Abstract
We analyzed five bacterial strains, designated 19982, 9194, 10457, 10790, and 12502, that were isolated from stool specimens of individuals with diarrheal illness by the International Centre for Diarrhoeal Disease Research in Dhaka, Bangladesh (M. J. Albert, S. M. Faruque, M. Ansaruzzaman, M. M. Islam, K. Haider, K. Alam, I. Kabir, and R. Robins-Browne, J. Med. Microbiol. 37:310–314, 1992). The strains were initially identified as Hafnia alvei with a commercial identification system and were reported to contain the eae gene of enteropathogenic Escherichia coli. Results of conventional biochemical analyses, testing of susceptibility to cephalothin, lysis by a Hafnia-specific phage, and amplification of the outer membrane protein gene phoE with species-specific primers support the identification of these strains as members of the genus Escherichia rather than Hafnia alvei. These strains varied from typical E. coli strains by their inability to produce acid from lactose or d-sorbitol and failure to elaborate the enzyme β-d-glucuronidase. PCR analysis confirmed previous findings that the strains were positive for the eae gene and negative for other virulence markers present among recognized categories of diarrheagenic E. coli. Our findings support the hypothesis that these strains are a new category of diarrheagenic isolates belonging to the genus Escherichia and illustrate the importance of using multiple methodologies when identifying new bacterial agents of diarrheal disease.
Over the past three decades the number of bacterial taxa proven to cause gastrointestinal disease has risen dramatically. In addition to bona fide enteric pathogens, a number of reputed agents of gastroenteritis which had previously been thought to be simple commensals of the gastrointestinal tract have been described (1). One such agent is Hafnia alvei. In 1991, John Albert and colleagues at the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDRB), reported on laboratory investigations concerning a strain of H. alvei (designated 19982) recovered from a 9-month-old girl seen at ICDDRB with vomiting, mild dehydration, fever, abdominal distention, and diarrhea of 72 h duration (2). This fecal isolate was found to induce diarrhea in 8 of 12 adult rabbits by using the removable intestinal tie-adult rabbit diarrhea model. Electron microscopic studies of the apical surface of the rabbit’s infected gastrointestinal epithelium revealed classic attachment-effacement lesions traditionally associated with enteropathogenic Escherichia coli (EPEC). Subsequent investigations by the same investigators at ICDDRB identified an additional six strains of H. alvei with similar properties (3). Like EPEC, all seven H. alvei strains were fluorescent actin staining positive and hybridized with the EPEC attaching-and-effacing (eae) probe (3). These findings led to the hypothesis that the eae locus is highly conserved among diverse species within the family Enterobacteriaceae (16).
On the basis of these investigations, several case reports and case-control studies followed, and they documented H. alvei as an enteric pathogen (18–20, 25). However, a 1996 Canadian study failed to detect virulence characteristics present in H. alvei 19982 in any of nine H. alvei isolates recovered from children with diarrhea at The Hospital for Sick Children in Toronto or in two strains previously implicated in outbreaks of gastroenteritis (12). That study also noted that the outer membrane, plasmid, and pulsed-field gel electrophoresis (PFGE) profiles of Canadian isolates differed significantly from those of H. alvei 19982. Finally, recent collaborative studies between ICDDRB and the University of Helsinki found a low level of 16S rRNA sequence homology (92%) between eae-positive and eae-negative H. alvei strains, raising concerns about the correct taxonomic position of these strains (20). Here we report on the results for five of these strains and provide data indicating that these isolates are, in fact, unusual biotypes of E. coli or represent a new species in the genus Escherichia.
MATERIALS AND METHODS
H. alvei 19982, 9194, 10457, 10790, and 12502 were provided by M. John Albert (Dhaka, Bangladesh). The enterovirulence characteristics of these strains have been described previously (2, 3). Isolates were kept as working cultures on motility deeps, with permanent stock cultures maintained at −70°C. Conventional biochemical tests were performed as described previously (13), and susceptibility to cephalothin was determined by the AB Biodisk E test (Remel, Lexena, Kans.) according to the manufacturer’s instructions.
