Summary
Quiescence is a dynamic process of reversible cell cycle arrest. High-level persistent expression of the HES1 transcriptional repressor, which oscillates with an ultradian periodicity in proliferative neural stem cells (NSCs), is thought to mediate quiescence. However, it is not known whether this is due to a change in levels or dynamics. Here, we induce quiescence in embryonic NSCs with BMP4, which does not increase HES1 level, and we find that HES1 continues to oscillate. To assess the role of HES1 dynamics, we express persistent HES1 under a moderate strength promoter, which overrides the endogenous oscillations while maintaining the total HES1 level within physiological range. We find that persistent HES1 does not affect proliferation or entry into quiescence; however, exit from quiescence is impeded. Thus, oscillatory expression of HES1 is specifically required for NSCs to exit quiescence, a finding of potential importance for controlling reactivation of stem cells in tissue regeneration and cancer.
Subject areas: molecular biology, cell biology, stem cells research
Graphical abstract
Highlights
-
•
HES1 oscillates in proliferative, quiescent, and reactivated NSCs
-
•
Ectopic UbC-driven HES1 expression overrides the endogenous HES1 oscillations
-
•
Persistent UbC-HES1 expression impedes reactivation of quiescent NSCs
-
•
Total HES1 level remains within physiological range in NSCs expressing UbC-HES1
Molecular biology; Cell biology; Stem cells research
Introduction
Quiescence is defined as a state of reversible growth arrest in which cells are not actively dividing but they retain the capacity to re-enter the cell cycle when exposed to the appropriate signals. It is a common feature adapted by many stem cell populations that enables them to retain a “reservoir” of cells in an idling state that can respond to changes affecting the actively dividing stem cells (van Velthoven and Rando, 2019). As such, dysregulation of this state has huge implications in the health of an organism as in tissue homeostasis and regeneration (Marescal and Cheeseman, 2020). In the adult vertebrate brain, neural stem cells (NSCs) exist primarily in two main brain regions, the subgranular zone (SGZ) of the dentate gyrus in the hippocampus and the ventricular-subventricular zone (V-SVZ) that lines the lateral ventricles, where they remain largely in a quiescent state (Doetsch et al., 1999; Doetsch, 2003; Seri et al., 2001; Urban et al., 2019; Obernier and Alvarez-Buylla, 2019). Although adult NSCs continuously generate new neurons in vivo, this does not seem to be a sufficient mechanism for regeneration and brain repair. It appears that the extracellular environment can restrict the activation of quiescent NSCs or their full differentiation potential upon activation (Magnusson and Frisen, 2016). Therefore, understanding how quiescent cells can be reactivated is the first key step to regeneration.
Although mostly studied in the adult brain, quiescent stem cells also exist in the embryonic brain, in spite of the overall extensive proliferation in the embryo. In particular, it was shown that a subpopulation of embryonic neural progenitors is ‘set aside’ during early embryonic development and remains in a quiescent state that later contributes to the adult NSC pool (Fuentealba et al., 2015; Furutachi et al., 2015). The Notch signaling pathway, and especially its effectors and members of the bHLH family Hes1 and Ascl1, are known to play an important role during embryonic neural development and in the adult brain (Cau et al., 2002; Pattyn et al., 2006; Hatakeyama et al., 2004; Baek et al., 2006; Sueda et al., 2019), which makes them attractive candidates for regulating the state of quiescence.
HES1 is a transcriptional repressor that inhibits its own transcription and causes HES1 to oscillate with a 2–3h periodicity in NSCs. In turn, the proneural gene Ascl1, which is a downstream target of HES1 (Chen et al., 1997), also oscillates with a similar period and it is inversely correlated with HES1 expression (Imayoshi et al., 2013). ASCL1 oscillations are needed to maintain NSCs in a proliferative state while switching to sustained ASCL1 expression at similar level promotes neurogenesis (Imayoshi et al., 2013). Similarly, in the adult brain, the ASCL1 oscillations control the transition from NSC quiescence to activation and differentiation toward neurons. ASCL1 expression is mostly lost in quiescent NSCs, however induced oscillatory Ascl1, but not sustained, more effectively activates quiescent NSCs to generate neurons (Sueda et al., 2019). These findings highlight the importance of dynamically expressed genes where the pattern of protein expression on its own, and not level, can dictate cell fate. This provides a new perspective of studying cell–state transitions, and especially the state of quiescence which is inherently dynamic.
Within the context of quiescence, HES1 is expressed at high level in slow dividing boundary regions in the roof and floor plate of the embryonic mouse spinal cord and was found to oscillate at high level in quiescent embryonic and adult NSCs (Baek et al., 2006; Sueda et al., 2019). Moreover, forced expression of sustained HES1 has been shown to reduce cell proliferation (Shimojo et al., 2008; Baek et al., 2006) and maintain NSCs in a quiescent state (Sueda et al., 2019). Therefore, high sustained or high oscillatory HES1 expression inhibits NSC proliferation. Unlike ASCL1, the role of HES1 dynamics on the ability of NSCs to enter and/or exit quiescence still remains unknown and it is not clear whether the dynamic expression of HES1 itself, rather than the high level, can drive this phenotype.
Here, we provide a detailed characterization of HES1 expression in embryonic NSCs as they transit in and out of quiescence in vitro and we explore the role of HES1 dynamics during this transition. We find that upon induction of NSC quiescence with BMP4 the total HES1 level does not increase while the HES1 oscillations are retained. To assess the role of HES1 dynamics in driving entry or exit from quiescence we first generated a new fluorescent HES1mScarlet-I/mScarlet-I transgenic mouse line, where the gene encoding for the mSCARLET-I fluorophore has been fused to the endogenous Hes1 locus, and we established NSC cultures from homozygote (HOM) mice where mSCARLET-I represents total endogenous HES1 expression. We then introduced a persistent mVENUS:HES1 input under a moderate strength UbC promoter (UbC-mVENUS:HES1) and assessed its effect on endogenous HES1 dynamics and HES1 level. We found that upon expression of UbC-mVENUS:HES1 the endogenous HES1 oscillations were abolished and endogenous HES1 level decreased by x3.5fold. Absolute quantification of the HES1 concentration with fluorescence correlation spectroscopy (FCS) showed that although the total HES1 concentration was increased in UbC-mVENUS:HES1 expressing cells, this was still within the range of HES1 concentrations found in cells not expressing the persistent input. Interestingly, under these conditions NSCs continue to proliferate, and they can be induced into quiescence; however, exit from quiescence is impeded. We, therefore, provide evidence that oscillatory HES1 dynamics are required specifically for reactivation of NSCs from quiescence.
