Abstract
Hypodiploidy with < 40 chromosomes is associated with poor prognosis in B cell precursor acute lymphoblastic leukemia. In some patients, the hypodiploid clone undergoes endoreduplication, resulting in doubling of the number of chromosomes and masquerades as a high hyperdiploid BCP-ALL. Karyotyping reveals metaphases with 50–79 chromosomes masking the hypodiploid clone. Identifying hypodiploidy in such cases requires awareness of non random alterations of chromosomal copy numbers found in hypodiploid BCP-ALL. We used a systematic strategy to identify masked hypodiploidy integrating targeted fluorescence in situ hybridization (FISH) analysis directed towards identifying monosomies of chromosomes 7, 15 and 17 and flow cytometry-based ploidy analysis (FCPA). Of 445 patients diagnosed as BCP ALL, 2.9% (13/445) were classified as hypodiploid including patients with masked hypodiploidy. Karyotype analysis showed hypodiploidy in 3 patients, near triploidy in 4 patients and normal karyotype in 6 patients. Four patients with near triploid clone on karyotype showed either bimodal peak (2 patients) or single low hypodiploid peak (1 patient) or only near triploid peak (1 patient) on FCPA. All 6 patients with normal karyotype revealed either bimodal peak (4 patients) or hypodiploid peak (2 patients) on FCPA. Targeted FISH analysis unmasked hypodiploid clone showing monosomies of chromosomes 7, 15 and 17 in all ten patients. Our algorithm successfully identified masked hypodiploidy in patients, including those with endoreduplication (4 patients) and normal karyotype (6 patients). Integrating FCPA with targeted FISH analysis provides a practical, sensitive and specific approach to identify masked hypodiploidy in low resource settings.
Keywords: Hypodiploidy, Endoreduplication, Near haploidy, FISH, BCP-ALL, Flow cytometry based ploidy analysis
Introduction
Precursor B cell acute lymphoblastic leukaemia (BCP-ALL) is characterized by recurring chromosomal abnormalities that include aneuploidies and rearrangements. Modern treatment protocols use cytogenetic characteristics of ALL blasts to risk stratify and modify therapy. Among the aneuploidies, high hyperdiploidy (> 50 chromosomes) is associated with a favorable outcome whereas hypodiploidy is associated with poor prognosis [1–3].
Hypodiploidy is further classified into high hypodiploidy (40–45 chromosomes), low hypodiploidy (30–39 chromosomes) and near haploidy (25–29 chromosomes), with the latter two being associated with extremely poor prognosis [1, 2, 4–7]. Hypodiploidy has been reported in approximately 1% of childhood and 3–4% of adult BCP-ALL [1, 2, 4].
In some patients with hypodiploidy, the hypodiploid clone duplicates the number of chromosomes through a process called endoreduplication, masking the hypodiploid clone. These patients maybe erroneously categorized as having high hyperdiploidy as karyotyping reveals only metaphases of the endoreduplicated clone with 50–79 chromosomes [2, 4, 7–9]. The presence of the masked hypodiploid clone in these patients is suspected based on the characteristic patterns of chromosomal gains from cytogenetic studies (karyotyping and/or FISH) and/or presence of two peaks (related to the hypodiploid and endoreduplicated clones) on flow cytometry-based ploidy analysis (FCPA) in ALL blasts and by techniques that assess loss of heterozygosity (LOH) i.e. single nucleotide polymorphism(SNP) arrays [10].
Near haploid and low hypodiploid karyotypes commonly retain two copies of chromosomes X, 14, 18 and 21while chromosomes 3, 4, 13, 15, 16, 7 and 17 are commonly monosomic [2, 3, 6]. The endoreduplicated clone classically shows tetrasomies of the disomic chromosomes and disomies of the monosomic chromosomes. However, in some cases the duplication may not be exact resulting in trisomies of chromosomes. This phenomenon of near exact duplication is more common in masked low hypodiploidy than near haploidy [7]. Based on the characteristic pattern of losses and gains of chromosomes in hypodiploid ALL, we have developed a systematic approach to diagnose masked hypodiploidy that uses a combination of targeted FISH analyses using commonly available FISH probes in the laboratory (i.e. probes used to identify ETV6/RUNX1, PML/RARA fusions and additionally, deletion 7(q) and FCPA in lymphoblasts.
Materials and Methods
All consecutive patients included in the study were diagnosed as BCP-ALL based-on morphology and immunophenotyping studies (October 2016–June 2020). Ethical clearance was obtained from Institutional Ethical Committee. Karyotyping and FISH analysis together with FCPA were performed in all the patients. They were managed, on a risk-stratified, standardized, ethically-approved institutional protocol after written informed consent.
FCPA was performed using FxCycle™ Violet (FCV)(assay sensitivity 0.01%) on BD FACS Canto™ II flow cytometer (Becton Dickinson, San Jose, CA). Post immunophenotypic characterization of the tumor cells, the left over labeled and fixed cells were processed for DNA ploidy analysis as per previously described guidelines [11]. 10,000–30,000 events were acquired for each sample at a low rate (approximately 200 events/s). Ploidy data was analyzed on Kaluza 1.3 software using overlay plots. DNA index (DI) was calculated as a ratio of geometric mean (GM) of FCV in G0/G1 peak of blasts (CD19/CD10 and/or CD34 positive) to the GM of FCV in G0/G1 peak of normal lymphocytes. Ploidy was categorized as near-haploid, low-hypodiploid, high hypodiploid, diploid, low-hyperdiploid, high-hyperdiploid, near-triploid, near-tetraploid for DI 0.55– 0.69, 0.70– 0.88, 0.89– 0.95, 0.96– 1.05, 1.06–1.15, 1.16–1.39, 1.40–1.79, and 1.80–2.28, respectively based on published literature (Table 1).
