Abstract
mRNA is a blooming technology for vaccination and has gained global attention during the SARS-CoV-2 pandemic. However, the recent clinical trials have highlighted increased reactogenicity when using high mRNA doses. Intending to increase the potency of mRNA therapeutics and to decrease the therapeutic dose, we designed a mRNA backbone and optimized the mRNA purification process. We used the enhanced green fluorescent protein (eGFP) reporter gene flanked by one 5′ untranslated region (UTR) and two 3′ UTRs of the human β-globin as a reference mRNA and identified the most promising mRNA sequence using in vitro and in vivo models. First, we assessed the impact of different poly(A) sizes on translation and selected the most optimal sequence. Then, we selected the best 5′ UTR among synthetic sequences displaying a high ribosome loading. Finally, we evaluated the transfection efficiency of our standard mRNA template after two capping strategies and purification using either double-stranded RNA (dsRNA) depletion or dephosphorylation of 5′PPP RNA or both combined. Double purification was shown to give the best results. Altogether, the use of a newly defined 5′ UTR coupled to post-transcriptional treatments will be of great interest in the mRNA vaccine field, by limiting the amount of the antigen-coding transcript and subsequently the formulation components needed for an efficient vaccination.
Keywords: mRNA translation efficiency, mRNA untranslated sequence, dsRNA, dendritic cells, mRNA-LNP complex, zebrafish injection
Graphical abstract

To increase the potency of mRNA therapeutics and decrease their reactogenicity, we have combined several strategies to optimize mRNA expression in transfected target cells. Our research arrives timely in the field, providing the “know-how” to achieve high-potency mRNA, with a 3-fold more efficient mRNA translation into antigen-presenting cells.
Introduction
The use of mRNA as a vaccine candidate is transforming the field of vaccinology and has the potential to initiate a new era in the prevention of infectious diseases.1 In addition to their safety profile, the principal advantages of mRNAs as a vaccination platform are speed and versatility during their development and cost and scalability in their production, which makes them the perfect platform to tackle pandemics. Several candidates based on mRNA are in development against SARS-CoV-2. Two of these candidates have provided exceptional preliminary results (>90% efficiency) in phase III clinical trials in a record time and are currently used worldwide. However, lessons learned from phase I of the clinical trials emphasize the importance to increase the potency of mRNA therapeutics to decrease the reactogenicity.2 Indeed, the use of high doses of mRNA formulated with lipid nanoparticles (LNPs) resulted in some side effects in the candidates mRNA-1273 and BNT162b1.3,4 Moreover, increasing the potency of mRNA vaccines is crucial for dose sparing, which will decrease the production cost and increase the number of doses of potential life-saving vaccines, extremely needed in the current and future pandemics.
To improve the potency of mRNA vaccines and optimize mRNA stability, researchers focused early efforts on modifications of the untranslated sequences. Indeed, because of their long half-life and efficient protein production, the use of highly stable untranslated regions (UTRs) from the α- and β-globin genes from human or Xenopus laevis improved mRNA stability.5,6 Later, the use of two copies of the 3′ UTR of β-globin in tandem improved the stability and the translational efficiency of mRNAs. Recent studies provided promising results such as optimizing the 3′ UTR region by selecting stable sequences and using two copies in tandem.7,8 Another strategy to improve mRNA stability was to tailor the poly(A) region. Initially, Holtkamp et al.9 found that 120 adenosines were optimal for translation in human dendritic cells. However, the impact of poly(A) size has been recently re-evaluated, demonstrating optimal sizes of longer tails (∼300 A).10 Furthermore, apart from mRNA stability, balancing the innate and adaptive immune systems is critical in mRNA vaccines.11 A breakthrough to achieve this balance was accomplished by the impairment of the innate immune system by blocking the activation of pattern recognition receptors (PRRs) through different approaches. Double-stranded RNA (dsRNA) generated during in vitro transcription (IVT) synthesis is the major contributor in the immunogenicity.12 dsRNA contaminants can be purified by high-performance liquid chromatography (HPLC) or cellulose chromatography13,14 or diminished during the IVT reaction with thermostable T7 polymerase or through Mg2+ concentration lowering.12,15 Others strategies aim to impair the PRR recognition of IVT mRNA with the use of modified nucleosides,16 uridine depletion in the coding sequence,17 or dephosphorylation of 5′PPP mRNAs.18
This study aimed to improve overall mRNA expression and therefore its potency for future vaccine or therapeutic purposes. To do so, we improved the state-of-the-art sequence, production, and purification methods to provide the “know-how” to achieve high-potency mRNA vaccines. First, we set our standard mRNA by flanking the enhanced green fluorescent protein (eGFP) coding sequence with the 5′ UTR and two copies in tandem of the 3′ UTR of the human β-globin gene. Then, we evaluated in two different cell lines the impact of poly(A) length into this standard mRNA backbone. We used the same approach after exchanging the 5′ UTR of the human β-globin by high-ribosome loading synthetic sequences. Additionally, this study aimed to improve the mRNA purification based on cellulose chromatography by including a phosphatase treatment and following its impact on the protein expression in the same cell lines as well as primary human monocyte-derived dendritic cells (DCs) (moDCs). Those results were then confirmed in vivo with the embryonic zebrafish model. Finally, the potency of the optimized mRNA and mRNA-LNP complexes was explored in adult zebrafish via intramuscular injection. Altogether, these results showed a difference in mRNA expression patterns, and this difference was consistent in all models. This allowed the determination of 5′ UTR sequences positively impacting the translation efficiency of mRNA. Moreover, adequate treatment of the IVT mRNA (vaccinia capping, dsRNA, and 5′PPP depletions) increased eGFP expression by 3-fold.
Results
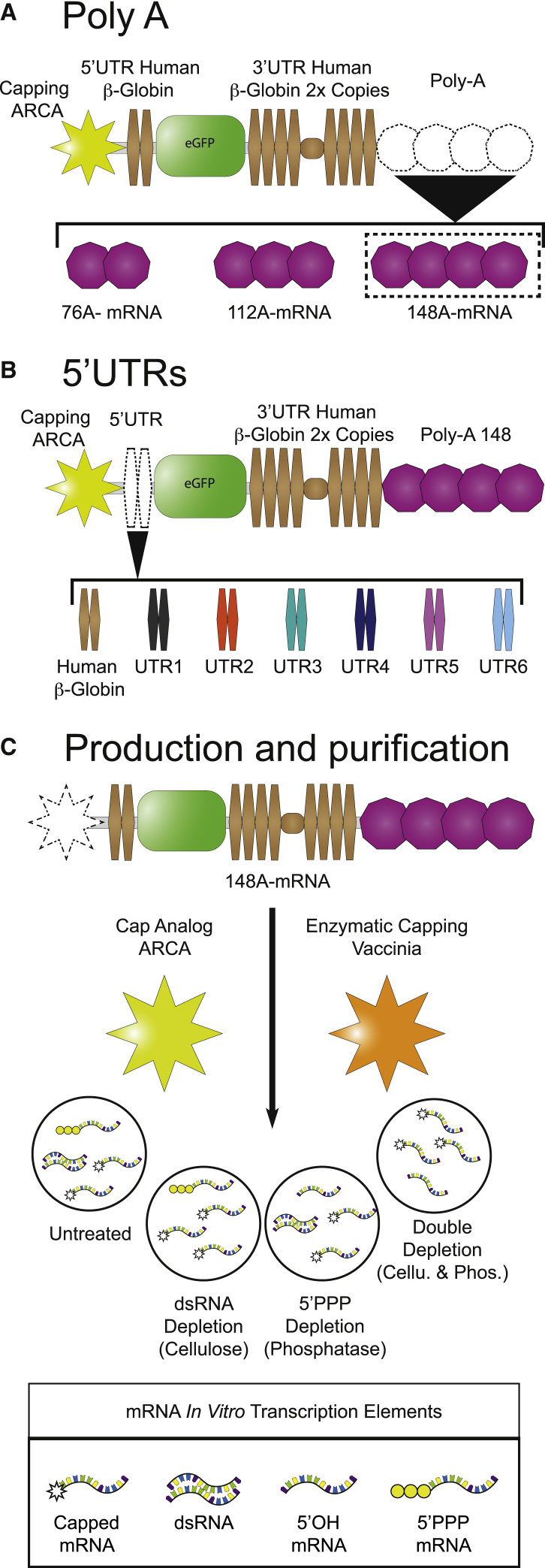
A standard IVT expression plasmid (Figures 1A and S1) was built, consisting of the eGFP reporter gene flanked by the 5′ UTR and two copies in tandem of the 3′ UTR of the long-half-life human β-globin RNA. This expression plasmid was used (1) as mRNA expression positive control and (2) as a template for the sequence modifications realized to increase the mRNA translation.
