Abstract
Recent studies have shown that during apoptosis protein synthesis is inhibited and that this is in part due to the proteolytic cleavage of eukaryotic initiation factor 4G (eIF4G). Initiation of translation can occur either by a cap-dependent mechanism or by internal ribosome entry. The latter mechanism is dependent on a complex structural element located in the 5′ untranslated region of the mRNA which is termed an internal ribosome entry segment (IRES). In general, IRES-mediated translation does not require eIF4E or full-length eIF4G. In order to investigate whether cap-dependent and cap-independent translation are reduced during apoptosis, we examined the expression of c-Myc during this process, since we have shown previously that the 5′ untranslated region of the c-myc proto-oncogene contains an IRES. c-Myc expression was determined in HeLa cells during apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand. We have demonstrated that the c-Myc protein is still expressed when more than 90% of the cells are apoptotic. The presence of the protein in apoptotic cells does not result from either an increase in protein stability or an increase in expression of c-myc mRNA. Furthermore, we show that during apoptosis initiation of c-myc translation occurs by internal ribosome entry. We have investigated the signaling pathways that are involved in this response, and cotransfection with plasmids which harbor either wild-type or constitutively active MKK6, a specific immediate upstream activator of p38 mitogen-activated protein kinase (MAPK), increases IRES-mediated translation. In addition, the c-myc IRES is inhibited by SB203580, a specific inhibitor of p38 MAPK. Our data, therefore, strongly suggest that the initiation of translation via the c-myc IRES during apoptosis is mediated by the p38 MAPK pathway.
The cellular proto-oncogene c-myc is involved in very disparate cellular processes including proliferation, transformation, and cell death (apoptosis) (17, 22). It encodes a transcription factor of the helix-loop-helix/leucine zipper class and binds, in conjunction with its partner Max, to E-box sequences (7, 8, 9). Myc-Max heterodimers are potent activators of transcription, and it has been suggested elsewhere that Myc acts by activating a critical set of target genes (1, 2, 13). The regulation of c-myc expression is complex and occurs at multiple levels, including control of transcription (38), stability of both the mRNA and the protein (8, 35, 36, 51, 56), and by control of translation (11, 43, 46, 53, 59).
In eukaryotic cells, the majority of the control of translation occurs at the initiation stage by a scanning mechanism. This involves the binding of eukaryotic initiation factor 4F (eIF4F), a complex of proteins which includes eIF4E (the cap binding protein), eIF4G [a large protein which acts as a scaffold for the proteins in the complex and has binding sites for eIF4E, eIF4A, eIF3, and poly(A) binding protein], and eIF4A (RNA helicase) to the m7 GpppN cap structure, recruitment of a 40S ribosomal subunit, and scanning to the first AUG codon in the correct context (45). Much of the control of translation initiation is mediated by changes in the phosphorylation states of proteins in the eIF4F complex and their binding partners, e.g., 4EBP1 and 4EBP2 (49). Messages that have large structured 5′ untranslated regions (UTRs) such as c-myc are, under normal cellular circumstances, only poorly translated, and it has been suggested elsewhere that this is because the levels of eIF4E are normally limiting and so restrict the formation of the active eIF4F complexes which are required for translation initiation (15, 33). In agreement with this, in cells overexpressing eIF4E there is an increase in the expression of c-myc (14, 15). We have shown that the control of cap-dependent translational regulation of c-myc is mediated by changes in the phosphorylation of 4EBP1 via the FRAP/mTOR signaling pathway (62).
Translation initiation can also occur by a mechanism that does not require the cap structure, and in this case, ribosomes enter at a region termed an internal ribosome entry segment (IRES), which can be up to 1,000 nucleotides from the 5′ end of the RNA (24–26). IRESs were originally identified in picornaviral RNAs, and upon picornaviral infection, there is often a switch of translation from host-encoded cellular mRNAs to viral transcripts. In some picornaviruses, this is in part mediated by the proteolytic cleavage of eIF4G into an N-terminal and a C-terminal domain serving to bifurcate the eIF4E and eIF4A binding functions (34). This C-terminal domain has been shown to be sufficient to support cap-independent translation, in the absence of eIF4E, providing a rationale for the preferential translation of viral mRNAs due to their internal mechanism of ribosome entry (44, 47). There are now several examples of mammalian mRNAs which contain IRESs, and interestingly, many of them are associated with proteins that are involved in the control of cell growth. These include fibroblast growth factor 2, platelet-derived growth factor, vascular endothelial growth factor, and c-myc (6, 41, 43, 58–60).
One of the main areas of interest in the study of eukaryotic IRESs is the situations in which they are required, especially since initiation of translation of certain IRES-containing mRNAs, e.g., c-myc and fibroblast growth factor 2, can also be cap dependent (14, 31). Given that many viral IRESs function when the host cell cap-dependent translation is severely compromised, one hypothesis is that eukaryotic IRESs will also be used by a cell where the normal scanning cap-dependent mechanism of translation is inactive. In agreement with this, the vascular endothelial growth factor IRES is active during hypoxia when protein synthesis is inhibited (58) and the platelet-derived growth factor IRES is more active during cell differentiation, where protein synthesis rates are also reduced (6).
During apoptosis induced by Fas/CD95L, cap-dependent translation is decreased due to the cleavage of eIF4G by caspase 3 (39, 42). Induction of apoptosis by members of the tumor necrosis factor (TNF) transmembrane receptor family (which includes Fas/CD95) which results from binding to their cognate ligands, e.g., TNF and CD95L, has been well studied (44, 55). Apoptosis induced by TNF-related apoptosis-inducing ligand (TRAIL) has only recently been investigated and is more complex due to the existence of multiple TRAIL receptors (20, 37). The intracellular domains of this family of proteins contain highly conserved “death domains” which aggregate upon induction of apoptosis and recruit a group of proteins which form the death-inducing signaling complex (40, 55). Recruitment of the initiator caspase, caspase 8, into the death-inducing signaling complex results in its activation and in turn leads to activation of downstream effector caspases that are responsible for many of the morphological and biochemical changes associated with apoptosis. Overexpression of c-myc in the absence of the correct survival factors leads to apoptosis (18), and it has been shown previously that this mechanism of apoptotic induction is downstream of CD95 (23). It has been therefore proposed that c-myc could promote the efficacy with which CD95 and its ligand engage the apoptotic machinery of the cell (23). More recently, it has been shown that activation of c-myc triggers the release of cytochrome c from the mitochondria (28).