Table 1.
Flow cytometric DNA Index and corresponding modal chromosomal number [11]
| Ploidy group | Flow cytometric DNA ploidy | Modal chromosome number |
|---|---|---|
| Near haploid | 0.55–0.69 | 24–29 |
| Low-hypodiploid | 0.70–0.88 | 31–39 |
| High-hypodiploid | 0.89–0.95 | 40–45 |
| Diploid | 0.96–1.05 | 46 |
| Low-hyperdiploid | 1.06–1.15 | 47–50 |
| High-hyperdiploid | 1.16–1.39 | 51–65 |
| Near-triploid | 1.40–1.79 | 66–80 |
| Near-tetraploid | 1.80–2.28 | 81–102 |
Conventional karyotyping was performed on baseline bone marrow aspirate samples using standard cytogenetic protocols [12]. For each sample, 10–20 GTG banded (G banding with trypsin using Giemsa stain) metaphases were obtained from at least two unstimulated overnight bone marrow cultures (with and without Colcemid). An automated karyotyping system (Metasystems, GmbH, Altlussheim, Germany) was used for analysis.
Routine FISH analysis using the ETV6/RUNX1 dual-colour extra signal fusion probe (Abbott, Illinois, USA), the BCR/ABL1 dual-colour dual-fusion probe and the KMT2A dual-colour break-apart probe (both from ZytoVision GmbH, Bremerhaven, Germany) was performed in all patients using triple probe screening strategy as reported earlier [13].
The karyotyping and FISH results in all the patients were reported as per International System for Human Cytogenomic nomenclature 2016 (ISCN 2016) [14].
The karyotype, FISH findings and FCPA values were analyzed and integrated in all patients. Cases revealing the hypodiploid clone on karyotyping were diagnosed as hypodiploid BCP-ALL.
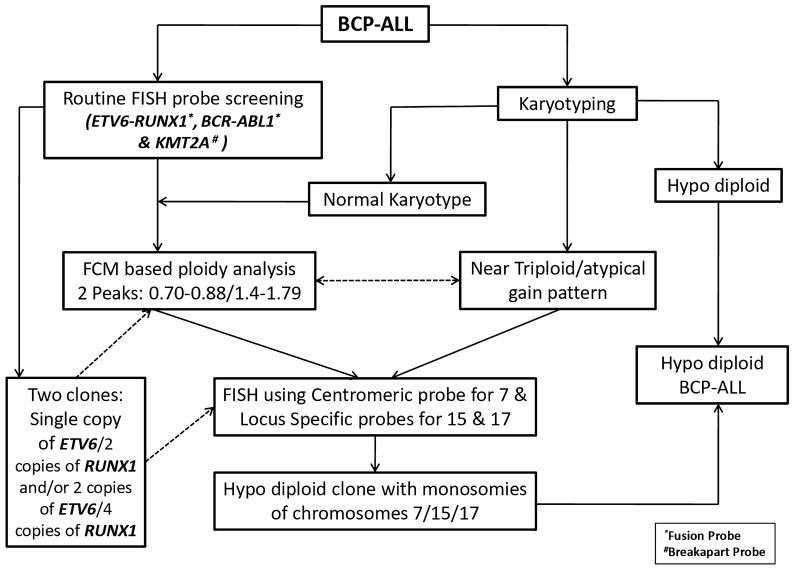
A diagnostic algorithm (Fig. 1) based on a targeted FISH approach to detect monosomies of chromosomes 7, 15 and 17 was used to identify the masked hypodiploid clone in patients whose karyotype, routine FISH analyses and FCPA were suggestive of presence of masked hypodiploidy. The criteria that were considered together to screen for masked hypodiploid clone included (a) near triploidy on karyotyping (b) two clones on ETV6/RUNX1 FISH analysis: single copy of the ETV6 gene with 2 copies of RUNX1or 2 copies of ETV6 gene with 4 copies of RUNX1 gene (c) FCPA showing either a hypodiploid peak, near triploid peak or bimodal peak distribution. Some of these patients had a normal karyotype. Targeted FISH studies were performed using the -7/del(7q) deletion probe (Abbott, Illinois, USA) for monosomy 7 and the PML/RARA dual-colour dual-fusion probe (ZytoVision GmbH, Bremerhaven, Germany).The loss of a single copy of PML or RARA was interpreted as a surrogate marker for monosomy of chromosomes 15 and 17 respectively.
Fig. 1.
Diagnostic algorithm integrating flow cytometry- based ploidy analysis, targeted FISH analysis to identify masked hypodiploid clone
Details of the treatment administered to the patients and follow up course were also documented for the cohort.
Results
Of 445 patients diagnosed as BCP ALL, 2.9% (13/445) were classified as hypodiploid including patients with masked hypodiploidy. Of these, 7 were adults and 6 were children with a median age of 19.5 years (range 4–48 years) and a female predominance (male: female, 1:1.6). The laboratory findings of the patients are provided in Table 2.
Table 2.