Results
1. HES1 oscillations persist through quiescence and reactivation, and are of better quality in quiescent NSCs
To monitor the HES1 dynamics in active and quiescent NSCs, we first established NSC lines from the telencephalic lateral ganglionic eminence (LGE) of E13.5 mouse embryos (Figure 1A). The embryos were derived from HES1 reporter mice where the firefly Luciferase 2 (LUC2) cDNA had been inserted in-frame upstream of the Hes1 gene in a bacterial artificial chromosome (BAC), to generate a LUC2-HES1 fusion protein that mimics the endogenous HES1 expression (Imayoshi et al., 2013) (Figure 1A). Embryonic LGEs were chosen as their descendants contribute to the pool of adult NSCs that reside in the ventral aspect of the subventricular zone (SVZ) (Young et al., 2007).
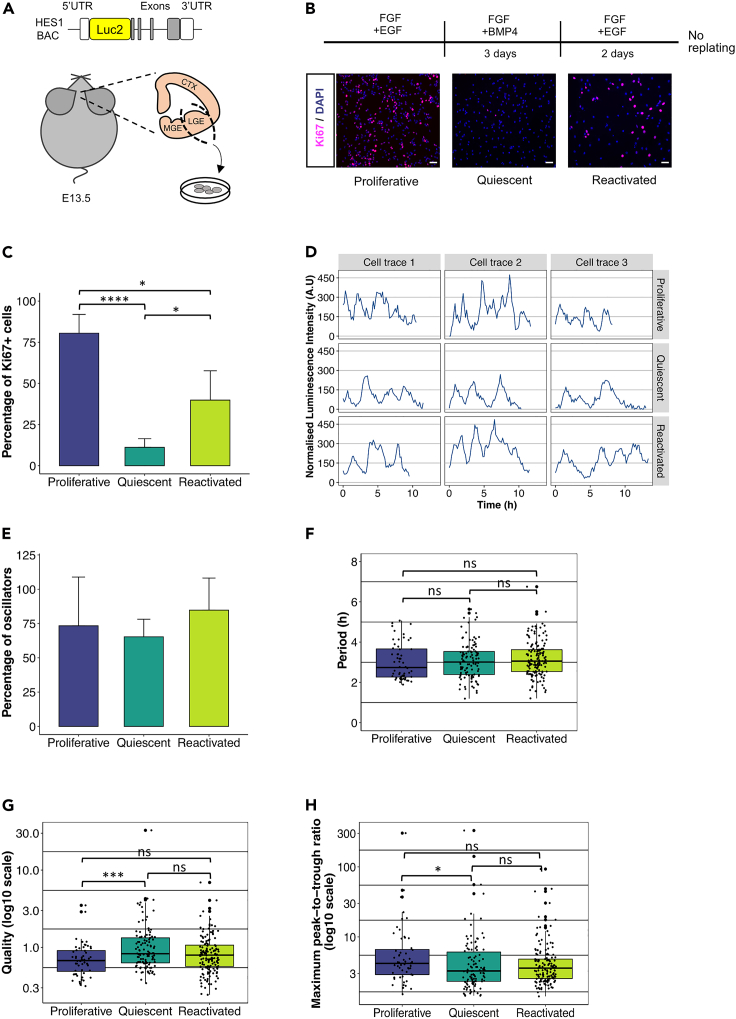
Figure 1.
Characteristics of HES1 dynamic expression in proliferative, quiescent and reactivated conditions
(A) Schematic structure of LUC2:HES1 BAC used to generate transgenic mice. NSC lines were established from the LGE region of E13.5 LUC2:HES1 embryos (CTX = cortex, LGE = lateral ganglionic eminence, MGE = medial ganglionic eminence).
(B) Experimental design for induction of quiescence and reactivation (upper panel). Example images from the analysis of cell proliferation by Ki67 staining at proliferative, quiescent and reactivated conditions. Cells were counterstained with DAPI (bottom panels) (scale bars = 30μm).
(C) Percentage of Ki67 positive cells in proliferative, quiescent and reactivated conditions (error bars represent standard deviation, n = 9 biological experiments, One-way ANOVA with Uncorrected Dunn’s multiple comparison test, proliferative versus quiescent ∗∗∗∗p < 0.0001, quiescent versus reactivated ∗p = 0.04, proliferative versus reactivated ∗p = 0.04).
(D) Example cell traces of E13.5 LUC2:HES1 NSCs cultured in proliferative, quiescent and reactivated conditions (A.U. = arbitrary units).
(E) Percentage of oscillators in proliferative, quiescent and reactivated conditions (error bars represent standard deviation, number of 10-h cell traces analyzed: 75 proliferative from n = 8, 171 quiescent from n = 4, 182 reactivated from n = 5 biological experiments).
(F–H) Box plots representing analysis of 10h bioluminescence traces for period of oscillations (F), quality of oscillations (G) and maximum peak-to-trough ratio per oscillatory trace (H). Note that the first 10h of imaging of individual cells have a random starting point from the start of the imaging of the culture (dots represent individual oscillatory cell traces, black horizontal lines represent median, number of 10-h oscillatory cell traces analyzed: 56 proliferative from n = 7, 110 quiescent from n = 4, 149 reactivated from n = 5 biological experiments, Kruskal-Wallis test with Dunn’s multiple comparison test, ∗p = 0.038, ∗∗∗p = 0.001, ns = not significant)
E13.5 LUC2-HES1 LGEs were cultured in the presence of epidermal growth factor (EGF) and basic fibroblast growth factor (FGF) to promote their proliferation, as indicated by the expression of the cell proliferation marker Ki67 (Figures 1B and 1C), and they uniformly expressed the stem cells markers SOX2 and PAX6 suggesting that they retain their stem cell characteristics in culture (Figures S1A–S1C and S1E–S1G). To induce quiescence, EGF was replaced by BMP4 for 3 days upon which the NSCs lost their Ki67 expression and they were cell-cycle arrested (Figures 1B and 1C). It has been previously shown that addition of BMP4 in the presence of FGF activates a quiescence gene expression program (Martynoga et al., 2013). The main mediators of BMP signaling were found to be ID proteins (inhibitors of differentiation), and not SMADS, although Id genes were not considered to be the main mechanism by which quiescence was established (Martynoga et al., 2013). Accordingly, we find in our cultures that upon addition of BMP4 Id genes (Id1,2,4) are upregulated with Id4 upregulated the most (Figure S2A). To reactivate the cells, BMP4 was replaced by EGF, enabling the cells to re-enter the cell cycle and start proliferating again (Figures 1B and 1C) (Martynoga et al., 2013). The NSCs retained expression of the SOX2 and PAX6 stem cells markers throughout quiescence and reactivation, and irrespectively of their Ki67 status (Figures S1A–S1H), suggesting that their stem cell identity was not lost during this manipulation. To further ensure that upon reactivation we are not promoting the proliferation of the minority of cells that failed to enter quiescence, we reactivated the cells without replating and only by replacing the culture media (Figure 1B).