Testing of individual bacterial strains for Hafnia-specific phage susceptibility (9) and for E. coli-specific genes and virulence characteristics were determined by multiplex PCR through the courtesy of the Centers for Communicable Disease Control by previously described methods (5, 7, 8, 11, 14, 17, 22–24). The PFGE profiles of selected strains were determined with a CHEF II system (Bio-Rad, Hercules, Calif.) according to the manufacturer’s instruction with the restriction endonuclease XbaI.
RESULTS AND DISCUSSION
All five ICDDRB strains were oxidase and indole negative, nonmotile, lysine and ornithine decarboxylase positive, o-nitrophenyl-β-d-galactopyranoside positive, and methyl red positive. Key differential tests useful in the separation of hafniae from E. coli are listed in Table 1. Several reactions, including failure to produce aceytlmethycarbinol (Voges-Proskauer), lack of growth in KCN broth, and failure to utilize acetate, suggested that the ICDDRB strains were not H. alvei. Among the common sugars, only d-glucose, d-mannitol, l-arabinose, trehalose, and maltose were fermented. The inability to produce acid from lactose and d-sorbitol and the failure to elaborate the enzyme β-d-glucuronidase (MUG) did not, however, fit the typical pattern exhibited by most E. coli strains.
TABLE 1.
Comparison of properties of Albert’s diarrheal isolates to characteristic properties of H. alvei and E. coli strains
| Test | Result for the following strainsa:
|
||||||
|---|---|---|---|---|---|---|---|
| E. colib | H. alveib | ICDDRB strains
|
|||||
| 19982 | 9194 | 10457 | 10790 | 12502 | |||
| Voges-Proskauer | − | + | − | − | − | − | − |
| KCN | − | + | − | − | − | − | − |
| Acetate | + | − | + | + | + | + | + |
| Acid from: | |||||||
| Lactose | + | − | − | − | − | − | − |
| d-Sorbitol | + | − | − | − | − | − | − |
| MUG | + | − | − | − | − | − | − |
| Cephalothin | S | R | I | I | S | I | S |
| Hafnia phage 1672 | − | + | − | − | − | − | − |
| phoE gene probe | + | − | + | + | + | + | + |
Abbreviations: +, ≥85% positive; −, ≤15% positive; S, susceptible; I, intermediate; R, resistant.
Typical profiles of each species (active E. coli, H. alvei) are from Farmer (6).
By PCR, all five of the ICDDRB strains were positive for the eaeA gene, as reported previously (14), but were negative by multiplex PCR (5, 7, 8, 11, 17, 22, 23) for the following virulence characteristics: enterohemolysin (18), bundle-forming pilus A (bfpA) (20, 25), EAF plasmid (21), Shiga toxins stx1 and stx2 (22, 24), E. coli invasion plasmid antigen H (ipaH) (23), and enterotoxigenic E. coli heat-labile and heat-stable enterotoxins (22).
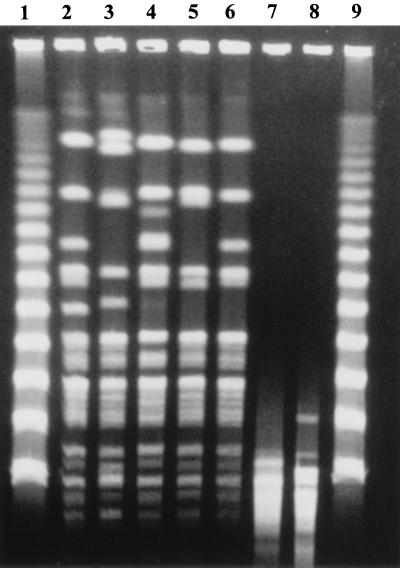
Evidence indicating that these strains were in fact members of the genus Escherichia comes from several lines of evidence. Almost all H. alvei strains are resistant to cephalothin (6), but all of these strains were partially or completely susceptible to cephalosporin by the E test. A genus-specific Hafnia phage, originally described by Guinée and Valkenburg in 1968 (9), failed to lyse any of these strains. Furthermore, a PCR probe encoding the phoE outer membrane protein gene, which is found only in E. coli and Shigella species, hybridized to all five strains but not to true H. alvei isolates (24). Finally, when all five of the ICDDRB strains were typed by PFGE with XbaI they generated a typical series of 10 to 15 large DNA fragments since only a few restriction sites are available (Fig. 1). However, when H. alvei ATCC 13337 and ATCC 29927 were typed by PFGE they produced a large series of small DNA bands, indicating that many XbaI restriction sites were present in true hafniae.