Complete blood counts and immunophenotype findings of the patients described in the study
| UPN | Age | Sex | Hb [g/dl] | TLC[× 109/L] | Platelets [× 109/L] | Immunophenotye (IPT) |
|---|---|---|---|---|---|---|
| 1 | 4 | F | 7.9 | 3.5 | 13 | Pre B ALL |
| 2 | 16 | F | 6.2 | 1.8 | 23 | Pre B ALL |
| 3 | 27 | M | 8.3 | 5.2 | 67 | Pre B ALL |
| 4 | 45 | M | 9.2 | 3.6 | 66 | Pre B ALL |
| 5 | 19 | F | 9.9 | 4.2 | 67 | Pre B ALL |
| 6 | 16 | F | 5.5 | 14.6 | 12 | Pre B ALL |
| 7 | 14 | F | 7.7 | 2.1 | 21 | Pre B ALL |
| 8 | 20 | M | 7 | 2.8 | 58 | Pre B ALL |
| 9 | 21 | F | 9.9 | 7.7 | 30 | Pre B ALL |
| 10 | 22 | M | 9 | 1.3 | 53 | Pre B ALL |
| 11 | 48 | F | 5 | 4.1 | 41 | Pre B ALL |
| 12 | 15 | F | 7.5 | 2.5 | 29 | Pre B ALL |
| 13 | 13 | M | 5.5 | 2.7 | 18 | Pre B ALL |
Karyotype analysis showed hypodiploidy in 3 patients (1 near haploid; 2 low hypodiploid), near triploidy in 4 patients and normal karyotype in 6 patients (Table 3).
Table 3.
Table depicting details of karyotyping, flow cytometry-based ploidy analysis results, routine diagnostic FISH panel findings and targeted FISH analysis results
| UPN* | Modal Number | Karyotype | Flow Cytometry-based Ploidy Analysis | FISH Panel (ETV6-RUNX1, BCR-ABL1, KMT2A, TCF3) | ETV6-RUNX1 | BCR-ABL1 |
|---|---|---|---|---|---|---|
| 1 | Near haploid(25) | 25, X, -X, − 2, − 3, − 6, − 7, − 9, − 10, − 11, − 12, − 13, − 15, − 16, − 17, − 19, − 20, − 22[2]/46, XX [13] | 0.58 (Near haploid) | Hypodiploid clone (ETV6, BCR, ABL1, KMT2A single copy) | nuc ish (ETV6 × 1),(RUNX1 × 2)—170 [85%]nuc ish (ETV6 × 2), (RUNX1 × 2)—30 [15%] | nuc ish (BCR × 1), (ABL1 × 1)—148 [74%] nuc ish (BCR × 2), (ABL1 × 2)—52 [26%] |
| 2 | Near triploid (79 ~ 80) | 79 ~ 80, XXX, + X, + 1, + 3, + 5, + 6, + 7, + 10, + 11, + 11, + 12, + 17, + 21, + 22, + 3 ~ 4mar, inc [cp2]/46, XX [18] | 0.79 /1.55 (Low hypodiploid/Near triploid) | Hypodiploid (ETV6 single copy) + duplicated clone | nuc ish (ETV6 × 2),(RUNX1 × 4)—134 [67%] nuc ish (ETV6 × 1),(RUNX1 × 2)—16 [08%] nuc ish (ETV6 × 2),(RUNX1 × 2)—50 [25%] | nuc ish (BCR × 4), (ABL1 × 3)—122 [61%] nuc ish (BCR × 2), (ABL1 × 2)—78 [39%] |
| 3 | Near triploid (64 ~ 77) | 64 ~ 77,XXY, + 1,-2,-3,-4, + 6,-7, + 8,-9, + 10,-12, + 14,-15,-16,-17, + 18, + 19, + 21, + 22, + mar[cp10]/46,XY[8] | 1.43 (Near triploid) | Hypodiploid(ETV6,BCR,ABL1,KMT2A single copy) + duplicated clone | nuc ish (ETV6 × 2),(RUNX1 × 4)—82 [41%] nuc ish (ETV6 × 3),(RUNX1 × 3)—10 [5%] nuc ish (ETV6 × 1), (RUNX1 × 2)—10 [5%] nuc ish (ETV6 × 2),(RUNX1 × 2)—98 [49%] | nuc ish (BCR × 1), (ABL1 × 1)—22 [11%] nuc ish (BCR × 3), (ABL1 × 3)—12 [6%] nuc ish (BCR × 2), (ABL1 × 2)—166 [83%] |
| 4 | Normal Karyotype (46) | 46, XY [2] | 0.80 (Low hypodiploid) | Hypodiploid clone (ETV6 single copy) | nuc ish (ETV6 × 1), (RUNX1 × 2)—48 [24%] nuc ish (ETV6 × 2), (RUNX1 × 2)—152 [76%] | nuc ish (BCR × 2), (ABL1 × 2)—200 [100%] |
| 5 | Near triploid (69 ~ 70) | 69 ~ 70, XXX, + 1, − 2, − 3, − 4, − 7, + 8, − 10, + 13, + 14, − 15, − 16, + 18, + 19, + 20, + 21, + 22, + mar [cp5]/46, XX [14] | 0.73 (Low hypodiploid) | Hypodiploid (ETV6 single copy) + duplicated clone | nuc ish (ETV6 × 1),(RUNX1 × 2)—124[62%] nuc ish (ETV6 × 2),(RUNX1 × 4)—24[12%] nuc ish (ETV6 × 2),(RUNX1 × 2)—52[26%] | nuc ish (BCR × 3), (ABL1 × 3)—16 [8%] nuc ish (BCR × 4), (ABL1 × 3)—12 [6%] nuc ish (BCR × 2), (ABL1 × 2)—172 [86%] |
| 6 | Low hypodiploid (37 ~ 39) | 37 ~ 39, X, -X, − 3, − 7, − 13, − 15, − 16, − 17, − 19, − 20, + 1 ~ 2mar [CP5]/40, X, -X, − 3, − 7, − 16, − 17, − 19, − 20, + mar [1]/46, XX [8] | 0.88 (Low hypodiploid) | Diploid clone | nuc ish (ETV6 × 2),(RUNX1 × 2)—200 [100%] | nuc ish (BCR × 2), (ABL1 × 2)—200 [100%] |