We next performed bioluminescent single-cell live imaging of NSCs cultured in proliferative, quiescent, or reactivated conditions. HES1 was expressed and expression was dynamic under all three conditions (Figure 1D). Proliferating NSCs are characterized by increased cell motility, which restricts the amount of time individual cells can be tracked for before they exit the field of view, whereas quiescent cells are less motile and therefore can be tracked for longer (Figures S2B and S2C). To therefore avoid any bias introduced by the length of tracking we truncated all cell traces to have a maximum length of 10h, to match the average cell trace length in proliferating conditions (Figure S2C). For example, traces that were 30h long were split into 3 independent 10-h traces so that no information was lost.
We first sought to determine whether the HES1 dynamic expression was oscillatory by using a previously described Gaussian processes (GP)-based method (see STAR Methods) that can classify cell traces as periodic or not, with statistical significance (Phillips et al., 2017). Our analysis revealed that across all different conditions ∼70% of cell traces were oscillatory (Figure 1E). HES1 was found to oscillate in proliferative conditions with a median period of 2.7h, similarly to what has been previously described (Imayoshi et al., 2013), whereas in quiescent and reactivated conditions the HES1 median period was slightly increased by ∼20min, although it was not statistically significant (Figure 1F).
We also assessed and compared the quality of oscillations, which is the ratio of the frequency of oscillations to the timescale of damping (Phillips et al., 2017). Oscillations with high quality better match a sine wave whereas low quality oscillations tend to have higher peak-to-peak variability. We found that HES1 oscillations in quiescent cells are of better quality (Figure 1G). Finally, we compared the amplitude of HES1 oscillations and found that proliferative cells tend to have a higher peak-to-trough ratio (Figure 1H). Oscillations in reactivated cells showed a mixed ‘phenotype’ with intermediate levels of quality and amplitude, indicative of their tendency to return to a proliferative state (Figures 1G and 1H). Finally, to ensure that these comparisons are not affected by potential changes in the HES1 behavior over long periods of time, we compared the first 10h traces to the remaining 10-h blocks within each condition and found no differences for any of the parameters tested (Figure S2D). Overall, our results show that HES1 continues to oscillate in quiescent and reactivated NSCs, similarly to proliferative conditions, with HES1 oscillations being of better quality in quiescent NSCs.
2. HES1 level does not increase in BMP4 induced quiescent NSCs
High HES1 expression has been reported to associate with slow-dividing or quiescent cells whereas enforced high sustained expression prevents cell proliferation (Baek et al., 2006; Shimojo et al., 2008; Sueda et al., 2019) suggesting that high HES1 is potentially required for quiescence to be established. To assess how HES1 level is affected upon induction of quiescence with BMP4 in our system, we performed bioluminescent live imaging of LUC2-HES1 NSCs as they transition from proliferation into quiescence. We began imaging NSCs in proliferating conditions and then replaced the media to induce quiescence while continuing to image. Following media replacement, the cells were imaged for at least three more days, by the end of which the vast majority of the cells had become quiescent (Figures 1B and 1C). Individual cells expressing luminescence were tracked before and after induction of quiescence to record HES1 levels in three independent experiments (Figure 2A). To assess the HES1 level we first estimated the median luminescence expression from all cell traces at every time point and then compared the median expression of all sampling intervals between proliferative and quiescent cells. We found that shortly after the media change luciferase expression was temporarily lost, however, this did not seem to be associated with the content of the media, but it was rather a response to the media change itself (Figure S3A). Our analysis revealed that overall there was no significant difference in HES1 level between proliferative and quiescent NSCs, although the responses were variable with quiescent cells either retaining similar HES1 level or showing a decrease (Figure 2B). Similar results were obtained when we compared the median luminescent expression per cell trace in proliferative versus quiescent conditions (Figure S3B). Quantitative analysis of the Hes1 mRNA also showed no significant differences between proliferative and quiescent cells, but similarly to the luciferase-HES1 expression the response was variable (Figure S3C). Induction of quiescence was efficient in all cases, as judged by analysis of the Ki67 mRNA expression, and therefore unlikely for it to have contributed to the Hes1 level variability (Figure S3C).
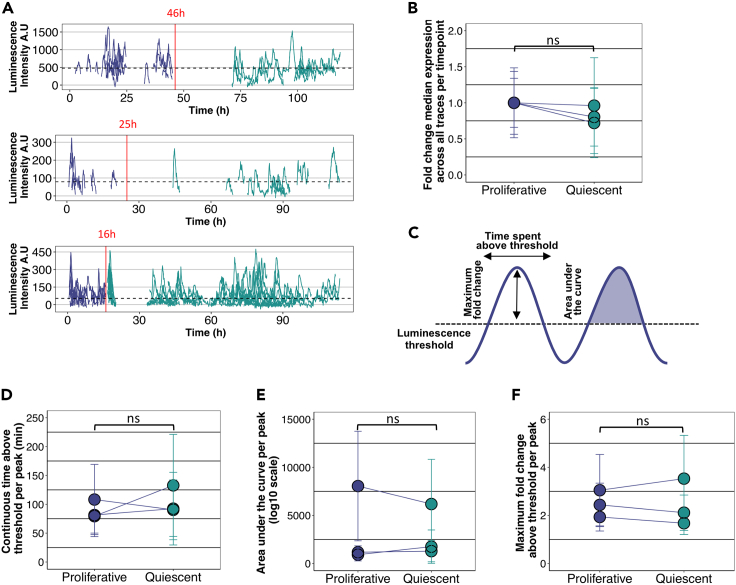
Figure 2.
HES1 level does not increase as NSCs transition from proliferation into quiescence
(A) Graphs showing luminescence expression of LUC2:HES1 NSCs as they transition from proliferation into quiescence from 3 independent experiments. Red vertical lines indicate the time at which proliferation media was replaced by quiescence media and horizontal dotted lines represent the luminescence thresholds defined as the median luminescence expression across proliferative and quiescence conditions per experiment, estimated by the median luminescence expression at each sampling interval (A.U. = arbitrary units).