FIG. 1.
PFGE profile (XbaI) of Albert’s strains and reference strains of H. alvei. Lanes 1 and 9, bacteriophage lambda markers; lanes 2, Albert’s strain 12502; lane 3, Albert’s strain 10790; lane 4, Albert’s strain 10457; lane 5, Albert’s strain 9194; lane 6, Albert’s strain 19982; lane 7, H. alvei ATCC 13337; and lane 8, H. alvei ATCC 29927.
Cumulative results from the present investigation indicate that the diarrheagenic H. alvei isolates originally described by Albert and associates (2, 3) either are unusual biotypes of E. coli or represent a new species in the genus Escherichia. This conclusion is based upon biochemical and antimicrobial susceptibility differences between true hafniae and the ICDDRB “H. alvei-like” strains and the failure of the latter group to be lysed by Hafnia-specific phage and homology between the phoE genes of the latter group and that of E. coli. Recent sequence data on a 353-bp fragment from the 5′ end of the 16S rRNA sequences of 10 eae-positive H. alvei strains indicates homology closest to an EPEC strain designated E2348/69 (20), further supporting the identification of these isolates as E. coli.
When these strains were initially characterized as H. alvei they were identified on the basis of API 20E (bioMérieux Vitek, Hazelwood, Mo.) reactions. Although the likelihood of a correct identification was not stated (e.g., excellent identification, acceptable identification), several reactions, notably a negative Voges-Proskauer reaction and the inability to ferment l-rhamnose, could have suggested the possibility that these strains were not true hafniae. Because of these initial misidentifications, the association of “H. alvei” with bacterial gastroenteritis was subsequently reinforced and was disseminated in a series of case reports, clinical studies, and a review (10, 18, 19, 21, 25).
Although many new potential enteric pathogens have been described over the past decade, current data do not strongly support a role for hafniae in infectious gastroenteritis. Furthermore, the present report points out the critical importance of bacterial taxonomy and the complete biochemical characterization of isolates in this process (4, 15). Without such analysis the impact of molecular characterization may be lost or provide incorrect information if the isolate is not initially placed in the correct taxa. Therefore, infrequently isolated groups, particularly those with aberrant properties, should receive close systematic scrutiny prior to the reporting of new disease associations or virulence properties linked to such groups.
ACKNOWLEDGMENTS
We thank Nancy Strockbine, Evan Sowers, and Caroline O’Hara for fruitful discussions and for assistance in the genotyping of our strains.