| 7 | Near triploid(61 ~ 70) | 61 ~ 70, XXX, + 1, − 3, + 4, + 5, + 6, − 7, + 8, − 9, + 10, − 11, − 12, − 13, − 14, − 15, − 16, − 17, + 18, − 20, + 21, + 1 ~ 2mar [cp7]/46, XX [13] | 0.84/1.46 (Low hypodiploid/Near triploid) | Duplicated clone only | nuc ish (ETV6 × 2),(RUNX1 × 4 ~ 6) -96 [48%] nuc ish (ETV6 × 2),(RUNX1 × 3)- 24[12%] nuc ish (ETV6 × 2),(RUNX1 × 2)- 80[40%] | nuc ish (BCR × 4),(ABL1 × 2 ~ 3) -90[45%] nuc ish (BCR × 2),(ABL1 × 2) -110[55%] |
| 8 | Normal Karyotype (46) | 46, XY [10] | 0.83/1.56 (Low hypodiploid/Near triploid) | Hypodiploid (ETV6 single copy) + duplicated clone | nuc ish (ETV6 × 1),(RUNX1 × 2)—64[32%] nuc ish (ETV6 × 2),(RUNX1 × 4)—88[44%] nuc ish (ETV6 × 2),(RUNX1 × 2)—48[24%] | nuc ish (BCR × 4),(ABL1 × 4)—78 [39%] nuc ish (BCR × 2),(ABL1 × 2)—122 [61%] |
| 9 | Normal Karyotype (46) | 46, XX [6] | 0.82/1.58 (Low hypodiploid/Near triploid) | Hypodiploid (ETV6 single copy) + duplicated clone | nuc ish (ETV6 × 1),(RUNX1 × 2)—124[62%] nuc ish (ETV6 × 2),(RUNX1 × 4)—40[20%] nuc ish (ETV6 × 2),(RUNX1 × 2)—36[18%] | nuc ish (BCR × 3 ~ 4), (ABL1 × 3 ~ 4)—40 [20%] nuc ish (BCR × 2), (ABL1 × 2)—160 [80%] |
| 10 | Normal Karyotype (46) | 46,XY[20] | 0.86/1.65 (Low hypodiploid/ Near triploid) | Hypodiploid (ETV6, ABL1 single copy) + duplicated clone | nuc ish (ETV6 × 1),(RUNX1 × 2)—06[03%] nuc ish (ETV6 × 3),(RUNX1 × 4)—42[21%] nuc ish (ETV6 × 2),(RUNX1 × 2)—152[76%] |
nuc ish (BCR × 4),(ABL1 × 2)—70[35%] nuc ish (BCR × 2),(ABL1 × 1)—14[07%] nuc ish (BCR × 2),(ABL1 × 2)—116[58%] |
| 11 | Normal Karyotype (46) | 46, XX [20] | 0.91/1.61 (High hypodiploid/Near triploid) | Duplicated clone only | nuc ish (ETV6 × 2),(RUNX1 × 3)-14[07%] nuc ish (ETV6 × 3),(RUNX1 × 6)-12[06%] nuc ish (ETV6 × 2),(RUNX1 × 2)-174[87%] | nuc ish (BCR × 3 ~ 4),(ABL1 × 3 ~ 4)-18[09%] nuc ish (BCR × 2),(ABL1 × 2)-182[91%] |
| 12 | Low hypodiploid (37) | 37, XX, − 3, − 4, − 5, − 7, − 8, − 9, − 13, − 16, − 17, − 19, − 20 [cp8]/46, XX [12] | 0.83 (Low hypodiploid) | Hypodiploid clone (ABL1 single copy) | nuc ish (ETV6 × 2),(RUNX1 × 2)—200[100%] | nuc ish (BCR × 2),(ABL1 × 1)—34 [17%] nuc ish (BCR × 2),(ABL1 × 2)—166 [83%] |
| 13 | Normal Karyotype (46) | 46, XY [20] | 0.81 (Low hypodiploid) | Hypodiploid clone (ETV6 single copy) | nuc ish (ETV6 × 1),(RUNX1 × 2)—172[86%] nuc ish (ETV6 × 2),(RUNX1 × 2)—28[14%] | nuc ish (BCR × 2),(ABL1 × 2)—200[100%] |
| UPN* | KMT2A | Targeted FISH Panel | MONOSOMY 7 | MONOSOMY 15 | MONOSOMY 17 | Final Diagnosis |
|---|---|---|---|---|---|---|
| 1 | nuc ish (MLL × 1)—134 [67%] nuc ish (MLL × 2)—66 [33%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—132 [66%] nuc ish (CEP7 × 2),( D7S486 × 2)—68[34%] | nuc ish (PML × 1), (RARa × 1)—142 [71%] nuc ish (PML × 2), (RARa × 2)—66 [33%] | nuc ish (PML × 1), (RARa × 1)—142 [71%] nuc ish (PML × 2), (RARa × 2)—66 [33%] | Near haploid |
| 2 | nuc ish (MLL × 5)—156 [78%] nuc ish (MLL × 2)—44 [22%] | Monsomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—18 [09%] nuc ish (CEP7 × 2), (D7S486 × 2)—182 [91%] | nuc ish (PML × 1), (RARa × 1)—14 [07%] nuc ish (PML × 4), (RARa × 2)—186 [93%] | nuc ish (PML × 1), (RARa × 1)—14 [07%] nuc ish (PML × 4), (RARa × 2)—186 [93%] | Low hypodploid/Near triploid |
| 3 | nuc ish (MLL × 1)—20 [10%] nuc ish (MLL × 3)—20 [10%] nuc ish (MLL × 2)—160 [80%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—28 [14%] nuc ish (CEP7 × 2), (D7S486 × 2)—172 [86%] | nuc ish (PML × 1),(RARa × 1)—16 [08%] nuc ish (PML × 2),(RARa × 2)—184 [92%] | nuc ish (PML × 1), (RARa × 1)—16 [08%] nuc ish (PML × 2),(RARa × 2)—184 [92%] | Low hypodploid/Near triploid |
| 4 | nuc ish (KMT2A × 2) – 200 [100%] | Monosomy 7 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—20 [10%] nuc ish (CEP7 × 2), (D7S486 × 2)—180 [90%] | ND# | nuc ish (CEP17 × 1) (TP53 × 1)—26 [13%] nuc ish (CEP17 × 2) (TP53 × 2)—174 [87%] | Low hypodiploid |
| 5 | nuc ish (KMT2A × 3 ~ 4)—44[22%] nuc ish (KMT2A × 2)—156[78%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—38 [19%] nuc ish (CEP7 × 2), (D7S486 × 2)—162 [81%] | nuc ish (PML × 1), (RARa × 1)—46 [23%] nuc ish (PML × 3), (RARa × 3)—08 [4%] nuc ish (PML × 2), (RARa × 2) -146 [73%] | nuc ish (PML × 1), (RARa × 1)—46 [23%] nuc ish (PML × 3), (RARa × 3)—08 [4%] nuc ish (PML × 2), (RARa × 2) -146 [73%] | Low hypodploid/Near triploid |
