Abstract
Patients with chronic hepatitis B virus (HBV) infection are at risk of developing hepatocellular carcinoma (HCC), and serum markers reflecting viral replication are potential predictors for HCC development. Besides the levels of serum HBV DNA and hepatitis B surface antigen (HBsAg), hepatitis B core-related antigen (HBcrAg) quantification is an emerging serological marker for viral replication. Unlike HBV DNA and HBsAg, HBcrAg is a covalently closed circular DNA-derived protein marker, consisting of hepatitis B e antigen (HBeAg), p22cr, and hepatitis B core antigen. In treatment-naïve HBV patients, higher HBcrAg levels are shown to be associated with an increased risk of HCC in several studies. More importantly, HBcrAg may complement HBV DNA level to predict HCC development. For example, an Asian treatment-naïve cohort study’s data showed that HBcrAg level of 4 log U/mL was effective to stratify HCC risk in HBeAg-negative patients with intermediate viral loads, who may not need antiviral therapy because of the low to moderate risk of HCC. In patients receiving prolonged nucleos(t)ide analogue with profound viral suppression, most data indicated that HBV DNA and HBsAg levels no longer serve as HCC predictors. However, several studies suggested on-treatment HBcrAg levels may remain as an HCC predictor. In summary, HBcrAg level can be a useful biomarker for treatment-naïve patients, but its value in on-treatment patients needs validation. The next challenge is how to combine HBcrAg with the other viral markers to construct a better HCC prediction model, optimizing the management of HBV patients.
Keywords: Hepatitis B virus, Hepatocellular carcinoma, Biomarker
INTRODUCTION
Chronic hepatitis B virus (HBV) infection continues to be a major public health issue worldwide, although safe and effective vaccines are available for more than 3 decades. Recent data estimated that more than 257 million individuals worldwide are positive for hepatitis B surface antigen (HBsAg) [1]. These individuals with chronic hepatitis B (CHB) infection are at an increased risk of developing liver cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC); 15% to 40% of these individuals will develop these serious sequelae during their lifetime.
Nucleos(t)ide analogues (NAs) are the most adopted antiviral treatment for patients with CHB. NAs effectively suppress HBV replication to undetectable levels through the inhibition of viral reverse transcriptase [2]. It not only stops the progression of liver fibrosis but also reduces the risk of HCC. However, rebound of viremia frequently occurs after the discontinuation of NA, primarily because of the persistence of the active transcriptional template of HBV covalently closed circular DNA (cccDNA). Therefore, prolonged antiviral therapy is usually necessary until the clearance of HBsAg.
Identifying CHB patients with high HCC risk for early antiviral therapy is an urgent need because more than half of these patients will suffer from serious liver sequelae during their lifetime. So far, there are several serum quantitative markers for viral replication, including HBV DNA, HBsAg, and hepatitis B core-related antigen (HBcrAg). Previous studies showed and validated the role of HBV DNA and HBsAg in predicting HCC development in treatment-naïve patient [3-5]. Recently, HBcrAg level has been increasingly recognized as an emerging predictor for HCC development [6-8]. In this review, the evolutionary roles of these three biomarkers in predicting HCC development among CHB patients will be summarized and discussed.
WHAT IS HBcrAg?
HBcrAg consists of 3 precore/core protein products sharing an identical 149 amino acid sequence: hepatitis B core antigen (HBcAg), hepatitis B e antigen (HBeAg), and a 22-kDa precore protein (p22cr) (Fig. 1) [9]. HBcAg is the nucleocapsid that encloses the viral DNA. HBeAg is a circulating protein derived from the core gene, then modified and secreted from liver cells. It usually serves as a marker of active viral replication [10]. The p22cr is the dominant precore/core protein in HBV DNA-negative particles. For both HBeAg-positive and -negative patients, HBcAg only accounts for 3.1–37.4% (median 10.5%) of HBcrAg in HBsAg-positive particles [11].
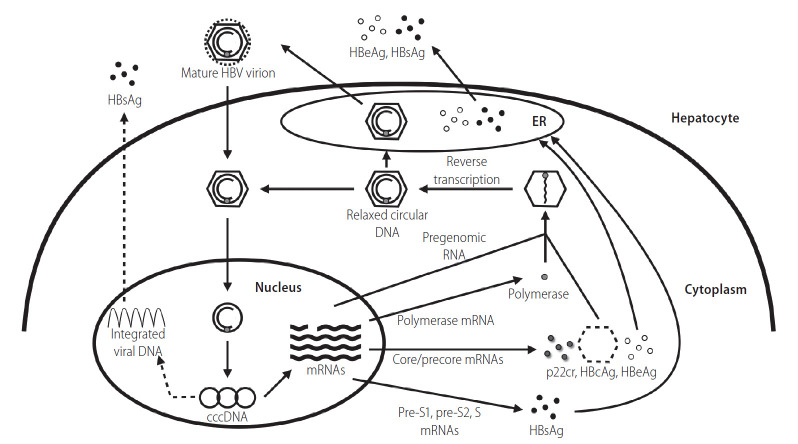
Figure 1.
The HBV replication cycle. HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HBeAg, hepatitis B e antigen; ER, endoplasmic reticulum; cccDNA, covalently closed circular DNA; p22cr, 22-kDa precore protein; HBcAg, hepatitis B core antigen.