Figure 1.
Strategies used to increase mRNA potency
(A) Poly(A) tail. The ARCA-capped mRNA backbone consisted of an eGFP reporter gene flanked by the 5′ UTR and two copies in tandem of the 3′ UTRs of the human β-globin and ended with a 76, 112, or 148 polyadenylated tail. (B) Synthetic 5′ UTRs. In the mRNA backbone containing the optimal poly(A)148 tail (148A-mRNA), the 5′ β-globin UTR region was replaced by synthetic sequences with high ribosome loading to generate UTR1 to UTR6 mRNAs. (C) Optimization of IVT mRNA production and purification steps. The standard IVT 148A-mRNA was produced in the presence of ARCA or capped with the vaccinia virus enzyme. Then, the mRNAs could be treated by cellulose chromatography for dsRNA depletion and/or Antarctic phosphatase for the dephosphorylation of 5′PPP-mRNAs.
Tailoring the poly(A) tail
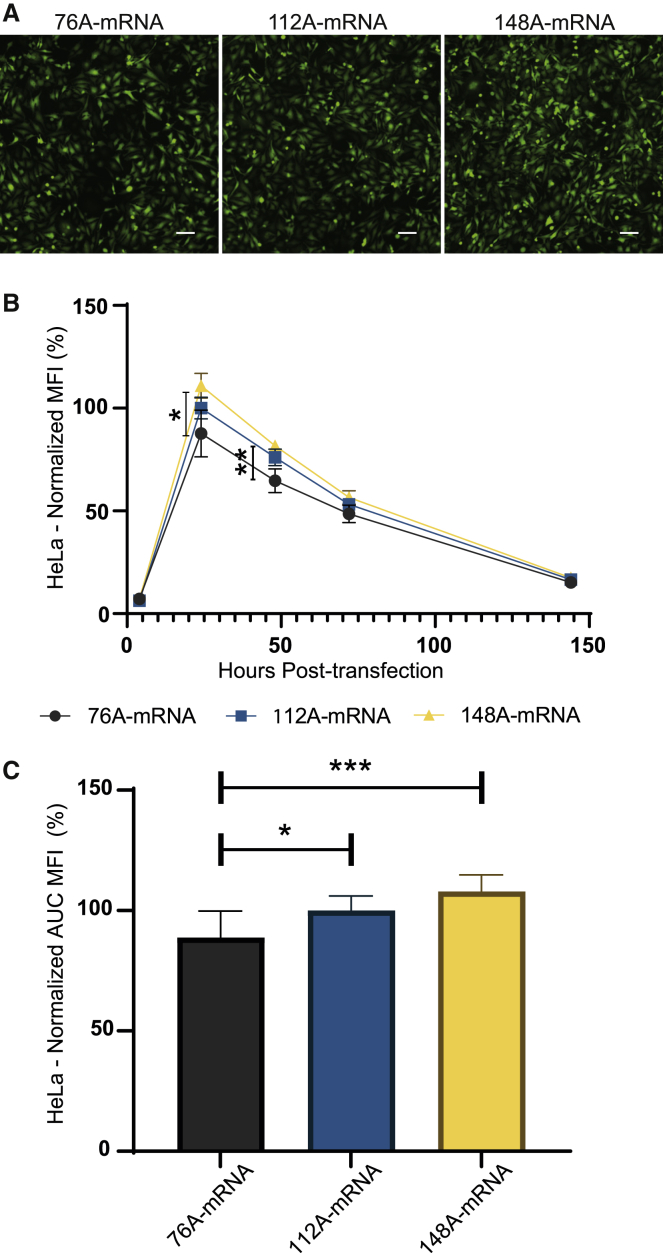
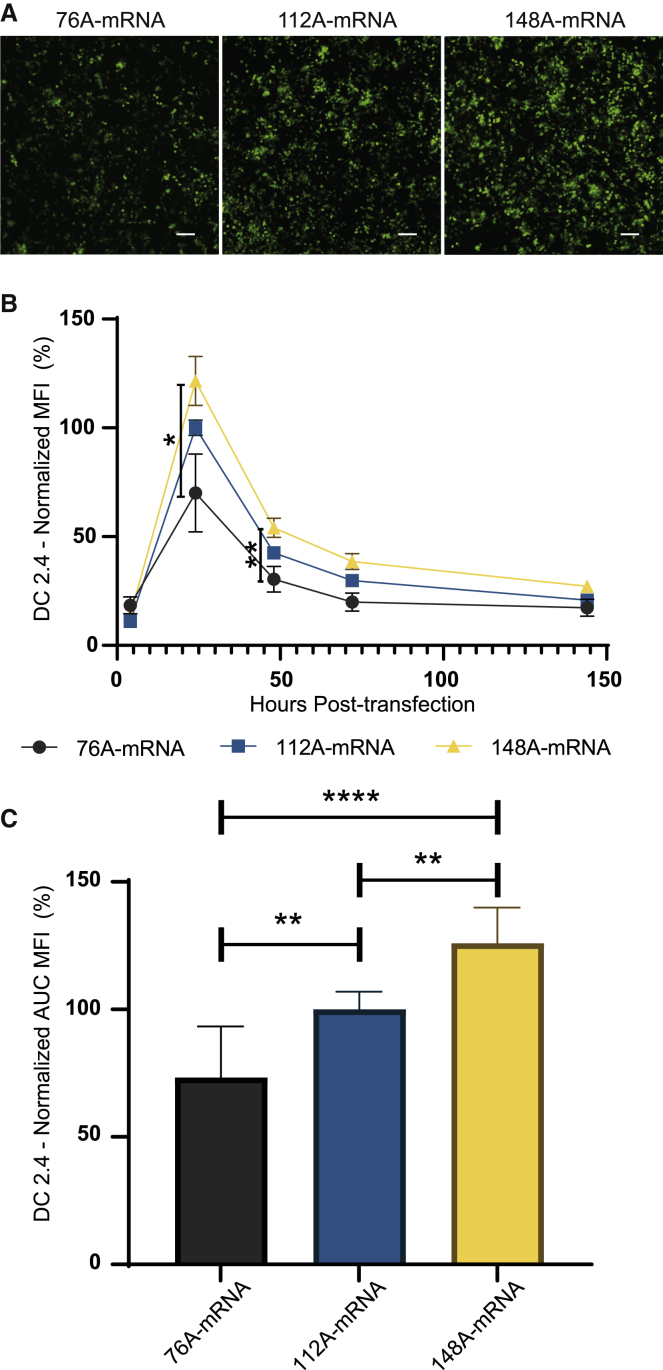
The poly(A) is a critical region that contributes to translation efficiency, mRNA half-life, and therefore mRNA potency.19 A poly(A) tail of 120 units is considered the standard size for mRNA vaccines.9,20,21 To unravel the impact of poly(A) stretch size, different plasmid templates (Figure 1A) harboring distinct poly(A) lengths were generated, with one poly(A) shorter than the standard (76A-mRNA), one with similar size (112A-mRNA), and another one longer (148A-mRNA). The different mRNAs were produced in vitro with anti-reverse cap analog (ARCA) and were then transfected into HeLa cells. At 24 h post-transfection, qualitative fluorescence microscopy analysis showed that the 148A-mRNA provided the highest eGFP expression (Figure 2A) for the same transfection efficiency and cell viability for all assays (Figures S2A and S2B). To better characterize the kinetics of eGFP expression, HeLa cells were analyzed by flow cytometry at different time points after transfection. A similar eGFP expression kinetic was observed with all the poly(A) mRNA, with a maximal eGFP expression at 24 h post-transfection followed by a gradual decrease of eGFP over time (Figure 2B). At 24 h, the intensity of expression was significantly higher with 148A-mRNA than with 76A-mRNA. After assessment of the area under the curve (AUC) (Figure 2C), the 148A-mRNA significantly increased the overall eGFP expression in comparison to 76A- and 112A-mRNAs. With the aim to optimize the IVT mRNA as a vaccine platform, the impact of the poly(A) size was evaluated with the murine DC line DC2.4. Akin to HeLa cells, fluorescence microscopy analysis showed the highest expression with 148A-mRNA (Figure 3A). Quantitative time course analysis of eGFP expression in DC2.4 transfected cells revealed similar kinetics, with a maximal fluorescence at 24 h post-transfection (Figure 3B). However, this peak of expression is followed by a faster decrease of eGFP over time than for HeLa cells (Figure 2B). The 148A-mRNA resulted in higher eGFP expression compared with 76A- and 112A-mRNAs at 24 and 48 h post-transfection (Figure 3B) and also considering the complete time course study (Figure 3C). Thus, a 65% and a 25% increase in eGFP expression were obtained with 148A-mRNA in comparison to 76A- and 112A-mRNAs, respectively (Figure 3C). There was no significant difference in cell viability between the different assays, but a slight decrease of transfection efficiency was observed between 76A- and 148A-mRNAs (p < 0.05). To get closer to physiological conditions, the different polyadenylated mRNAs were microinjected into one-cell-stage zebrafish embryos. Fluorescence analysis of zebrafish embryos at 24 and 48 h post-injection by microscopy (Figure S3) showed that 148A-mRNA provided the highest eGFP expression. This qualitative study confirmed the results obtained in the in vitro cell assays.
