Abstract
Intestinal functions demonstrate circadian rhythms thought to be entrained, in part, by an organisms’ intrinsic feeding and fasting periods as well as by the intestinal microbiome. Circadian disruption as a result of ill-timed nutrient exposure and obesogenic feeding poses an increased risk to disease. As such, the aim of this study was to assess the relationships between dietary timing, composition, and the microbiome with regard to rhythmic small intestinal structure and mucosal immunity. Rodent chow (RC)-mice exhibited time-dependent increases in small intestinal weight, villus height, and crypt depth as well as an increased proportion of CD8αα+ cells and concomitant decrease in CD8αβ+ cells at the onset of the feeding period (p < 0.05–0.001). Western diet (WD)-animals displayed disrupted time-dependent patterns in intestinal structure and lymphocyte populations (p < 0.05–0.01). Antibiotic-induced microbial depletion abrogated the time- and diet-dependent patterns in both RC- and WD-mice (p < 0.05–0.001). However, although germ-free-mice displayed altered rhythms, fecal microbial transfer from RC-mice was generally unsuccessful in restoring structural and immune changes in these animals. This study shows that adaptive changes in the small intestine at the onset of the feeding and fasting periods are disrupted by WD-feeding, and that these changes are dependent, in part, on the intestinal microbiome.
Subject terms: Physiology, Gastroenterology
Introduction
While circadian rhythms have developed over evolutionary time as an adaptive response to external light–dark cycles, peripheral tissues, including the gastrointestinal (GI) tract, exhibit diurnal rhythms in the absence of direct exposure to these cycles1,2. Hence, although the GI tract does receive input from the master clock expressed within the suprachiasmatic nuclei, the gut intestinal epithelial cells (IECs) also demonstrate self-sustained rhythmic expression of the circadian clock genes that are entrained by extrinsic cues originating from both ingested nutrients and the microbiome3–11. These rhythms are established by a transcriptional-translational feedback loop between the positive (i.e. Arntl (aka Bmal1)/Clock) and negative (i.e. Per1-3/Cry1-2) arms of the core clock12, and have been estimated to affect the expression of 43% of the genes in the mammalian genome13.
Over several decades, circadian misalignment due to the disruption of clock gene expression has been linked to impaired human health. Hence, ill-timed exposure to light and/or nutrients leads to an increased risk for metabolic disease, as documented in individuals who conduct shift work and in normal rodents exposed to constant light or obesogenic diets12,14–16. In parallel, clock gene mutant rodent models also demonstrate impaired metabolic homeostasis17–21. However, both GI dysfunction and microbial dysbiosis have been also reported for humans undergoing shift work and rodents subjected to food-, light- or genetically-induced circadian disruption5,7,8,22–25. Consistent with these findings, IEC proliferation also follows a 24-h rhythm, which is abolished in Clock mutant and Arntl knockout mice26,27. Furthermore, important digestive and absorptive functions of the intestine, as well as barrier integrity, all follow a circadian rhythm as a result of clock-controlled expression of key enzymes, transporters, and tight junction proteins11.
While the majority of the small intestinal epithelium is comprised of nutrient-absorbing enterocytes, intraepithelial lymphocytes (IELs) dispersed between the IECs function to establish mucosal immune responses against invading pathogens and to maintain epithelial integrity9,28. The other major intestinal lymphocyte population resides in the lamina propria, deemed lamina propria lymphocytes (LPLs). Like IELs, LPLs are critical for defense against intestinal pathogens, but they also serve a role in establishing the IEL cell population through specific changes in homing markers28. Diurnal patterns in the immune system have been demonstrated in several peripheral tissues, including the intestine, whereby the number of immune cells is reduced during the fasting period through the induction of apoptosis29. The circadian regulation of immune cell populations in the intestine is believed to result, at least in part, from immune responsiveness to dietary antigen exposure following food intake9,29. However, the factors that regulate the daily patterns in IELs and LPLs remain of great interest, as the SI must maintain dietary antigenic tolerance while simultaneously providing immune protection during the rhythmic cycles of feeding and fasting.
Recent findings have demonstrated that both the IECs and the intestinal immune system are profoundly influenced by the microbiome4,5,8,9. Germ-free (GF) mice display reduced IEC proliferation in association with a thinning of the intestinal mucosa30. Intestinal microbes have been further demonstrated as essential for intestinal repair, as mice deficient for innate immune toll-like receptors exhibit reduced IEC proliferation, survival, and barrier function31. Moreover, the intestinal microbiome has been well documented as a fundamental regulator of intestinal lymphocyte localization and function32. Furthermore, recent evidence supports the notion that disruption of the microbiome through dietary changes or antibiotic use leads to a perturbation in overall SI homeostasis, including disrupted gene expression, barrier integrity, and cytokine production5,6,9.
While there appears to be an essential interaction between the circadian rhythms expressed by the intestinal epithelium, the intestinal lymphocytes, and the intestinal microbiome, the exact relationship between these key determinants of GI function remains unclear. The goal of the present study was, therefore, to interrogate the effects of both diet and microbial dysbiosis on circadian intestinal homeostasis. Thus, at the onset of the active/feeding and rest/fasting periods, regular chow (RC)- and high-fat/high-sucrose Western diet (WD)-fed mice, as well as RC- and WD-animals with antibiotic-induced microbial disruption (AIMD) and GF-mice with and without fecal microbial transfer (FMT) from RC-mice were used to characterize the intimate relationship between an intact microbiome, intestinal structure and mucosal immunity. The findings indicate that time- and diet-dependent changes in SI gravimetric and morphometric parameters as well as in intestinal lymphocyte populations are, at least in part, dependent on an intact intestinal microbiome.
Results
The intestinal microbiome regulates time- and diet-dependent structural changes in the small intestine
To characterize time-dependent changes in the SI of normal mice, RC-fed, male and female mice were examined at the onset of the established rodent fasting and feeding periods, Zeitgeber times 2 and 14 (ZT; hours after lights-on), respectively. SI wet weight increased by 19% at ZT14 as compared ZT2, with no differences by time observed in SI length (Fig. 1a and b). To account for the time-of-day differences in SI weight independent of length, ileal villus height and crypt depth were also determined. Villus height as well as crypt depth were significantly increased at ZT14 as compared to ZT2 by 12% and 11%, respectively (Fig. 1c and d; Supplementary Figure 1). Together, the increases in intestinal weight, villus height, and crypt depth at ZT14 suggest anticipatory changes in the SI mucosa in preparation for the onset of the animals’ feeding period.
Figure 1.
WD-feeding and microbial disruption alter time-dependent patterns in SI gravimetric and morphometric parameters. (a) SI weight, (b) SI length, (c) ileal villus height, and (d) ileal crypt depth in RC- and WD-fed animals with and without AIMD measured at ZT2 and ZT14. n = 3–6 male and 3–6 female mice. *p < 0.05 **p < 0.01 ***p < 0.001.
