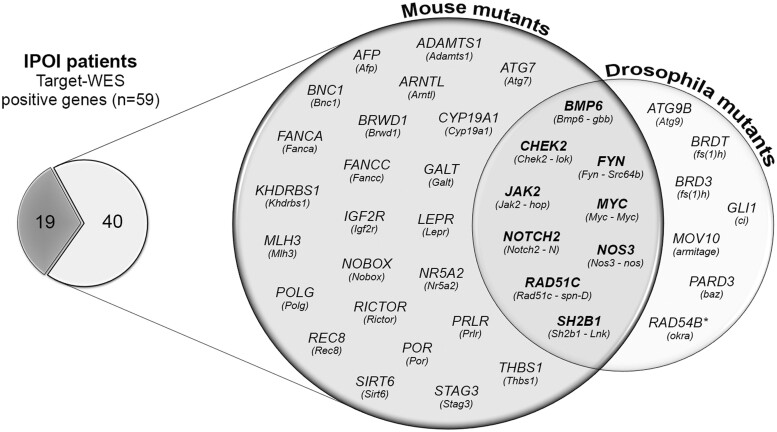
We outlined the mouse and Drosophila genes homologous to those identified in the POI patients to trace the maintenance of their fertility-related function. Among the 59 validated and significant genes disclosed in the Italian cohort, 45 and 25 have a mouse and Drosophila orthologue, respectively, that plays a role during ovarian development, oocyte maturation or DNA repair, hence confirming a conserved function between the three species. Indeed 40 out of 59 orthologue mutants present infertile or sub-fertile phenotypes; the Venn diagram in Fig. 3 shows the 33 mice and the 16 Drosophila orthologous with 9 genes shared by the two species (Supplementary Table SV).
Figure 3.
Diagrams showing the IPOI positive genes with a mouse and/or Drosophila orthologue whose mutants present an infertile/sub-fertile phenotype (n = 40). The shared genes among the two species are shown in bold in the Venn intersection. POI, primary ovarian insufficiency.
Functional studies of MOV10 and DMRT3 in D. melanogaster
In order to support the role of MOV10 and DMRT3 in female fertility and during ovary development, we performed an analysis of the behaviour of D.melanogaster mutants for armitage and in RNAi strains for doublesex-Mab-related 93B, the orthologues of the mammalian MOV10 and DMRT3, respectively.
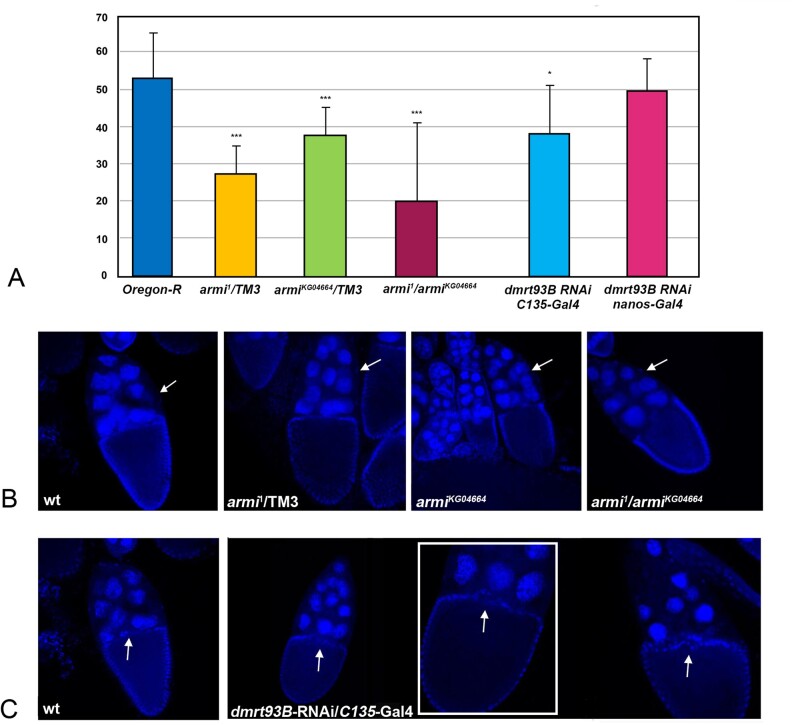
The armi1 and armiKG04664 mutations are ‘loss of function’ mutant alleles characterised by a reduced amount of armitage mRNA. We tested armi1/TM3, armiKG04664/TM3 heterozygote females, and armi1/armiKG04664 trans-heterozygotes. The results of the fertility tests of armi alleles are shown in Fig. 4A where a significant reduction in female fertility is evident in armi1 (51.5% of the control: 27.5 progeny in armi1 heterozygote females vs. 53.4 progeny in control females; P = 0.000013), in armiKG04664 heterozygote females (70.5% of the control: 37.7 progeny in armiKG04664 heterozygote females vs. 53.4 progeny in control females; P = 0.0022) and in armi1/armiKG04664 trans-heterozygotes (37.6% of the control: 21.2 progeny in armi1/armiKG04664 vs. 53.4 progeny in control females; P = 0.00038). We were also interested in investigating the structure of the ovaries in armi alleles armi1, armiKG04664 in heterozygote females and in armi1/armiKG04664 trans-heterozygotes, in order to investigate for alterations and possible cause of failure events during oogenesis. For this purpose, dissected ovaries from heterozygote and trans-heterozygotes mutant females were labelled with DAPI to highlight egg chamber organisation. The analysis demonstrated that the number and structure of ovarioles are apparently the same as in the control, but some of the egg chambers are defective in the number of nurse cells that are fewer (<13) compared to the control (40% in armi1/TM3; 35% in armiKG04664/TM3; 55% in armi1/armiKG04664 trans-heterozygotes) with respect to the control egg chambers (Fig. 4B). In order to count the nurse cells, the images that we analysed report the sum of all the planes throughout the egg chambers.
Figure 4.