There are two start codons in the HBV’s precore/core open reading frame (Fig. 2A). The first ATG encodes the entire amino acid sequence of HBcAg plus a 29 amino acids precore sequence at the N-terminal end [12]. There are two major products derived from post-translational modifications of precore protein. HBeAg is generated after the first cleavage of the first 19 amino acids, a signal sequence that allows translocation into the lumen of endoplasmic reticulum (ER), and second cleavage of up to 34 amino acids from the arginine-rich C-terminal end. HBeAg is then secreted through the ER and Golgi apparatus [12]. Another product is p22cr, containing the uncleaved signal peptide and lacking the arginine-rich domain involved in binding the RNA pregenome or the DNA genome. In contrast to the first start codon, the second ATG specifically encodes HBcAg only.
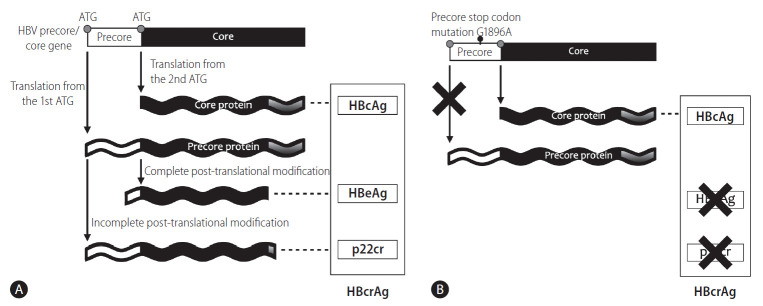
Figure 2.
Precore/core protein translation is initiated from (A) different start codons of the precore/core mRNA and (B) the translation of precore protein, including HBeAg and p22cr, is abolished by emergence of precore stop codon mutation (G1896A). HBV, hepatitis B virus; HBcAg, hepatitis B core antigen; HBeAg, hepatitis B e antigen; p22cr, 22-kDa precore protein; HBcrAg, hepatitis B core-related antigen.
The viral mutations of the precore region highly affect the ratio between HBeAg/p22cr and HBcAg. The precore stop codon mutation (G1896A) is a single-base substitution of G-to-A at nucleotide position 1896 located in the precore gene, which creates a stop codon that prevents the translation of the precore open reading frame and terminates the production of HBeAg/p22cr, but not HBcAg. Therefore, emergence of the mutation lowers the expression level of HBeAg, which has been shown to be associated with HBeAg-negative disease and is expected to cause a lower HBcrAg level in serum [13-17]. This variant may explain the different HBcrAg levels between HBeAg-negative and -positive patients [18,19].
A sensitive enzyme immunoassay specific for HBcAg and HBeAg was first introduced in 2002, and Kimura et al. [20] designated these proteins translated from the precore/core gene as HBcrAg. With the pretreatment by detergents, this assay can detect HBcAg and HBeAg even in anti-HBc or anti-HBe positive specimens. Not only HBcAg and HBeAg but also p22cr can be measured by the serological testing. So far, there is only one commercial HBcrAg assay available (Lumipulse G HBcrAg assay; Fujirebio, Tokyo, Japan). Its linear range for quantification spans from 3 to 7 log U/mL and its lowest sensitivity limit could reach 2 log U/mL.
HBcrAg levels well correlate with HBV DNA levels. [20] Yoshida et al. [21] reported good performance of the HBcrAg assay in identifying high viremic individuals among the treatment-naïve CHB patients using systematic review and meta-analysis. A positive correlation between HBcrAg and HBsAg levels was also observed in Asian and European cohorts [18,19]. A Taiwanese cohort study including 2,666 patients showed a high correlation between HBcrAg and HBV DNA levels (r=0.83, P<0.001), while a moderate correlation between HBcrAg and HBsAg levels (r=0.59, P<0.001) [8]. It is plausible that HBcrAg is better correlated with HBV DNA than HBsAg because both HBcrAg and HBV DNA are only derived from cccDNA, whereas HBsAg may also be translated from the integrated viral genome [22]. In addition, serum HBcrAg levels have been shown to correlate well with cccDNA level and its transcription activity [19,23-25]. Suzuki et al. [23] first reported the correlation between HBcrAg and other viral markers, including HBeAg, HBsAg, serum HBV DNA, and intrahepatic cccDNA. They enrolled a total of 57 patients with CHB, and HBcrAg was observed to correlate positively with cccDNA (r=0.692, P<0.001). This correlation was also observed in another study of HBV re-infection after liver transplantation [24]. Conducted by Wong et al. [19], a study enrolling 138 patients with CHB reported that HBcrAg correlated positively with cccDNA not only in overall patients (r=0.70, P<0.001) but also in patients achieving undetectable HBV DNA level after antiviral therapy (r=0.42, P<0.001).
VIRAL REPLICATION MARKERS TO PREDICT HCC RISK IN TREATMENT-NAÏVE PATIENTS
To optimally investigate the causal relationship of dynamic factors, cohort studies are preferred over cross-sectional studies. Therefore, the role of viral factors in predicting HCC is only reviewed in cohort studies (Table 1).
Table 1.