To establish the effects of dietary alterations on the gravimetric and morphometric changes in the SI with time-of-day, mice were fed a WD for 16 weeks. Unexpectedly, the increase in SI weight between ZT2 and ZT14 that was observed in RC-animals was lost in the WD-fed mice, as a result of increased weight at ZT2 (Fig. 1a). Furthermore, an overall decrease in SI length was observed in these animals at both time points (Fig. 1b). In keeping with loss of the rhythm in SI weight, villus height in WD-animals was also increased at ZT2, with no differences observed in crypt depth (Fig. 1c and d; Supplementary Figure 1). These findings suggest the existence of diet-driven temporal effects on SI structure.
As one potential regulator of the time- and diet-dependent changes in the SI is the intestinal microbiome, depletion of the microbiome was accomplished by administration of antibiotics (AIMD). As previously reported at the phyla level in these animals33, microbial dysbiosis in the RC AIMD-animals was characterized by a massive increase in the relative abundance of colonic Proteobacteria as compared to RC mice. Parallel analysis at the family level confirmed microbial disruption as RC AIMD mice were predominantly colonized by Burkholderiaceae (Supplementary Figure 2). RC-fed AIMD-mice were also found to have increased SI weight and length at both ZT2 and 14, along with abrogation of their normal diurnal rhythm in SI weight (Fig. 1a and b). Interestingly, this was accompanied by a loss in the normal time-dependent difference in villus height but, again, with no changes in crypt depth (Fig. 1c and d; Supplementary Figure 1). In contrast, antibiotic-treated WD-fed mice, the colons of which were colonized almost exclusively by the Streptococcaceae family (Supplementary Figure 2), a member of the Firmicutes phylum33, did not demonstrate the increases in SI weight and length that were observed in RC AIMD-animals (Fig. 1a and b) and, indeed, demonstrated decreased villus height and crypt depth at ZT14 as compared to WD-fed mice (Fig. 1c and d; Supplementary Figure 2). Together, these findings suggest a role for the microbiome in both the time- and diet-dependent changes in SI structure.
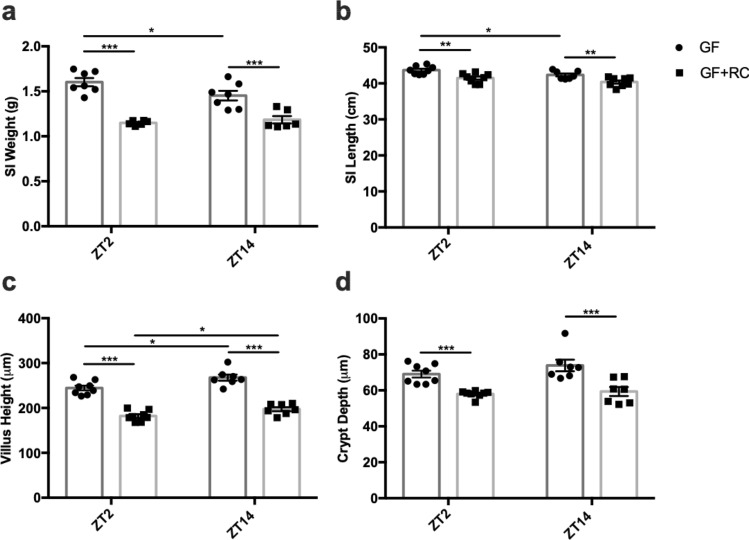
To establish the essentiality of the microbiome in the structural changes in the SI with time, GF-mice were examined without (control) and with transplantation of feces from conventional RC-mice (GF + FMT). Successful recolonization was achieved as evidenced by the presence of major phyla33 and families (Supplementary Figure 3) that were observed in normal mice. However, it is important to note that the relative abundance of select families differed between the RC and GF + FMT mice (Supplementary Figure 2 and 3). GF-mice displayed SI weights and lengths that were comparable to those observed in RC-AIMD animals (Fig. 2a and b vs Fig. 1a and b) and, as found in the antibiotic-treated mice, did not demonstrate the normal time-dependent changes in SI weight. Interestingly, FMT into the GF-mice reduced both SI weight and length to levels that were comparable to those observed in conventional RC-animals; however, SI weight did not regain the time-dependent difference between ZT2 and ZT14. Consistent with the findings made for SI weight, GF-mice demonstrated villi and crypts that appeared slightly longer than normal (Fig. 2c and d vs. Figure 1c and d; Supplementary Figure 4), although the normal time-dependent pattern in villus height was maintained. Furthermore, both villus height and crypt depth were significantly reduced in GF + FMT mice, to levels that were comparable to those observed in the RC-animals, with maintenance of the temporal pattern in villus height. When taken with findings in the AIMD animals, these results demonstrate a key role for the microbiome in the establishment of temporal patterns in SI structure.
Figure 2.
FMT reduces SI gravimetric and morphometric parameters independent of time. (a) SI weight, (b) SI length, (c) ileal villus height, and (d) ileal crypt depth in GF and GF-mice following FMT with RC-microbiome (GF + RC) measured at ZT2 and ZT14. n = 3–4 male and 3–4 female mice. *p < 0.05 **p < 0.01 ***p < 0.001.
The intestinal microbiome regulates time- and diet-dependent changes in small intestinal gene expression
Given that both diet and the intestinal microbiome have been shown to alter the intestinal epithelial circadian clock and immune homeostasis4–9,33, expression of the core clock genes, Arntl and Per2 as well as several markers of intestinal inflammation (Tnf, Ifng, Il6, Il10,Tgfb1) were examined in the SI mucosa. In line with previous evidence33,34, Arntl and Per2 exhibited anti-phasic expression in normal mouse SI, with greater expression of Arntl at ZT2 as compared to ZT14, and of Per2 at ZT14 versus ZT2 (Fig. 3a and b). Although previous reports indicate that obesogenic diets disrupt metabolic tissue clock gene expression35,36, including the intestinal enteroendocrine L cell33, WD-feeding did not significantly alter the levels of, or patterns in mucosal clock gene expression. However, microbial dysbiosis disrupted SI clock gene expression in both RC- and WD-fed mice, in a diet-dependent manner (Fig. 3a and b). Hence, RC-AIMD animals demonstrated increased Arntl expression at ZT2 and a complete loss of rhythmic Per2 expression, whereas WD-AIMD mice lost rhythmic Arntl expression (Fig. 2a and b).
Figure 3.
Intestinal mucosal gene expression is altered by WD-feeding and microbial disruption. (a) Arntl, (b) Per2, (c) Tnf, (d) Ifng, (e) Il6, (f) Il10, and (g) Tgfb mRNA expression measured from SI mucosa isolated at ZT2 and ZT14 from RC- and WD-fed animals with and without AIMD. n = 2–6 male and 2–6 female mice. *p < 0.05 **p < 0.01 ***p < 0.001.