| 6 | nuc ish (KMT2A × 2) – 200 [100%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—146[73%] nuc ish (CEP7 × 2), (D7S486 × 2) – 54 [27%] | nuc ish (PML × 1), (RARa × 1)—138 [69%] nuc ish (PML × 2), (RARa × 2)—62 [31%] | nuc ish (PML × 1),(RARa × 1)—138 [69%] nuc ish (PML × 2),(RARa × 2)—62 [31%] | Low hypodiploid |
| 7 | nuc ish (KMT2A × 3 ~ 4) – 80 [40%] nuc ish (KMT2A × 2) – 120 [60%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1) – 40 [20%] nuc ish (CEP7 × 2), (D7S486 × 2) – 160 [80%] | nuc ish (PML × 1), (RARa × 1)—40 [20%] nuc ish (PML × 2), (RARa × 2)—160 [80%] | nuc ish (PML × 1), (RARa × 1)—40 [20%] nuc ish (PML × 2), (RARa × 2)—160 [80%] | Low hypodploid/Near triploid |
| 8 | nuc ish (KMT2A × 3 ~ 6) – 112 [56%] nuc ish (KMT2A × 2) – 88 [44%] | Monosomy 7 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—82 [41%] nuc ish (CEP7 × 2), (D7S486 × 2)—118 [59%] | nuc ish (PML × 2), (RARa × 1)—86 [43%] nuc ish (PML × 4), (RARa × 2)—60 [30%] | nuc ish (PML × 2), (RARa × 1)—86 [43%] nuc ish (PML × 4), (RARa × 2)—60 [30%] | Low hypodploid/Near triploid |
| 9 | nuc ish (KMT2A × 3 ~ 4) – 80 [40%] nuc ish (KMT2A × 2) – 120 [60%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—116[58%] nuc ish (CEP7 × 2), (D7S486 × 2) – 84 [42%] | nuc ish (PML × 1) (RARa × 1)—130[65%] nuc ish (PML × 2) (RARa × 2) -70 [35%] | nuc ish (PML × 1) (RARa × 1)—130[65%] nuc ish (PML × 2) (RARa × 2) -70 [35%] | Low hypodploid/Near triploid |
| 10 | nuc ish (KMT2A × 3 ~ 4) – 58 [29%] nuc ish (KMT2A × 2) – 142 [71%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—24[12%] nuc ish (CEP7 × 2), (D7S486 × 2)—176[88%] | nuc ish (PML × 1), (RARa × 1)—28[14%] nuc ish (PML × 2), (RARa × 2)—172[86%] | nuc ish (PML × 1), (RARa × 1)—28[14%] nuc ish (PML × 2), (RARa × 2)—172[86%] | Low hypodploid/Near triploid |
| 11 | nuc ish (KMT2A × 4 ~ 6)—24 [12%] nuc ish (KMT2A × 2)—176 [88%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)-12[06%] nuc ish (CEP7 × 2), (D7S486 × 2)-188[94%] | nuc ish (PML × 1), (RARa × 1)-10[05%] nuc ish (PML × 2), (RARa × 1)-12[06%] nuc ish (PML × 2), (RARa × 2)-178 [89%] | nuc ish (PML × 1), (RARa × 1)-10[05%] nuc ish (PML × 2), (RARa × 1)-12[06%] nuc ish (PML × 2), (RARa × 2)-178 [89%] | Low hypodploid/Near triploid |
| 12 | nuc ish (KMT2A × 2)—200 [100%] | Monosomy 7 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1)—46[23%] nuc ish (CEP7 × 2), (D7S486 × 2)—46[77%] | nuc ish (PML × 2), (RARa × 1)—50 [25%] nuc ish (PML × 2), (RARa × 2)—150 [75%] | nuc ish (PML × 2), (RARa × 1)—50 [25%] nuc ish (PML × 2), (RARa × 2)—150 [75%] | Low hypodiploid |
| 13 | nuc ish (KMT2A × 2)—200 [100%] | Monosomy 7, 15 and 17 detected | nuc ish (CEP7 × 1), (D7S486 × 1) – 146 [73%] nuc ish (CEP7 × 2), (D7S486 × 2) – 54 [27%] | nuc ish (PML × 1), (RARa × 1)—170 [85%] nuc ish (PML × 2), (RARa × 2)—30 [15%] | nuc ish (PML × 1), (RARa × 1)—170 [85%] nuc ish (PML × 2), (RARa × 2)—30 [15%] | Low hypodiploid |
*UPN unique patient number
#ND not done
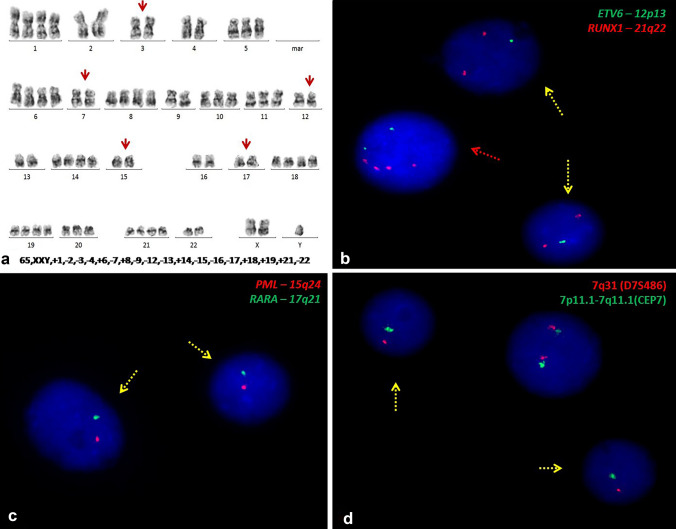
Of the 4 patients with near triploidy on karyotyping, the FCPA showed the presence of a hypodiploid clone in three patients. Two of these showed bimodal peaks ranging in near triploid and hypodiploid values (0.79 & 1.55; 0.84 & 1.46) indicating the presence of endoreduplication and one patient showed only the low hypodiploid peak (0.73) on FCPA. Both patients with bimodal peaks on FCPA showed monosomies of chromosomes 7, 12, 15 and 17 and 4 copies of RUNX1, BCR and KMT2A genes on FISH analysis. The patient with a single low hypodiploid peak (UPN-5) had monosomies of 7, 15 and 17 on targeted FISH analysis. The one remaining patient (UPN-3, Fig. 2) with a near triploid karyotype showed only a near triploid peak on FCPA (1.43). Masked low hypodiploidy was confirmed in this patient based on FISH analysis which demonstrated the presence of monosomies of chromosome 7, 15 and 17. Single copies of ETV6, BCR, ABL1 and KMT2A genes were also recorded together with 4 copies of the RUNX1 gene.