Figure 2.
Impact of poly(A) size on eGFP expression in HeLa cells
(A) 76A-, 112A-, and 148A-mRNAs were transfected (125 ng mRNA per well, 48-well plates). Fluorescence microscopy analysis was performed 24 h post-transfection. Images were obtained with equal acquisition parameters. Scale bar, 100 μm. (B) Time course analysis of eGFP expression in cells transfected with the different poly(A) mRNAs was performed by flow cytometry. The measurements were acquired at 4, 24, 48, 72, and 144 h post-transfection, and results are expressed as the mean ± SD of 3 independent experiments with four technical replicates. Results are represented as the percentage of change with respect to normalized eGFP expression of 112A-mRNA at 24 h post transfection. (C) Total eGFP expression of the different mRNAs was represented by calculating the area under the curve (AUC). The relative eGFP expression levels were normalized to that of 112A-mRNA assay. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure 3.
Impact of poly(A) size on eGFP expression in DC2.4 cells
(A) 76A-, 112A-, and 148A-mRNAs were transfected (250 ng mRNA per well, 48-well plate). Fluorescence microscopy analysis was performed 24 h post-transfection. Images were obtained with equal acquisition parameters. Scale bar, 100 μm. (B) Time course analysis of eGFP expression in cells transfected with the different poly(A) mRNAs was performed by flow cytometry. The measurements were acquired at 4, 24, 48, 72, and 144 h post-transfection and MFIs normalized with eGFP expression of 112A-mRNA at 24 h post-transfection. Data were determined as the mean ± SD of 3 independent experiments with four technical replicates. (C) Total eGFP expression of the different mRNAs was represented by calculating the area under the curve (AUC). The relative eGFP expression levels were normalized to that of 112A-mRNA assay. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
Enhanced mRNA translation with high-ribosome loading 5′ UTR sequences
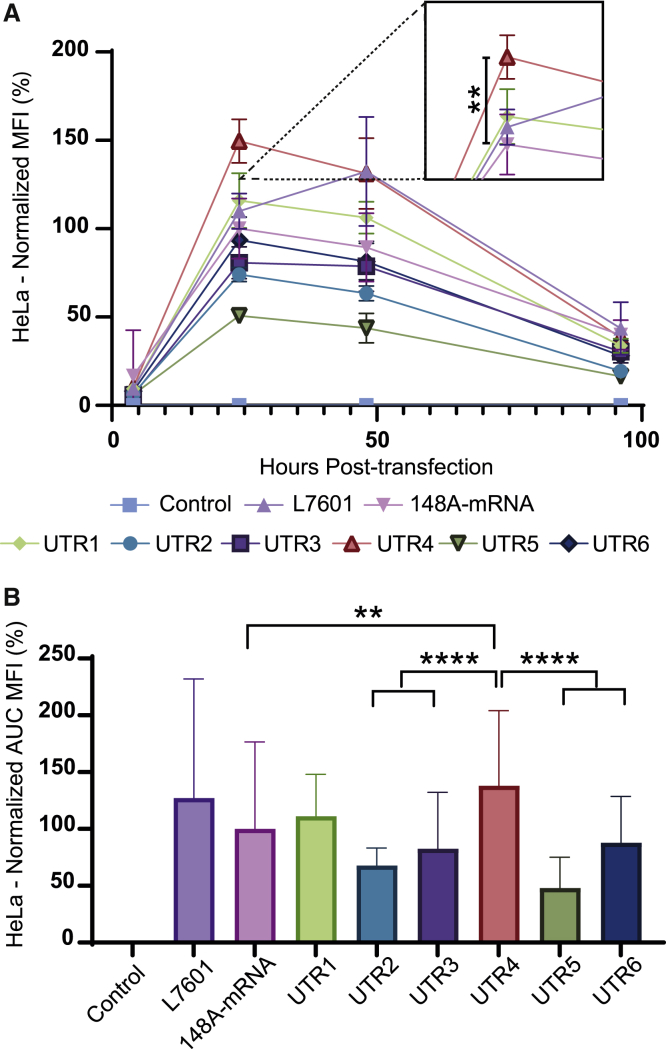
The 5′ UTR is a critical region where the mRNA translation begins after recognition by the ribosomes and therefore is essential for translation efficiency. Some attempts to optimize the 5′ UTR have focused on the use of these long mRNA half-lives.7 Recently, Sample et al.22 generated a library of 300,000 random synthetic 5′ UTR IVT mRNAs containing the eGFP sequence. They obtained the ribosome profiling of each sequence after transfecting HEK293T cells. With the aim to optimize the translation efficiency of our standard 148A-mRNA, 6 different synthetic 5′ UTRs with high ribosome loading and high number of reads were selected from this library. As shown in Figure 1B, the 5′ UTR of 148A-mRNA was exchanged by six new synthetic 5′ UTR sequences, allowing the formation of UTR1 to UTR6 mRNAs. HeLa cells were transfected with the new synthetic 5′ UTRs, the human β-globin (148A-mRNA), and a commercial eGFP (L7601) mRNA, and the eGFP expression was analyzed both by fluorescence microscopy (Figure S4) and flow cytometry (Figure 4). At 24 h post-transfection, fluorescence microscopy revealed a higher eGFP expression with UTR4 mRNA compared with other mRNAs. As for previous time course analyses (Figures 2B and 3B), similar flow cytometry kinetics were obtained, except for the L7601 mRNA, which presented an eGFP expression peak at 48 h post-transfection (Figure 4A). At 24 h post-transfection, the intensity of expression was significantly more important with UTR4 mRNA than other mRNAs (see magnification square in Figure 4A). When considering the overall fluorescence detected during this time course analysis, the UTR4 mRNA presented significantly higher eGFP expression than other mRNAs (Figure 4B), except the L7601 mRNA. Moreover, the UTR4 mRNA generated 33% more eGFP compared with the standard β-globin 148A-mRNA. Interestingly, the high-ribosome loading UTR2 and UTR5 showed a low level of eGFP production. Interestingly, among the lowest eGFP-expressing mRNAs, UTR3 and UTR5, presented a significantly lower percentage of viability at 24 h post-transfection (Figure S2A). Although the origin of this cell toxicity is unknown, the presence of a common 10 bp near the Kozak sequence on both 50-bp UTRs could participate in this toxicity (Figure S1, GTGTCAGTGA).
Figure 4.
Quantitative eGFP expression of HeLa cells transfected with eGFP mRNAs displaying distinct 5′ synthetic UTRs
Transit reagent alone (negative control), L7601 eGFP mRNA (external control), and eGFP mRNA displaying either the 5′ UTR of human β-globin or a synthetic 5′ UTR (UTR1 to UTR6) were transfected into HeLa cells (125 ng of mRNA per well). (A) Time course analysis of eGFP expression was performed by flow cytometry. The measurements were acquired at 4, 24, 48, and 96 h post-transfection and MFIs normalized with eGFP expression of 148A-mRNA at 24 h post-transfection. Data were determined as the mean ± SD of 3 independent experiments with four technical replicates. (B) Total eGFP expression of the different mRNAs was represented by calculating the area under the curve (AUC). The relative eGFP expression levels were normalized to that of 148A-mRNA assay. Statistical comparisons were only based on the UTR4 mRNA. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; NS, non-significant.