Relationship between HBcrAg level and HCC development in treatment-naïve cohort studies
| Study | Country | Study design | Disease status | Number of subjects | Number of HCC cases | Finding | Median follow-up time | Note |
|---|---|---|---|---|---|---|---|---|
| Tada et al. [6] (2016) | Japan | Retrospective | All HBV carriers with or without cirrhosis | 1,031 (only 711 patients had HBcrAg level) | 78 | Higher HBcrAg level (>2.9 log U/mL vs. ≤2.9 log U/mL) was independently associated with the incidence of HCC (adjusted HR, 5.05; 95% CI, 2.40–10.63). | 10.7 years | Remaining treatment-naïve during the whole follow-up |
| To et al. [7] (2019) | Hong Kong | Retrospective | HBeAg seroconverters with or without cirrhosis | 207 | 14 | Higher HBcrAg level (>5.21 log U/mL vs. <5.21 log U/mL) at HBeAg seroconversion was associated with HCC development (adjusted HR, 1.75; 95% CI, 1.06–2.90). | 13.1 years | Treatment-naïve at enrolment but nearly half of the patients started to receive treatment at a median of 5.5 years after HBeAg seroconversion |
| Tseng et al. [8] (2019) | Taiwan | Retrospective | All HBV carriers without cirrhosis | 2,666 | 209 | 1. For the overall patients, HBcrAg was positively associated with HCC with doseresponse relationship. | 16.0 years | Remaining treatment-naïve during the whole follow-up |
| 2. Among the HBeAg-negative patients with HBV between 2,000–20,000 IU/mL and normal baseline ALT level, a higher HBcrAg level (≥4 log U/mL vs. <4 log U/mL) was associated with an increased HCC risk (adjusted HR, 6.29; 95% CI, 2.27–17.48). |
HBcrAg, hepatitis B core-related antigen; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; HR, hazard ratio; CI, confidence interval; HBeAg, hepatitis B e antigen; ALT, alanine aminotransferase.
The positive relationship between serum HBV DNA level and HCC risk was first shown by a prospective community-based cohort study known as Risk Evaluation of Viral Load Elevation and Associated Liver Disease/Cancer-Hepatitis B Virus (REVEAL-HBV), which followed 3,653 adult Taiwanese HBsAg seropositive patients over a mean follow-up period of 11.4 years [3]. In particular, the HCC risk started to increase when serum viral load ≥2,000 IU/mL but increased dramatically when viral load ≥20,000 IU/mL. The HBV DNA cutoff levels of 2,000 IU/mL and 20,000 IU/mL are thus recommended to categorize the patients into low, intermediate, and high-risk groups.
The role of HBsAg level was first shown by a hospital-based cohort study with the acronym of Elucidation of Risk fActors for DIsease Control or Advancement in Taiwanese hEpatitis B carriers (ERADICATE-B), which enrolled 2,688 Taiwanese HBV carriers who did not have evidence of cirrhosis at baseline and remained treatment-free during the follow-up period [4]. The mean follow-up period was 14.7 years. The data showed that HBsAg level could complement HBV DNA level in predicting HCC risk. In patients with viral load <2,000 IU/mL, serum HBsAg level of 1,000 IU/mL further stratified the risk, which is also validated by the REVEAL-HBV cohort [5].
The association between serum HBcrAg level and the development of HCC was first reported by Kumada et al [26]. They selected 117 NA-treated and 117 treatment-naïve patients using the propensity score matching, which was designed to explore whether the NA treatment lowers the HCC risk. The multivariable analysis showed that serum HBcrAg level greater than 3.0 log U/mL was associated with HCC development.
The predictive value of serum HBcrAg level for the development of HCC in treatment-naïve patients was first reported by Tada et al. [6] in a Japanese cohort. The hospital-based retrospective cohort recruited a total of 711 treatment-naïve patients with available HBcrAg levels. After a median follow-up period of 10.7 years, patients with an HBcrAg level greater than 2.9 log U/mL were associated with more than 5-fold increase in the risk of HCC development than those below the level.
Another retrospective cohort study, enrolling 207 spontaneous HBeAg seroconverters from Hong Kong, was conducted to explore the relationship between HBcrAg level and HCC development [7]. The median follow-up period was 13.1 years. The data showed that a higher baseline serum HBcrAg level at the time of HBeAg seroconversion was associated with increased risk of HCC development. The adjusted hazard ratio (HR) was 1.75 when stratifying the patients by baseline HBcrAg level of 5.21 log U/mL. To be noted, it is not a treatment-free cohort as nearly half of the study patients received NAs at a median of 5.5 years after the spontaneous HBeAg seroconversion.
A shared limitation of both studies is relatively small patient numbers, which make it difficult to explore rare events, such as HCC development. The ERADICATE-B cohort study from Taiwan actually overcomes these challenges [8]. It showed a linear relationship between serum log10 HBcrAg level and HCC risk. The multivariable analysis revealed that, compared to the patients with the HBcrAg less than 4 log U/mL, those with the HBcrAg between 4 and 5 log U/mL, 5 and 6 log U/mL, and greater or equal to 6 log U/mL had higher HCC risk with adjusted HRs of 2.83 (95% confidence interval [CI], 1.67–4.80), 3.73 (95% CI, 2.08–6.70), and 5.33 (95% CI, 2.77–10.26), respectively [8]. Furthermore, the data showed that HBcrAg served as a better HCC predictor than HBsAg did in HBeAg-negative patients with intermediate HBV viral load and normal alanine aminotransferase (ALT) level at baseline, who fell into the grey zone of the HBV management, The serum HBcrAg level greater or equal to 4 log U/mL was associated with more than 6-fold increase in the risk of HCC development than those with HBcrAg level below this cutoff.