(B) Graph showing the relative fold change of median luminescence expression between proliferative and quiescent conditions per experiment. Luminescence expression was estimated by the median luminescence expression at each sampling interval across proliferative and quiescent cell traces (error bars represent standard deviation, two-tailed paired ttest, ns = not significant).
(C) Schematic showing the different parameters tested for each peak from all cell traces. These include the maximum fold change of luminescence expression per peak relative to the threshold, the continuous time spent above the threshold for every peak and the area under the curve per peak.
(D–F) Graphs comparing the time spend above threshold per peak (D), the area under the curve (E) and the maximum fold change above threshold per peak (F) between proliferative and quiescent conditions per experiment (circles represent mean values and error bars represent standard deviation, two-tailed paired ttest, ns = not significant)
To avoid any bias introduced by the fact that quiescent cells can be tracked for longer, where potentially more low expressing intervals can be captured, we performed additional analysis to overcome this limitation. For each replicate experiment, we calculated the median luminescence expression across both proliferative and quiescent conditions, which was estimated based on the median HES1 expression at each sampling interval, and we used it as a threshold (Figures 2A and 2C). We then explored for how long the HES1 level was detected above the threshold, continuously. That is, for every trace we would estimate continuous blocks of time intervals during which HES1 was expressed above the threshold. Each block of time interval represents an oscillatory peak and it was treated as an independent component. Blocks of intervals of 30 min or less were excluded from any downstream analysis as they were too short to be considered oscillatory peaks. Analysis of the continuous time spent above threshold between proliferative and quiescent conditions showed variable behavior but not statistically significant differences (Figure 2D). We also measured the area under the curve for each peak that had crossed the threshold, to assess how much HES1 protein was expressed (Figure 2E). Our results showed again no consistent requirements for HES1 level to increase or decrease in order for the cells to enter quiescence (Figure 2E). Finally, we compared the maximum fold change of each peak over the equivalent threshold value, where we found again no statistically significant differences (Figure 2F). Together these findings show that upon induction of quiescence with BMP4 the HES1 level does not increase and that high HES1 level is not required for cells to enter quiescence.
3. Ectopic persistent HES1 expression overrides the endogenous HES1 oscillations
To further gain insight into the functional role of the HES1 dynamics, we next sought to establish a system to manipulate the HES1 oscillatory expression. To this end, we first generated a transgenic mouse HES1 reporter line to mark endogenous HES1 expression. We manipulated the C-terminus of the endogenous HES1 protein by knocking in, in frame, a linker sequence followed by a 3xFlag epitope and the mSCARLET-I fluorophore directly after the last Hes1 exon and upstream of the 3′UTR (Figures 3A and S4). E13.5 LGEs were further isolated from HES1mScarlet-I/mScarlet-I mice (or HOM HES1:mSCARLET-I mice) to establish NSCs lines where mSCARLET-I expression represents the total expression of endogenous HES1 in the cells. To assess whether fusion of the HES1 protein with the mSCARLET-I fluorophore has not affected the function of the protein, we compared total Hes1 mRNA level and the level of known HES1 targets like Ascl1 and Dll1 (Chen et al., 1997; Kobayashi et al., 2009), in wild type (WT) vs HOM HES1:mSCARLET-I cells and found no statistical differences in their level of expression (Figure S5).
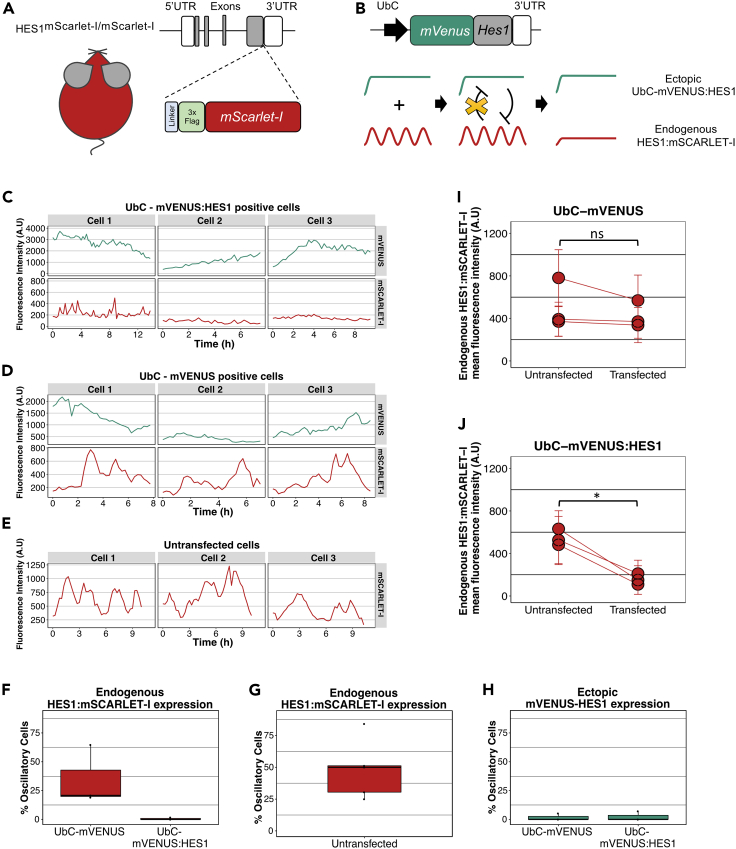
Figure 3.
Endogenous HES1 protein does not oscillate in the presence of an ectopic HES1 persistent input
(A) Schematic structure of the Hes1 locus in the HES1mScarlet-I/mScarlet-I transgenic mice. A DNA sequence encoding for a linker protein, a 3xFlag epitope and the mSACRLET-I protein has been inserted downstream of the last Hes1 exon and before the 3′UTR.
(B) Schematic showing the structure of UbC-mVENUS:HES1 reporter and the hypothesis for altering the endogenous HES1:mSCARLET-I oscillatory expression and level. mVenus has been fused to Hes1 cDNA followed by the Hes1 3′UTR and it is expressed under the constitutive UbC promoter. Persistent HES1 expression is expected to inhibit the endogenous HES1 level and override the endogenous HES1 oscillations.