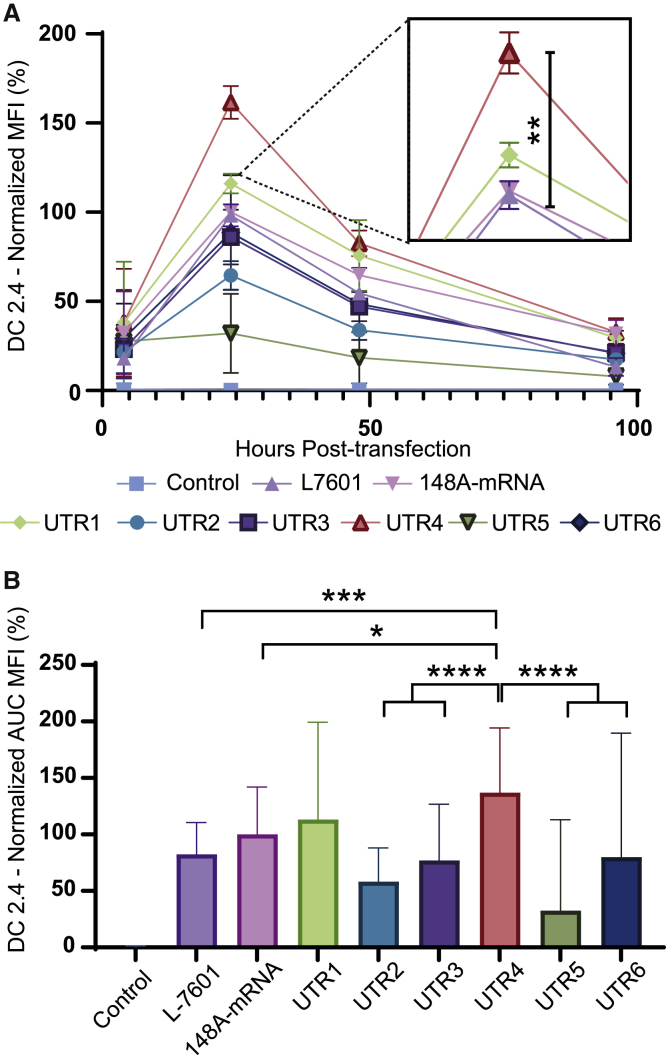
The same analyses were performed with DC2.4 cells. Fluorescence images of DC2.4 cells realized 24 h post-transfection showed that UTR4 mRNA presented the highest eGFP production (Figure S5). When the eGFP expression was assessed by flow cytometry (Figure 5A), at 24 h post-transfection, the intensity of expression was significantly more important with UTR4 mRNA than with other mRNAs. The total amount of fluorescence detected during the time course analysis (Figure 5B) confirmed that the UTR4 mRNA resulted in a significantly higher expression compared with other mRNAs. The total eGFP expression in DC2.4, calculated with the AUC (Figure 5B), was similar to the expression obtained in HeLa cells (Figure 4B). Thus, a 37% increase in eGFP expression was obtained with the UTR4 mRNA in comparison to 148A-mRNA, illustrating the added value of this specific 5′ UTR. Another transfection reagent was used to transfect DC2.4 cells with 148A and UTR4 mRNAs (Figure S6), and the results were similar.
Figure 5.
Quantitative eGFP expression of DC2.4 cells transfected with eGFP mRNAs displaying distinct 5′ synthetic UTRs
Transit reagent alone (negative control), L7601 eGFP mRNA (external control), and eGFP mRNA displaying either the 5′ UTR of human β-globin or a synthetic 5′ UTR (UTR1 to UTR6) were transfected into DC2.4 cells (125 ng of mRNA per well). (A) Time course analysis of eGFP expression was performed by flow cytometry. The measurements were acquired at 4, 24, 48, and 96 h post-transfection and MFIs normalized with eGFP expression of 148A-mRNA at 24 h post-transfection. Data were determined as the mean ± SD of 3 independent experiments with four technical replicates. (B) Total eGFP expression of the different mRNAs was represented by calculating the area under the curve (AUC). The relative eGFP expression levels were normalized to that of 148A-mRNA assay. Statistical comparisons were based only on the UTR4 mRNA. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
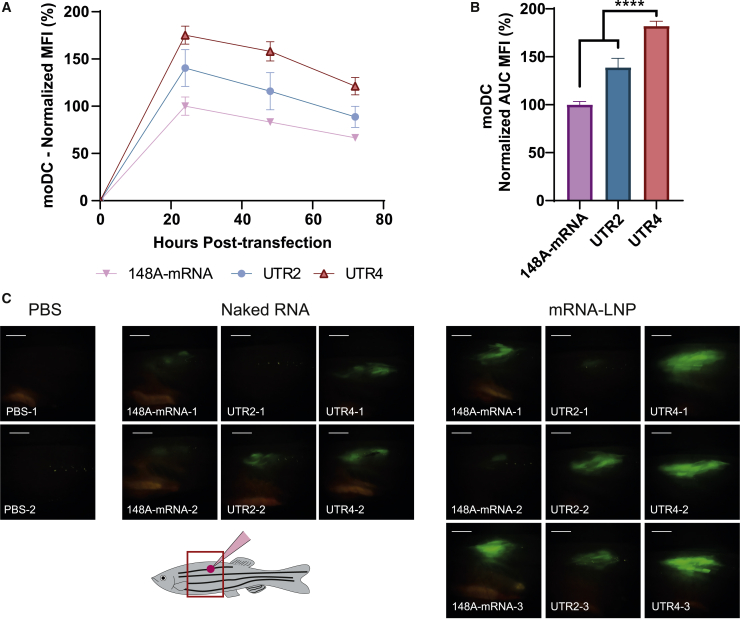
To further characterize the translational efficiency of UTR4 mRNA, primary DCs derived from human monocytes (moDCs) were transfected with 148A, UTR2, and UTR4 mRNAs and the eGFP expression was evaluated by flow cytometry 72 h after transfection (Figures 6A and 6B). As for HeLa and DC2.4 cell lines, the eGFP expression was significantly more important in primary cells transfected with UTR4 mRNA. An 80% increase of eGFP production was observed with UTR4 mRNA compared with 148A-mRNA. Contrarily to the results obtained with HeLa and DC2.4 cell lines, the UTR2 mRNA also showed a significantly better translation efficiency in moDCs compared with 148A-mRNA. As shown in Figures S2E and S2F, cell transfection efficiency and cell viability were lower than for HeLa and DC2.4 cell lines, which is expected when using a suspension of primary cells from hematopoietic origin. Moreover, the percentage of transfected moDCs was significantly higher with UTR4 than with 148A and UTR2 mRNAs.
Figure 6.
UTR4 versus UTR2 and 148A-mRNA expression into moDCs and adult zebrafish
UTR2, UTR4, and 148A-mRNAs were transfected into moDCs (125 ng of mRNA per well). (A) Time course analysis of eGFP expression was performed by flow cytometry. The measurements were acquired at 24, 48, and 96 h post-transfection and MFIs normalized with eGFP expression of 148A-mRNA at 24 h post-transfection. Data were determined as the mean ± SD of 3 independent experiments with four technical replicates. (B) Total eGFP expression of the different mRNAs was represented by calculating the area under the curve (AUC). The relative eGFP expression levels were normalized to that of 148A-mRNA assay. Statistical comparisons were based only on the UTR4 mRNA. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. (C) Two or three adult zebrafish were injected intramuscularly with 10 μL of naked mRNA (20 ng/μL) or 10 μL of mRNA-LNP complexes (200 ng of mRNA), respectively. eGFP expression observed with a Leica fluorescent stereoscope on anesthetized animals at 48 h post-injection is shown. The site of injection and the location where the picture was taken (red square) are presented in the drawing. Comparable results were obtained at 24 h post-injection (data not shown). Scale bar, 2 mm.