Analysis of select immune cytokines also revealed time-dependent expression of genes that was differentially affected by diet and AIMD. RC SI mucosal Tnf gene expression demonstrated decreased expression at ZT14 as compared to ZT2, a pattern that was lost with both WD-feeding and AIMD (Fig. 3c). Expression of other pro-inflammatory cytokines, Ifng and Il6, did not display time-dependent differences in RC-mice and was not significantly altered by feeding or AIMD (Fig. 3d and e). In contrast to Tnf, the anti-inflammatory cytokine, Il10, had elevated expression at ZT14 compared to ZT2 in RC-fed mice only, which was reversed by AIMD (Fig. 3f). Furthermore, Tgfb1 expression was largely unaffected by diet, but was significantly reduced in RC-AIMD mice at the ZT2 time point, with a similar trend at ZT14 (Fig. 3g).
Interestingly, in the complete absence of the microbiome, GF-animals maintained rhythmic anti-phasic Arntl and Per2 expression, which was not altered following FMT (Fig. 4a and b). In contrast, GF-mice did not demonstrate the normal patterns in Tnf and Il10 that were observed in RC-mice (Fig. 4c–g), although Il10 did regain rhythmic expression in GF + FMT mice, with increased levels at ZT14 (Fig. 4f). The minimal changes in gene expression seen following FMT in GF-mice may speak to the need for a microbial-immune interaction from birth32.
Figure 4.
Intestinal mucosal gene expression is partially restored following FMT. (a) Arntl, (b) Per2, (c) Tnf, (d) Ifng, (e) Il6, (f) Il10, and (g) Tgfb mRNA expression measured from SI mucosa isolated at ZT2 and ZT14 from GF and GF-mice following FMT with RC-microbiome (GF + RC) measured at ZT2 and ZT14. n = 3–4 male and 3–4 female mice. *p < 0.05 **p < 0.01 ***p < 0.001.
Differential effects of obesogenic feeding and microbial disruption on the diurnal patterns in intestinal lymphocytes
To characterize time-, diet- and microbiome interactions within the intestinal immune cell population, the IEL and LPL compartments were characterized in RC- and WD-mice, with and without antibiotic treatment. Examination of the IEL profile revealed both time- and diet-dependent changes, which appeared to be largely dependent on the existence of an intact intestinal microbiome. Thus, the proportion of CD8αβ+ IELs was elevated at ZT2 as compared to ZT14 in RC-mice, whereas WD-mice lacked a rhythm, having significantly reduced CD8αβ+ at ZT2 (Fig. 5a). AIMD significantly increased the proportion of CD8αβ+ IELs in both RC- and WD-animals at both ZT2 and 14, resulting in a loss of the normal rhythm. Furthermore, the highest proportions of CD8αβ+ IELs were observed in the WD-AIMD animals. In contrast, time-dependent differences in CD8αα+ IELs in RC-mice were opposite to those of the CD8αβ+ IELs, showing increased proportions at ZT14 (Fig. 5b). WD-feeding resulted in loss of rhythm in CD8αα+ IELs as a result of increased proportions observed at ZT2 (Fig. 5b). In addition, both RC- and WD-AIMD animals lost the rhythm in CD8αα+ IELs in association with a reduction in this cell population in WD-AIMD mice at both time points (Fig. 5b). As found for the CD8αβ+ IELs, the proportion of CD4+ cells was also decreased at ZT14 as compared to ZT2 in RC-mice, whereas WD-fed animals lost this rhythm in combination with increased CD4+ cells at ZT14 (Fig. 5c). AIMD in RC-mice also demonstrated reduced proportions of CD4+ cells at ZT2 while, in WD-fed AIMD-animals, this cell population was reduced at ZT14 (Fig. 5c).
Figure 5.

Time, diet, and microbial disruption alter proportions of SI IEL cell populations. Proportions of (a) CD8αβ+, (b) CD8αα+, and (c) CD4+ on TCRβ+ cells acquired from the SI epithelium at ZT2 and ZT14 from RC- and WD-fed animals with and without AIMD. n = 3–6 male and 3–6 female mice. *p < 0.05 **p < 0.01 ***p < 0.001.
Analysis of CD8αβ+ IELs in GF-mice revealed no time-dependent changes, although the overall proportions of these cells were greater than observed in either normal RC-mice or in RC-AIMD animals (Fig. 6a vs. Fig. 5a). Although, FMT failed to re-establish the rhythm in this cell population, the presence of a microbiome did reduce the overall proportion of these IELs at both time points (Fig. 6a). Interestingly, the time-dependent changes in the proportion of CD8αα+ IELs were maintained in both GF-mice and in GF-mice with FMT (Fig. 6b). However, the CD4+ IEL population lacked a significant rhythm in GF-mice, and this was restored following FMT (Fig. 6c).
Figure 6.

Time-dependent changes in SI IEL cell populations are partially restored following FMT in GF-mice. Proportions of (a) CD8αβ+, (b) CD8αα+, and (c) CD4+ on TCRβ+ cells acquired from the SI epithelium at ZT2 and ZT14 from GF and GF-mice following FMT with RC-microbiome. n = 2–3 male and 2–3 female mice. *p < 0.05 **p < 0.01.
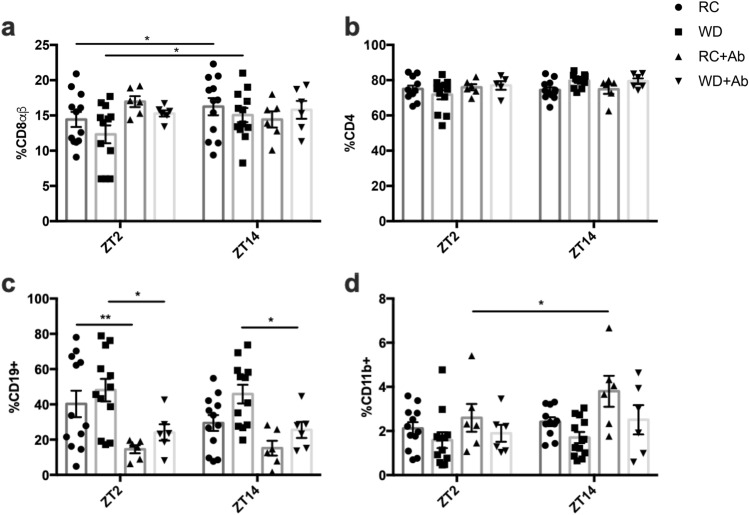
In contrast to the patterns observed in the IELs, the proportion of CD8αβ+ cells in the LPL compartment revealed opposing time-dependent changes in RC-mice, with an increase at ZT14 that was maintained upon WD-feeding (Fig. 7a). Interestingly, AIMD in both RC- and WD-fed mice resulted in loss of this rhythm (Fig. 7a). The CD4+ LPLs remained constant irrespective of time, diet, and microbial disruption (Fig. 7b). However, while analysis of the LPL B-cell (CD19 +) population revealed no time- or diet-dependent changes, consistent with previous reports37, antibiotic administration decreased the cell proportion under both diet conditions (Fig. 7c). Furthermore, no time-, diet-, or treatment-dependent differences were observed in the proportion of myeloid cells (CD11b +) within the LPL (Fig. 7d). Germ-free animals did not display time-dependent differences in CD8αβ+ IELs (Fig. 8a). However, as also observed for the proportion of CD8αβ+ IELs, FMT in GF-animals failed to re-establish the normal RC rhythm in CD8αβ+ LPLs, but did reduce the proportion of this cell population (Fig. 8a). CD4+ LPLs in GF and GF-mice following FMT remained unchanged, as also seen in RC- and WD-mice with and without microbial depletion (Fig. 8b vs Fig. 7b). Interestingly, the proportion of LPL B-cells was increased at the ZT14 time point in both GF and GF-FMT animals (Fig. 8c), despite no time-dependent patterns in RC-mice (Fig. 7c). In contrast, minimal changes were observed in the proportions of LPL myeloid cells in these animals (Fig. 8d).