Fertility tests and analysis of the egg chambers of armitage mutants and dmrt93B-RNAi. (A) Female fertility in Oregon-R control females, and in armi1/TM3, armiKG04664/TM3, armi1/armiKG04664 trans-heterozygotes, dmrt93B-RNAi/C135-Gal4 and dmrt93B-RNAi/nanos-Gal4 females (data are represented as a mean ± standard deviation, n = 20 females; ***P < 0.001, *P < 0.05: armi1/TM3 P = 0.000013; armiKG04664/TM3 P = 0.0022; armi1/armiKG04664P = 0.00038; dmrt93B-RNAi/C135-Gal4 P = 0.012; dmrt93B-RNAi/nanos-Gal4 P = 0.41). (B) wild type and armi mutants DAPI stained egg chambers, 40× magnification (images correspond to the sum of all planes that guarantees the possibility to count the nurse cells, whose position is indicated by white arrows); (C) wild type and dmrt93B-RNAi/C135-Gal4 DAPI stained egg chambers, 40× magnification (images correspond to a single plan, in order to show the border cells, whose position is indicated by white arrows, at stage 10). The zoom of the egg chamber is showed in the white panel.
The doublesex-Mab-related 93B gene, encoding a transcription factor, might, as recently suggested, be a good candidate for the correct development of female gonads (Lobell et al., 2017). In order to test if dmrt93B gene may be implicated in female fertility, we also analysed dmrt93B-RNAi after crossing the flies with two different Gal4 drivers, the C135-Gal4 and the nanos-Gal4, expressing Gal4 protein in follicle cells and in germ cells, respectively, activating the RNAi construct in these tissues specifically. The results of the fertility tests are shown in Fig. 4A, where a slight reduction of the fertility is evident in C135-Gal4/dmrt93B-RNAi flies (71.5% compared to control females: 38.2 progeny C135/dmrt93B-RNAi vs. 53.4 of the control; P = 0.012) whereas the effect on the fertility of nanos-Gal4/dmrt93B-RNAi females is not evident (92.6% compared to control females: 49.5 vs. 53.4 of the control; P = 0.41). Due to the reduction in fertility reported above, we also analysed the structure of the ovaries of females in which the dmrt93B gene was silenced in somatic cells of the gonads by the C135-Gal4 driver. Interestingly, some (almost 30%) of the developing egg chambers at stage 10 exhibit defects compared to the control (Fig. 4C). The main defect was in the anterior part of the oocyte, where a specific small group of follicle cells, the border cells (arrowed in Fig. 4C which shows a normal egg chamber at stage 10), localises after migration during the earlier stages of development contributing to define the anterior identity to the developing oocyte. We analysed the images throughout all plans of the egg chambers, even though, only a single plan is reported in the figures in order to see the region in which the border cells localise in the wild type.
Discussion
Targeted WES and Drosophila comparative analysis
Integrated genome-wide approaches such as array-CGH and WES have been recently used for POI diagnosis and molecular research (Tšuiko et al., 2016), improving our knowledge of the molecular basis of POI. Furthermore, 17 out of the 41 IPOI patients analysed in this study by means of targeted-WES were previously screened by array-CGH (Bestetti et al., 2019). The combined approach revealed in some cases the co-occurrence of both rare CNVs and SNVs/Indels, confirming the oligogenic inheritance and high genetic heterogeneity of POI disorder, and highlighting the need for complementary genome-wide approaches to identify the molecular basis of POI (Supplementary Fig. S1).
In this framework, our previous array-CGH study turned out to be a valid tool to identify new POI genes in patients with a severe phenotype. Indeed, 44 candidate genes disclosed by array-CGH and 18 genes encoding for interactors of the most significant candidates were included in the gene list used for targeted-WES with the aim of detecting other pathogenic variants and supporting their involvement in severe POI. There were 26 SNVs identified in array-CGH genes among the patients analysed, highlighting a significant enrichment in rare variants in five of them. This approach fostered the research of additional genes that had never previously been investigated, leading to an overall detection rate of 73%, which is significantly higher than those reported in recent studies, 48% (Patiño et al., 2017) and 30% (Yang et al., 2019).
Besides the identification of pathogenic or likely pathogenic variants in OMIM disease genes for POI (GALT, NOBOX, POLG and STAG3) and in genes already POI-associated thus further proving their pathogenetic contribution (AFF2, ATG7, CYP17A1, CYP19A1, NOTCH2, PMM2, WT1 and XPNPEP2), 10 extremely rare SNVs (13%), were detected in 8 array-CGH disclosed genes, namely BNC1, MOV10, PARD3, RYR3, SIRT6, WHAMM plus CHEK2 and FYN which code interactors of the candidate TP63 and VLDLR genes, respectively (Bestetti et al., 2019). Interestingly, out of these genes, BNC1 and WHAMM are in the 15q25.2 POI-specific region that also contains CPEB1, which is involved in synaptonemal complex formation during oocyte maturation (Tay and Richter, 2001) and has been already associated with POI (McGuire et al., 2011; Hyon et al., 2016; Tšuiko et al., 2016; Bestetti et al., 2019). BNC1 (basonuclin1) is a recently confirmed POI causative gene (MIM#618723, POF16) whose haploinsufficiency inhibits human oocyte meiosis by a higher oocyte degradation percentage (Zhang et al., 2018a). Little is known about the ovarian function of WHAMM (WAS protein homologue associated with actin, golgi membranes and microtubules), however, in mouse oocytes, it is important for meiotic spindle migration and asymmetric cytokinesis (Huang et al., 2013).