Taken together, serum HBcrAg level could be used to evaluate the risk of HCC development in treatment naïve CHB patients. On top of the risk groups stratified by HBV DNA level, HBsAg and HBcrAg levels further categorize their HCC risk in those with low-and intermediate-risk levels. Such a risk stratification system may help physicians optimize the clinical management of HBV patients.
VIRAL REPLICATION MARKERS TO PREDICT HCC DEVELOPMENT IN ON-TREATMENT PATIENTS
Oral antiviral therapy is effective in inhibiting reverse transcription of pregenomic RNA, thus serum HBV DNA could be lowered to an undetectable level in most of the on-treatment patients. In contrast, HBsAg is translated from the viral mRNA directly, thus it remained high and stable. However, the HBcrAg kinetic was different from both as it consists of the protein products translated from the precore/core mRNA.
A decline in serum HBcrAg level in CHB patients after NA therapy was first reported by a study from Hong Kong [27]. They found that HBcrAg level declined faster (-0.244 log KU/mL/year) than HBsAg level did (-0.107 log IU/mL/year). HBcrAg level dropped more rapidly in the first year of follow-up compared with the reduction in subsequent years (-0.815 vs. -0.117 log KU/mL/year, P<0.001). In addition, only about 32.0% of the patients had undetectable HBcrAg at the end of follow-up although most of the patients cleared their serum HBV DNA levels. The rapid decline was also validated in a Japanese study [28]. They reported a decline in the serum HBcrAg level of >1 log U/mL in both the HBeAg-positive and -negative cohorts after NA treatment.
The mechanism of the decline in the serum HBcrAg level after NA treatment remains vague. In the HBV replication cycle, HBcrAg comes from the transcription of the HBV precore/core gene, in which NA treatment has no direct effect. A drop of serum HBcrAg level could be attributed to the prevention of new infection, which may further reduce the intrahepatic cccDNA pool. This inference needs more longitudinal data of intrahepatic cccDNA and serum HBcrAg levels during different time points of the treatment course.
The viral factors impact the development of HBV-related HCC differently between the patients with and without antiviral treatment. Most of the studies suggested that HBV DNA and HBsAg levels did not predict HCC development after NA treatment. However, several reports suggested a different role of HBcrAg in predicting HCC development. The positive association between HBcrAg level and HCC risk was first reported by Honda et al. [29] in a Japanese cohort study. The hospital-based retrospective cohort enrolled 109 CHB patients receiving NA therapy for more than 2 years. About half the patients received potent antiviral treatment. After a mean follow-up duration of 6.5 years, 36 patients developed HCC. The HBcrAg positivity before treatment was shown not to predict HCC development. In contrast, at the end of follow-up, the patients with HCC development were more likely to have a detectable HBcrAg than those without HCC development. Ando et al. [30] also explored the relationship between HBcrAg and HCC development in another Japanese treatment cohort. A total of 133 CHB patients who received NA therapy and achieved undetectable serum HBV DNA were enrolled. Most of them received potent NA therapy. The median follow-up duration after the disappearance of HBV DNA was 4.8 years and 13 patients subsequently developed HCC. The data showed that patients with higher serum HBcrAg levels when the serum HBV DNA was cleared were associated with an increased HCC risk.
A small patient number is a common limitation for both studies. In a recent Japanese treatment cohort on a large number of patients, Hosaka et al. [28] found a positive correlation between on‐ treatment HBcrAg level and HCC risk. A total of 1,268 CHB patients were enrolled and about half of them received potent NA therapy. The median follow-up duration was 8.9 years. The data showed that a higher HBcrAg level at 1 year after antiviral therapy was associated with higher HCC risk and the cutoff values were 4.9 log U/mL and 4.4 log U/mL for the HBeAg-positive and HBeAg-negative patients, respectively. The multivariable analysis revealed that, compared to the patients with low HBcrAg levels after 1-year treatment, those with high HBcrAg levels had a higher risk of HCC, with adjusted HRs of 6.15 (95% CI, 1.89–20.0) for the HBeAg-positive patients, and 2.54 (95% CI, 1.40–4.60) for the HBeAg-negative patients. The role of pre-treatment HBcrAg level could not be evaluated in the HBeAg-positive patients because most HBcrAg levels at baseline were beyond the assay’s upper detection limit. The pre-treatment HBcrAg level was not associated with HCC development in the HBeAg-negative patients.
In addition to the Japanese cohort studies, a recent Hong Kong study enrolling 1,400 NA-treated CHB patients was conducted to explore the relationship between HBcrAg level and HCC development [31]. All the patients received potent NA therapy. During a median follow-up duration of 45 months, 85 patients developed HCC. High serum HBcrAg levels were defined as >2.9 log U/mL in the HBeAg-negative patients and >4.9 log U/mL in the HBeAg-positive patients (cutoffs adopted from Tada’s and Hosaka’s reports, respectively) [6,28]. They found that high on-treatment HBcrAg levels were associated with an increased risk of HCC development in the overall cohort and HBeAg-negative patients (n=1,042), but not in HBeAg-positive patients (n=358). In addition, serum HBsAg levels were not associated with HCC risk. However, 92.6% of the patients had received NA before the enrollment with various duration (median: 44 months, interquartile range: 18–71 months), and around 70% had already achieved undetectable HBV DNA when HBcrAg was determined. All these factors might influence the interpretation of the findings and thus should be cautious.