Figure 7.
Time-dependent changes are lost in LPL %CD8αβ+ cells following microbial disruption. Proportions of (a) CD8αβ+and (b) CD4+ on TCRβ+ cells, as well as (c) CD19 + B-cells and (d) CD11b + myeloid cells acquired from the SI lamina propria at ZT2 and ZT14 from RC- and WD-fed animals with and without AIMD. n = 3–6 male and 3–6 female mice. * p < 0.05.
Figure 8.
FMT in GF-mice alters proportion of CD8αβ+ but not CD4+ SI LPL cell populations. Proportions of (a) CD8αβ+and (b) CD4+ on TCRβ+ cells, as well as (c) CD19 + B-cells and (d) CD11b + myeloid cells acquired from the SI lamina propria at ZT2 and ZT14 from GF and GF-mice following FMT with RC-microbiome. n = 2–3 male and 2–3 female mice. *p < 0.05.
Discussion
The SI is the major site of dietary exposure, carrying out the critical roles of nutrient digestion and absorption11. Given the short lifespan of the IECs (3–5 days), the intestine has been deemed the most highly regenerative organ in the body, accounting for 20–30% of whole-body protein synthesis38. At the same time, the balance between IEL and LPL immune cell populations ensures that the SI can conduct immunosurveillance while maintaining an immunotolerant environment following dietary challenge. The findings of the present study demonstrate that both structure and immune functions of the SI exhibit patterns that are not only influenced by time-of-day, but also by diet and the presence of a normal intestinal microbiome.
Intestinal growth is affected not only by crypt cell proliferation, but also by IEC use of dietary nutrients as an energy source38. Hence, changes in intestinal growth, as determined by analyses of intestinal weight and crypt-villus height, have been observed in response to changes in nutrient intake in murine models of fasting and re-feeding39,40. Indeed, crypt-villus height is reduced by a remarkable 19% in 24-h fasted mice, and refeeding for 24-h completely restores the SI architecture38. Furthermore, an approximately 18% increase in small intestinal villus height within a 6-h period has been reported in piglets following feeding of medium chain triacylglycerol41. These studies are consistent with the 12% difference in villus height observed during the 12-h between the onset of fasting and onset of feeding observed herein. Similarly, following an extreme fasting period, food intake in snakes is associated with a doubling in intestinal mass as a result of a six-fold increase in microvillus length42. The concomitant increase in intestinal weight and villus height observed in RC-animals at ZT14, the onset of the feeding period in rodents, is therefore consistent with SI growth being induced by the increase in food intake.
The IEC that line the villi constitute the absorptive surface of the small intestine, and small intestinal surface area is thus the critical physiological determinant of the absorption of orally ingested substances, in rodents as well as humans43–45. For example, in the rat jejunum, the surface area of the villi amplifies that of the mucosa by 5-fold43,44. It can therefore be estimated that the 12% increase in villus height, as observed between ZT2 and ZT14, represents a 12% increase in absorptive capacity, even without taking into account any additional contributions by the microvilli, which amplify the absorptive surface area by an additional 21-fold43,44. Such analyses have largely been conducted to determine the translatability of studies on oral drug absorption from rodents to humans44,45 but, indeed, villus height positively correlates with nutrient absorption in a variety of different animals models46,47 as well as following administration of long-acting agonists of the intestinal growth factor, glucagon-like peptide-2 (GLP-2) in rodents and humans48–50. Furthermore, reductions in villus height, such as observed in congenital-, environmental- and infection-induced enteropathies are associated with malabsorption and diarrhea, some of which can be reversed by restoration of villus height51–54. Finally, a recent study has demonstrated using both gain- and loss-of-function approaches that a 25% increase in villus length and the associated expansion of intestinal surface area leads to enhanced absorption of dietary fat55. Therefore, in keeping with the evolutionary presentation of the circadian system, it can be hypothesized that the 12% increase in villus height observed herein during the transition from ZT2 to ZT14 is anticipatory, to enhance the absorptive capacity of SI and thus, ensure increased energy availability during the normal rodent feeding period.
Of importance, the increased intestinal weight, villus height, and crypt depth in RC-mice at ZT14 corresponds to the previously identified peak in circadian secretion of intestinal incretin hormone Glucagon-like Peptide-1 (GLP-1)33. GLP-1 receptor agonists are known to enhance SI weight through the induction of crypt fission56. However, co-secreted alongside GLP-1 from the enteroendocrine L-cell is the intestinotrophic hormone, GLP-2, which has been shown to stimulate crypt cell proliferation and inhibit IEC apoptosis, resulting in increased SI villus height and weight49,57. Furthermore, the disrupted rhythms in intestinal weight and villus height observed in WD-fed mice, coincide not only with a disrupted food intake pattern in these animals, but also with parallel changes in L-cell hormone secretion33. The larger SI observed in both GF- and RC-mice following antibiotic administration not only parallels the previously reported dramatic increase in basal L-cell secretion found in these animals33, but also regulation of intestinal hormone secretion. The microbiome has also previously been reported to impact growth related endocrine pathways such as the somatotrophic axis, altering the production and activity of insulin-like growth factor-158. Hence, circadian- and diet-induced changes in intestinal L-cell secretion of GLP-1 and/or GLP-2 may contribute to the rhythmic changes in SI growth observed in normal mice, as well as in animals fed a high-fat diet and GF-mice.
SI mucosal expression of the core clock genes Arntl and Per2 in RC-mice mimicked that of other known metabolic tissues, with a higher expression of Arntl at ZT2 as compared to ZT14 that was anti-phasic to that of Per234. While obesogenic feeding has been shown to alter metabolic tissue clock gene expression33,35,36, the SI rhythm in Arntl was largely unaffected by either WD-feeding or disruption/absence of the microbiome as in the AIMD- and GF-mice, respectively. Therefore, clock gene expression in the heterogenous cell population that constitutes the intestinal mucosa appears to be regulated differentially as compared to metabolic tissues. It is therefore also possible that the observed changes in SI growth are independent of the IEC clock. However, a limitation of this conclusion is that changes in mucosal clock gene expression may not reflect those in the proliferating crypt base stem cells, which are also known to exhibit circadian rhythms59.