(C–E) Example cell traces showing mVENUS and mSCARLET-I fluorescence expression over time in the same cell where mVENUS represents expression of either UbC-mVENUS:HES1 (C) or the control UbC-mVENUS (D) and mSCARLET-I represents endogenous HES1 expression. In untransfected cells only the mSCARLET-I endogenous HES1 expression has been recorded (E) (A.U. = arbitrary units). Oscillatory period of mSCARLET-I expression in panel (D) Cell 1 = 2.4h, Cell 2 = 2.4h, Cell 3 = 3h and in panel (E) Cell 1 =3.2h, Cell 2 =3.1h, Cell 3 = 5.3h.
(F and G) Box plots showing the percentage of cells that have oscillatory endogenous HES1:mSCARLET-I expression in the presence of ectopic UbC-mVENUS or UbC-mVENUS:HES1 (F) or in untransfected cells (G) (dots represent means of biological replicates, black horizontal lines represent median, n = 3 biological experiments).
(H) Box plot showing the percentage of cells where UbC-mVENUS or UbC-mVENUS:HES1 exhibit oscillatory expression (dots represent means of biological replicates, black horizontal lines represent median, n = 3 biological experiments).
(I and J) Graphs showing the endogenous HES1:mSCARLET-I expression level in untransfected vs transfected cells with UbC-mVENUS (I) or UbC-mVENUS:HES1 reporter (J) per experiment (circles represent mean values and error bars represent standard deviation, A.U. = arbitrary units). Number of cells analyzed for (I) untransfected = 191, transfected = 135, n = 3 biological experiments, two-tailed paired ttest, ns = not significant. Number of cells analyzed for (J) untransfected = 222, transfected = 88, n = 3 biological experiments, two-tailed paired ttest, ∗p = 0.015.
To manipulate the endogenous HES1 dynamics we introduced into the HOM HES1:mSCARLET-I NSCs an mVENUS:HES1 reporter driven by the ubiquitously expressed and moderate strength UbC promoter. The notion behind this is that ectopic expression of a persistent HES1 input (i.e., UbC-mVENUS:HES1) will continuously suppress the endogenous HES1 expression (due to HES1 autorepression) while the endogenous HES1 protein will not be able to bind and inhibit the UbC-mVENUS:HES1 reporter, as it lacks the HES1 binding sites (Figure 3B). Therefore, we hypothesized that ectopic persistent HES1 expression will override the endogenous HES1 oscillations.
To assess whether this is the case, we performed dual-color fluorescence live imaging of HOM HES1:mSCARLET-I NSCs expressing the UbC-mVENUS:HES1 reporter, to record the endogenous HES1 dynamics in the presence of a HES1 persistent input. We then compared the results against HOM HES1:mSCARLET-I NSCs expressing a control UbC-mVENUS reporter or no reporter at all. We found that HES1:mSCARLET-I endogenous expression continued to be dynamic in untransfected cells or cells expressing the control UbC-mVENUS whereas the oscillations were completely abolished in cells expressing the UbC-mVENUS:HES1 (Figures 3C–3E). Note that untransfected cells are cells from the same culture, treated with the same transfection reagents but they do not express the UbC-mVENUS or UbC-mVENUS:HES1 protein. Oscillatory analysis of the traces, using the GP based method previously described, confirmed that endogenous HES1:mSCARLET remains oscillatory in untransfected and control UbC-mVENUS transfected cells, whereas almost no oscillations were found in cells expressing UbC-mVENUS:HES1 (Figures 3F and 3G). We found that the percentage of oscillators in UbC-mVENUS was lower than the one in untransfected samples, suggesting that transfection of the plasmid on its own may have an effect and, therefore, including an UbC-mVENUS control is always necessary to account for this effect. Finally, as expected, neither the ectopic persistent UbC-mVENUS:HES1 expression nor the control UbC-mVENUS expression were found to be oscillatory as expression of these reporters is driven by the constitutive UbC promoter (Figure 3H).
Our dual-color fluorescence live-imaging also showed a reduction in the endogenous HES1 levels upon ectopic expression of the UbC-mVENUS:HES1 (Figure 3C). We therefore estimated the mean endogenous HES1:mSCARLET-I fluorescence intensity per cell trace in the presence or absence of an ectopic reporter. In particular, we found that in control UbC-mVENUS expressing cells the endogenous HES1 expression was not significantly altered when compared to untransfected cells however, in the presence of ectopic persistent HES1 the endogenous HES1 level was decreased by a x3.5-fold (Figures 3I and 3J). These results confirm our expectation that endogenous HES1 expression would be repressed by UbC-mVENUS:HES1.
Examining the correlation of mVENUS vs mSCARLET expression for the different levels of mVENUS expression in single cells showed that endogenous HES1 expression is inhibited for medium and even low level expression of UbC-mVENUS:HES1 (Figures S6A–S6C). This suggests that inhibition of endogenous expression is efficient even with minimal level of ectopic HES1 expression. In high UbC-mVENUS:HES1 expressing cells the endogenous HES1 increased, which may be due to an indirect effect from the high level of HES1 expression in these cells (Figure S6C).
In summary, we conclude that a low or medium ectopic persistent HES1 expression suppresses the endogenous HES1 level and overrides the endogenous HES1 oscillations, thus, it can be used as a means to manipulate the endogenous HES1 dynamics.
4. Persistent HES1 expression impedes reactivation of quiescent NSCs
Having established that we can override the endogenous oscillations by introducing ectopically a persistent HES1 input, we next sought to determine the effect of altering the HES1 dynamics on the ability of NSCs to proliferate as well as to enter and exit quiescence. To address that, we employed a similar method as before whereby the UbC-mVENUS:HES1 reporter was transfected into WT NSCs to create a mosaic population of UbC-mVENUS:HES1 positive and negative cells. We then cultured the cells either in proliferative conditions or we induced quiescence for 3 days, followed by reactivation into proliferating conditions again for 2 days. At each condition, we fixed the cells and performed immunofluorescent staining against the Ki67 proliferation marker and mVENUS (Figure 4A). As a control, we applied the same process to NSCs transfected with the UbC-mVENUS reporter (Figure 4B).