REFERENCES
- 1.Abbott S L, Janda J M. Bacterial gastroenteritis. I. Incidence and etiologic agents. Clin Microbiol Newsl. 1992;14:17–21. [Google Scholar]
- 2.Albert M J, Alam K, Islam M, Montanaro J, Rahman A S M H, Haider K, Hossain M A, Kibriya A K M G, Tzipori S. Hafnia alvei, a probable cause of diarrhea in humans. Infect Immun. 1991;59:1507–1513. doi: 10.1128/iai.59.4.1507-1513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert M J, Faruque S M, Ansaruzzaman M, Islam M M, Haider K, Alam K, Kabir I, Robins-Browne R. Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol. 1992;37:310–314. doi: 10.1099/00222615-37-5-310. [DOI] [PubMed] [Google Scholar]
- 4.Brenner D J. Phylogenetic classification of bacteria and recent developments in bacterial classification and nomenclature: impact on clinical microbiology. Clin Microbiol Newsl. 1988;10:153–156. [Google Scholar]
- 5.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;32:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer J J., III . Enterobacteriaceae: introduction and identification. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 438–449. [Google Scholar]
- 7.Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler L H, Balijer G, Beutin L, Karch H. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol. 1994;32:2460–2463. doi: 10.1128/jcm.32.10.2460-2463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fratamico P M, Sackitey S K, Wiedmann M, Yi Deng M. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guinée P A M, Valkenburg J J. Diagnostic value of a Hafnia-specific bacteriophage. J Bacteriol. 1968;96:564. doi: 10.1128/jb.96.2.564-.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Günthard H, Pennekamp A. Clinical significance of extraintestinal Hafnia alvei isolates from 61 patients and review of the literature. Clin Infect Dis. 1996;22:1040–1045. doi: 10.1093/clinids/22.6.1040. [DOI] [PubMed] [Google Scholar]
- 11.Gunzburg S T, Tornieporth N G, Riley L W. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismaili A, Bourke B, de Azavedo J C S, Ratnam S, Karmali M A, Sherman P M. Heterogeneity in phenotypic and genotypic characteristics among strains of Hafnia alvei. J Clin Microbiol. 1996;34:2973–2979. doi: 10.1128/jcm.34.12.2973-2979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda J M, Abbott S L, Cheung W K W, Hanson D F. Biochemical identification of citrobacteria in the clinical laboratory. J Clin Microbiol. 1994;32:1850–1854. doi: 10.1128/jcm.32.8.1850-1854.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louie M, De Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcon M J. The medical relevance of molecular taxonomy and associated name changes, or “What’s in a name?”. Clin Microbiol Newsl. 1996;18:123–125. [Google Scholar]
- 16.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsvik O, Strockbine N A. Polymerase chain reaction detection of heat-stable, heat-labile, and Shiga-like toxin genes in Escherichia coli. In: Persing D H, Tenover F C, Smith T F, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: American Society for Microbiology; 1993. pp. 271–276. [Google Scholar]
- 18.Ratnam S. Etiologic role of Hafnia alvei in human diarrheal illness. Infect Immun. 1991;59:4744–4745. doi: 10.1128/iai.59.12.4744-4745.1991. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reina J, Hervas J, Borrell N. Acute gastroenteritis caused by Hafnia alvei in children. Clin Infect Dis. 1993;16:443. doi: 10.1093/clind/16.3.443. . (Correspondence.) [DOI] [PubMed] [Google Scholar]
- 20.Ridell J, Siitonen A, Paulin L, Lindroos O, Korkeala H, Albert M J. Characterization of Hafnia alvei by biochemical tests, random amplified polymorphic DNA PCR, and partial sequencing of 16S rRNA gene. J Clin Microbiol. 1995;33:2372–2376. doi: 10.1128/jcm.33.9.2372-2376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridell J, Siitonen A, Paulin L, Mattila L, Korkeala H, Albert M J. Hafnia alvei in stool specimens from patients with diarrhea and healthy controls. J Clin Microbiol. 1994;32:2335–2337. doi: 10.1128/jcm.32.9.2335-2337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–705. doi: 10.1128/jcm.33.3.701-705.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sethabutr O, Venkatesan M, Murphy G S, Eampokalap B, Hoge C W, Echeverria P. Detection of shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis. 1992;167:458–461. doi: 10.1093/infdis/167.2.458. [DOI] [PubMed] [Google Scholar]
- 24.Spierings G, Ockhuijsen C, Hofstra H, Tommassen J. Polymerase chain reaction for the specific detection of Escherichia coli/Shigella. Res Microbiol. 1993;144:557–564. doi: 10.1016/0923-2508(93)90005-m. [DOI] [PubMed] [Google Scholar]
- 25.Westblom T U, Milligan T W. Acute bacterial gastroenteritis caused by Hafnia alvei. Clin Infect Dis. 1992;14:1271–1272. doi: 10.1093/clinids/14.6.1271. . (Correspondence.) [DOI] [PubMed] [Google Scholar]