Fig. 2.
a GTG banded karyogram (UPN-3)showing an endoreduplicated with tetrasomies/trisomies of chromosomes 1, 5, 6, 8, 10, 11, 14, 18, 19, 20 and 21 and gain of sex chromosome, X. b Interphase FISH analysis using ETV6-RUNX1single fusion extra signal probe shows two clones: one clone with single copy of ETV6 gene and two copies of RUNX1gene (1G2R) depicting monosomy of chromsome 12 and another clone showing two copies of ETV6 gene with four copies of RUNX1 gene (2G4R) indicating duplicated clone. c Interphase FISH analysis using PML-RARadual fusion dual colour probe reveals cells with single copies of both PML and RARa (clone with modal number 65 showing disomies of chromsomes 2, 3, 4, 7, 9, 12, 13, 15, 16, 17 and 22 along 1R1G) genes suggestive of monosomies of chromosomes 15 and 17 respectively. d Interphase FISH examination using -7/del(7q) probe shows nuclei with 1G1R pattern suggestive of monosomy of chromosome 7
Of the six patients with a normal karyotype, 4 showed bimodal peaks on FCPA including low hypodiploidy/near triploidy (0.83 and 1.56; 0.82 and 1.58; 0.86 and 1.65) in three patients and high hypodiploidy/near triploidy in one (0.91 and 1.61). Monosomies of chromosomes 7, 15 and 17 were detected in all four on targeted FISH analysis. Single copies of ETV6 and ABL1 genes were seen in one of these patients (UPN-10).The remaining two patients demonstrated hypodiploidy with a single hypodiploid peak (DNA index of 0.8 and 0.81 respectively). Monosomies of chromosomes 7 and 17 was noted in one case (UPN- 4) while another one (UPN-13) had monosomies of chromosomes 7, 15 and 17 along with a single copy of the ETV6 gene on FISH analysis.
All the thirteen patients in cohort received treatment as per Indian Childhood Collaborative Leukaemia Group (ICiCLe) ALL-14 high risk protocol or Berlin-Frankfurt-Munster (BFM 95) or (BFM 2002) protocol. Post induction complete morphological remission (CMR) was achieved in all the patients however, FCM based measurable residual disease (MRD) analysis was positive in 3 out of 12 patients (MRD ≥ 10–3 cells). The overall median follow-up was 12 months (range 2–61 months) during which six patients relapsed, the majority of them were late relapse (duration > 6 months). Three out of six patients who relapsed succumbed to death owing to the severity of disease and/or bacterial or fungal infections within 4 months after relapse. Two of the relapsed patients did not continue therapy and opted for supportive treatment. Single patient was on maintenance therapy post Hyper CVAD regimen for relapsed disease during last follow up. Two patients underwent allogenic stem cell transplant post induction while the rest five are on either consolidation or maintenance phase of their treatment regimen (2- consolidation; 3- maintenance).
Discussion
Aneuploidy in BCP ALL arising out of loss of chromosomes is subdivided into near-haploidy (25–29 chromosomes), low-hypodiploidy (30–39 chromosomes)and high-hypodiploidy (40–45 chromosomes)[1, 7, 15]. Using our algorithm and integrated analysis, we identified hypodiploidy in 1·8%(6/329) of paediatric BCP ALL patients and 6.0% (7/116) of adult BCP- ALL patients, proportions that are slightly higher than reported in western literature [1, 2]. Patients with nearhaploid and low hypodiploid ALL usually have low total leukocyte count (< 50 × 109/L) at presentation [1, 2, 4, 16]. Similar findings were observed in our patients. Nine out of thirteen patients had pancytopenia with a median leukocyte count of 3.5 × 109/L at presentation.