The role of c-Myc during apoptosis, the short half-life of this protein, and the two alternative mechanisms of c-myc mRNA translation initiation led us to investigate the synthesis of this protein in apoptotic cells. In this paper, we show that c-Myc protein expression in apoptotic HeLa cells, initiated with TRAIL, remains constant for up to 8 h. We demonstrate that c-Myc protein synthesis under these circumstances is initiated from the c-myc IRES, and this is the first example of a specific function which has been ascribed to this region of RNA. We investigated events that lie upstream of IRES-mediated c-Myc protein synthesis and show that signaling through the p38 mitogen-activated protein kinase (MAPK) pathway is required. Hence, c-myc internal initiation was stimulated by overexpression of MKK6, whereas the p38 kinase inhibitor SB203580 inhibited both c-Myc protein expression and internal ribosome entry on dicistronic mRNAs containing the c-myc IRES in apoptotic cells.
MATERIALS AND METHODS
Cell culture.
HeLa cells were maintained on 90-mm-diameter plates in culture at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (GIBCO-BRL) and 10% fetal calf serum (Advanced Protein Products). To induce apoptosis, cells were treated with 0.25 μg of recombinant human TRAIL (37) per ml or 1 μM staurosporine. For treatment with signaling inhibitors, cells were preincubated with medium containing rapamycin (20 nM), SB203580 (40 μM), or an equivalent dilution of solvent (dimethyl sulfoxide) for 1 h before the addition of TRAIL.
Immunoprecipitation.
Cells were labeled and immunoprecipitations were performed as described previously (36). Briefly, 2 × 106 cells, either untreated or used 4 h after the addition of 0.25 μg of TRAIL per ml, were labeled with 250 μCi of [35S]methionine in 1 ml of methionine-free medium for 30 min. After addition of fresh complete medium, cell samples were harvested at 0, 20, 30, and 50 min. Cells were solubilized in antibody buffer (36) and disrupted by passage through a syringe attached to a 21-gauge needle. The samples were precleared by incubation for 1 h at 4°C with mouse immunoglobulin G and protein A/G-agarose (Santa Cruz Biotechnology, Inc.). Myc proteins were immunoprecipitated overnight at 4°C using Myc monoclonal antibody C-33 (Santa Cruz Biotechnology, Inc.). Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the amount of radiolabel incorporated was visualized on a PhosphorImager (Molecular Dynamics). Experiments were performed on three independent occasions.
DNA transfections and reporter gene analysis.
HeLa cells were transfected using the calcium phosphate DNA coprecipitation method as described previously (3). Cells (2 × 105) were transfected with 2 μg of pGL3Rutr (59), 0.5 μg of the MKK6 vectors (50), and 0.2 μg of β-galactosidase construct pcDNA3.1/HisB/LacZ (Invitrogen) as a transfection control. Cells were harvested after 48 h, luciferase expression was determined using the dual-luciferase assay system (Promega), and β-galactosidase expression was determined using a Galactolight Plus system (Tropix). Both activities were measured in an Opticomp-1 luminometer (MGM Instruments). Variations in transfection efficiency were corrected by normalizing luciferase activity to β-galactosidase activity. All assays were performed in triplicate on three independent occasions.
p38 kinase assays.
HeLa cells with or without preincubation with 40 μM SB203580 for 1 h were then treated with 0.25 μg of TRAIL per ml. Cells were harvested at predetermined times, washed in ice-cold phosphate-buffered saline (PBS), and lysed in 250 μl of Triton lysis buffer (20 mM HEPES [pH 7.5]; 137 mM NaCl; 25 mM β-glycerolphosphate; 2 mM NaPPi; 2 mM EDTA; 10% glycerol; 1% Triton X-100; 1 mM phenylmethylsulfonyl fluoride; 2.5 μg of pepstatin, antipain, and leupeptin per ml; 2 mM benzamidine; 0.5 mM dithiothreitol; 1 mM Na3VO4). After centrifugation, the supernatants were incubated with anti-p38 antibody and 4 mg of protein A-Sepharose for 3 h. After this period, the pellets were washed three times with Triton lysis buffer and one time with kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerolphosphate, 25 mM MgCl2, 0.5 mM Na3VO4, 0.5 mM EDTA, 0.5 mM dithiothreitol). Pellets were then resuspended in 30 μl of kinase assay buffer–5 μg of glutathione S-transferase–ATF2 (1–109)–50 μM [γ–32P]ATP (2,000 cpm/pmol) for 30 min at 30°C. Samples were electrophoresed on an SDS–10% polyacrylamide gel, and incorporation of 32P into glutathione S-transferase–ATF2 protein was determined by phosphorimager analysis.
Determination of protein synthesis rates.
For determination of protein synthesis rates in the presence of TRAIL, the method used was that described previously (62).
Cell viability.
Apoptosis was determined using propidium iodide staining of nuclei. Cells (2 × 105 per time point) were washed three times in PBS and then fixed in a solution of 1:1 methanol-acetone for 10 min. Following five washes in PBS, the cells were incubated with 900 μl of PBS–100 μl of propidium iodide (50 μg/ml) at 37°C for 30 min before fluorescence analysis. Apoptosis was assessed by determining the percentage of cells with condensed nuclei relative to the total population of nuclei counted.
SDS-PAGE and Western blotting.