In short, on-treatment levels and kinetic of HBcrAg are potential predictors for HCC development in NA-treated patients. Most data are coming from Asian countries, especially from Japan (Table 2). We need more data from other countries for validation.
Table 2.
Relationship between HBcrAg level and HCC development in cohort studies of patients with oral antiviral treatment
| Study | Country | Study design | HBcrAg measurement time points | Number of subjects | Number of HCC cases | Finding | Median follow-up time | Treatment drugs |
|---|---|---|---|---|---|---|---|---|
| Honda et al. [29] (2016) | Japan | Retrospective | Before treatment and at the end of follow-up | 109 | 36 | 1. The HBcrAg positivity (≥3.0 log U/mL vs. <3.0 log U/mL) before treatment was not associated with HCC development. | 6.5 years | Patients with NA therapy for >2 years were enrolled; LAM: 12, LAM to LAM+ADV: 25, LAM to ETV: 17, ETV: 55 |
| 2. At the end of follow-up, the patients with HCC development had higher detectable rates of HBcrAg than those without HCC development. | ||||||||
| Ando et al. [30] (2018) | Japan | Retrospective | At the time of HBV DNA disappearance | 133 | 13 | Higher HBcrAg level (≥3.4 log U/mL vs. <3.4 log U/mL) at the time of HBV‐DNA disappearance was associated with HCC development (adjusted HR, 13.532; 95% CI, 1.683–108.815). | 4.8 years (after HBV‐DNA disappearance) | Patients with NA and achieved HBV DNA disappearance were enrolled; LAM: 14, LAM+ADV: 18, ETV/ET-V+ADV/TDF: 101 |
| Hosaka et al. [28] (2019) | Japan | Retrospective | At baseline and 1 year after receiving NA therapy | 1,268 | 113 | 1. Higher HBcrAg levels at 1 year after treatment were significantly associated with HCC development, for both HBeAg-positive (≥4.9 log U/mL vs. <4.9 log U/mL; adjusted HR, 6.15; 95% CI, 1.89–20.0), and HBeAg-negative patients (≥4.4 log U/mL vs. <4.4 log U/mL; adjusted HR, 2.54; 95% CI, 1.40–4.60). | 8.9 and 8.4 years for HBeAgpositive and -negative patients, respectively | Patients with NA therapy for >1 year were enrolled; LAM: 683, ETV: 585 |
| 2. Pre-treatment HBcrAg levels could not predict the development of HCC in HBeAg-negative patients. They could not analyze the role of HBcrAg in HBeAg-positive patients as most of the HBcrAg data are greater than upper limit of quantification. | ||||||||
| Liang et al. [31] (2020) | Hong Kong | Retrospective | Baseline, defined as the earliest serum samples available for HBcrAg | 1,400 | 85 | Higher baseline HBcrAg level (>2.9 log U/mL vs. ≤2.9 log U/mL) was an independent factor for HCC in HBeAg-negative patients (adjusted HR, 2.13; 95% CI, 1.10–4.14). | 45 months | Patients with NA therapy were enrolled; ETV: 77%, TDF: 24% |
HBcrAg, hepatitis B core-related antigen; HCC, hepatocellular carcinoma; NA, nucleos(t)ide analogue; LAM, lamivudine; ADV, adefovir; ETV, entecavir; HBV, hepatitis B virus; HR, hazard ratio; CI, confidence interval; TDF, tenofovir disoproxil fumarate; HBeAg, hepatitis B e antigen.
CHALLENGES AHEAD AND THE WAY FORWARD
Current data have suggested that HBcrAg level serves as an HCC predictor in treatment-naïve patients and on-treatment level or kinetic of HBcrAg may stratify HCC risk in NA-treated patients. However, several fundamental and essential questions remain to be answered.
First, HBcrAg assay detects HBeAg, p22cr and HBcAg, and HBcAg represents around 10.5% of the precore/core proteins in HBsAg-positive particles only [11]. As HBeAg and p22cr share the first start codon of the precore/core region, production of both proteins could be highly affected by the precore stop codon (G1896A) mutation (Fig. 2B). In other words, serum HBcrAg levels will be reduced when precore stop codon mutation (G1896A) emerges. This may raise two issues: 1) although HBcrAg is now considered as a surrogate marker of intrahepatic cccDNA level, the proportion of precore stop codon mutation (G1896A) should be included as an adjusting variable in the prediction formula. And 2) although the REVEAL-HBV cohort study has shown that the precore stop codon mutation (A1896) was associated with a lower HCC risk compared to the precore wild-type (G1896) [32], the relationship between the viral variant and HCC has not been well validated. If the relationship is confirmed, the viral variants may confound the relationship between HBcrAg level and HCC as it also affects the production of HBcrAg. In addition, basal core promoter mutations are another potential confounder as they are HCC-associated variants that may influence HBcrAg production [33-36]. More studies are needed to clarify whether HBcrAg levels remain an independent risk factor after adjusting these viral variants (Fig. 3).