Endoreduplicated hypodiploid clones pose a diagnostic challenge in distinguishing from true high hyperdiploid ALL. In high hyperdiploid ALLs, gain of chromosomes occurs mostly as trisomies while the endoreduplicated clone in hypodiploid cases harbors tetrasomies and trisomies of disomic chromosomes from the hypodiploid clone together with whole chromosome uniparental isodisomies of the monosomic chromosomes when analyzed usingSNP arrays [2, 8, 10, 15, 17]. It is not clear whether there is any additional advantage of chromosomes doubling in a hypodiploid clone. A few authors have described differential expression of genes present on heterodisomic chromosomes [2, 16, 17]. It has been hypothesized that endo-reduplication may be an attempt by cancer cells to restore/normalise the gene copy numbers thereby increasing the viability of the cell [2].
In the absence of SNP array studies, a systematic scrutiny of patterns of gains and losses of chromosomes helps identify cases of masked (i.e. either with normal karyotype or endoreduplication) hypodiploidy. Chromosomes X/Y, 8, 10, 14, 18, and 21 are retained (disomies) in near haploid ALL while chromosomes X/Y, 1, 5, 6, 8, 10, 11, 14, 18, 19, 21, and 22 are retained (disomies) in low hypodiploid ALL [1, 2, 7, 18]. In both low hypodiploidy and near haploidy ALL, chromosomes 3, 7, 15 and 17 are almost always monosomic and doubling of chromosomes 3, 4, 13, 16, 2 and 12 occurs only rarely [4, 7, 8, 18].
In our case series, 4 of 10 cases revealed near triploidy on karyotype. The karyotype analysis showed disomies for chromosomes 3, 7, 15 and 17. However, targeted FISH analysis revealed small clones showing monosomies of chromosomes 7, 15 and 17 in these cases of masked hypodiploidy. Rest of the 6 cases presented with normal karyotype on chromosomal analysis, however, our integrated approach (combined FCPA and targeted FISH analyses) helped us to identify masked hypodiploid clone in all the cases.
Discrepancies between Cytogenetics and DNA index by flow cytometry have been decribed in the literature [19]. We observed discordant results in FCPA (hypodiploid with DI of 0.80 and 0.81 respectively) and karyotyping (diploid) in UPN 4 and UPN 13, and hypodiploidy was diagnosed based on the targeted FISH analysis. Similarly, UPN 8, 9, 10 and 11 showed bimodal peak distribution on FCPA, and a diploid clone on karyotyping, targeted FISH based analysis identified monosomies of chromosomes 7, 15 and 17 confirming the diagnosis of hypodiploidy.
In our cohort, UPN 3 would have been simply misdiagnosed as Heh/NT, had it not been further examined using targeted FISH strategy which identified hypodiploid clone showing monosomies of chromosome 7, 15 and 17 retreating the fact that FCPA, karyotyping and targeted FISH analysis are complimentary techniques which help in identifying hypodiploid as well as duplicated clone in cases of BCP-ALL.
It has already been well established that hypodiploid BCP-ALL confers an extremely unfavorable prognosis in pediatric as well as adult population. Both near haploidy and low hyodiploid childhood BCP-ALL are associated with a dismal outcome, with 5–8-year event-free survival rates ranging from25 to 50% [2, 5–7, 20]. Event free survival (EFS) rate further plunges down to 9% in relapsed childhood hypodiploid BCP-ALLs treated under ALL REZ BFM 2002 trial [21].Similarly, adult patients with low hypodiploidy have an extremely poor EFS of 0–20% despite been treated upfront with high risk protocols [4, 5]. On the other hand, high hyperdiploidy is associated with standard risk. Therefore, it is critical to differentiate between the two entities considering genetic (TP53 alterations) and prognostic implications [10, 18]. Also, an optimal risk stratification will lead to timely intensification of treatment in hypodiploid patients.
Children’s Oncology Group (COG) reported that around 25% of hypodiploidy in children with BCP-ALL may be masked [8]. Microsatellite panel analysis along with FCPA was used in the COG study to identify masked hypodiploidy. In our case series, 50% of the masked hypo diploid paediatric patients (3/6 patients) were identified using the targeted FISH approach which revealed the minor hypodiploid clone. In one patient (UPN-3), the FCPA showed only the near triploid peak and the masked hypodiploid clone was detected by identifying monosomy of chromosome 7 and single copies of ETV6, BCR, ABL1, KMT2A, PML and RARA genes clearly. The caveat to the approach is that in some patients, the monosomic clone cannot be identified and the only way to diagnose the hypodiploid clone is through SNP array analysis.
SNP microarrays or microsatellite panel analysis reveal copy neutral loss of heterozygosity in patients with masked hypodiploidy [1, 8, 15, 17, 22]. In limited resource countries, SNP array analysis is not available and even if the infrastructure is present, the cost of the tests is prohibitive. Hence a cytogenetic strategy based on an integrated FCPA and targeted FISH analyses using FISH probes commonly available in diagnostic laboratories provides a practical, sensitive and specific method to identify masked hypodiploidy, facilitating optimal risk stratification in patients with BCP-ALL.
Acknowledgements
Prof. Vaskar Saha, Prof. Mammen Chandy and Prof. Reena Nair for their support, guidance and scientific inputs. Mr. Harshit Rao for assisting with the figures.
Funding
This work was partially funded by the Department of Biotechnology Govt of India (DBT) project under Grant No. BT/PR12046/MED/12/665/2014.