A special focus should be given to the MOV10 gene, which was found disrupted or altered in two POI patients (IPOI-09 and POI-25; Bestetti et al., 2019). In mouse testis its orthologue, Mov10, is highly expressed in spermatogonia and acts in the maintenance of gene expression by regulating splicing and mRNA processing of spermatogonia progenitor cells (Fu et al., 2019). The function of MOV10 in germ cells is highly conserved, as inferred by validation of its mutant effect in the fruit fly. During germline development Piwi-interacting RNAs (piRNAs) lead a host safeguarding system that transcriptionally and post-transcriptionally silences transposons. Transposons can activate and prompt mutations, DNA breaks and chromosome rearrangements and their silencing is fundamental in the germline, which is dedicated to the transmission of the inherited genome. The Drosophila orthologue, armitage (armi; Cook et al., 2004) is involved in the RNAi pathway, required for genome stability in the ovary, where silencing is needed to establish the oocyte cytoskeleton polarity during early oogenesis. The main role of Armi is in the biogenesis of primary piRNAs in the somatic cells of the gonads (Lim and Kai, 2007; Olivieri et al., 2010), where it interacts with several components of the piRNA pathway, thus being crucial for the genome stability and fertility of both sexes (Megosh et al., 2006; Saito et al., 2010; Bozzetti et al., 2015; Specchia et al., 2017; Cusumano et al., 2019; Durdevic and Ephrussi, 2019; Ge et al., 2019a; Ishizu et al., 2019; Specchia et al., 2019). Indeed, armitage mutants support the early stages in the RNAi pathway but fail in the production of active RNA-induced silencing complex (RISC), which mediates target RNA destruction in RNAi (Tomari et al., 2004). It is also known that mutations affecting piRNA pathways in Drosophila cause infertility/subfertility in both females and males (Atikukke et al., 2014). In addition to literature data, our analysis of two heterozygote alleles of armitage (armi1, armiKG04664) and of trans-heterozigote armi1/armiKG04664 flies support the role of the gene in female gonads function. The Drosophila ovary is composed of strings of developing egg chambers and each egg chamber contains an oocyte and 15 nurse cells. The analysed mutant armi alleles, both heterozygotes and trans-heterozygote, display defects in the number of nurse cells in the egg chambers and a consequent significant reduction of female fertility, supporting the role of genes involved in the piRNA pathway and in DNA repair in the POI phenotype in humans.
Interestingly, one of the genetic interactors of armitage in Drosophila is the loki gene (lok) coding for the orthologue of the human CHEK2, known to be an interactor of the previously identified TP63 array gene (Bestetti et al., 2019) and found mutated in a new familial case of POI (Mathorne et al., 2020). CHEK2, defined as a guardian of female germline integrity (Amelio et al., 2012; Xian and McKeon, 2018), is a multifunctional kinase involved in cell cycle checkpoint regulation, DNA repair and apoptosis (Gebel et al., 2020). After DNA double-stranded breaks (DSBs), it is required to phosphorylate TP63 leading to its activation through the formation of a tetrameric complex that triggers pro-apoptotic factors to induce oocyte apoptosis. Thus, in the context of POI, it is hypothesised that a super active CHEK2 might lead to a premature loss of oocytes thus explaining the effect of secondary POI (Tuppi et al., 2018). Our IPOI-20 patient carries a reported missense variant in CHEK2 (rs200928781, c.A1169C, p.Y390S) classified as pathogenic according to the computational analysis and ACMG/AMP guidelines. This variant localises in exon 10 within the t-loop activation segment in the protein kinase domain, where phosphorylation takes place to trigger its functional activity and currently it appears more likely to inhibit the function of CHEK2 than to make it super active. A possible explanation could be that CHEK2 is also important for inducing DNA repair (Zannini et al., 2014). If this repair mechanism is less efficient, due to the lower activity of CHEK2, more DNA DSBs might persist, likely resulting in TP63-dependent elimination of the oocytes. In this regard, IPOI-20 might have a smaller pool of oocytes to start with, which will be exhausted earlier in life. The mutant females of the Drosophila orthologue, lok, further support its role in ovarian function since they display a reduced fertility. Moreover, in addition to armitage, lok interacts with other piRNA and DNA repair genes (Klattenhoff et al., 2009; Orsi et al., 2010; Specchia et al., 2017), suggesting a link between the piRNA and the DNA repair pathways in both females and males.
Another gene identified in the screening reported in our study is DMRT3 (Doublesex and Mab-3 Related Transcription Factor 3). This is a not surprising finding because in Drosophila other genes involved in sex determination like doublesex affect female fertility. Thus, we also tested flies in which the dmrt93B gene was silenced in the somatic cells of the ovary and demonstrated that female fertility is slightly reduced. Intriguingly these flies show specific defects in the structure of some egg chambers at stage 10, in the region where the border cells normally localise. Border cells are a group of follicular cells that detach from the anterior part of the egg chamber in the early stages of development and migrate towards the anterior boundary of the oocyte, playing an important role in the specification of the antero-posterior axis of the oocyte (He et al., 2011).
The array-CGH appointed gene, FYN, encoding a VLDLR interactor (Bestetti et al., 2019) acts in the Reelin pathway. Reelin is highly expressed in the ovary, mainly in the granulosa cells of growing follicles (Meseke et al., 2018), and the identification of significant unreported variants in POI patients in more than one gene of this pathway (FYN and VLDLR) highlights the contribution of FYN to POI aetiology, supporting its involvement in ovarian maturation. The FYN protein is a tyrosine kinase that, by the way of the Ras pathway and ERK activation, triggers the PI3K pathway in granulosa cells to ensure good quality and maturation of the oocytes. In mammalian oocytes, a role of Fyn in the first meiotic division, in particular at the metaphase checkpoint, has been highlighted (Levi et al., 2010a,b) and Fyn pivotal function in meiotic resumption has recently been demonstrated (Grossman et al., 2017). Furthermore, the male mouse model presents oligospermia and abnormalities of the genital apparatus, while knock-out female mice were found to be sub-fertile due to the quality of their oocytes (Luo et al., 2010): they failed to mature, were small, less competent and did not perform a correct asymmetric division. In addition, Drosophila females mutant for Src64B, the orthologue of FYN, are semi-sterile with oogenesis disruption due to a compromised interaction between nurse cells and the oocyte (Djagaeva et al., 2005).