Immune function has been well established to be circadian in nature, as immune cell trafficking and cytokine levels differ over the course of the day11,29. Circulating levels of proinflammatory cytokines have been demonstrated to peak during the active phase in mice, whereas circulating leukocytes are highest during the rest phase and are recruited to tissues at the onset of the active phase29,60. As previously demonstrated in the intestine27, assessment of select cytokine genes in SI revealed elevated Tnf at the onset of the fasting/rest period in normal mice. While IECs have been shown to be influenced by changes in intestinal cytokine levels, no clear patterns were observed in the selected cytokine profile in relation to gravimetric and morphometric SI parameters27,61. Specifically, Tnf was found to change in a time-, diet-, and microbiome-dependent manner; however, expression did not parallel SI changes despite the fact that epithelial-derived Tnf has been speculated to boost Wnt signalling to promote IEC proliferation62. Furthermore, while obesogenic diets have been associated with an increased proinflammatory phenotype63, Tnf expression was reduced at ZT2 in WD-fed animals, disrupting the rhythm found in RC-fed mice. Short-chain fatty acids (SCFA) have been also been reported to modify cytokine production by IECs and, interestingly, previous analyses in RC-fed mice revealed a reduction in total cecal SCFA at ZT14, as well as decreases at both ZT2 and ZT14 following WD-feeding, providing a potential mechanism for altered Tnf expression64. This is further supported by the reduction in Tnf expression in AIMD-mice, which have an impaired ability to generate microbial metabolites. Furthermore, GF-mice lacked the rhythms in cytokine levels seen in normal mice, although FMT failed to restore these rhythms, a finding that requires further investigation.
Circadian rhythms observed within the immune system include those of leukocyte trafficking to peripheral tissues, which peak during the active/feeding period, likely to maintain adequate immunosurveillance during the period of maximal bacterial exposure29,60. Given that the active period is also associated with increased exposure to nutrient-derived luminal antigens, inappropriate immune responses are thought to be inhibited by the upregulation of CD8αα on intestinal lymphocytes28,65. Previous reports have demonstrated diurnal changes in SI IELs, wherein the ratio of CD8αα homodimer- to CD8αβ heterodimer-expressing cells is greater in the rodent active phase66. In keeping with these findings, the increase in CD8αα+ cells in association with the decrease in proportion of CD8αβ+ cells at the onset of the feeding period in RC-mice reported here, may play a physiological role in maintaining immunological tolerance in the SI. Furthermore, the disrupted pattern in WD-IELs, is likely a response to their disruption in food intake pattern33. Interestingly, recent studies have identified the GLP-1 receptor on IELs and have demonstrated their role regulating GLP-1 bioavailability67. Consistently, parallel changes were observed in the proportion of CD8αβ+ cells and the previously-reported stimulation of GLP-1 secretion in both RC- and WD-mice with or without AIMD and in GF-mice with or without FMT33. Finally, CD8αβ+ LPLs displayed an opposing pattern to the IELs, likely indicative of lymphocyte homing from the lamina propria to the epithelium28.
Given the unique interaction between the microbiome and the intestinal immune system, as well as the differential microbial composition and function along the length of the GI tract68, analysis of the SI microbiome would be warranted to infer specific microbial-driven changes in intestinal immunity. Furthermore, while GF-mice were employed to study the rhythmic nature of select parameters of intestinal homeostasis in the absence of a microbiome, in general, FMT failed to re-establish patterns observed in normal RC-mice. A limitation of the present study is the lack of FMT experiments using WD-fed mice microbiome, as well as the use of pooled RC fecal samples, which precludes interrogation of the effects of the normal day vs. night pattern in microbial composition; both of these issues warrant future consideration. Furthermore, the overall inability of the RC FMT to restore the physiologic rhythms in the observed intestinal parameters may be a result of the variation in the relative abundance of major families between normal RC-mice and the recolonized GF-animals. The use of aerobic- rather than anaerobically-reduced PBS to resuspend the RC feces may have contributed to this variation through reductions in strict anaerobes. However, while an important tool for microbiome research, GF-animals have been demonstrated to have impairments in both early development and education of the intestinal immune compartment32. As such, conducting FMT studies using AIMD adult mice might further support of the role of the microbiome in maintaining SI responses over the 24-h day and in response to WD-feeding.
In addition, time-restricted feeding, as well as the implementation of reverse light/dark cycle studies would also provide further insight into the relationship between feeding patterns and intestinal homeostasis. Notwithstanding, the findings of this study enhance our current understanding of diurnal SI changes in both structure and immune cell populations, showing that these are driven by dietary composition and timing, as well as by the presence of a normal microbiome. Such insights may explain the increased risk for intestinal dysfunction in individuals suffering from circadian disruption or obesity.
Methods
Animal models
Male and female C57Bl6/J mice (5–7 weeks) were purchased from Jackson Laboratories, and randomized into treatment groups upon receipt and allowed to acclimate to the University of Toronto animal facility for one week prior to the beginning of the experimental conditions. For the duration of the experiment, mice were housed in a 12/12-h light/dark cycle at constant room temperature with free access to water, and were fed either regular chow (RC (n = 24 with 12 mice used per time point); 18%, 58%, and 24% calories from fat, carbohydrate and protein, respectively; overall caloric density of 3.1 kcal/g [2018 Envigo]) or a high-fat/high-sucrose Western diet (WD (n = 24 with 12 mice used per time point); 41%, 43%, and 17% calories from fat, carbohydrate [29% sucrose, 14% other], and protein, respectively; overall caloric density of 4.7 kcal/g [D120798, Research Diets]) for 16 weeks to establish a model of diet-induced obesity. To achieve antibiotic-induced microbial depletion (AIMD), mice (n = 12 with 6 mice used per time point) were orally gavaged twice-daily with either vehicle (water) or an antibiotic/antifungal cocktail (ampicillin (100 mg/kg), vancomycin (50 mg/kg), metronidazole (100 mg/kg), neomycin (100 mg/kg), amphotericin B (1 mg/kg)) during the final 2 weeks of feeding33,69. Male (n = 7–8 with 3–4 mice used per time point) and female (n = 7–8 with 3–4 mice used per time point) RC germ-free mice on a C57Bl/6 N background were bred under standard germ-free conditions prior to transfer to a sterile 12/12-h light/dark cycle for fecal microbiome transfer (FMT). In brief, fecal pellets from RC-animals were collected over 24-h and resuspended in normal, oxygenated PBS (200 μL; 5 g/mL) prior to oral gavage daily for 2 days into 8-wk old germ-free mice, followed by a 3-week rest period. The sample size for all experiments was determined based on previous studies of the same nature conducted in our laboratory33. All animal studies were approved by the University of Toronto Animal Care Committee and conformed to the Canadian Council on Animal Care guidelines. All of the protocols, as well as the results of the fecal 16S rRNA gene sequencing, cecal bile acid and short chain fatty acid, and metabolic data in these mice have been previously reported33.