A final in vivo experiment was realized by injecting 8-month-old zebrafish intramuscularly with 148A, UTR2, and UTR4 naked mRNA or mRNA-LNP complexes (Figure 6C). This qualitative study indicated a higher eGFP expression with UTR4 than with 148A and UTR2 mRNAs and confirmed the results obtained in vitro. Moreover, a better eGFP expression was obtained with mRNA-LNP complexes than with naked mRNA.
Capping and purification of IVT mRNA
The adequate production and purification of IVT mRNA can drastically affect antigen expression. Indeed, a high capping efficiency in the adequate Cap-1 conformation evades host responses by interferon (IFN)-induced protein with tetratricopeptide repeats (IFIT) family members and favors mRNA translation.23 Moreover, it is also essential to purify the IVT mRNA from the contaminants generated during the reaction. Indeed, those contaminants, such as dsRNA or to a lesser extent 5′PPP uncapped mRNA, activate PRRs and block the cellular protein synthesis.12
Here, the standard 148A-mRNA was produced with either the ARCA or the vaccinia virus capping enzyme. Then, the IVT mRNAs were depleted of dsRNA by cellulose chromatography and/or dephosphorylated with Antarctic phosphatase. An illustration of the purification strategy and mRNA recovery yields is presented in Figure S7. All these mRNAs were analyzed by agarose gel electrophoresis, and the presence of sharp bands confirmed their overall integrity (Figure S8A).
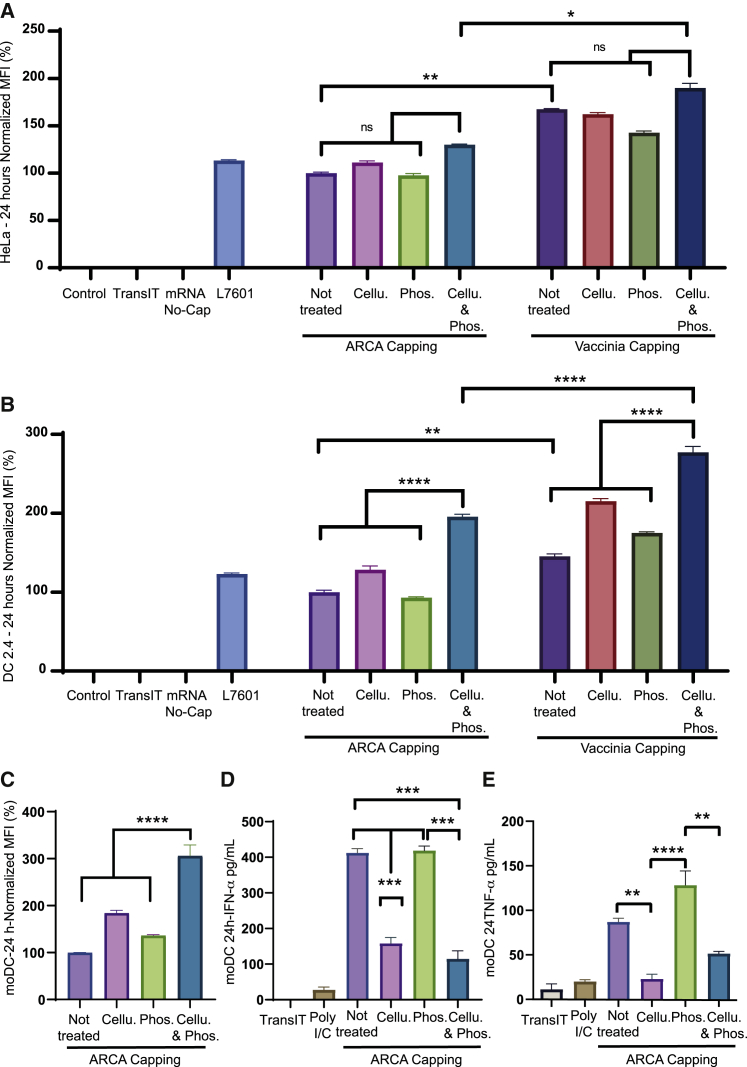
After cellulose chromatography, dsRNA depletion was confirmed by dot blot using the J2 anti-dsRNA antibody (Figure S8B). Interestingly, the amount of dsRNA depended on the method used for mRNA production, being higher in the ARCA than the vaccinia capping system. HeLa or DC2.4 cells were transfected with the different types of 148A-mRNAs and their eGFP expression evaluated by fluorescence microscopy 24 h post-transfection (Figure S9). Vaccinia virus-capped mRNAs provided a higher eGFP expression compared with ARCA-capped mRNA in both cell lines. Moreover, among all the transfected types of 148A-mRNAs in DC2.4 cells, those depleted of dsRNA and submitted to a dephosphorylation step showed the highest eGFP expression independently of the capping procedure. Cellulose purification alone offered an increase in protein expression compared with untreated 148A-mRNA, especially in DC2.4 cell assays. The eGFP expression was then quantified by flow cytometry 24 h after mRNA transfection in both HeLa and DC2.4 cells (Figures 7A and 7B, respectively). The results showed that vaccinia virus capping is more efficient than ARCA capping in both cell lines. As presented in Figure 7A, the different treatments had no significant impact either in the translation of ARCA-capped mRNA or in vaccinia-capped mRNA in HeLa cells. On the other hand, the phosphatase treatment alone provided a nonsignificant detrimental effect on eGFP expression. DC2.4 cells showed different patterns of expression among some of the purification treatments compared with HeLa cells (Figure 7B). Hence, the eGFP expression of ARCA-capped mRNA was significantly increased (∼2-fold) after cellulose purification and dephosphorylation. The same pattern of expression was observed for vaccinia virus-capped mRNA, suggesting the beneficial impact of the combined treatment, i.e., dsRNA depletion plus dephosphorylation of 5′PPP mRNAs. Finally, a 3-fold increase in eGFP expression was observed between dephosphorylated/dsRNA-depleted vaccinia mRNA and untreated ARCA mRNA in DC2.4 cells.
Figure 7.
Vaccinia capping followed by cellulose dsRNA depletion and dephosphorylation of 5′PPP mRNA promoted high eGFP expression
ARCA- or vaccinia-capped 148A-mRNAs were submitted to dsRNA depletion by cellulose chromatography and/or a phosphatase treatment. These mRNAs were transfected into HeLa (A) or DC2.4 (B) cells, and eGFP expression was monitored 24 h later by flow cytometry. (C) ARCA-capped 148A-mRNAs submitted to dsRNA depletion by cellulose chromatography and/or a phosphatase treatment were transfected into moDCs, and eGFP expression was determined 24 h later by flow cytometry. Transfection were realized in 48-well plates with 125 ng or 250 ng of mRNA for HeLa cells and DC2.4 and moDC cells. The different types of 148A-mRNAs were transfected into moDCs, and IFN-α (D) and TNF-α (E) were measured in the supernatant 24 h later. Control, untransfected cells; TransIT, TransIT reagent without mRNA; mRNA No-Cap, uncapped 148A-mRNA; L7601, eGFP mRNA from commercial company. The relative eGFP expression levels were normalized to that of 148A-mRNA ARCA non-treated. Statistical significance was assessed by nested one-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; NS, non-significant.
To evaluate the gain of translation of these purification procedures in human primary cells, human moDCs were transfected with the different types of ARCA-capped 148A-mRNAs and their eGFP expression quantified by flow cytometry 24 h after transfection (Figure 7C). The results obtained were comparable with those generated with DC2.4 cells but revealed a higher increase of eGFP expression between the dephosphorylated/dsRNA depleted mRNA and untreated mRNA (3-fold). moDCs being a better tool for analyzing innate immune responses via RNA sensors,13 IFN-α and tumor necrosis factor alpha (TNF-α) productions were analyzed 24 h post-transfection with ARCA-capped 148A-mRNA treated or not with phosphatase and cellulose chromatography (Figures 7D and 7E). The lowest IFN-α and TNF-α amounts were detected in the supernatant of moDCs transfected with dsRNA-depleted mRNA. Phosphatase treatment did not decrease the levels of IFN-α and TNF-α in comparison to untreated mRNA. Altogether, these results indicate that, at 24 h post transfection, the dsRNA has an important impact on innate immune response. However, the double treatment of mRNA allows the best eGFP production.