Among the ovary-related genes herein investigated, special attention should be given to the variants identified in BMP6 and REC8 genes. BMP6 encodes a secreted ligand of the TGF-beta (transforming growth factor-beta) superfamily of proteins, similarly to the OMIM POI gene BMP15, and has a critical role in the regulation of follicular development (Zhang et al., 2018b). The identified variant in exon 1 encodes for the protein segment that needs to be cleaved to perform its proper activity. Several pathogenic prediction tools have classified this variant as damaging and its homozygous state, never previously reported, is strongly suggestive of a pathogenic effect impairing the correct activation of the protein. Dysfunctional BMP6 might be at the basis of the patient phenotype but further studies are needed to confirm this assumption. Mutations of BMP6 in Drosophila are lethal and some studies have revealed that the BMP signal is required for maintaining male and females germ stem cells (Kawase et al., 2004).
A never reported SNV in exon 13 of REC8, a gene involved in meiosis encoding for a member of the cohesin complex (Ishiguro and Watanabe, 2016), and a deletion on the other allele (also never reported) were identified in IPOI-02. Until now only two REC8 heterozygous missense variants have been reported in three patients with POI (Bouilly et al., 2016). Among them, two affected sisters presented with a maternally inherited REC8 variant and a paternal mutation in the OMIM POI gene GDF9. Our compound heterozygous patient, together with the reported data, allow us to speculate that an REC8 single hit might not be sufficient to produce the overt phenotype, which requires an additional hit. Our case is the first evidence of a double-hit event in the gene and further studies are warranted to clarify the pathogenic role of REC8 in POI. As in humans, the Drosophila REC8 orthologue, verthandi (vtd), encoding a subunit of the cohesin complex, has a role during meiosis. In addition, it is also involved in the repair of DSBs (Gaudet et al., 2011) but no specific information regarding the effect of mutant alleles on the female fertility is reported.
Functional classification of the identified genes
As recently postulated (França and Mendonca, 2019), genes involved in meiosis and DNA repair play key roles in POI development and our functional classification of the identified genes is consistent with this assumption. Indeed, the above-mentioned genes BNC1, WHAMM, FYN, MOV10, BMP6 and REC8 can be assigned to the first class as being primarily involved in meiosis, namely in synaptonemal complex formation, asymmetric division and oocyte maturation. PARD3, another array-CGH disclosed gene, can be added to this class as it encodes a member of the PARD protein family involved in the asymmetrical cell division to direct polarised cell growth (Bultje et al., 2009; Chen et al., 2017). In mouse ovaries, Pard3 was found to surround the condensing chromosomes and to associate with the meiotic spindles defining the future site of polar body emission (Duncan et al., 2005). RYR3 can be also assigned to this group of genes as it contributes to the Ca2+ release which is crucial for meiosis resumption and oocyte maturation (Díaz-Muñoz et al., 2008; Toranzo et al., 2014).
Among the DNA repair genes, CHEK2 is definitely to be included, as its defect may also impact on oocyte apoptosis by means of the perturbed interaction with TP63. Moreover, its phosphorylation is facilitated by RAD51C, an early player in DNA repair, thereby leading to the protection of genomic integrity by cell cycle arrest and homologous recombination activation (Badie et al., 2009). Similarly, the MLH3 gene is also implicated in maintaining genomic integrity during DNA replication and after meiotic recombination (Manhart et al., 2017). In addition, the array-CGH SIRT6 gene clusters to this class as it has been recently identified to be a sensor of DSBs activating the DNA damage response to prevent genomic instability (Onn et al., 2020). In particular, it has a role in modulating telomere function in ovaries during oocyte maturation, ensuring a good quality control of oocytes (Ge et al., 2019b) and preventing both spindle morphology defects and chromosome misalignment in mouse oocytes during meiosis (Han et al., 2015). FANCA and FANCC are also well-known to be involved in this process (Niedzwiedz et al., 2004; Benitez et al., 2018); moreover, recent studies have highlighted the implication of JAK2 activating mutations in the development of DNA damage (Karantanos and Moliterno, 2018).
Burden test identification of POI candidate genes
To make further progress in the validation and ranking of the genes identified by the integrated array-CGH and WES approaches on the Italian cohort, we processed a wider French-American POI cohort by targeted WES analysis. We confirmed the presence of significant and new variants in 31 shared genes, highlighting that there are common deregulated pathways in the two cohorts. To identify the most constrained candidate genes, a Burden test was performed and showed five significant genes shared by the two cohorts: FYN, AFP, MYC, DMRT3 and MOV10 (Fig. 2C and Supplementary Table SIV). We have discussed the pre-eminent ovarian roles of FYN, MOV10 and DMRT3 above and have underlined that their function is maintained through evolution, in mammals (Levi et al., 2010a,b; Grossman et al., 2017; Fu et al., 2019) and also in Drosophila (Cook et al., 2004; Djagaeva et al., 2005; Ge et al., 2019a).
As regards the other significant genes, according to the literature, AFP is involved in oocyte maturation as Afp knock-out mice present defects in the reproductive tract, with no ovulation, infertility and anomalies in oocyte morphology, with hyperplasia of the endometrium (MGI: 2450139; Gabant et al., 2002). Indeed, AFP has been shown to be relevant for female fertility being involved in estrogen uptake thus inhibiting the estrogen response and exerting an anti-estrogenic role. In mice, the perinatal excessive exposure to estrogens leads to sterility in the adult without ovulation (De Mees et al., 2007). Accordingly, a defective AFP protein might not properly sequestrate estrogens, leading to poor feminisation in humans too.
Concerning the well-known MYC proto-oncogene, its animal models had already highlighted its role in ovarian fertility. The myc mouse model displays reduced female fertility and abnormality of the ovary (MGI: 2182124; Davis et al., 1993); a similar scenario is observed in the Drosophila model where a loss of activity of the orthologue Myc, with its key role in oogenesis, delays oocyte maturation, growth and DNA replication with size reduction (Maines et al., 2004). Likewise, it may be assumed that in female patients, MYC defects lead to a reduction of ovarian reserve, with oocyte loss and premature ageing.