For analysis of c-Myc, eIF4G, and poly(ADP-ribose) polymerase (PARP), cell pellets were solubilized in electrophoresis buffer (50 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 1 mM EDTA, 10% glycerol, and 0.01% bromophenol blue) by sonication. Cell extracts (106 cells per lane) were then analyzed by SDS-PAGE on 7.5 or 10% polyacrylamide 16-cm gels (Bio-Rad), and proteins were transferred to nitrocellulose (Schleicher and Schuell) by electroblotting in transfer buffer (0.2 M glycine, 20 mM Tris, 20% [vol/vol] methanol) for 1.5 h at 85 V. Equal loading of protein was determined on all blots by staining with Ponceau S. Blots were blocked by incubation in 5% skimmed milk in Tris-buffered saline–Tween for 1 to 2 h and then probed with the relevant antibodies for 1 h at room temperature. c-Myc protein was detected using the mouse monoclonal antibody 9E10 (generated by T. Harrison) at a 1:400 dilution, and α-tubulin proteins were detected using a mouse monoclonal antibody (Sigma) at a 1:10,000 dilution. Rabbit polyclonal antibodies used to detect eIF4G were kindly provided by S. Morley (Sussex University) at dilutions of 1:7,000. A mouse monoclonal antibody which was used to detect PARP was obtained from G. Poirier, Laval University, Quebec, Canada. Blots were then incubated with peroxidase-conjugated secondary antibodies raised against mouse or rabbit immunoglobulins and developed using the chemiluminescence reagent Illumin 8 (generated by M. Murray, Department of Genetics, Leicester University).
Northern blot analysis.
Total cellular RNA and poly(A)+-selected (using DynaBeads) mRNA were prepared and analyzed by Northern blotting exactly as described previously (63). DNA probes used for the detection of c-myc and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA species were also as described previously (63).
RESULTS
c-Myc protein levels remain constain during apoptosis when protein synthesis is inhibited.
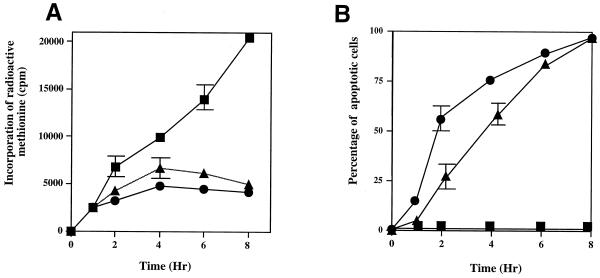
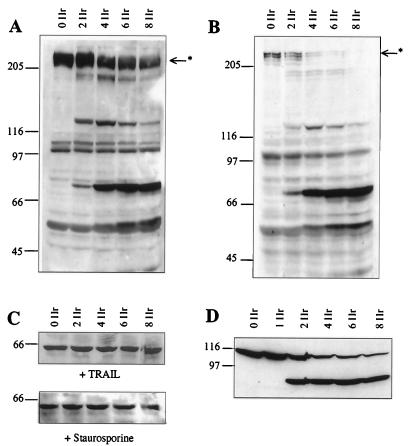
In HeLa cells in which apoptosis has been triggered with either staurosporine or TRAIL, protein synthesis is inhibited within 2 h (Fig. 1). This inhibition of protein synthesis correlates with the cleavage of eIF4G (Fig. 2A and B) that occurs at the same time as cleavage of PARP (Fig. 2D), known to be cleaved by effector caspases. Thus, there is a rapid cleavage of eIF4G into fragments of 150 and 76 kDa and a corresponding appearance of the specific 89-kDa cleavage fragment of PARP (Fig. 2D). The degradation of eIF4G appears to be a general phenomenon during the onset of apoptosis and has also been shown to occur in Jurkat cells treated with anti-CD95 (42), in BJAB cells during serum starvation (12), and in HeLa cells treated with etoposide or cisplatin (39). Since protein synthesis rates are very much decreased during apoptosis, it would be expected that the abundance of proteins that have a short half-life would be greatly reduced a few hours after the onset of apoptosis. The initiation of synthesis of the c-Myc protein (which has a relatively short half-life of between 20 and 40 min [21]) can occur both in a cap-dependent manner and by internal ribosome entry (43, 59, 62). Thus, to determine the steady-state levels of c-Myc protein in HeLa cells undergoing apoptosis, when cap-dependent translation is compromised, Western blots were probed with anti-c-Myc antibodies (Fig. 2C). Interestingly, we found that the levels of this protein remained constant during apoptosis (Fig. 2C). Thus, even 8 h after the onset of apoptosis, when 95% of cells were apoptotic as assessed by propidium iodide staining of condensed nuclei, c-Myc protein levels were unaltered (Fig. 1B and 2C). Consistent with the intimate role of the c-myc gene product in the regulation of cellular proliferation and differentiation, the expression of this proto-oncogene is controlled at multiple levels (38). Thus, there are a number of possible explanations for these data: firstly, levels of c-myc RNA are increased during apoptosis; secondly, there is a change in the stability of c-Myc protein during this process; and finally, the c-Myc protein is translated by an alternative mechanism during apoptosis which does not require full-length eIF4G. To distinguish between these possibilities, the following series of experiments were carried out.
FIG. 1.
Treatment of HeLa cells with either TRAIL or staurosporine causes a general inhibition of translation and a decrease in cell viability. HeLa cells, in triplicate, were incubated in either the absence (closed squares) or the presence of 0.25 μg of TRAIL (closed triangles) per ml or 1 μM staurosporine (closed circles). (A) Protein synthesis was estimated by labeling the cells with [35S]methionine before harvesting at the times indicated. (B) Parallel cultures were used to determine apoptosis by staining with propidium iodide.
FIG. 2.