Discussion
The use of mRNA in vaccinology is a revolutionary technology that has exploded in recent years. Although two mRNA-based COVID-19 vaccines are already on the market, the clinical trials have reported more adverse events in patients injected with high doses of mRNA vaccines. Thus, to lower the injection dose, it is important to increase the stability and translation efficiency of the mRNA and its release into the cytosol. In this work, we aimed to enhance the mRNA potency by improving the 5′ UTR and poly(A) regions and by optimizing the capping and purification steps. Hence, the use of synthetic 5′ UTRs in an eGFP reporter mRNA harboring a 148(A) tail allows us to increase their translation efficiency in either HeLa cells or DC2.4 cells, a DC model, in comparison to a human β-globin UTR-containing and a commercial eGFP mRNA control. Moreover, the depletion of dsRNA conjugated to a phosphatase treatment increases the translational potential of mRNA, especially in DCs, which are an important target for eliciting a strong immune response.
Poly(A) tail plays an important function in the stability and regulation of translation efficiency of the mRNA.24 However, because of its instability during bacterial plasmid production, the poly(A) tail has been limited to 120 units. An alternative strategy has been developed to avoid this instability, consisting of including an internal 10-nucleotide linker into the poly(A) stretch.25,26 By using highly stable bacteria (New England Biolabs [NEB] stable competent Escherichia coli strain), we have been able to propagate and maintain a plasmid containing a 148 A tail and showed that this mRNA offers the best mRNA translation efficiency into DC2.4 cells. These results are in line with the results obtained by Grier et al.,10 who showed that the translational efficiency of mRNA was improved in conjunction with increasing poly(A) size until a length of 300 A, a value in agreement with those of newly transcribed mRNA in Metazoa.24 Interestingly, these authors used the linear plasmid technology that allowed the propagation of longer poly(A) stretch, which could be an interesting alternative to conventional circular plasmids producing 120–148 poly(A) or segmented poly(A) tail mRNAs.25,26
Besides the poly(A) tail, numerous works have developed specific UTRs to increase the translational efficiency of mRNAs, as the use of one or two copies of the 3′ UTR of the globin or other known stable mRNA8,9 or short 14-nucleotide 5′ UTR.27 Here, we improved the mRNA translation by using a new set of 5′ UTR synthetic sequences based on the selection of synthetic 5′ UTRs with high ribosome loading.22 Among these synthetic 5′ UTRs, UTR4 provided the highest translation efficiency, with a 33%, 37%, and 80% increase in eGFP expression compared with the standard 5′ UTR human β-globin in HeLa cells, DC2.4 cells, and moDCs, respectively (Figures 4, 5, and 6). The eGFP expression level of the mRNAs tested is comparable between HeLa and DC2.4 cell lines, except for the L7601 commercial mRNA. Hence, only UTR4 mRNA allowed us to produce a significantly higher level of eGFP than L7601 mRNA in DC2.4 cells. Since the untranslated (origin) and translated (codon optimization) sequences of this commercial mRNA are unknown, the discrepancy observed between these two cell lines cannot be discussed. From this study, we showed that the selection of high-ribosome loading 5′ UTR sequence is a promising tool to improve mRNA potency, although it is not always correlated with high protein expression. Hence, a low level of eGFP production is obtained with UTR2 and UTR5 mRNAs in both cell lines (Figures 4 and 5), whereas UTR2 presented a higher eGFP expression in moDCs (Figures 6A and 6B). Therefore, it is important to validate the selected 5′ UTR and 3′ UTR used for the gene of interest and the target cell types. Here, we have shown that UTR4 allows a high level of eGFP expression in human and mouse cells as well as in vivo after intramuscular injection of adult zebrafish. As shown in Figure 6C, incorporation of naked mRNA into zebrafish cells and subsequent GFP expression in the zebrafish tissues was monitored. A comparable result had been previously observed after intramuscular injection of plasmid DNA into adult zebrafish and showed the nucleic acid entering the cells (myocytes and antigen-presenting cells [APCs]) at the site of administration.28 Other strategies can be used to improve mRNA translation, for example, the depletion of strong secondary structures preventing ribosome scanning within the 5′ UTR, a codon optimization of the coding sequences, or the reduction of miRNA sites.
Apart from the untranslated sequences, other parameters can modulate the translation process, such as secondary structures or innate immunity induction. For instance, a successful approach to avoid overactivation of the innate immunity and increase translation of mRNA is the use of modified nucleosides such as methyl-pseudo uridine.29 This strategy is the approach followed by the two currently used worldwide SARS-CoV-2 vaccines. However, other factors like the presence of uncapped mRNA and dsRNA are still a source of innate immunity activation. As shown in Figure 7, a better efficiency is achieved with enzymatic capped mRNA compared with ARCA-capped mRNA in the two cell lines used (+67% in HeLa, +45% in DC2.4). This can be due to the difference in capping efficiency (70%–80% with ARCA versus 98%–100% with vaccinia virus system), the cap conformation (Cap-0 in ARCA versus Cap-1 in vaccinia virus system), and the amount of dsRNA, which is more important in mRNA samples produced with the ARCA capping method (Figure S8B). It has been shown that dsRNA removal through HPLC or cellulose methods drastically enhances the translational efficiency of IVT mRNAs.13,14 This dsRNA depletion leads to a decrease of type I IFN and a translational enhancement of the mRNA into transfected DC cells. As shown in Figures 7D and 7E, depletion of dsRNA decreases the production of IFN-α and TNF-α in moDCs in comparison to untreated mRNA. The dsRNA production might be related to the buffer composition, as shown by Mu et al.,12 a reduction in the MgCl2 concentration decreasing the synthesis of dsRNA. This illustrates the strong added value of vaccinia virus capping. However, despite its advantages, vaccinia virus capping presents some limitations compared with ARCA capping. First, it is a more complex process requiring extra steps of production and purification. Second, the capping reaction should be validated for every batch, therefore being a more time-consuming and laborious process. From a more general view, our results demonstrated a great synergic improvement in mRNA potency by combining dsRNA depletion and 5′PPP dephosphorylation in our APC model, the DC2.4 cells, and even more in human moDCs. Activation by dsRNA leads to a detrimental strong innate immune activation, impairing the cellular translational machinery, and therefore the eGFP expression. In phosphatase-treated samples, the presence of dsRNA masks any PRR activation by 5′PPP mRNA. After cellulose chromatography and phosphatase treatment, dsRNA contaminants are depleted; thus the decrease of 5′PPP mRNA is beneficial for eGFP expression. The positive effect of the double purification is more pronounced in DCs (moDCs and DC2.4 cells) than in HeLa cells, possibly because of the higher expression of PRRs into DCs and therefore their sensing capacity to contaminants. This highlights the importance of dsRNA depletion and 5′PPP dephosphorylation of mRNA used for vaccine purposes, since immune cells are more sensitive to pathogen-associated molecular patterns (PAMPs). Finally, although we provide promising results for the optimization of the IVT mRNA platform with up to a 3-fold expression increase, our study contains some limitations. Although this study is informative about the high potency and versatility (human and mouse cell lines, human DCs derived from monocytes, and intramuscular injection in adult zebrafish) of UTR4, it will be interesting to compare the added value of this UTR relative to 5′ β-globin UTR in mouse and non-human primate models.
There are no doubts that mRNA vaccine platform will revolutionize medicine. Most of the recent research efforts are focused on the production of mRNA formulations exhibiting an efficient delivery into immune cells and target tissues. However, increasing potency and diminishing reactogenicity of mRNA species are also key elements to improve mRNA therapeutics, especially in the vaccinology field.