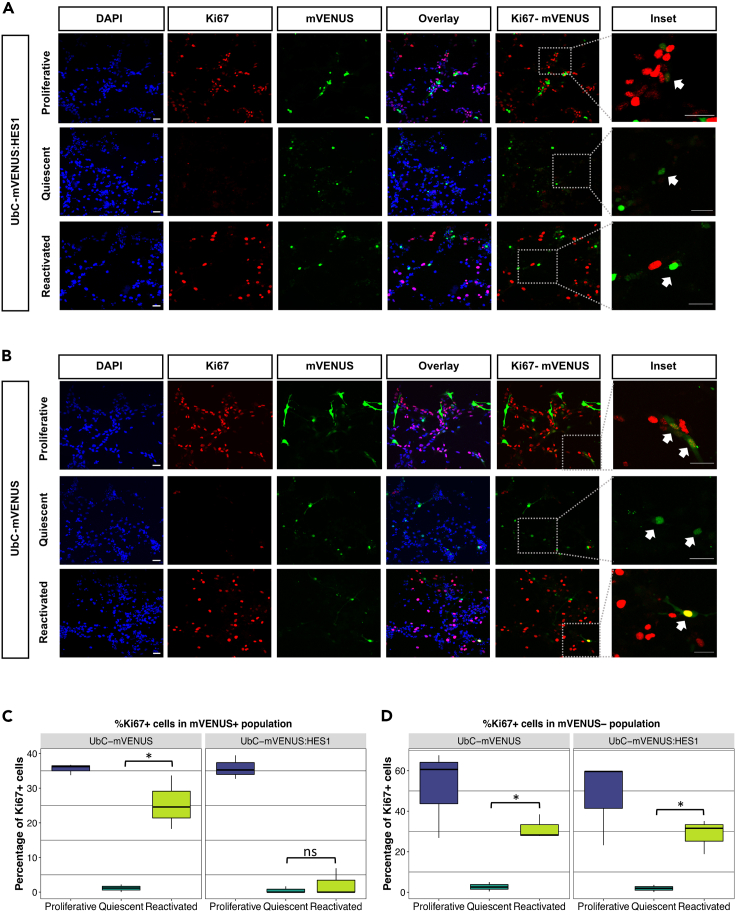
Figure 4.
Loss of HES1 oscillations in NSCs impedes reactivation from quiescence
(A and B) Immunofluorescence staining for the proliferation marker Ki67 and mVENUS. E13.5 NSCs were transfected either with the UbC-mVENUS:HES1 (A) or the control UbC-mVENUS reporter (B). Cells were fixed and stained in proliferative (1d or 2d after transfection in proliferation media), quiescent (4d after transfection, 3d in quiescent culture media) or reactivated (6d after transfection, 3d in quiescent media, and 2d in proliferation media) culture conditions. Cells were also counterstained with DAPI. The insets depict the dotted square areas magnified. White arrows in the insets in proliferative conditions in (A) and (B) mark cells that stain for both Ki67 and mVENUS whereas in quiescent conditions in (A) and (B) white arrows mark mVENUS positive cells that are negative for Ki67. In reactivated conditions in (A) white arrow marks a cell that stains positive only for mVENUS but it is negative for Ki67 whereas in (B) the arrow marks a cell that stains positive for both mVENUS and Ki67 (scale bars = 30μm).
(C and D) Box plots showing the percentage of Ki67 positive cells in UbC-mVENUS or UbC-mVENUS:HES1 positive (C) and negative cells (D) in proliferative, quiescent, and reactivated conditions. Note that the percentage of Ki67 positive cells for UbC-mVENUS:HES1 in proliferative cells in (C) ranges from 35 to 39%, meaning that fixation 1d or 2d after transfection produced similar results (black horizontal lines represent median, n = 3 biological experiments, C left panel: two-tailed paired ttest, ∗p = 0.028, C right panel: Wilcoxon test, ns = not significant, D left panel: two-tailed paired ttest, ∗p = 0.016, D right panel: two-tailed paired ttest, ∗p = 0.022).
We estimated the percentage of Ki67 positive cells at each condition and within the mVENUS positive and mVENUS negative populations. We found that within the mVENUS positive population (i.e., transfected cells), in proliferative conditions, UbC-mVENUS:HES1 expressing cells were proliferating at the same rate as the control UbC-mVENUS expressing cells, suggesting that switching from oscillatory to stable HES1 expression does not prevent cells from cycling (Figure 4C). Equally, upon induction of quiescence, almost all UbC-mVENUS:HES1 expressing cells lost Ki67 expression, similar to control, indicating that dynamic expression of HES1 is not required for cells to enter quiescence (Figure 4C). Interestingly, when UbC-mVENUS:HES1 expressing quiescent cells were exposed to proliferating media they did not resume proliferation (Figure 4C). This impediment in reactivation was only present in UbC-mVENUS:HES1 expressing cells but not in their control counterparts. Accordingly, we found that within the mVENUS negative population (i.e., untransfected cells in the same culture), in both the control UbC-mVENUS and the UbC-mVENUS:HES1 treated cultures, the NSCs were able to proliferate when cultured in proliferating conditions and they would undergo quiescence when exposed to BMP4 (loss of Ki67 expression) whereas upon reactivation they resumed proliferation capacity (Figure 4D). Differences observed in the percentage of Ki67 positive cells in proliferative conditions between the mVENUS positive and mVENUS negative population, are potentially due to the effect of transfection itself which is taken into account by comparing UbC-mVENUS to UbC-mVENUS:HES1 expressing cells (Figures 4C and 4D).
Although we did not detect any differences in the percentage of Ki67 positive cells expressing either UbC-mVENUS or UbC-mVENUS:HES1 in proliferative conditions (Figure 4C), to further examine how variable levels of UbC-mVENUS:HES1 may affect expression of Ki67, we looked at the correlation of mVENUS and Ki67 protein expression intensity in the immunofluorescence data. We found that the mVENUS intensity in cells expressing UbC-mVENUS:HES1 at either high or low level was not correlated with the Ki67 expression intensity, similar to control UbC-mVENUS positive cells (Figure S7A). Additionally, quantitative analysis of the Ki67 mRNA in proliferative cells, sorted for either high or low expression of UbC-mVENUS and UbC-mVENUS:HES1, showed no statistically significant differences between control and experimental cells (Figures S7B and S7C).
Together, these findings suggest that oscillatory HES1 expression is required specifically for cells to be reactivated from quiescence.
5. Total HES1 level remains within physiological level range in cells expressing persistent HES1
To determine whether the failure of UbC-mVENUS:HES1 expressing NSCs to reactivate is due to the change in HES1 dynamic expression rather than the HES1 protein level, it is imperative to assess how the total HES1 level is affected in the presence of the ectopic persistent HES1 input.