Inhibition of translation correlates with cleavage of eIF4G and PARP. Cells were incubated with 0.25 μg of TRAIL (A) per ml or 1 μM staurosporine (B) and harvested at the time points shown. Cell extracts (106 cells) were separated by SDS-PAGE, Western blotted, and probed with antibodies to eIF4G and then with the c-Myc antibody 9E10 (C). Samples from the blot shown in panel A were rerun on an SDS–7.5% polyacrylamide gel and then probed with antibodies to PARP (D). Results are representative of at least three independent experiments. The asterisk marks the full-length eIF4G. Numbers to the left of each panel indicate molecular masses in kilodaltons.
During apoptosis in HeLa cells, there is no increase in the level of c-myc mRNA.
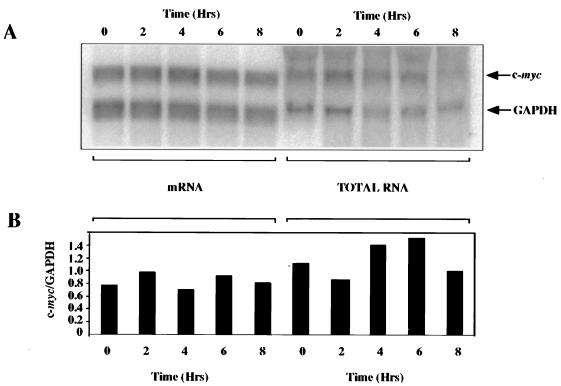
An increase in the levels of c-myc mRNA could occur either by an increase in the transcription of c-myc or by an increase in the stability of the message. To investigate these possibilities, the steady-state level of c-myc mRNA was analyzed during apoptosis. HeLa cells were incubated with TRAIL, total and poly(A)+ mRNA was isolated over 8 h, and Northern blot analysis was performed. There was no change in the ratio of c-myc to GAPDH during apoptosis (Fig. 3), and in the absence of a large induction of mRNA expression, it is evident that increased transcription rates and/or increased mRNA stability is not the key mechanism involved in the expression of c-Myc protein described here. Since c-Myc protein expression has been shown to be subject to regulation at the level of both protein stability and translation (36, 56, 57), it was then necessary to distinguish between these two possibilities.
FIG. 3.
c-myc mRNA levels are unchanged during apoptosis. Poly(A)+ mRNA or total cellular RNA was prepared from samples taken over an 8-h period after the addition of TRAIL and then analyzed by Northern blotting for levels of c-myc mRNA, and levels of the control message GAPDH were determined using specific probes (A). The Northern blot was analyzed using a Molecular Dynamics PhosphorImager, and c-myc mRNA levels were normalized to those of GAPDH (B).
During apoptosis, there is no change in the half-life of c-Myc protein.
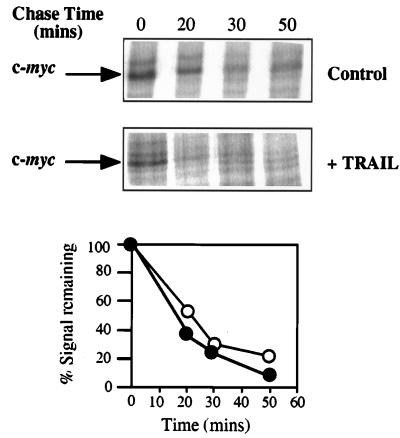
Studies of the rate of degradation of the c-Myc protein using pulse-chase analysis revealed that there was no increase in the stability of the protein 4 h after the addition of TRAIL, a point at which more than 65% of the cells were undergoing apoptosis (Fig. 1C and 4). Indeed, the Myc protein appeared to be slightly less stable during apoptosis, with a half-life of approximately 17 min, compared to the control samples, which had a half-life of approximately 23 min, a value consistent with published data (21, 62). In addition, the amounts of c-Myc protein synthesized during the 30-min labeling time (t = 0) were also similar, demonstrating that Myc is still synthesized from the endogenous message during apoptosis.
FIG. 4.
c-Myc protein stability is unaltered during apoptosis. Determination of the half-life of the c-Myc protein in control HeLa cells and at 4 h post-addition of TRAIL is shown. Pulse-chase analysis and immunoprecipitation were performed as described in Materials and Methods. Cells were labeled for 30 min and then harvested following the chase times indicated. Samples were subjected to PAGE, and the amount of radiolabel incorporated into each band was determined by using a phosphorimager. (A) Representative gels of pulse-chase–immunoprecipitations. (B) Phosphorimager analysis of the gels shown in panel A. Open circles, untreated HeLa cells; closed circles, HeLa cells incubated with TRAIL for 4 h.
Further experiments performed were aimed at determining the mechanism of c-myc translation initiation in apoptosis.
The c-myc IRES is functional during apoptosis.
We and others have shown that the c-myc 5′ UTR contains an IRES and that translation initiation of this message can therefore occur in a cap-independent manner (43, 59). In addition, it has been shown that c-myc is still translated in cells which have been infected with polio virus (27). In this situation, cap-dependent translation of most host cell mRNAs is blocked due to the specific cleavage of eIF4G.