Materials and methods
Production of DNA templates
The initial plasmid (called p40A-IVT) contained the T7 promoter and the eGFP reporter gene flanked by the 5′ UTR and two copies in tandem of the 3′ UTR of the human β-globin mRNA (HBB gene). The end of the 3′ UTR contains a poly(A) tail region (A)40, and a unique BspQI site downstream of the poly(A). This type IIS restriction enzyme will allow the linearization of the plasmid after the last adenosine of the poly(A) stretch. Another synthetic gene (called poly-A), consisting of a 40 A sequence flanked by BfuAI and BtgZI sites, was used to elongate the poly(A) tail. GenScript (Netherlands) produced both synthetic genes. To get a longer poly(A) tail, the BfuAI/BtgZI restriction fragment of the poly-A plasmid was inserted into the BspQI site of the p40A-IVT vector, generating the p76A-IVT construct with a poly(A) stretch of 76 units. Two more rounds of digestion/insertion allowed the production of the p112A-IVT and p148A-IVT plasmids. Finally, a new set of plasmids was generated by exchanging the 5′ β-globin UTR with the new synthetic sequences. For that, synthetic 5′ UTR regions with high ribosome loading were chosen from a database (NCBI Gene Expression Omnibus: GSE114002). The original library ID sequences used for this study are UTR1, 33372; UTR2, 324103; UTR3, 241769; UTR4, 317915; UTR5, 215240; UTR6, 100382. The sequences were slightly modified to include in the 5′ and 3′ ends a HindIII site and a NcoI site that is part of the Kozak sequence, respectively. The synthetic 5′ UTR inserts were generated with two complementary primers (Table S1). Briefly, an equimolar amount of oligonucleotide pairs was heated for 5 min at 95°C and then slowly cooled to room temperature to allow their hybridization. The different ds 5′ UTRs were inserted between the HindIII and NcoI sites of the IVT-148A plasmid, allowing the formation of the IVT-UTR1 to IVT-UTR6 constructs.
All plasmids were propagated with NEB stable competent E. coli cells (NEB, France), and sequences were checked by Sanger sequencing. The FASTA sequences are available (Figure S1).
Production of IVT mRNA
For IVT, the different plasmids were linearized with the type IIS restriction enzyme BspQI and purified with the Monarch PCR & Cleanup Kit (NEB). Capped or uncapped mRNAs were produced with the T7 ULTRA mMESSAGE mMACHINE Kit (Ambion, Life Technologies, France) or the HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB), respectively. The reactions were done according to the manufacturer’s recommendations. Then, the IVT mRNAs were precipitated with LiCl and their integrity evaluated by agarose gel electrophoresis. The enzymatic mRNA capping was performed with the Vaccinia Capping System Kit (NEB) according to the manufacturer’s recommendations. Vaccinia virus-capped mRNAs were precipitated by adding 1 volume of isopropanol 100% and 0.1 volume of 3 M sodium acetate, pH 5.5. After 30 min at −20°C, the RNA solution was centrifuged (19,000 × g, 4°C, 45 min) and the pellet resuspended in nuclease-free water. The enzymatic capping efficiency was validated by fluorescence microscopy after mRNA transfection into HeLa cells.
Post-transcriptional mRNA treatments
To eliminate dsRNA, the IVT mRNAs were subjected to a cellulose-based purification method, as described by Baiersdörfer et al.14 The dsRNA elimination was checked by dot-blotting. Briefly, 1 μg of cellulose-purified mRNA was plotted into a Whatman Nytran SuPerCharge (SPC) nylon blotting membrane (Sigma, France), and the membrane was blocked for 1 h in TBS-T buffer (20 mM Tris [pH 7.4], 150 mM NaCl, and 0.1% [v/v] Tween 20) with 5% (w/v) nonfat dry milk. The mouse monoclonal antibody anti-dsRNA antibody J2 (Scicons, Hungary) was diluted at 1:5,000 in 1% (w/v) milk TBS-T buffer, and the membrane was incubated overnight under agitation at 4°C. The secondary antibody used was the horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) (H+L) (Jackson ImmunoResearch) diluted at 1:5,000 in 1% (w/v) milk TBS-T buffer. The positive control contained 20 ng of the dsRNA ladder (NEB), and negative controls contained nuclease-free water or 1 μg of plasmid DNA. The phosphatase treatment was performed with 15 units of Antarctic phosphatase (NEB) for 40 μg of IVT mRNA. The reaction was then incubated at 37°C for 30 min, and the mRNA was isopropanol purified as previously described. Finally, the mRNA was resuspended in nuclease-free water, and the integrity of the mRNA was evaluated by agarose gel electrophoresis. For the double mRNA treatment, we first purified the mRNA with cellulose and subsequently treated it with Antarctic phosphatase.
Cell culture
HeLa cells were purchased from ATCC (ATCC CCL-2) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies, France) supplemented with 10% fetal bovine serum (Gibco) at 37°C and 5% CO2. DC2.4 cells, which are immortalized murine bone marrow DCs, were kindly provided by Jacqueline Marvel (CIRI, France).30 They were cultured in RPMI 1640 medium supplemented with GlutaMAX (Gibco), 10% fetal bovine serum, 50 μM β-mercaptoethanol, and 10 mM HEPES. Both cell lines were propagated in 48-well plates, and the medium was renewed every 72 h. Cells were used with a low passage number (<10). For the production of human moDCs, venous blood samples were purchased from Etablissement Français du Sang (Décines-Charpieu) and treated as previously described.31 First, density gradient centrifugations using Ficoll-Paque Plus (Cytiva, France) and Percoll (Sigma) allowed isolation of peripheral blood mononuclear cells (PBMCs) from blood samples. Then, final depletion of erythrocytes and natural killer (NK), B, and T cells was realized with anti-glycophorin, anti-CD56, anti-CD19, anti CD3 (Beckman Coulter, France), and anti-CD235a (BD Biosciences, France) antibodies, respectively, and Dynabeads Pan mouse IgG (Invitrogen, France). The differentiation of purified monocytes into moDCs was realized by cultivating the cells for 5 days in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum, gentamycin (50 U/mL), 100 U/mL penicillin, and 100 mg/mL streptomycin in the presence of human interleukin-4 (62.5 ng/mL; Miltenyi Biotec, France) and human granulocyte macrophage colony-stimulating factor (75 ng/mL; Miltenyi Biotec, France).
“Monitoring” of mRNA translation efficiency
The translation efficiency of the different mRNAs was evaluated through the detection of the eGFP reporter protein by fluorescence microscopy or by flow cytometry. One day before the transfection, HeLa cells, DC2.4 cells, or moDCs were seeded in 48-well plates at a density of 3.75 × 105 cells/well. Transfections were realized with the TransIT-mRNA Transfection Kit (Mirus Bio) according to the manufacturer’s recommendations. The transfection complex was prepared by adding 1 μL of mRNA (125 and 250 ng/μL for HeLa cells and DC2.4 cells and moDCs, respectively) into 26 μL of serum-free culture medium and then adding 0.5 μL of each transfecting reagent. Then, each transfection complex solution was added dropwise into the wells, and cells were incubated at 37°C and 5% CO2 until analysis. Some transfections were also realized with Lipofectamine 2000 (Thermo Fisher Scientific, France) according to the manufacturer’s recommendations. The eGFP mRNA (#L-7601) from Trilink BioTechnologies (Tebu, France) was used as a positive commercial control. This mRNA has been capped with their proprietary cap analog Clean-Cap. eGFP expression was (1) qualitatively assessed in adherent cells (i.e., HeLa and DC2.4 cells) by fluorescence microscopy using the Eclipse TE-300 inverted fluorescence microscope (Nikon) and (2) quantitatively measured in all cells (adherent and in suspension) by flow cytometry LSRII (BD Biosciences; AniRA platform in Lyon) with the following protocol. The cells were harvested, centrifugated and resuspended in phosphate-buffered saline (PBS) (Gibco), and kept on ice until their analysis. Cell suspension was incubated with 3 μM propidium iodide (PI) and analyzed 10 min later by flow cytometry analysis. The cells were then gated with DIVA software based on side scatter height (SSC-H), and side scatter area (SSC-A), excluding cell doublets and larger aggregates from the acquisition gate. A SSC-H versus forward scatter area (FSC-A) density plot was used to eliminate debris from the acquisition gate. Dead cells were excluded with the PI (710/750 nm) channel, and eGFP expression was measured with the (525/550 nm) eGFP channel. The compensation between both colors was set up with the auto-compensation tool from the acquisition software. The threshold between eGFP-positive and eGFP-negative cells was defined so that >99.5% of non-transfected cells are considered not expressing eGFP. Data were determined as mean of three independent experiments, each of them including four technical replicates. The results were analyzed with FlowJo software (FlowJo version 10.6.2.). The mean fluorescence intensity (MFI) corresponded to the geometric mean of alive positive eGFP cells.