Due to the heterogeneity of the POI disorder, other genes were found, as expected, to be significant in individual Burden analyses of the two cohorts (Fig. 2C). Among them CYP19A1, involved in the production of estrogens (Kim et al., 2011), BNC1 (Tšuiko et al., 2016; Bestetti et al., 2019), RAD54L and PRIM1, the latter two implicated in menopause, hence linking the onset and end of the reproductive lifespan (Day et al., 2017), and STAG3, encoding a subunit of the cohesin complex essential for proper pairing and segregation of chromosomes during meiosis, which is responsible for Premature Ovarian Failure 8 (MIM#615723). All these genes safeguard oocyte maturation, including RAD54L and PRIM1, which act in basic pathways of DNA repair (Stolk et al., 2012; Ghamrasni et al., 2016).
Conclusions
In conclusion, defects in multiple genes involved in common cell maintenance pathways could lead to the POI phenotype, making it crucial to elucidate the molecular networks involved in ovarian and follicular growth. The omics approaches used in this study and the analysis of some of the genes altered in POI patients in the Drosophila model led to the discovery of new candidate genes and new pathways (piRNA and DNA repair pathways) that affect female fertility and may be strongly implicated in the oligogenic origin of POI. However, further functional studies are needed to clarify and prove the involvement of the identified variants on POI phenotype. Likewise, the contribution of each of the discussed genes to POI, and their epidemiological prevalence require additional studies on larger patient cohorts to be assessed and defined. Hopefully, further research on mammal gonadal defects and human POI should validate these data and elucidate the molecular basis of female fertility.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
The datasets for the Italian cohort generated during the current study are publicly available at ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/): accession numbers SCV001364312 to SCV001364375.
Supplementary Material
Acknowledgements
The authors thank the patients for their participation in this research study on the genetics of primary ovarian insufficiency.
Authors’ roles
I.B.: study design, WES data analysis, statistical analyses, interpretation of data and manuscript draft preparation. C.B.: WES data analysis and statistical analyses. A.S.: variants validation analysis and interpretation of data. V.S.: functional studies in D. melanogaster. S.A.Y.: WES experiments. M.D.D.D.: functional studies in D. melanogaster. C.C.: variants validation analysis. D.G.: bioinformatic processing of WES raw data files. M.C.: variants validation analysis. A.M.: study design, variants validation analysis, comparative analysis of mouse models and critical revision of the manuscript. L.L.: critical revision of the manuscript. A.R.: recruitment of FPOI, PPOI and BPOI cohorts, WES experiments and critical revision of the manuscript. D.T.: recruitment of IPOI cohort, clinical data collection, study design and critical revision of the manuscript. M.P.B.: comparative analysis and functional studies in D. melanogaster, data interpretation, manuscript preparation and critical revision of the manuscript. P.F.: study design and coordination, interpretation of data and critical revision of the manuscript. All authors read and approved the manuscript for submission.
Funding
This study was supported by the Italian Ministry of Health grants ‘Ricerca Corrente’ (08C621_2016 and 08C924_2019) provided to IRCCS Istituto Auxologico Italiano, and by the ‘Piano Sostegno alla Ricerca’ (PSR2020_FINELLI_LINEA_B) provided by University of Milan; M.P.B. was supported by Telethon-Italy (grant number GG14181).
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR.. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboura A, Dupas C, Tachdjian G, Portnoi MF, Bourcigaux N, Dewailly D, Frydman R, Fauser B, Ronci-Chaix N, Donadille B. et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab 2009;94:4540–4546. [DOI] [PubMed] [Google Scholar]
- Alvarez‐Mora MI, Todeschini AL, Caburet S, Perets LP, Mila M, Younis JS, Shalev S, Veitia RA.. An exome-wide exploration of cases of primary ovarian insufficiency uncovers novel sequence variants and candidate genes. Clin Genet 2020;98:293–298. [DOI] [PubMed] [Google Scholar]
- Amelio I, Grespi F, Annicchiarico-Petruzzelli M, Melino G.. p63 the guardian of human reproduction. Cell Cycle 2012;11:4545–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atikukke G, Albosta P, Zhang H, Finley RL Jr. A role for Drosophila Cyclin J in oogenesis revealed by genetic interactions with the piRNA pathway. Mech Dev 2014;133:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie S, Liao C, Thanasoula M, Barber P, Hill MA, Tarsounas M.. RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J Cell Biol 2009;185:587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez A, Liu W, Palovcak A, Wang G, Moon J, An K, Kim A, Zheng K, Zhang Y, Bai F. et al. FANCA promotes DNA double-strand break repair by catalyzing single-strand annealing and strand exchange. Mol Cell 2018;71:621–628.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestetti I, Castronovo C, Sironi A, Caslini C, Sala C, Rossetti R, Crippa M, Ferrari I, Pistocchi A, Toniolo D. et al. High-resolution array-CGH analysis on 46,XX patients affected by early onset primary ovarian insufficiency discloses new genes involved in ovarian function. Hum Reprod 2019;34:574–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouilly J, Beau I, Barraud S, Bernard V, Azibi K, Fagart J, Fèvre A, Todeschini AL, Veitia RA, Beldjord C. et al. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab 2016;101:4541–4550. [DOI] [PubMed] [Google Scholar]
- Bozzetti MP, Specchia V, Cattenoz PB, Laneve P, Geusa A, Sahin HB, Di Tommaso S, Friscini A, Massari S, Diebold C. et al. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J Cell Sci 2015;128:2070–2084. [DOI] [PubMed] [Google Scholar]