Figure 3.
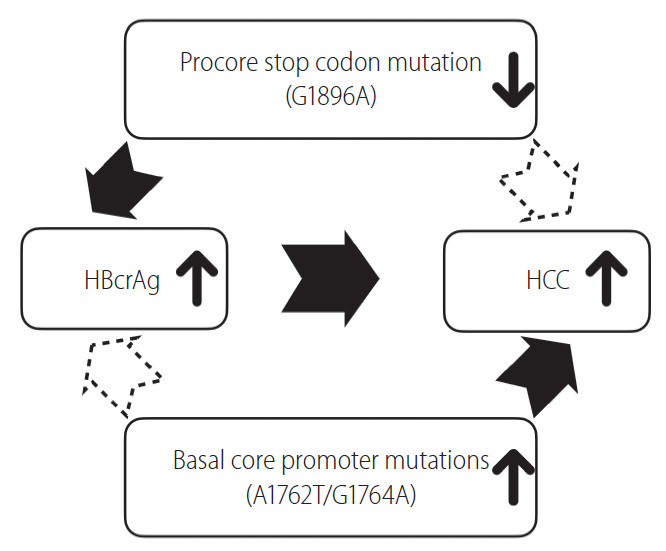
Precore stop codon mutation (G1896A) and basal core promoter mutations may confound the relationship between hepatitis B core-related antigen (HBcrAg) level and risk of hepatocellular carcinoma (HCC).
Second, the underlying mechanism of how HBcrAg levels affect HCC development is unclear. As HBcrAg is a viral protein translated from cccDNA level, a common hypothesis is that higher HBcrAg levels indicate more replication competent viral templates and less controlled viral replication, which could lead to more liver damage followed by higher risk of cirrhosis and HCC development. The data from ERADICATE-B study has supported the hypothesis by showing the positive relationship between HBcrAg and the risk of developing pre-HCC adverse events in treatment-naïve patients [37]. A Japanese cohort study also had a similar finding. These data confirmed the hypothesis in treatment-naïve patients [38]. In contrast, once patients receive NA treatment, it prevents not only the HBV-related liver necroinflammation but also the progression of liver fibrosis. That is why most of the data suggest that HBV DNA and HBsAg levels are no more HCC predictors after NA treatment [39,40]. It is still unknown whether HBcrAg level plays a different role from HBV DNA and HBsAg levels in predicting the residual HCC risk in patients after prolonged NA therapy. More clinical data and mechanistic studies are awaited to address this issue.
The last challenge is how to apply the HBcrAg quantification in clinical practice. Current lines of evidence show that it could not replace HBV DNA level to predict HCC development or to serve as the criteria for initiating antiviral treatment [8], but whether HBcrAg levels may serve as a biomarker to optimize the management of HBeAg-negative patients at grey zone, who have either high HBV DNA levels or mildly elevated ALT levels, deserves further investigation [41]. Conflicting results about the HCC risk in the grey zone patients existed in recent studies, suggesting this is a heterogeneous population [42-44]. As HBcrAg level is another biomarker for viral replication, it may play a given role in stratifying the risk of HCC development in this special clinical setting, which may help physicians optimize the management of the so-called “grey zone” patients.
CONCLUSION
In summary, the viral markers representative of HBV replication are useful predictors for HBV-related HCC in treatment-naïve patients. The next key question is how to combine all of them to achieve a more accurate HCC prediction in treatment-naïve patients. In those receiving prolonged antiviral therapy, more conclusive studies are needed to clarify the relationship between HBcrAg level and the HCC risk.
Acknowledgments
This work was supported by the grants from the National Taiwan University Hospital (107-N4041,108-N4157,109-N4644,109-P05), the Ministry of Science and Technology, Executive Yuan, Taiwan (MOST 106-2314-B-002-136), the National Health Research Institutes (NHRI-EX108-10807BC).
Abbreviations
- ALT
alanine aminotransferase
- cccDNA
covalently closed circular DNA
- CHB
chronic hepatitis B
- CI
confidence interval
- ER
endoplasmic reticulum
- ERADICATE-B
Elucidation of Risk fActors for DIsease Control or Advancement in Taiwanese hEpatitis B carriers
- HBcAg
hepatitis B core antigen
- HBcrAg
hepatitis B core-related antigen
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- NA
nucleos(t)ide analogue
- p22cr
22-kDa precore protein
- REVEAL-HBV
Risk Evaluation of Viral Load Elevation and Associated Liver Disease/Cancer-Hepatitis B Virus
Footnotes
Authors’ contributions
Conceptualization: Jer-Wei Wu and Tai-Chung Tseng; Drafting of the manuscript: Jer-Wei Wu and Tai-Chung Tseng; Critical review and editing of the manuscript: Jia-Horng Kao and Tai-Chung Tseng
Conflicts of Interest
The authors have no conflicts to disclose.