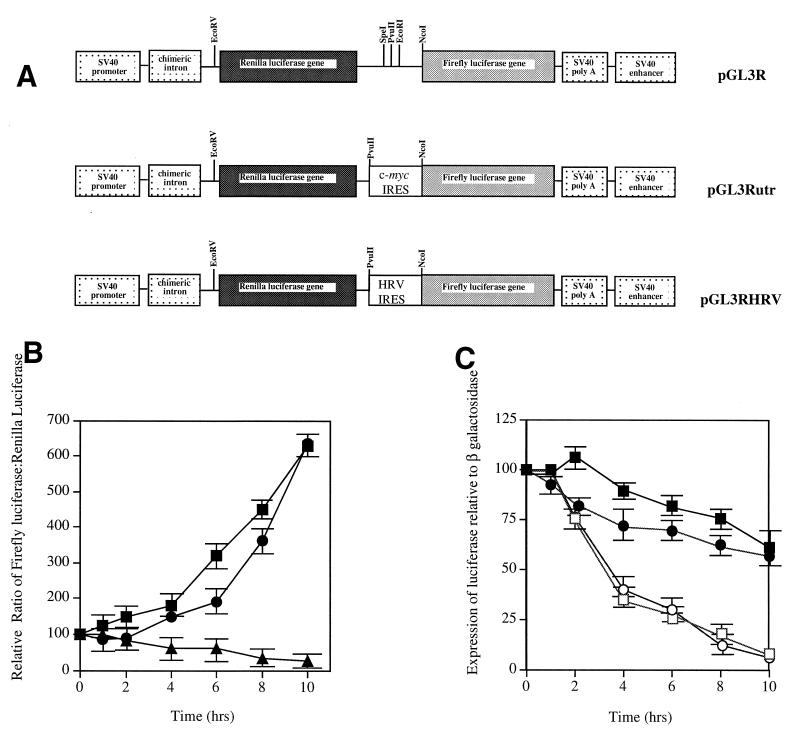
To determine whether the c-myc IRES was active during apoptosis, HeLa cells were transiently transfected with either the control dicistronic plasmid containing the Renilla luciferase gene upstream and the firefly luciferase gene downstream (pGL3R [Fig. 5A]), the dicistronic construct containing the c-myc IRES fused in frame with the firefly luciferase gene (pGL3Rutr [Fig. 5A]) (59), or a plasmid construct containing the human rhinovirus IRES (HRV-IRES, pGL3RHRV [Fig. 5A]). This was used to determine whether a similar effect was observed using an IRES of viral origin (Fig. 5A) (59). Apoptosis was initiated with TRAIL, and the amount of luciferase produced from each cistron was measured over 10 h (Fig. 5B and C). When firefly luciferase activity was normalized to Renilla luciferase activity, there was an apparent increase in firefly luciferase activity following stimulation with TRAIL (Fig. 5B). This suggests that, during apoptosis, cap-dependent translation from the Renilla luciferase cistron was down modulated whereas initiation of translation by internal ribosome entry was maintained. The difference between the synthesis of these proteins cannot be accounted for by a difference in half-life between the firefly luciferase and the Renilla luciferase, since we have found that in HeLa cells they have similar half-lives of approximately 2.5 h, in agreement with previously published data (data not shown and reference 10). Indeed, this interpretation was confirmed when the firefly luciferase activity (IRES mediated) was calculated relative to that of β-galactosidase, which is a more stable protein with a half-life of approximately 20 h (Fig. 5C). In this case, the firefly luciferase actually decreased to approximately 70% 10 h after apoptosis had been triggered, whereas the Renilla luciferase activity decreased to 11%. These data are consistent with the IRES sustaining c-Myc protein expression, as opposed to inducing it, during apoptosis when cap-dependent translation is compromised.
FIG. 5.
The c-Myc IRES is active during apoptosis. (A) HeLa cells were cotransfected (in triplicate) with pcDNA3.1/HisB/LacZ and either pGL3Rutr, pGL3RHRV, or pGL3R. Twenty-four hours after transfection, apoptosis was induced by the addition of 0.25 μg of TRAIL per ml, and samples were harvested at the time points indicated. (B) The expression of firefly luciferase generated from the IRES (by a cap-independent mechanism) was calculated relative to the expression of the upstream Renilla luciferase (cap-dependent expression), pGL3Rutr (closed circles), pGL3RHRV (closed squares), or pGL3R (closed triangles). (C) The expression of firefly luciferase generated from translation initiation of the IRES was also determined relative to that of the transfection control β-galactosidase, pGL3Rutr firefly luciferase (closed circles), pGL3RHRV firefly luciferase (closed squares), pGL3Rutr Renilla luciferase (open circles), and pGL3RHRV Renilla luciferase (open squares). All experiments were performed in triplicate on three independent occasions. SV40, simian virus 40; HRV, human rhinovirus.
To gain further information about the cellular consequences of c-myc expression during apoptosis, the effects of compounds which have been shown to specifically inhibit translation were investigated.
Proteins required for initiation of protein synthesis by the c-myc IRES are downstream of the MAPK pathway.
We have shown recently that cap-dependent translational regulation of c-myc is blocked by rapamycin, suggesting that regulation of this type of translation is mediated by signaling through the FRAP/mTOR pathway (62). Cap-dependent translation is also regulated by both the p38 and extracellular signal-related kinase MAPK pathways, the downstream substrates of which include MNK1 and MNK2, which have been shown to phosphorylate the cap-binding protein eIF4E in vivo and thus modulate its activity (19, 61). However, during apoptosis initiated by either anti-CD95 or TNF alpha, there is a large transient induction of p38 kinase (29, 52), and yet this does not result in the phosphorylation of eIF4E (42). Experiments were therefore performed to address whether cap-independent translation of c-myc via the IRES during TRAIL-induced apoptosis is downstream of either of these signaling pathways.
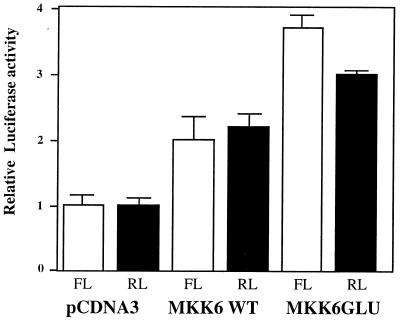
Preincubation of transfected cells with rapamycin before the addition of TRAIL had no effect on the luciferase activity produced from the downstream cistron (our unpublished data); therefore, we investigated whether the MAPK pathways were involved in IRES-mediated translation initiation during apoptosis. Dicistronic plasmid constructs harboring the c-myc IRES were cotransfected with plasmids expressing either wild-type MKK6 (MKK6WT), an immediate upstream activator of p38 MAPK, or a constitutively active form, MKK6Glu (50). Both the wild-type and the constitutively active version of MKK6 cause an increase in the activity of both the Renilla luciferase (cap dependent) and the firefly luciferase (cap independent) (Fig. 6). For the Renilla luciferase, this is not surprising, since downstream substrates in this pathway include MNK1 and MNK2, which phosphorylate the cap-binding protein of eIF4E. However, these data also suggest that the p38 MAPK pathway activates a protein(s) which is required for cap-independent translation.