We had previously found that ectopic UbC-mVENUS:HES1 expression reduces the endogenous HES1:mSCARLET-I level by x3.5-fold (Figure 3J). However, this decrease in the endogenous HES1 level does not inform on how the total HES1 level is affected in UbC-mVENUS:HES1 expressing cells compared to untransfected. This is because in untransfected cells total HES1 protein is represented only by mSCARLET-I expression whereas in UbC-mVENUS:HES1 treated cells total HES1 is represented by both mSCARLET-I and mVENUS expression, which cannot be compared to just mSCARLET-I expression alone. To this end, we employed FCS to perform absolute quantification of HES1 protein molecules. FCS is an imaging technique used to estimate absolute numbers of fluorescent protein molecules based on their fluorescence intensity and irrespective of the type of fluorophore (Elson, 2011). During FCS analysis, the intensity of fluorescent proteins within a minute volume (also known as confocal volume) is collected over time and temporally autocorrelated to infer the concentration of the protein.
To measure the HES1 concentration we established mosaic cultures of HOM HES1:mSCARLET-I NSCs expressing persistent UbC-mVENUS:HES1 and performed a dual-color FCS for mSCARLET-I and mVENUS (Figure 5Ai). Absolute quantification of mSCARLET-I fluorescent particles per confocal volume is a direct estimation of the endogenous HES1 concentration whereas quantification of mVENUS particles is a direct estimation of mVENUS:HES1 concentration. To estimate the total concentration of HES1, the number of mVENUS and mSCARLET-I HES1 molecules per confocal volume were combined in UbC-mVENUS:HES1 expressing cells and compared against the HES1:mSCARLET-I molecules in untransfected cells (Figure 5Aii). We found that in transfected cells the endogenous HES1 protein expression was dramatically decreased in the vast majority of the cells, preventing the accurate measurement of HES1:mSCARLET-i molecules by FCS. To therefore ensure that we are not underestimating the endogenous HES1 expression in those cells, by assuming zero contribution when accurate measurements cannot be obtained, we decided instead to infer the HES1 concentration from the mSCARLET-I fluorescence intensity. For this purpose we constructed standard curves of mSCARLET-I and mVENUS fluorescence intensity versus mSCARLET-I concentration (determined by FCS in unstransfected cells) and mVENUS concentration (determined by FCS in UbC-mVENUS:HES1 expressing cells) respectively in the same cell, from 3 independent experiments (Figure S8A). In each case we found a strong correlation of 0.8 or greater, suggesting that mSCARLET-I and mVENUS fluorescent intensities can be used as a proxy to estimate protein concentration without performing any further FCS.
Figure 5.
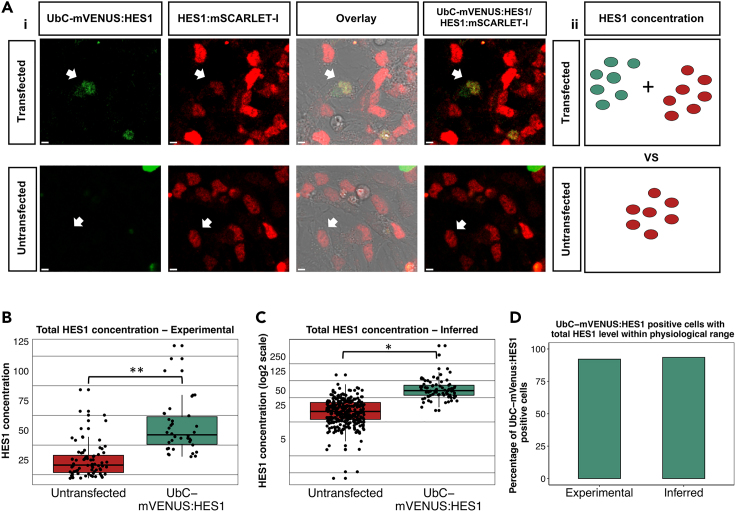
Ectopic persistent HES1 expression does not increase total HES1 level above physiological range
(A) Example pictures of E13.5 HES1:mSCARLET-I NSCs transfected with UbC-mVENUS:HES1 reporter. White arrows mark transfected (top panels) or untransfected (bottom panels) cells where FCS was performed (1). Schematic showing how total HES1 concentration is compared between transfected and untransfected cells. Green dots represent UbC-mVENUS:HES1 concentration and red dots represent HES1-mSCARLET-I concentration (2) (scale bars = 5μm).
(B and C) Box plots showing total HES1 concentration in untransfected and UbC-mVENUS:HES1 transfected E13.5 HOM HES1:mSCARLET-I NSCs. In the reporter transfected cells, the total HES1 concentration is the sum of UbC-mVENUS:HES1 concentration and HES1:mSCARLET-I concentration. In (B) all concentrations have been estimated experimentally by FCS (apart from the HES1:mSCARLET-I concentration in transfected cells which was inferred). In (C) all concentrations have been inferred (black horizontal lines represent median, dots represent individual cells, number of cells analyzed in B) untransfected= 75, transfected = 38, n = 3 biological replicates, in (C) untransfected = 316, transfected = 77, n = 3 biological experiments, two-tailed paired ttest, ∗p = 0.047, ∗∗p = 0.008).
(D) Bar graph showing the percentage of E13.5 HOM HES1:mSCARLET-I NSCs transfected with UbC-mVENUS:HES1 that have a total HES1 concentration (either experimentally estimated or inferred) within the range of total HES1 concentration values in their untransfected counterparts.
We performed two types of analyses in order to estimate the total amount of HES1 protein. One analysis was based on the experimental values obtained by FCS (called experimental analysis), with the exception of the endogenous HES1:mSCARLET-I particles in the UbC-mVENUS:HES1 expressing cells, which had to be inferred from the mSCARLET-I fluorescence intensity. For the second analysis we inferred all HES1 concentration based on mSCARLET-I and mVENUS fluorescence intensity (called inferred analysis). The aim of the latter analysis was to increase the number of cells tested in order to get a better representation of the HES1 level heterogeneity in the culture.
Overall, we found that total HES1 levels in UbC-mVENUS:HES1 expressing cells increased by either x2.1-fold (experimental analysis) or x2.9-fold (inferred analysis) (Figures 5B and 5C). However, over 90% of the UbC-mVENUS:HES1 expressing cells had a total HES1 concentration still within the range of HES1 concentrations recorded in untransfected cells (which we consider to represent the physiological range of HES1 level) (Figure 5D). These results suggest that despite the overall increase in total HES1 level, cells expressing a HES1 persistent input operate within normal HES1 level. We also calculated the contribution of endogenous HES1:mSCARLET-I and ectopic mVENUS:HES1 concentration in the total HES1 level of UbC-mVENUS:HES1expressing cells (in both the experimental and inferred analysis) and found an approximate x4fold decrease in endogenous HES1 levels compared to untransfected cells (Figures S8B and S8C), similar to what we had found before based on the fluorescence intensity comparison (Figure 3J). Altogether these findings reveal that ectopic expression of persistent HES1 under a weak ubiquitous promoter does not increase total HES1 level above normal values, revealing that the impediment of reactivation in UbC-mVENUS:HES1 expressing cells is due to the change in HES1 dynamics and not levels.