Compliance with Ethical Standards
Ethical Approval
All procedures performed in the study were in accordance with the ethical standards of the Institutional Research Committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moorman AV. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 2012;26:123–135. doi: 10.1016/j.blre.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Safavi S, Paulsson K. Near-haploid and low-hypodiploid acute lymphoblastic leukemia: two distinct subtypes with consistently poor prognosis. Blood. 2017;129:420–423. doi: 10.1182/blood-2016-10-743765. [DOI] [PubMed] [Google Scholar]
- 3.Heerema NA, Raimondi SC, Anderson JR, et al. Specific extra chromosomes occur in a modal number dependent pattern in pediatric acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46:684–693. doi: 10.1002/gcc.20451. [DOI] [PubMed] [Google Scholar]
- 4.Charrin C. A report from the LALA-94 and LALA-SA groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2 possible expressions of a sole entity conferring poor prognosis in adult acute lymphoblastic leukemia (ALL) Blood. 2004;104:2444–2451. doi: 10.1182/blood-2003-04-1299. [DOI] [PubMed] [Google Scholar]
- 5.Nachman JB, Heerema NA, Sather H, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110:1112–1115. doi: 10.1182/blood-2006-07-038299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raimondi SC, Zhou Y, Mathew S, et al. Reassessment of the prognostic significance of hypodiploidy in pediatric patients with acute lymphoblastic leukemia. Cancer. 2003;98:2715–2722. doi: 10.1002/cncr.11841. [DOI] [PubMed] [Google Scholar]
- 7.Harrison CJ, Moorman AV, Broadfield ZJ, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol. 2004;125:552–559. doi: 10.1111/j.1365-2141.2004.04948.x. [DOI] [PubMed] [Google Scholar]
- 8.Carroll AJ, Shago M, Mikhail FM, et al. Masked hypodiploidy: Hypodiploid acute lymphoblastic leukemia (ALL) mimicking hyperdiploid ALL in children: a report from the children’s oncology group. Cancer Genet. 2019;238:62–68. doi: 10.1016/j.cancergen.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma SK, Chan GCF, Wan TSK, et al. Near-haploid common acute lymphoblastic leukaemia of childhood with a second hyperdiploid line: a DNA ploidy and fluorescence in-situ hybridization study. Br J Haematol. 1998;103(3):750–755. doi: 10.1046/j.1365-2141.1998.01044.x. [DOI] [PubMed] [Google Scholar]
- 10.Inaba H, Mullighan CG. Pediatric acute lymphoblastic leukemia. Haematologica. 2020 doi: 10.3324/haematol.2020.247031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta N, Parihar M, Banerjee S, et al. FxCycleTM based ploidy correlates with cytogenetic ploidy in B-cell acute lymphoblastic leukemia and is able to detect the aneuploid minimal residual disease clone. Cytom B Clin Cytom. 2019;96:359–367. doi: 10.1002/cyto.b.21765. [DOI] [PubMed] [Google Scholar]
- 12.Parihar M, Kumar JA, Sitaram U, et al. Cytogenetic analysis of acute myeloid leukemia with t(8;21) from a tertiary care center in India with correlation between clinicopathologic characteristics and molecular analysis. Leuk Lymphoma. 2012;53:103–109. doi: 10.3109/10428194.2011.603447. [DOI] [PubMed] [Google Scholar]
- 13.Parihar M, Singh MK, Islam R, et al. A triple-probe FISH screening strategy for risk-stratified therapy of acute lymphoblastic leukaemia in low-resource settings. Pediatr Blood Cancer. 2018;65:e27366. doi: 10.1002/pbc.27366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGowan-Jordan J, Simons A, Schmid M. An International system for human cytogenomic nomenclature (ISCN 2016). Reprint of: cytogenetic and genome research. Basel, New York: Karger; 2016. [Google Scholar]
- 15.Choi SM, Papenhausen P, Wertheim G, King RL. Near-haploid B lymphoblastic leukemia with an apparent hyperdiploid karyotype: the critical role of SNP analysis in establishing proper diagnosis. J Hematop. 2014;7:27–32. doi: 10.1007/s12308-013-0189-5. [DOI] [Google Scholar]
- 16.Safavi S, Olsson L, Biloglav A, et al. Genetic and epigenetic characterization of hypodiploid acute lymphoblastic leukemia. Oncotarget. 2015;6:42793–42802. doi: 10.18632/oncotarget.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mühlbacher V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%: characterization of Low Hypodiploid ALL. Genes Chromosomes Cancer. 2014;53:524–536. doi: 10.1002/gcc.22163. [DOI] [PubMed] [Google Scholar]
- 19.Almozain N, Mashi A, Al-Sweedan S, et al. Discrepancies between DNA index by flow cytometry and cytogenetic studies in childhood B-lymphoblastic leukemia. J Appl Hematol. 2018;9(2):45–50. doi: 10.4103/joah.joah_14_18. [DOI] [Google Scholar]
- 20.Heerema NA, Nachman JB, Sather HN, et al. Hypodiploidy with less than 45 chromosomes confers adverse risk in childhood acute lymphoblastic leukemia: a report from the children’s cancer group. Blood. 1999;94(12):4036–4045. [PubMed] [Google Scholar]
- 21.Groeneveld-Krentz S, Schroeder MP, Reiter M, et al. Aneuploidy in children with relapsed B-cell precursor acute lymphoblastic leukaemia: clinical importance of detecting a hypodiploid origin of relapse. Br J Haematol. 2019;185:266–283. doi: 10.1111/bjh.15770. [DOI] [PubMed] [Google Scholar]
- 22.Creasey T, Enshaei A, Watts K, et al. Single nucleotide polymorphism array-based signature of genetic ploidy groups in acute lymphoblastic leukemia. Blood. 2019;134:1473–1473. doi: 10.1182/blood-2019-122556. [DOI] [Google Scholar]