FIG. 6.
Proteins which mediate internal ribosome entry of c-myc are downstream of MKK6. HeLa cells were cotransfected with the dicistronic c-myc IRES-containing plasmid pGL3Rutr and either pcDNA3 (control empty vector), pcDNA3 containing DNA encoding MKK6WT, or pcDNA3 containing the constitutively active version of this kinase, MKK6Glu. Cells were harvested and assayed for luciferase activity to determine whether coexpression of the dicistronic plasmid with MKK6 increased cap-dependent (Renilla luciferase [RL] expression) and/or cap-independent (firefly luciferase [FL] expression) translation initiation. All experiments were performed in triplicate on three independent occasions.
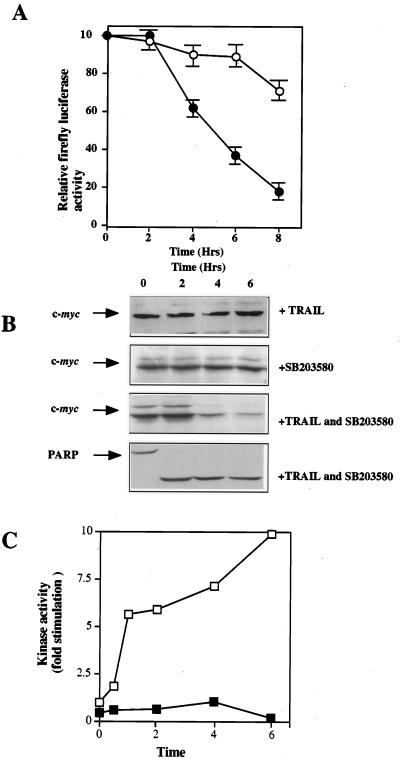
To investigate these observations further, cells (pGL3Rutr) were transfected with the dicistronic plasmid construct incubated with TRAIL either with or without pretreatment with SB203580, and harvested over an 8-h period. As before, (Fig. 5C), relative to the expression of β-galactosidase, the level of firefly luciferase decreased to approximately 70% of the original value by 8 h (Fig. 7A). However, in cells pretreated with SB203580 (which inhibits p38 MAPK [Fig. 7C]), the expression of firefly luciferase derived from the c-myc IRES is very much reduced (Fig. 7A). Hence, after 4 h the expression is decreased to 60%, compared to 93% in the untreated control cells, and by 8 h, the levels of firefly luciferase are reduced to 18% of their original value. To determine whether a similar effect was observed with c-Myc protein expression, HeLa cells were pretreated with SB203580 before the addition of TRAIL and Western blot analysis was performed as before using an anti-c-Myc antibody. Pretreatment with SB203580 alone had no effect on c-Myc expression; however, this inhibitor had a very marked effect on the expression of c-Myc protein during apoptosis; 4 h after the addition of TRAIL, the levels of the c-Myc protein decreased by approximately 70% (Fig. 7B). Taken together, these results add credence to our hypothesis that the c-myc IRES is responsible for maintaining expression of this protein during apoptosis. We posit that the switch from cap-dependent translation initiation to internal ribosome entry is likely to be an early event in the apoptotic pathway. In agreement with previously published data with anti-CD95 and TNF (42, 52), we have found that in cells preincubated with SB203580 apoptosis occurs slightly more rapidly (data not shown). Therefore, the decrease in c-myc expression does not appear to affect the efficiency with which cells initiate apoptosis.
FIG. 7.
The c-myc IRES is downstream of p38 MAPK. (A) cells were cotransfected with dicistronic reporter plasmids containing the c-myc IRES (pGL3Rutr) and β-galactosidase (pcDNA3.1/HisB/LacZ), and half the samples were preincubated for 1 h with 40 μM SB203580. Open circles, absence of inhibitor; closed circles, presence of inhibitor. Cells were then treated with 0.25 μg of TRAIL per ml, and samples were harvested at the time points shown and assayed for firefly luciferase and β-galactosidase activity. The relative ratio of firefly luciferase to β-galactosidase is shown. (B) Cells (106) were incubated with TRAIL with or without a 1-h preincubation with 40 μM SB203580. Samples were harvested at the time points shown, analyzed by PAGE and Western blotting with an anti-c-Myc antibody, and then stripped and reprobed with an anti-PARP antibody. (C) p38 MAPK activity was assayed in the presence (closed squares) or absence (open squares) of SB203580 (preincubation for 1 h) followed by the addition of 0.25 μg of TRAIL per ml. As expected, the activity of this enzyme is induced up to 10-fold during apoptosis, and this induction is inhibited by the presence of the inhibitor.
These data strongly suggest that proteins which are responsible for IRES-mediated translation initiation during apoptosis are downstream of p38 MAPK.
DISCUSSION
Protein synthesis of the proto-oncogene c-myc can be initiated by two mechanisms. Evidence for a cap-dependent scanning mechanism of translation initiation comes from the observations that the translation of c-myc mRNA is increased in cells which overexpress eIF4E (14) and that an increase in the degree of phosphorylation of 4EBP1 (which causes the dissociation of eIF4E from this binding partner) results in an increase in c-Myc expression (62). More recently, we and others have shown that the c-myc 5′-UTR contains an IRES and that therefore c-myc mRNA can be translated in a manner which is independent of the cap structure (43, 59). The c-myc gene encodes four different transcripts, P0, P1, P2, and P3, of approximately 3.1, 2.4, 2.25, and 2.0 kb, respectively (4, 5, 64), and most of this heterogeneity is in the length of the 5′ UTR. The IRES is contained within the 5′ UTR of mRNAs which initiate from P2 (59); thus (as these transcripts represent 75 to 90% of the c-myc mRNA, in most cells), the majority of c-myc mRNA in a cell has the potential to initiate translation via internal ribosome entry.