REFERENCES
- 1.World Health Organization (WHO) Global Hepatitis Report 2017. Geneva: WHO; 2017. [Google Scholar]
- 2.Tseng TC, Kao JH. Elimination of hepatitis B: is it a mission possible? BMC Med. 2017;15:53. doi: 10.1186/s12916-017-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 4.Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC, Chen CL, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology. 2012;142:1140–1149.e3. doi: 10.1053/j.gastro.2012.02.007. quiz e13-e14. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Yang HI, Lee MH, Jen CL, Batrla-Utermann R, Lu SN, et al. Serum levels of hepatitis B surface antigen and DNA can predict inactive carriers with low risk of disease progression. Hepatology. 2016;64:381–389. doi: 10.1002/hep.28552. [DOI] [PubMed] [Google Scholar]
- 6.Tada T, Kumada T, Toyoda H, Kiriyama S, Tanikawa M, Hisanaga Y, et al. HBcrAg predicts hepatocellular carcinoma development: an analysis using time-dependent receiver operating characteristics. J Hepatol. 2016;65:48–56. doi: 10.1016/j.jhep.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 7.To WP, Mak LY, Wong DK, Fung J, Liu F, Seto WK, et al. Hepatitis B core-related antigen levels after HBeAg seroconversion is associated with the development of hepatocellular carcinoma. J Viral Hepat. 2019;26:1473–1480. doi: 10.1111/jvh.13191. [DOI] [PubMed] [Google Scholar]
- 8.Tseng TC, Liu CJ, Hsu CY, Hong CM, Su TH, Yang WT, et al. High level of hepatitis B core-related antigen associated with increased risk of hepatocellular carcinoma in patients with chronic HBV infection of intermediate viral load. Gastroenterology. 2019;157:1518–1529. doi: 10.1053/j.gastro.2019.08.028. e3. [DOI] [PubMed] [Google Scholar]
- 9.Inoue T, Tanaka Y. Novel biomarkers for the management of chronic hepatitis B. Clin Mol Hepatol. 2020;26:261–279. doi: 10.3350/cmh.2020.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dandri M, Locarnini S. New insight in the pathobiology of hepatitis B virus infection. Gut. 2012;61 Suppl 1:i6–i17. doi: 10.1136/gutjnl-2012-302056. [DOI] [PubMed] [Google Scholar]
- 11.Kimura T, Ohno N, Terada N, Rokuhara A, Matsumoto A, Yagi S, et al. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J Biol Chem. 2005;280:21713–21719. doi: 10.1074/jbc.M501564200. [DOI] [PubMed] [Google Scholar]
- 12.Ganem D, Schneider RJ. In: Fields Virology. J Biol Chem. 4th ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. The Molecular Biology of the Hepatitis B Viruses; pp. 2923–2970. [Google Scholar]
- 13.Lok AS, Akarca US, Greene S. Predictive value of precore hepatitis B virus mutations in spontaneous and interferon-induced hepatitis B e antigen clearance. Hepatology. 1995;21:19–24. [PubMed] [Google Scholar]
- 14.Chang MH, Hsu HY, Ni YH, Tsai KS, Lee PI, Chen PJ, et al. Precore stop codon mutant in chronic hepatitis B virus infection in children: its relation to hepatitis B e seroconversion and maternal hepatitis B surface antigen. J Hepatol. 1998;28:915–922. doi: 10.1016/s0168-8278(98)80337-1. [DOI] [PubMed] [Google Scholar]
- 15.Chan HL, Hussain M, Lok AS. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology. 1999;29:976–984. doi: 10.1002/hep.510290352. [DOI] [PubMed] [Google Scholar]
- 16.Yuen MF, Sablon E, Yuan HJ, Hui CK, Wong DK, Doutreloigne J, et al. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J Infect Dis. 2002;186:1335–1338. doi: 10.1086/344327. [DOI] [PubMed] [Google Scholar]
- 17.Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, et al. Prevalence of HBV precore/core promoter variants in the United States. Hepatology. 2003;38:619–628. doi: 10.1053/jhep.2003.50352. [DOI] [PubMed] [Google Scholar]
- 18.Maasoumy B, Wiegand SB, Jaroszewicz J, Bremer B, Lehmann P, Deterding K, et al. Hepatitis B core-related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin Microbiol Infect. 2015;21:606.e1–e10. doi: 10.1016/j.cmi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Wong DK, Seto WK, Cheung KS, Chong CK, Huang FY, Fung J, et al. Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 2017;37:995–1001. doi: 10.1111/liv.13346. [DOI] [PubMed] [Google Scholar]
- 20.Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, et al. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J Clin Microbiol. 2002;40:439–445. doi: 10.1128/JCM.40.2.439-445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida K, Desbiolles A, Feldman SF, Ahn SH, Alidjinou EK, Atsukawa M, et al. Hepatitis B core-related antigen to indicate high viral load: systematic review and meta-analysis of 10,397 individual participants. Clin Gastroenterol Hepatol. 2021;19:46–60.e8. doi: 10.1016/j.cgh.2020.04.045. [DOI] [PubMed] [Google Scholar]
- 22.Wooddell CI, Yuen MF, Chan HL, Gish RG, Locarnini SA, Chavez D, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9:eaan0241. doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki F, Miyakoshi H, Kobayashi M, Kumada H. Correlation between serum hepatitis B virus core-related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J Med Virol. 2009;81:27–33. doi: 10.1002/jmv.21339. [DOI] [PubMed] [Google Scholar]