- Bramble MS, Goldstein EH, Lipson A, Ngun T, Eskin A, Gosschalk JE, Roach L, Vashist N, Barseghyan H, Lee E. et al. A novel follicle-stimulating hormone receptor mutation causing primary ovarian failure: a fertility application of whole exome sequencing. Hum Reprod 2016;31:905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH.. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron 2009;63:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z, García-Tuñón I. et al. Mutant cohesin in premature ovarian failure. N Engl J Med 2014;370:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlosama C, Elzaiat M, Patiño LC, Mateus HE, Veitia RA, Laissue P.. A homozygous donor splice-site mutation in the meiotic gene MSH4 causes primary ovarian insufficiency. Hum Mol Genet 2017;26:3161–3166. [DOI] [PubMed] [Google Scholar]
- Chen X, An Y, Gao Y, Guo L, Rui L, Xie H, Sun M, Lam Hung S, Sheng X, Zou J. et al. Rare deleterious PARD3 variants in the aPKC-binding region are implicated in the pathogenesis of human cranial neural tube defects via disrupting apical tight junction formation. Hum Mutat 2017;38:378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna V, Pistis G, Bomba L, Mona S, Matullo G, Boano R, Sala C, Viganò F, Torroni A, Achilli A. et al. Small effective population size and genetic homogeneity in the Val Borbera isolate. Eur J Hum Genet 2013;21:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE.. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 2004;116:817–829. [DOI] [PubMed] [Google Scholar]
- Cusumano P, Damulewicz M, Carbognin E, Caccin L, Puricella A, Specchia V, Bozzetti MP, Costa R, Mazzotta GM.. The RNA helicase BELLE is involved in circadian rhythmicity and in transposons regulation in Drosophila melanogaster. Front Physiol 2019;10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A.. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev 1993;7:671–682. [DOI] [PubMed] [Google Scholar]
- Day FR, Thompson DJ, Helgason H, Chasman DI, Finucane H, Sulem P, Ruth KS, Whalen S, Sarkar AK, Albrecht E. et al. ; The LifeLines Cohort Study. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet 2017;49:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mees C, Bakker J, Szpirer J, Szpirer C.. Alpha-fetoprotein: from a diagnostic biomarker to a key role in female fertility. Biomark Insights 2007;1:82–85. [PMC free article] [PubMed] [Google Scholar]
- de Vries L, Behar DM, Smirin-Yosef P, Lagovsky I, Tzur S, Basel-Vanagaite L.. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab 2014;99:E2129–E2132. [DOI] [PubMed] [Google Scholar]
- Desai S, Wood-Trageser M, Matic J, Chipkin J, Jiang H, Bachelot A, Dulon J, Sala C, Barbieri C, Cocca M. et al. MCM8 and MCM9 nucleotide variants in women with primary ovarian insufficiency. J Clin Endocrinol Metab 2017;102:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Muñoz M, de la Rosa Santander P, Juárez-Espinosa AB, Arellano RO, Morales-Tlalpan V.. Granulosa cells express three inositol 1,4,5-trisphosphate receptor isoforms: cytoplasmic and nuclear Ca2+ mobilization. Reprod Biol Endocrinol 2008;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djagaeva I, Doronkin S, Beckendorf SK.. Src64 is involved in fusome development and karyosome formation during Drosophila oogenesis. Dev Biol 2005;284:143–156. [DOI] [PubMed] [Google Scholar]
- Dudding TE, Lawrence O, Winship I, Froyen G, Vandewalle J, Scott R, Shelling AN.. Array comparative genomic hybridization for the detection of submicroscopic copy number variations of the X chromosome in women with premature ovarian failure. Hum Reprod 2010;25:3159–3160. [DOI] [PubMed] [Google Scholar]
- Duncan FE, Moss SB, Schultz RM, Williams CJ.. PAR-3 defines a central subdomain of the cortical actin cap in mouse eggs. Dev Biol 2005;280:38–47. [DOI] [PubMed] [Google Scholar]
- Durdevic Z, Ephrussi A.. Germ cell lineage homeostasis in Drosophila requires the Vasa RNA helicase. Genetics 2019;213:911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchereau F, Shalev S, Chervinsky E, Beck-Fruchter R, Legois B, Fellous M, Caburet S, Veitia RA.. A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency. Clin Genet 2016;89:603–607. [DOI] [PubMed] [Google Scholar]
- Fonseca DJ, Patiño LC, Suárez YC, de Jesús Rodríguez A, Mateus HE, Jiménez KM, Ortega-Recalde O, Díaz-Yamal I, Laissue P.. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril 2015;104:154–162.e2. [DOI] [PubMed] [Google Scholar]
- França MM, Mendonca BB.. Genetics of primary ovarian insufficiency in the next-generation sequencing era. J Endocr Soc 2019;4:bvz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu K, Tian S, Tan H, Wang C, Wang H, Wang M, Wang Y, Chen Z, Wang Y, Yue Q. et al. Biological and RNA regulatory function of MOV10 in mammalian germ cells. BMC Biol 2019;17:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabant P, Forrester L, Nichols J, Van Reeth T, De Mees C, Pajack B, Watt A, Smitz J, Alexandre H, Szpirer C. et al. Alpha-fetoprotein, the major fetal serum protein, is not essential for embryonic development but is required for female fertility. Proc Natl Acad Sci U S A 2002;99:12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD.. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform 2011;12:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, Zamore PD.. The RNA-binding ATPase, armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol Cell 2019a;74:982–995.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Li C, Li C, Huang Z, Zeng J, Han L, Wang Q.. SIRT6 participates in the quality control of aged oocytes via modulating telomere function. Aging (Albany, NY). 2019b;11:1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebel J, Tuppi M, Sänger N, Schumacher B, Dötsch V.. DNA damaged induced cell death in oocytes. Molecules 2020;25:5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamrasni SE, Cardoso R, Li L, Guturi KKN, Bjerregaard VA, Liu Y, Venkatesan S, Hande MP, Henderson JT, Sanchez O. et al. Rad54 and Mus81 cooperation promotes DNA damage repair and restrains chromosome missegregation. Oncogene 2016;35:4836–4845. [DOI] [PubMed] [Google Scholar]