IFN-α and TNF-α analyses
For IFN-α and TNF-α level quantification, moDC suspension was centrifuged after 24 h of culture and supernatant was analyzed by ELISA according to the manufacturer’s recommendations (Mabtech, Sweden). Poly(I:C) was purchased from InvivoGen (France).
mRNA-LNP production
mRNA-LNP complexes were produced by microfluidics (NanoAssemblr Spark; Precision Nanosystem). Lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (25 mM D-Lin-MC3-DMA, ClinicScience; 5 mM 1,2-distearoyl-sn-glycero-3-phosphocholine 18:0 DSPC, Avanti; 19.25 mM cholesterol, Sigma; 0.75 mM DMG-PEG-2000, Avanti). The self-assembly process of mRNA-LNP complexes was realized by mixing the lipid solution with an acidic aqueous mRNA solution (100 mM sodium acetate, pH 5.3) at a 1:2 volume ratio, with the Spark setting at “5.” An N/P ratio of 7:1 between the cationic amines in the lipids and the anionic phosphate on mRNA was used for the formulations. The LNPs were recovered and resuspended in PBS before being added to the diafiltration column (10-kDa cutoff Amicon Ultra-4 Centrifugal Filter Unit). The diafiltration was carried out at 2,000 × g, 8°C for 30 min. The LNPs were then recovered and resuspended in PBS before being stored at 4°C. The hydrodynamic diameter and the surface charge (zeta potential) of mRNA-LNP complexes were characterized by dynamic light scattering with the Zetasizer Nano ZS (Malvern Instruments, UK). Quantification of mRNA encapsulation into LNPs was performed with RiboGreen (Invitrogen) according to the manufacturer’s instructions. The hydrodynamic diameter, the polydispersity index, the surface charge, and the % of mRNA encapsulation of the produced LNPs-mRNA complexes were between 77 and 110 nm, 0.15 and 0.23, −2.8 and +2.8, and 89% and 91%, respectively.
Zebrafish injection
All animals provided by the PRECI platform (UMS 3444, SFR Biosciences) were maintained at 28°C under 14-h/10-h light/dark cycle. One-cell-stage casper zebrafish embryos were microinjected with 1 nL (20 ng/μL) of mRNA coding for eGFP.32 The embryos were then kept at 28°C until the evaluation of mRNA efficiency through detection of GFP intensity in each injected embryo by fluorescence microscopy and ImageJ analysis. The procedures on adult zebrafish were conducted in accordance with the guidelines of the European Union and French laws and approved by the local animal ethic committee under regulatory governmental authority (CECCAPP, Comité d’Evaluation Commun au Centre Léon Bérard, à l’Animalerie de transit de l’ENS, au PBES et au laboratoire P4 [no. C2EA15], APAFIS #16888-2018082321405041). Eight-month-old casper zebrafish were anesthetized with 0.02% 3-aminobenzoic acid ethyl ester (tricaine) solution (pH 7) and then injected intramuscularly with a micromanipulator in the dorsal muscle in the area in the front left of the dorsal fin with 10 μL of 20 ng/μL mRNA or mRNA-LNP complexes (20 ng/μL) as previously described.28 After injection, fishes were gently transferred into the recovery tank without tricaine until observation 24 and 48 h post-injection with a fluorescent stereoscope from Leica (MZ10F).
Statistical analysis
The eGFP MFI was analyzed with GraphPad Prism (version 8.4.2 for Windows, GraphPad Software, La Jolla CA, USA). The AUC was calculated in kinetic expression experiments. Data were tested for normality with the Shapiro-Wilk test.33 MFIs are represented as the mean ± standard deviation of the mean (SD), and experimental values were compared by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 10−4.
Acknowledgments
We would like to thank Stephane Paul from Gimap and Christophe Guillon for helpful discussion and Danielle Arruda and Pierre Libeau for interesting discussions on mRNA delivery and transfection experiments. We are indebted to Claire Monge and Fanny Charriaud for critically reading and editing the manuscript. We would also like to thank Paul J. Sample and Georg Seelig for providing early access to their 5′ UTR library. We acknowledge the contribution of SFR Biosciences (UMS3444/CNRS, US8/Inserm, ENS de Lyon, UCBL), AniRA-Cytométrie platform, for assistance with flow cytometry, the PLATIM-Microscopie platform, and the PRECI fish facility. This research was funded by HIVERA JTC 2014 (HIV NANOVA) to B.V., by ANR-16-CE20-0002-01 554 (FishRNAVax) to B.V., and by Ministère de la Recherche (MRT to S.L.-F). The doctoral contract of S.L.-F was funded by University Claude Bernard Lyon 1and the doctoral school EDISS 205 and by ANRS ECTZ 119388 to B.V.
Author contributions
S.L.-F., B.V., and J.-Y.E. participated in the conception and design of the study. S.L.-F. created the figures and, together with B.V. and J.-Y.E., wrote the manuscript. S.L.-F. produced the DNA plasmids, synthetized the IVT mRNAs, performed the cell transfection, mRNA purification, fluorescence microscopy, and flow cytometry experiments, and analyzed the samples. J.M. helped in the production of DNA plasmids, IVT mRNA synthesis, and purification. P.M.-G. was involved in moDC preparation and in cell culture experiments. L.V. set up the experiments on zebrafish embryos and adults. E.L. performed experiments on zebrafish embryos and adults and helped with the analysis of the data on zebrafish. E.L. reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.10.007.
Contributor Information
Bernard Verrier, Email: Bernard.verrier@ibcp.fr.
Jean-Yves Exposito, Email: jy.exposito@ibcp.fr.
Supplemental information
References
- 1.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wadman M. Fever, aches from Pfizer, Moderna jabs aren’t dangerous but may be intense for some. Science Insider. 2020 doi: 10.1126/science.abf7805. Published online November 18, 2020. [DOI] [Google Scholar]
- 3.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 5.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 6.Falcone D., Andrews D.W. Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell. Biol. 1991;11:2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferizi M., Aneja M.K., Balmayor E.R., Badieyan Z.S., Mykhaylyk O., Rudolph C., Plank C. Human cellular CYBA UTR sequences increase mRNA translation without affecting the half-life of recombinant RNA transcripts. Sci. Rep. 2016;6:39149. doi: 10.1038/srep39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 10.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M., Jacoby K. pEVL: A Linear Plasmid for Generating mRNA IVT Templates With Extended Encoded Poly(A) Sequences. Mol. Ther. Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M.Z., Asahara H., Tzertzinis G., Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26:345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Vaidyanathan S., Azizian K.T., Haque A.K.M.A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt A., Schwerd T., Hamm W., Hellmuth J.C., Cui S., Wenzel M., Hoffmann F.S., Michallet M.-C., Besch R., Hopfner K.-P. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachs A. The role of poly(A) in the translation and stability of mRNA. Curr. Opin. Cell Biol. 1990;2:1092–1098. doi: 10.1016/0955-0674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira C.C., McCarthy J.E.G. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 21.Oh S., Kessler J.A. Design, Assembly, Production, and Transfection of Synthetic Modified mRNA. Methods. 2018;133:29–43. doi: 10.1016/j.ymeth.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sample P.J., Wang B., Reid D.W., Presnyak V., McFadyen I.J., Morris D.R., Seelig G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019;37:803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U.T., Pestova T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5’ppp- mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson A.L., Pasquinelli A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019;29:191–200. doi: 10.1016/j.tcb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 26.Trepotec Z., Geiger J., Plank C., Aneja M.K., Rudolph C. Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA. 2019;25:507–518. doi: 10.1261/rna.069286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trepotec Z., Aneja M.K., Geiger J., Hasenpusch G., Plank C., Rudolph C. Maximizing the Translational Yield of mRNA Therapeutics by Minimizing 5′-UTRs. Tissue Eng. Part A. 2019;25:69–79. doi: 10.1089/ten.TEA.2017.0485. [DOI] [PubMed] [Google Scholar]
- 28.Myllymäki H., Niskanen M., Oksanen K., Rämet M. Immunization of Adult Zebrafish for the Preclinical Screening of DNA-based Vaccines. J. Vis. Exp. 2018;140:e58453. doi: 10.3791/58453. [DOI] [PubMed] [Google Scholar]
- 29.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinman R.M., Cohn Z.A. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coolen A.L., Lacroix C., Mercier-Gouy P., Delaune E., Monge C., Exposito J.Y., Verrier B. Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials. 2019;195:23–37. doi: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 32.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., Zon L.I. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro S.S., Wilk M.B. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.