Gravimetric and morphometric analysis
The small intestine was isolated, rinsed with ice-cold PBS, and both the weight and length (under constant tension using a 2.8 g weight) were measured. A two cm section from the ileum was collected into formalin for paraffin-embedding, sectioning and staining with hematoxylin & eosin (Pathology Research Program, University Health Network, Toronto, ON). Ileal villus height and crypt depth were measured in blinded fashion using a Zeiss microscope with AxioVision software (Zeiss Microscopy Canada), with at least 20 well-oriented crypts and villi measured from each animal, to make n = 1.
Gene expression analysis
RNA was isolated from ileal mucosal scrapes using the RNeasy Plus Mini Kit (Qiagen Inc.) and cDNA was generated using 5 × All-in-One RT Mastermix (Applied Biological Materials Inc.). Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed using the Taqman Gene Expression Assay (ThermoFisher) with primers (ThermoFisher) as listed in Table 1.
Table 1.
List of primer sequences.
| Target | Product number |
|---|---|
| Arntl | Mm00500226_m1 |
| Per2 | Mm00478113_m1 |
| Tnf | Mm00443258_m1 |
| Ifng | Mm01168134_m1 |
| Il6 | Mm00446190_m1 |
| Il10 | Mm01288386_m1 |
| Tgfb | Mm01178820_m1 |
| 18S | Hs99999901_s1 |
Lymphocyte collection and flow cytometry
In brief, IELs and LPLs were isolated by dividing the distal 10 cm of the small intestine into two cm segments and incubating in HBSS with 5 mM DTT to remove mucus. Two consecutive 10 min incubations with 5 mM EDTA were performed to collect the intestinal epithelial cells and associated IELs after debris removal using a tube-top 40 µm strainer. To collect the LPLs, 3 additional EDTA incubations were conducted followed by mincing of the tissue, enzymatic digestion with 0.2 U/ml Liberase TM (Roche) and 200 U/ml DNAse I (Roche) for 30 min, trituration of the digested tissue through a 21G needle and filtration to remove debris70.
Single cell suspensions were stained with LIVE/DEAD fixable aqua dead cell stain (ThermoFisher) to exclude dead cells from flow cytometric analysis. Suspensions were then incubated in flow cytometry staining buffer (0.3%FBS, 0.1% NaAzide) with Anti-Mo CD16/CD32 (ThermoFisher, Clone 93) to block non-specific binding of antibodies by Fc receptors. Antibodies (ThermoFisher) listed in Table 2 were used for staining of cell surface antigens, following which, the eBioscience Foxp3/Transcription Factor Staining Kit (ThermoFisher) was used for fixation. Analysis was conducted using the LSRFortessa Cell Analyzer (BD Biosciences) running FACSDiva acquisition software, and data were analyzed using FlowJo v1071.
Table 2.
List of antibodies.
| Target | Fluorophore |
|---|---|
| CD45 | PerCP-Cy5.5 |
| CD3 | PE-Cy7 |
| TCRb | eFluor 450 |
| CD8α | Alexa Fluor 700 |
| CD8β | APC |
| CD4 | APC-eFluor 780 |
| CD19 | PE-eFluor 610 |
| CD11b | Super Bright 780 |
| Ly6g | FITC |
Statistical analysis
Statistical analysis was conducted using GraphPad Prism. Data were analyzed for significance by 2-way ANOVA followed by appropriate post hoc analysis for 3 or more groups, or by Student’s t-test for two groups. Statistical outliers were determined using the Grubbs’ test. All data are expressed as mean ± SEM.
Supplementary Information
Acknowledgements
The authors are extremely grateful to D. White and N. Simard from the University of Toronto Faculty of Medicine Flow Cytometry Facility, A. Cao and L. Kent from the Division of Comparative Medicine, and Dr. A. Waller, Brock University, for the family-level microbiome analysis. S.E.M. was supported by graduate awards from the Ontario Graduate Scholarship program and the Banting and Best Diabetes Centre (University of Toronto), D.P by a postdoctoral award from the Canadian Institutes of Health Research, Canadian Association of Gastroenterology, and Crohn’s and Colitis Canada, A.M. by graduate awards from the Canadian Institutes of Health Research, the Ontario Graduate Scholarship, the Banting and Best Diabetes Centre, and the Biological Rhythms Training Program (University of Toronto), M.E.S. by summer studentships from the Banting and Best Diabetes Centre, and P.L.B. by a Tier I Canada Research Chair.
Abbreviations
- AIMD
Antibiotic-induced microbial disruption
- GF
Germ-free
- GI
Gastrointestinal
- GLP-1
Glucagon-like peptide-1
- IEC
Intestinal epithelial cell
- IEL
Intraepithelial lymphocytes
- LPL
Lamina propria lymphocytes
- RC
Regular chow
- SI
Small intestine
- WD
Western diet
Author contributions
S.E.M. researched data, analyzed data, and wrote the manuscript. D.P. researched data, analyzed data, and reviewed and edited the manuscript. A.M. researched data and reviewed and edited the manuscript. M.E.S. researched data and reviewed and edited the manuscript. D.J.P provided funding and reviewed and edited the manuscript. P.L.B provided funding, reviewed and edited the manuscript, and is the guarantor of the work.
Funding
This work was supported by an operating grant from the Canadian Institutes of Health Research (PJT-15308 to P.L.B.) and the Banting and Best Diabetes Centre (Pilot and Feasibility Grant, to P.L.B and D.J.P.). Some of the equipment was supported by the 3D (Diet, Digestive Tract and Disease) Centre funded by the Canadian Foundation for Innovation and Ontario Research Fund (project numbers 19442 and 30961).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98986-7.