- Grossman H, Har-Paz E, Gindi N, Levi M, Miller I, Nevo N, Galiani D, Dekel N, Shalgi R.. Regulation of GVBD in mouse oocytes by miR-125a-3p and Fyn kinase through modulation of actin filaments. Sci Rep 2017;7:2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Ge J, Zhang L, Ma R, Hou X, Li B, Moley K, Wang Q.. Sirt6 depletion causes spindle defects and chromosome misalignment during meiosis of mouse oocyte. Sci Rep 2015;5:15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wang X, Montell DJ.. Shining light on Drosophila oogenesis: live imaging of egg development. Curr Opin Genet Dev 2011;21:612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WB, Du J, Yang XW, Li W, Tang WL, Dai C, Chen YZ, Zhang YX, Lu GX, Lin G. et al. Novel inactivating mutations in the FSH receptor cause premature ovarian insufficiency with resistant ovary syndrome. Reprod Biomed Online 2019;38:397–406. [DOI] [PubMed] [Google Scholar]
- Huang X, Ding L, Pan R, Ma PF, Cheng PP, Zhang CH, Shen YT, Xu L, Liu Y, He XQ. et al. WHAMM is required for meiotic spindle migration and asymmetric cytokinesis in mouse oocytes. Histochem Cell Biol 2013;139:525–534. [DOI] [PubMed] [Google Scholar]
- Hyon C, Mansour-Hendili L, Chantot-Bastaraud S, Donadille B, Kerlan V, Dodé C, Jonard S, Delemer B, Gompel A, Reznik Y. et al. Deletion of CPEB1 gene: a rare but recurrent cause of premature ovarian insufficiency. J Clin Endocrinol Metab 2016;101:2099–2104. [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Watanabe Y.. The cohesin REC8 prevents illegitimate inter-sister synaptonemal complex assembly. EMBO Rep 2016;17:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Kinoshita T, Hirakata S, Komatsuzaki C, Siomi MC.. Distinct and collaborative functions of Yb and armitage in transposon-targeting piRNA biogenesis. Cell Rep 2019;27:1822–1835.e8. [DOI] [PubMed] [Google Scholar]
- Jaillard S, Akloul L, Beaumont M, Hamdi-Roze H, Dubourg C, Odent S, Duros S, Dejucq-Rainsford N, Belaud-Rotureau MA, Ravel C.. Array-CGH diagnosis in ovarian failure: identification of new molecular actors for ovarian physiology. J Ovarian Res 2016;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A, Bayram Y, Turan S, Aycan Z, Tos T, Abali ZY, Hacihamdioglu B, Coban Akdemir ZH, Hijazi H, Bas S. et al. Exome sequencing of a primary ovarian insufficiency cohort reveals common molecular etiologies for a spectrum of disease. J Clin Endocrinol Metab 2019;104:3049–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantanos T, Moliterno AR.. The roles of JAK2 in DNA damage and repair in the myeloproliferative neoplasms: opportunities for targeted therapy. Blood Rev 2018;32:426–432. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP. et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E, Wong MD, Ding BC, Xie T.. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 2004;131:1365–1375. [DOI] [PubMed] [Google Scholar]
- Kim S, Pyun JA, Kang H, Kim J, Cha DH, Kwack K.. Epistasis between CYP19A1 and ESR1 polymorphisms is associated with premature ovarian failure. Fertil Steril 2011;95:353–356. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A. et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009;138:1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauff EA, Blauw HM, Pearson PL, Kok K, Wijmenga C, Veldink JH, van den Berg LH, Bouchard P, Fauser BC, Franke L; Dutch Primary Ovarian Insufficiency Consortium. Copy number variants on the X chromosome in women with primary ovarian insufficiency. Fertil Steril 2011;95:1584–1588.e1. [DOI] [PubMed] [Google Scholar]
- Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, Massouras A.. VarSome: the human genomic variant search engine. Bioinformatics 2019;35:1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue P. Aetiological coding sequence variants in non-syndromic premature ovarian failure: from genetic linkage analysis to next generation sequencing. Mol Cell Endocrinol 2015;411:243–257. [DOI] [PubMed] [Google Scholar]
- Ledig S, Ropke A, Wieacker P.. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev 2010;4:225–232. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB. et al. ; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M, Maro B, Shalgi R.. The involvement of Fyn kinase in resumption of the first meiotic division in mouse oocytes. Cell Cycle 2010a;9:1577–1589. [DOI] [PubMed] [Google Scholar]
- Levi M, Maro B, Shalgi R.. Fyn kinase is involved in cleavage furrow ingression during meiosis and mitosis. Reproduction 2010b;140:827–834. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang K.. InterVar: clinical interpretation of genetic variants by ACMG-AMP 2015 guideline. Am J Hum Genet 2017;100:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T.. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A 2007;104:6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell AS, Kaspari RR, Serrano Negron YL, Harbison ST.. The genetic architecture of ovariole number in Drosophila melanogaster: genes with major, quantitative, and pleiotropic effects. G3 (Bethesda) 2017;7:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, McGinnis LK, Kinsey WH.. Role of Fyn kinase in oocyte developmental potential. Reprod Fertil Dev 2010;22:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines JZ, Stevens LM, Tong X, Stein D.. Drosophila dMyc is required for ovary cell growth and endoreplication. Development 2004;131:775–786. [DOI] [PubMed] [Google Scholar]
- Manhart CM, Ni X, White MA, Ortega J, Surtees JA, Alani E.. The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol 2017;15:e2001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathorne SW, Ravn P, Hansen D, Beck-Nielsen SS, Gjørup H, Sørensen KP, Fagerberg CR.. Novel phenotype of syndromic premature ovarian insufficiency associated with TP63 molecular defect. Clin Genet 2020;97:779–784. [DOI] [PubMed] [Google Scholar]
- McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A.. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril 2011;95:1595–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F.. The ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H.. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 2006;16:1884–1894. [DOI] [PubMed] [Google Scholar]