- 24.Matsuzaki T, Tatsuki I, Otani M, Akiyama M, Ozawa E, Miuma S, et al. Significance of hepatitis B virus core-related antigen and covalently closed circular DNA levels as markers of hepatitis B virus re-infection after liver transplantation. J Gastroenterol Hepatol. 2013;28:1217–1222. doi: 10.1111/jgh.12182. [DOI] [PubMed] [Google Scholar]
- 25.Testoni B, Lebossé F, Scholtes C, Berby F, Miaglia C, Subic M, et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615–625. doi: 10.1016/j.jhep.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 26.Kumada T, Toyoda H, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, et al. Effect of nucleos(t)ide analogue therapy on hepatocarcinogenesis in chronic hepatitis B patients: a propensity score analysis. J Hepatol. 2013;58:427–433. doi: 10.1016/j.jhep.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Lam YF, Seto WK, Wong D, Cheung KS, Fung J, Mak LY, et al. Sevenyear treatment outcome of entecavir in a real-world cohort: effects on clinical parameters, HBsAg and HBcrAg levels. Clin Transl Gastroenterol. 2017;8:e125. doi: 10.1038/ctg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosaka T, Suzuki F, Kobayashi M, Fujiyama S, Kawamura Y, Sezaki H, et al. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment Pharmacol Ther. 2019;49:457–471. doi: 10.1111/apt.15108. [DOI] [PubMed] [Google Scholar]
- 29.Honda M, Shirasaki T, Terashima T, Kawaguchi K, Nakamura M, Oishi N, et al. Hepatitis B virus (HBV) core-related antigen during nucleos(t)ide analog therapy is related to intra-hepatic HBV replication and development of hepatocellular carcinoma. J Infect Dis. 2016;213:1096–1106. doi: 10.1093/infdis/jiv572. [DOI] [PubMed] [Google Scholar]
- 30.Ando Y, Ishigami M, Ishizu Y, Kuzuya T, Honda T, Hayashi K, et al. Cumulative incidence and risk factors for the development of hepatocellular carcinoma in patients with chronic hepatitis B who achieved sustained disappearance of viremia by nucleos(t)ide analog treatment. Hepatol Res. 2018;48:E240–E251. doi: 10.1111/hepr.12976. [DOI] [PubMed] [Google Scholar]
- 31.Liang LY, Wong VW, Toyoda H, Tse YK, Yip TC, Yuen BW, et al. Serum hepatitis B core-related antigen predicts hepatocellular carcinoma in hepatitis B e antigen-negative patients. J Gastroenterol. 2020;55:899–908. doi: 10.1007/s00535-020-01700-z. [DOI] [PubMed] [Google Scholar]
- 32.Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29:946–953. doi: 10.1002/hep.510290336. [DOI] [PubMed] [Google Scholar]
- 34.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 35.Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, et al. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258–1265. doi: 10.1086/502978. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng TC, Liu CJ, Yang WT, Hsu CY, Hong CM, Su TH, et al. Serum hepatitis B core-related antigen level stratifies risk of disease progression in chronic hepatitis B patients with intermediate viral load. Aliment Pharmacol Ther. 2021;53:908–918. doi: 10.1111/apt.16266. [DOI] [PubMed] [Google Scholar]
- 38.Tada T, Kumada T, Toyoda H, Kobayashi N, Akita T, Tanaka J. Hepatitis B virus core-related antigen levels predict progression to liver cirrhosis in hepatitis B carriers. J Gastroenterol Hepatol. 2018;33:918–925. doi: 10.1111/jgh.13989. [DOI] [PubMed] [Google Scholar]
- 39.Papatheodoridis G, Dalekos G, Sypsa V, Yurdaydin C, Buti M, Goulis J, et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J Hepatol. 2016;64:800–806. doi: 10.1016/j.jhep.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 40.Tseng TC, Peng CY, Hsu YC, Su TH, Wang CC, Liu CJ, et al. Baseline mac-2 binding protein glycosylation isomer level stratifies risks of hepatocellular carcinoma in chronic hepatitis B patients with oral antiviral therapy. Liver Cancer. 2020;9:207–220. doi: 10.1159/000504650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Bisceglie AM, Lombardero M, Teckman J, Roberts L, Janssen HL, Belle SH, et al. Determination of hepatitis B phenotype using biochemical and serological markers. J Viral Hepat. 2017;24:320–329. doi: 10.1111/jvh.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou K, Wahed AS, Cooper S, Di Bisceglie AM, Fontana RJ, Ghany MG, et al. Phase transition is infrequent among north American adults with e-antigen-negative chronic hepatitis B and low-level viremia. Am J Gastroenterol. 2019;114:1753–1763. doi: 10.14309/ajg.0000000000000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonacci M, Lens S, Mariño Z, Londoño MC, Rodríguez-Tajes S, Mas A, et al. Anti-viral therapy can be delayed or avoided in a significant proportion of HBeAg-negative Caucasian patients in the grey zone. Aliment Pharmacol Ther. 2018;47:1397–1408. doi: 10.1111/apt.14613. [DOI] [PubMed] [Google Scholar]
- 44.Lee HW, Kim SU, Baatarkhuu O, Park JY, Kim DY, Ahn SH, et al. Progression of untreated minimally active chronic HBV infection compared to inactive infection. Clin Gastroenterol Hepatol. 2019;17:2808–2810.e2. doi: 10.1016/j.cgh.2019.01.002. [DOI] [PubMed] [Google Scholar]