We have investigated the circumstances in which the c-myc IRES is utilized and have found that it is active during apoptosis (Fig. 3 and 6) when the levels of cap-dependent translation are reduced due to cleavage of eIF4G (Fig. 2) (12, 39, 42). Our data suggest that cleaved eIF4G is sufficient to allow initiation of translation by internal ribosome entry by the c-myc IRES, since 8 h after the induction of apoptosis with staurosporine there is no remaining full-length eIF4G and yet c-Myc protein is still expressed (Fig. 2C). In addition, it has been shown recently that c-myc mRNA is still associated with the translational machinery in cells that have been infected with poliovirus, whereby eIF4G is cleaved subsequent to viral infection (25, 27).
p38 MAPK is activated during apoptosis (29, 52), and yet this is not accompanied by phosphorylation of eIF4E (42), which has been shown to be a downstream target in vivo (19, 61). Our data demonstrate that cotransfection of wild-type and constitutively active MKK6 increases the activity of IRES- mediated translation, suggesting that the c-myc IRES is also downstream of p38 MAPK (Fig. 6). The p38 inhibitor SB203580 blocks both the activity of the firefly luciferase downstream of the c-myc IRES and the expression of c-Myc during apoptosis (Fig. 6 and 7), again strongly suggesting that proteins which are required for internal ribosome entry are downstream of p38 MAPK.
Although c-Myc protein levels are maintained during apoptosis, a reduction in expression of this protein by preincubation with SB203580 does not block this process (data not shown and references 42 and 52). Thus, our data suggest that c-myc expression is not required for cell death in this system. Therefore, why are c-Myc protein levels selectively maintained during apoptosis? The dual hypothesis suggests that c-Myc promotes proliferation and apoptosis simultaneously through the modulation of appropriate target genes (17). Therefore, one possibility is that c-myc expression is required for the transcription of genes that are required at/for the end stage of apoptosis, i.e., engulfment. Cells undergoing apoptosis are normally cleared rapidly in vivo by phagocytes, and phagocytic recognition of “apoptotic self” is of the utmost importance to this process (54). CD14 and CD36 on phagocytic cells are directly involved in tethering to apoptotic cells, although the ligands with which they interact have yet to be defined (16). Therefore, c-myc expression during apoptosis could be required for the transcription of specific cell surface proteins required for phagocyte recognition, and it has been shown elsewhere that RNA synthesis still occurs during late-stage apoptosis (30). In agreement with this, it has been shown previously that c-Myc-overexpressing fibroblasts are more sensitive to the cytotoxic effects of natural killer cell-derived granules, and in coculture experiments natural killer cells were able to efficiently destroy only target cells which overexpressed c-Myc (32).
In conclusion, we show that c-Myc protein synthesis is initiated by internal ribosome entry during apoptosis when cap-dependent translation is reduced. The proteins which mediate internal entry are downstream of p38 MAPK, since inhibition of this kinase ablates expression both of the reporter enzyme and of c-Myc. The downstream function of c-Myc is unknown; however, one possibility which could be investigated is that c-Myc is required to transactivate genes involved in phagocytic recognition of apoptotic self.
ACKNOWLEDGMENTS
Thanks go to Mark Coldwell for help with initial experiments and for critically reading the manuscript.
This work was supported by grants from the Cancer Research Campaign (M.S.) and the Leukaemia Research Fund (S.A.C.). C.L.J. holds an MRC studentship.
REFERENCES
- 1.Amati B, Brooks M W, Levy N, Littlewood T D, Evan G I, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Littlewood T D, Evan G I, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel R M, et al. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1987. pp. 9.11–9.17. [Google Scholar]
- 4.Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, Lenoir G, Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983;34:779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- 5.Bentley D L, Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986;6:3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein J, Sella O, Le S, Elroy-Stein O. PDGF/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site. J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 8.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 9.Blackwood E M, Luscher B, Eisenman R N. Myc and Max associate in vivo. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 10.Bronstein I, Fortin J, Stanley P E, Stewart G S, Kricka L J. Chemiluminescent and bioluminescent reporter gene assays. Anal Biochem. 1994;219:169–181. doi: 10.1006/abio.1994.1254. [DOI] [PubMed] [Google Scholar]
- 11.Butnick N Z, Miyamoto C, Chizzonite R, Cullen B R, Ju G, Skalka A M. Regulation of the human c-myc gene: 5′ noncoding sequences do not affect translation. Mol Cell Biol. 1985;5:3009–3016. doi: 10.1128/mcb.5.11.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens M J, Bushell M, Morley S J. Degradation of eIF4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- 13.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Benedetti A, Joshi B, Graff J R, Zimmer S G. CHO cells are transformed by the translation initiation factor eIF-4E display increased levels of c-myc expression, but require concomitant overexpression of Max for tumorigenicity. Mol Cell Differ. 1994;2:347–371. [Google Scholar]
- 15.De Benedetti A, Rhoads R E. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci USA. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devitt A, Moffatt O C, Raykundalia C, Capra J D, Simmons D L, Gregory C D. Human CD14 mediates recognition and phagocytosis of apoptotic cells. Nature. 1998;392:505–509. doi: 10.1038/33169. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 18.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 19.Fukunaga F, Hunter T. MnK1, a new map kinase-activated protein kinase, isolated by novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin R, Smith C A. The TRAIL of death. Apoptosis. 1998;3:83–88. doi: 10.1023/a:1009640823621. [DOI] [PubMed] [Google Scholar]