References
- 1.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 2.Gachon F, Loizides-Mangold U, Petrenko V, Dibner C. Glucose homeostasis: Regulation by peripheral circadian clocks in rodents and humans. Endocrinology. 2017;158:1074–1084. doi: 10.1210/en.2017-00218. [DOI] [PubMed] [Google Scholar]
- 3.Hoogerwerf WA, Hellmich HL, Cornelissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. Clock gene expression in the murine gastrointestinal tract: Endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Thaiss CA, Zeevi D, Maayan L, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 6.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Fischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash M, Thaiss CA, Pinker EY, Ratiner K, Adlung L, Federici S, Kleimeyer C, Moresi C, Yamada T, Cohen Y, Zhang X, Massalha H, Massasa E, Kuperman Y, Koni PA, Harmelin A, Gao N, Itzkovitz S, Hona K, Shapiro H, Elinav E. Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell. 2020;182:1441–1459.e21. doi: 10.1016/j.cell.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez Y, Glotfelty LG, Blank N, Dohnalová L, Thaiss CA. The microbiome as a circadian coordinator of metabolism. Endocrinology 2020;161:bqaa059. [DOI] [PMC free article] [PubMed]
- 11.Martchenko A, Martchenko SE, Biancolin AD, Brubaker PL. Circadian rhythms and the gastrointestinal tract: Relationship to metabolism and gut hormones. Endocrinology 2020;161:bqaa167. [DOI] [PMC free article] [PubMed]
- 12.Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr. Rev. 2015;36:289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: Two prospective cohort studies in women. PLoS Med. 2011;8:e1001141. [DOI] [PMC free article] [PubMed]
- 15.Fonken LK, Nelson RJ. The effects of light at night on circadian clocks and metabolism. Endocr. Rev. 2014;35:648–670. doi: 10.1210/er.2013-1051. [DOI] [PubMed] [Google Scholar]
- 16.Qian J, Yeh B, Rakshit K, Colwell CS, Matveyenko AV. Circadian disruption and diet-induced obesity synergize to promote development of β-cell failure and diabetes in male rats. Endocrinology. 2015;156:4426–4436. doi: 10.1210/en.2015-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutsson A, Bøggild H. Gastrointestinal disorders among shift workers. Scand. J. Work Environ. Health. 2010;36:85–95. doi: 10.5271/sjweh.2897. [DOI] [PubMed] [Google Scholar]
- 23.Benedict C, Vogel H, Jonas W, Woting A, Blaut M, Schürmann A, Cedernaes J. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol. Metab. 2016;5:1175–1186. doi: 10.1016/j.molmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poroyko VA, Carreras A, Khalyfa A, Khalyfa AA, Leone V, Peris E, Almendros I, Gileles-Hillel A, Qiao Z, Hubert N, Farré R, Chang EB, Gozal D. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci. Rep. 2016;6:35405. doi: 10.1038/srep35405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds AC, Paterson JL, Ferguson SA, Stanley D, Wright KP, Jr, Dawson D. The shift work and health research agenda: Considering changes in gut microbiota as a pathway linking shift work, sleep loss and circadian misalignment, and metabolic disease. Sleep Med. Rev. 2017;34:3–9. doi: 10.1016/j.smrv.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida D, Aoki N, Tanaka M, Aoyama S, Shibata S. The circadian clock controls fluctuations of colonic cell proliferation during the light/dark cycle via feeding behavior in mice. Chronobiol. Int. 2015;32:1145–1155. doi: 10.3109/07420528.2015.1065415. [DOI] [PubMed] [Google Scholar]
- 27.Stokes K, Cooke A, Chang H, Weaver DR, Breault DT, Karpowicz P. The circadian clock Gene BMAL1 coordinates intestinal regeneration. Cell. Mol. Gastroenterol. Hepatol. 2017;4:95–114. doi: 10.1016/j.jcmgh.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutter L, Hoytema van Konijnenburg DP, Brand EC, Oldenburg B, van Wijk F. The elusive case of human intraepithelial T cells in gut homeostasis and inflammation. Nat. Rev. Gastroenterol. Hepatol. 15:637–649 (2018). [DOI] [PubMed]
- 29.Zheng D, Ratiner K, Elinav E. Circadian influences of diet on the microbiome and immunity. Trends Immunol. 2020;41:512–530. doi: 10.1016/j.it.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The intestinal epithelium: Central coordinator of mucosal immunity. Trends Immunol. 2018;39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martchenko SE, Martchenko A, Cox BJ, Naismith K, Waller A, Gurges P, Sweeney ME, Philpott DJ, Brubaker PL. Circadian GLP-1 secretion in mice is dependent on the intestinal microbiome for maintenance of diurnal metabolic homeostasis. Diabetes. 2020;69:2589–2602. doi: 10.2337/db20-0262. [DOI] [PubMed] [Google Scholar]
- 34.Cedernaes J, Huang W, Ramsey KM, Waldeck N, Cheng L, Marcheva B, Omura C, Kobayashi Y, Peek CB, Levine DC, Dhir R, Awatramani R, Bradfield CA, Wang XA, Takahashi JS, Mokadem M, Ahima RS, Bass J. Transcriptional basis for rhythmic control of hunger and metabolism within the AgRP neuron. Cell Metab. 2019;29:1078–191.e5. doi: 10.1016/j.cmet.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Prasai MJ, Mughal RS, Wheatcroft SB, Kearney MT, Grant PJ, Scott EM. Diurnal variation in vascular and metabolic function in diet-induced obesity: Divergence of insulin resistance and loss of clock rhythm. Diabetes. 2013;62:1981–1989. doi: 10.2337/db11-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang TWH, Chen HC, Chen CY, Yen CYT, Lin CJ, Prajnamitra RP, Chen LJ, Ruan SC, Lin JH, Lin PJ, Lu HH, Kuo CW, Chang CM, Hall AD, Vivas EI, Shui JW, Chen P, Hacker TA, Rey FE, Kamp TJ, Hsieh PCH. Loss of gut microbiota alters immune system composition and cripples postinfarction cardiac repair. Circulation. 2019;139:647–659. doi: 10.1161/CIRCULATIONAHA.118.035235. [DOI] [PubMed] [Google Scholar]
- 38.Nilaweera KN, Speakman JR. Regulation of intestinal growth in response to variations in energy supply and demand. Obes. Rev. 2018;19:61–72. doi: 10.1111/obr.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin ED, Estall JL, Izzo A, Drucker DJ, Brubaker PL. Mucosal adaptation to enteral nutrients is dependent on the physiologic actions of glucagon-like peptide-2 in mice. Gastroenterology. 2005;128:1340–1353. doi: 10.1053/j.gastro.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 40.Bahrami J, Yusta B, Drucker DJ. ErbB activity links the glucagon-like peptide-2 receptor to refeeding-induced adaptation in the murine small bowel. Gastroenterology. 2010;138:2447–2456. doi: 10.1053/j.gastro.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Chwen LT, Foo HL, Thanh NT, Choe DW. Growth performance, plasma fatty acids, villous height and crypt depth of preweaning piglets fed with medium chain triacylglycerol. Asian Australas. J. Anim. Sci. 2013;26:700–770. doi: 10.5713/ajas.2012.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Secor SM, Diamond J. A vertebrate model of extreme physiological regulation. Nature. 1998;395:659–662. doi: 10.1038/27131. [DOI] [PubMed] [Google Scholar]