- Meseke M, Pröls F, Schmahl C, Seebo K, Kruse C, Brandt N, Fester L, Zhou L, Bender R, Rune GM.. Reelin and aromatase cooperate in ovarian follicle development. Sci Rep 2018;8:8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki M, Moore B, Mosbruger T, Neklason DW, Yandell M, Jorde LB, Welt CK.. POLR2C mutations are associated with primary ovarian insufficiency in women. J Endocr Soc 2017;1:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Zeggini E.. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol 2010;34:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ.. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 2004;15:607–620. [DOI] [PubMed] [Google Scholar]
- Norling A, Hirschberg AL, Rodriguez-Wallberg KA, Iwarsson E, Wedell A, Barbaro M.. Identification of a duplication within the GDF9 gene and novel candidate genes for primary ovarian insufficiency (POI) by a customized high-resolution array comparative genomic hybridization platform. Hum Reprod 2014;29:1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J.. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 2010;29:3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn L, Portillo M, Ilic S, Cleitman G, Stein D, Kaluski S, Shirat I, Slobodnik Z, Einav M, Erdel F. et al. SIRT6 is a DNA double-strand break sensor. Elife 2020;9:e51636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Joyce EF, Couble P, McKim KS, Loppin B.. Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. J Cell Sci 2010;123:3515–3524. [DOI] [PubMed] [Google Scholar]
- Patiño LC, Beau I, Carlosama C, Buitrago JC, González R, Suárez CF, Patarroyo MA, Delemer B, Young J, Binart N. et al. New mutations in non-syndromic primary ovarian insufficiency patients identified via whole-exome sequencing. Hum Reprod 2017;32:1512–1520. [DOI] [PubMed] [Google Scholar]
- Qin Y, Guo T, Li G, Tang TS, Zhao S, Jiao X, Gong J, Gao F, Guo C, Simpson JL. et al. CSB-PGBD3 mutations cause premature ovarian failure. PLoS Genet 2015;11:e1005419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, Sargent CA, Affara NA.. Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF). Hum Reprod 2010;25:2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E. et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC.. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 2010;24:2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K.. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res 1999;9:677–679. [PubMed] [Google Scholar]
- Specchia V, D’Attis S, Puricella A, Bozzetti MP.. dFmr1 plays roles in small RNA pathways of Drosophila melanogaster. Int J Mol Sci 2017;18:1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Puricella A, D'Attis S, Massari S, Giangrande A, Bozzetti MP.. Drosophila melanogaster as a model to study the multiple phenotypes, related to genome stability of the fragile-X syndrome. Front Genet 2019;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolk L, Perry JR, Chasman DI, He C, Mangino M, Sulem P, Barbalic M, Broer L, Byrne EM, Ernst F. et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet 2012;44:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Richter JD.. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 2001;1:201–213. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD.. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 2004;116:831–841. [DOI] [PubMed] [Google Scholar]
- Toranzo GS, Bühler MC, Bühler MI.. Participation of IP3R, RyR and L-type Ca2+ channel in the nuclear maturation of Rhinella arenarum oocytes. Zygote 2014;22:110–123. [DOI] [PubMed] [Google Scholar]
- Tšuiko O, Nõukas M, Žilina O, Hensen K, Tapanainen JS, Mägi R, Kals M, Kivistik PA, Haller-Kikkatalo K, Salumets A. et al. Copy number variation analysis detects novel candidate genes involved in follicular growth and oocyte maturation in a cohort of premature ovarian failure cases. Hum Reprod 2016;31:1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker EJ, Grover SR, Robevska G, van den Bergen J, Hanna C, Sinclair AH.. Identification of variants in pleiotropic genes causing “isolated” premature ovarian insufficiency: implications for medical practice. Eur J Hum Genet 2018;26:1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppi M, Kehrloesser S, Coutandin DW, Rossi V, Luh LM, Strubel A, Hötte K, Hoffmeister M, Schäfer B, De Oliveira T. et al. Oocyte DNA damage quality control requires consecutive interplay of CHK2 and CK1 to activate p63. Nat Struct Mol Biol 2018;25:261–269. [DOI] [PubMed] [Google Scholar]
- Xian W, McKeon F.. TAp63 as a guardian of female germ line integrity. Nat Struct Mol Biol 2018;25:201–202. [DOI] [PubMed] [Google Scholar]
- Yang X, Touraine P, Desai S, Humphreys G, Jiang H, Yatsenko A, Rajkovic A.. Gene variants identified by whole-exome sequencing in 33 French women with premature ovarian insufficiency. J Assist Reprod Genet 2019;36:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannini L, Delia D, Buscemi G.. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol 2014;6:442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ.. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016;32:1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu Y, Zhang Z, Lv P, Liu Y, Li J, Wu Y, Zhang R, Huang Y, Xu G. et al. Basonuclin 1 deficiency is a cause of primary ovarian insufficiency. Hum Mol Genet 2018a;27:3787–3800. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Chang HM, Taylor EL, Liu RZ, Leung PCK.. BMP6 downregulates GDNF expression through SMAD1/5 and ERK1/2 signaling pathways in human granulosa-lutein cells. Endocrinology 2018b;159:2926–2938. [DOI] [PubMed] [Google Scholar]
- Zhen XM, Sun YM, Qiao J, Li R, Wang LN, Liu P. [Genome-wide copy number scan in Chinese patients with premature ovarian failure]. Beijing Da Xue Xue Bao Yi Xue Ban 2013;45:841–847. [PubMed] [Google Scholar]
- Zong X, Yang JX, Zhang Y.. Persistently elevated alpha-fetoprotein associated with chronic hepatitis B during chemotherapy for malignant ovarian germ cell tumors: a case series and a review of the literature. J Ovarian Res 2019;12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.
The datasets for the Italian cohort generated during the current study are publicly available at ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/): accession numbers SCV001364312 to SCV001364375.