- 21.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hueber A O, Evan G I. Traps to catch unwary oncogenes. Trends Genet. 1998;14:364–367. doi: 10.1016/s0168-9525(98)01520-0. [DOI] [PubMed] [Google Scholar]
- 23.Hueber A O, Zornig M, Lyon D, Suda T, Nagata S, Evan G I. Requirement for the CD95 receptor-ligand pathway in c-myc-induced apoptosis. Science. 1997;278:1305–1309. doi: 10.1126/science.278.5341.1305. [DOI] [PubMed] [Google Scholar]
- 24.Jackson R J, Hunt S L, Gibbs C L, Kaminski A. Internal initiation of translation of picornavirus RNAs. Mol Biol Rep. 1994;19:147–159. doi: 10.1007/BF00986957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 26.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 27.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, Bip, and eIF4F conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juin P, Hueber A, Littlewood T, Evan G. c-Myc-induced sensitization to apoptosis is mediated through cytochrome C release. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juo P, Kuo C J, Reynolds S E, Konz R F, Raingeaud J, Davis R J, Biemann H-P, Blenis J. Fas activationb of the p38 mitogen-activated protein kinase signalling pathway requires ICE/CED-3 family proteases. Mol Cell Biol. 1997;17:24–35. doi: 10.1128/mcb.17.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerkhoff E, Ziff E B. Deregulated messenger RNA expression during T cell apoptosis. Nucleic Acids Res. 1995;23:4857–4863. doi: 10.1093/nar/23.23.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kevil C, Carter P, Hu B, DeBenedetti A. Translational enhancement of FGF-2 by eIF-4 factors and alternate utilization of CUG and AUG codons for translation initiation. Oncogene. 1995;11:2339–2348. [PubMed] [Google Scholar]
- 32.Klefstrom J, Kovanen P E, Somersalo K, Hueber A-O, Littlewood T, Evan G I, Greenberg A H, Saksela E, Timonen T, Alitalo K. c-Myc and E1A induced cellular sensitivity to activated NK cells involves cytotoxic granules as death effectors. Oncogene. 1999;18:2181–2188. doi: 10.1038/sj.onc.1202546. [DOI] [PubMed] [Google Scholar]
- 33.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF4E. EMBO J. 1991;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 35.Lee C H, Leeds P, Ross R J. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J Biol Chem. 1998;273:25261–25271. doi: 10.1074/jbc.273.39.25261. [DOI] [PubMed] [Google Scholar]
- 36.Lüscher B, Eisenman R N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988;8:2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacFarlane M, Ahmad M, Srinivasula M, Fernandes-Alnemri T, Cohen G M, Alnemri E S. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J Biol Chem. 1997;272:25417–25420. doi: 10.1074/jbc.272.41.25417. [DOI] [PubMed] [Google Scholar]
- 38.Marcu K B, Bossone S A, Patel A J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 39.Marissen W, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medema J P, Scaffidi C, Kischkel F C, Shevchenko A, Mann M, Krammer P H, Peter M E. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller D, Dibbens J, Damert A, Risau W, Vadas M, Goodall G. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998;434:417–420. doi: 10.1016/s0014-5793(98)01025-4. [DOI] [PubMed] [Google Scholar]
- 42.Morley S J, McKendrick L, Bushell M. Cleavage of translation initiation factor 4G during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- 43.Nanbru C, Lafon I, Audiger S, Gensac G, Vagner S, Huez G, Prats A-C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 44.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF-4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 45.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 46.Parkin N, Darveau A, Nicholson R, Sonenberg N. cis-acting translational effects of the 5′ noncoding region of c-myc mRNA. Mol Cell Biol. 1988;8:2875–2883. doi: 10.1128/mcb.8.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pestova T, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peter M E, Krammer P H. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr Opin Immunol. 1998;10:545–551. doi: 10.1016/s0952-7915(98)80222-7. [DOI] [PubMed] [Google Scholar]
- 49.Proud C G, Denton R M. Molecular mechanisms for the control of translation by insulin. Biochem J. 1997;328:329–341. doi: 10.1042/bj3280329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raingeaud J, Whitmarsh A J, Barrett B, Derijard Y, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roulston A, Reinhard C, Amiri P, Williams L T. Early activation of c-jun N-terminal kinase and p38 kinase regulate cell survival response to tumour necrosis factor α. J Biol Chem. 1997;273:10232–10239. doi: 10.1074/jbc.273.17.10232. [DOI] [PubMed] [Google Scholar]
- 53.Saito H, Hayday A C, Wiman K, Hayward W S, Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci USA. 1983;80:7476–7460. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savill J, Fadok V, Henson P, Haslett C. Phagocyte recognition of cells undergoing apoptosis. Immunol Today. 1993;14:131–136. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 55.Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter M E. Apoptosis signaling by death receptors. Eur J Biochem. 1998;254:439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- 56.Shindo H, Tani E, Matsumuto T, Hashimoto T, Furuyama J. Stabilization of c-myc protein in human glioma-cells. Acta Neuropathol. 1993;86:345–352. doi: 10.1007/BF00369446. [DOI] [PubMed] [Google Scholar]
- 57.Spotts G D, Hann S R. Enhanced translation and increased turnover of c-myc proteins occur during differentiation of murine erythroleukemia cells. Mol Cell Biol. 1990;10:3952–3964. doi: 10.1128/mcb.10.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 60.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases MnK1 and MnK2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West M J, Stoneley M, Willis A E. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 63.West M J, Sullivan N F, Willis A E. Translational upregulation of the c-myc oncogene in Bloom's syndrome cell lines. Oncogene. 1995;13:2515–2524. [PubMed] [Google Scholar]
- 64.Yang J-Q, Bauer S R, Mushinski J F, Marcu K B. Chromosome translocations clustered 5′ of the murine c-myc gene qualitatively affect promoter usage: implications for the site of normal c-myc regulation. EMBO J. 1985;4:1441–1447. doi: 10.1002/j.1460-2075.1985.tb03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]