- 43.Mayhew TM, Middleton C. Crypts, villi and microvilli in the small intestine of the rat. A stereological study of their variability within and between animals. J. Anat. 1985;141:1–17. [PMC free article] [PubMed]
- 44.DeSesso JM, Lavin AL, Hsia SM, Mavis RD. Assessment of the carcinogenicity associated with oral exposures to hydrogen peroxide. Food Chem. Toxicol. 2000;38:1021–1041. doi: 10.1016/S0278-6915(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 45.Casteleyn C, Rekecki A, Van Der Aa A, Simoens P, Van Den Broeck W. Surface area assessment of the murine intestinal tract as a prerequisite for oral dose translation from mouse to man. Lab. Anim. 2010;44:176–183. doi: 10.1258/la.2009.009112. [DOI] [PubMed] [Google Scholar]
- 46.Greig CJ, Zhang L, Cowles, RA. Potentiated serotonin signaling in serotonin re-uptake transporter knockout mice increases enterocyte mass and small intestinal absorptive function. Physiol. Rep. 7:e14278 (2019). [DOI] [PMC free article] [PubMed]
- 47.Wang M, Yang C, Wang Q, Li J, Huang P, Li Y, Ding X, Yang H, Yin Y. The relationship between villous height and growth performance, small intestinal mucosal enzymes activities and nutrient transporters expression in weaned piglets. J. Anim. Physiol. Anim. Nutr. 2020;104:606–615. doi: 10.1111/jpn.13299. [DOI] [PubMed] [Google Scholar]
- 48.Jeppesen PB, Sanguinetti EL, Buchman A, Howard L, Scolapio JS, Ziegler TR, Gregory J, Tappenden KA, Holst J, Mortensen PB. Teduglutide (ALX-0600), a dipeptidyl peptidase IV resistant qlucagon-like peptide 2 analogue, improves intestinal function in short bowel syndrome patients. Gut. 2005;54:1224–1231. doi: 10.1136/gut.2004.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brubaker PL. Glucagon-like peptide-2 and the regulation of intestinal growth and function. Compr. Physiol. 2018;8:1185–1210. doi: 10.1002/cphy.c170055. [DOI] [PubMed] [Google Scholar]
- 50.Mouillot T, Boehm V, Treton X, Ferrandi E, Kapel N, Cazals-Hatem D, Joly F. Small-bowel adaptation: A case of morphological changes induced by teduglutide in short-bowel syndrome with intestinal failure. J. Parenter. Enter. Nutr. 2020;44:940–943. doi: 10.1002/jpen.1805. [DOI] [PubMed] [Google Scholar]
- 51.Davidson GP, Cutz E, Hamilton JR, Gall DG. Familial enteropathy: A syndrome of protracted diarrhea from birth, failure to thrive, and hypoplastic villus atrophy. Gastroenterology. 1978;75:783–790. doi: 10.1016/0016-5085(78)90458-4. [DOI] [PubMed] [Google Scholar]
- 52.Sherman PM, Mitchell DJ, Cutz E. Neonatal enteropathies: Defining the causes of protracted diarrhea of infancy. J. Pediatr. Gastroenterol. Nutr. 2004;38:16–26. doi: 10.1097/00005176-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Westergaard H. Tropical sprue. Curr. Treatm. Opt. Gastroenterol. 2004;7:7–11. doi: 10.1007/s11938-004-0020-6. [DOI] [PubMed] [Google Scholar]
- 54.Louis-Auguste J, Besa E, Zyambo K, Munkombwe D, Banda R, Banda T, Watson A, Mayneris-Perxachs J, Swann J, Kelly P. Tryptophan, glutamine, leucine, and micronutrient supplementation improves environmental enteropathy in Zambian adults: A randomized controlled trial. Am. J. Clin. Nutr. 2019;110:1240–1252. doi: 10.1093/ajcn/nqz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor SR, Ramsamooj S, Liang RJ, Katti A, Pozovskiy R, Vasan N, Hwang SK, Nahiyaan N, Francoeur NJ, Schatoff EM, Johnson JL, Shah MA, Dannenberg AJ, Sebra RP, Dow LE, Cantley LC, Rhee KY, Goncalves MD. Dietary fructose improves intestinal cell survival and nutrient absorption. Nature. 2021 doi: 10.1038/S41586-021-03827-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koehler JA, Baggio LL, Yusta B, Longuet C, Rowland KJ, Cao X, Holland D, Brubaker PL, Drucker DJ. GLP-1R agonists promote normal and neoplastic intestinal growth through mechanisms requiring Fgf7. Cell Metab. 2015;21:379–391. doi: 10.1016/j.cmet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Drucker DJ, Ehrlich P, Asa SL, Brubaker PL. Induction of intestinal epithelial proliferation by glucagon-like peptide 2. Proc. Natl. Acad. Sci. USA. 1996;93:7911–7916. doi: 10.1073/pnas.93.15.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol. 2019;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Parasram K, Karpowicz P. Time after time: circadian clock regulation of intestinal stem cells. Cell. Mol. Life Sci. 2020;77:1267–1288. doi: 10.1007/s00018-019-03323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat. Rev. Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Onyiah JC, Colgan SP. Cytokine responses and epithelial function in the intestinal mucosa. Cell. Mol. Life Sci. 2016;73:4203–4212. doi: 10.1007/s00018-016-2289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradford EM, Ryu SH, Singh AP, Lee G, Goretsky T, Sinh P, Williams DB, Cloud AL, Gounaris E, Patel V, Lamping OF, Lynch EB, Moyer MP, De Plaen IG, Shealy DJ, Yang GY, Barrett TA. Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J. Immunol. 2017;199:1886–1897. doi: 10.4049/jimmunol.1601066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Brooks RS, Ciappio ED, Kim SJ, Crott JW, Bennett G, Greenberg AS, Mason JB. Diet-induced obesity elevates colonic TNF-α in mice and is accompanied by an activation of Wnt signaling: A mechanism for obesity-associated colorectal cancer. J. Nutr. Biochem. 2012;23:1207–1213. doi: 10.1016/j.jnutbio.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venegas DP, et al. Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki H, Shibata S, Okutani T, Suzuki M, Nakayam M, Nishimura T, Doi K. Diurnal changes in intraepithelial lymphocytes (IELs) in the small intestine of mice. Exp. Anim. 1999;48:115–118. doi: 10.1538/expanim.48.115. [DOI] [PubMed] [Google Scholar]
- 67.He S, Kahles F, Rattik S, Nairz M, McAlpine CS, Anzai A, Selgrade D, Fenn AM, Chan CT, Mindur JE, Valet C, Poller WC, Halle L, Rotllan N, Iwamoto Y, Wojtkiewicz GR, Weissleder R, Libby P, Fernández-Hernando C, Drucker DJ, Nahrendorf M, Swirski FK. Gut intraepithelial T cells calibrate metabolism and accelerate cardiovascular disease. Nature. 2019;566:115–119. doi: 10.1038/s41586-018-0849-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes Environ. 2017;32:300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, Saghatelian A, Knight R, Panda S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018;9:2872. doi: 10.1038/s41467-018-05336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodyear AW, Kumar A, Dow S, Ryan EP. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods. 2014;405:97–108. doi: 10.1016/j.jim.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 71.Prescott D, Maisonneuve C, Yadav J, Rubino SJ, Girardin SE, Philpott DJ. NOD2 modulates immune tolerance via the GM-CSF–dependent generation of CD103 + dendritic cells. Proc. Natl. Acad. Sci. USA. 2020;117:10946–10957. doi: 10.1073/pnas.1912866117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.