To further gain an insight into how this change in HES1 dynamics, but not level, may affect gene expression, which ultimately impedes reactivation of quiescent cells, we performed quantitative PCR analysis in cells expressing UbC-mVENUS:HES1 vs control. We hypothesized that the main effect of persistent HES1 expression is exerted in quiescent cells hence, we sorted mVENUS positive quiescent NSCs expressing either control UbC-mVENUS or UBC-mVENUS:HES1, at either low or high levels (Figure S9A). We then compared mRNA expression for a panel of genes between control and experimental samples. In particular, we looked at expression of the direct target Ascl1 (Chen et al., 1997) and p21 (Castella et al., 2000), the indirect target CyclinD1 (regulator of cell cycle progression) (Shimojo et al., 2008), the Id genes (Id1-Id4), the cell cycle inhibitors Rb1, Rb2 and the tumor suppressor p53. When we looked within the mVENUS low expressing samples (which we hypothesize represent more physiological levels of persistent HES1 expression) we found that expression of Ascl1 was downregulated by x0.7 fold in cells expressing UbC-mVENUS:HES1 (although it was marginally not significant with a pvalue = 0.06) and expression of p21 was significantly upregulated by x3.3-fold suggesting that a change in HES1 dynamics directly affects HES1 downstream targets (Figure S9B). With regards to p21, regulation of this gene by HES1 has been found to be context-dependent with studies reporting both an upregulation and downregulation (Castella et al., 2000; Yu et al., 2006). Accordingly, we found that in proliferative cells, p21 was not differentially affected, supporting a context-dependent regulation by HES1, whereas Ascl1 was significantly reduced by x0.4-fold in UbC-mVENUS:HES1 low expressing cells, similar to quiescent conditions (Figure S9C).
Finally, within the high mVENUS expressors in quiescent cells, Ascl1 was not differentially affected whereas p21 showed an even greater increase of x12 fold in UbC-mVENUS expressing cells.
For the rest of the cell cycle regulator genes and the Ids we found no differential expression between control and experimental conditions in quiescent cells in both the low and high mVENUS expressors (where this was applicable) (Figure S9B). Therefore, a change in the HES1 dynamics, from oscillatory to sustained, can differentially affect downstream targets and also in a context-dependent manner.
Discussion
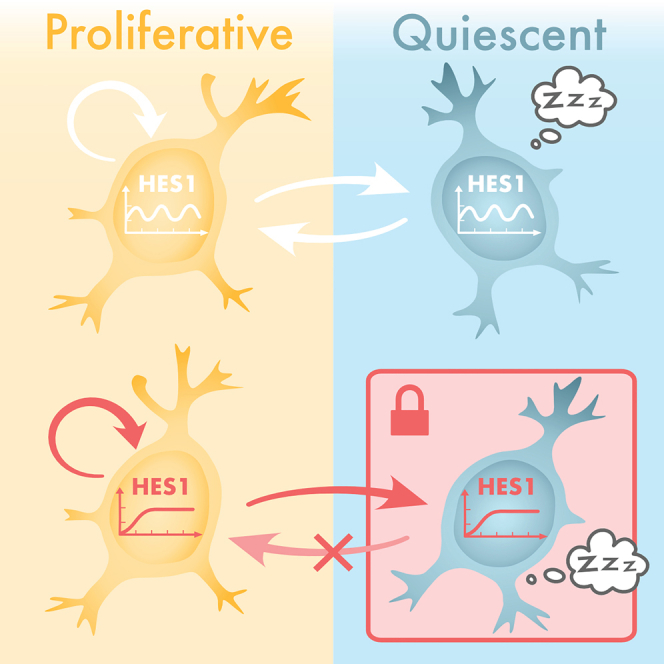
Here we have explored the role of HES1 ultradian oscillations in controlling the entry and exit of NSCs from quiescence in vitro. We showed that by altering the HES1 dynamic expression from oscillatory to persistent, while keeping the total HES1 level within physiological range, reactivation from quiescence is impeded while NSC proliferation and entry into quiescence is not affected.
Our findings highlight the importance of ultradian gene expression dynamics regulating cell fate, a concept that has been evolving over the last several years (Isomura and Kageyama, 2014) as evidence linking the temporal regulation of ultradian oscillations with distinct cellular outcomes emerges (Imayoshi et al., 2013; Shimojo et al., 2016; Santos et al., 2007; Purvis et al., 2012). For example, the Notch ligand Delta-like1 (DLL1) and the transcriptional repressors HES1 and HES7, exhibit oscillatory expression in the mouse presomitic mesoderm. Elongation or shortening of the Dll1 gene dampens the DLL1 oscillations, followed by a dampening of HES1 and HES7 oscillations, which lead to severe fusion of somites and their derivatives (Shimojo et al., 2016). Accordingly, the tumor suppressor protein P53 exhibits oscillatory expression (with a period of ∼5.5h) upon exposure to γ-irradiation (Lev Bar-Or et al., 2000; Lahav et al., 2004), which allows for cells to recover from DNA damage and survive, while induction of sustained P53 signaling leads to senescence (Purvis et al., 2012). Therefore, understanding how gene expression dynamics dictate cell fate is essential for being able to manipulate cell-state transitions at will.
High sustained HES1 expression has been previously shown to associate with slow-dividing cells in vivo or with preventing NSC proliferation both in vitro and in vivo, while quiescent NSCs were found to express high oscillatory HES1 (Baek et al., 2006; Shimojo et al., 2008; Sueda et al., 2019). However, in these cases, HES1 was expressed at a high level which did not allow to dissect out how each factor (HES1 level vs HES1 dynamic expression) contributes to this phenotype. Here we set out to explore the role of HES1 expression dynamics in the context of NSC quiescence. We first investigated how is the HES1 dynamic expression affected when proliferative NSCs are induced in and out of quiescence in vitro with the addition and subsequent removal of BMP4. Similarly to previously published data (Sueda et al., 2019) we found that HES1 oscillatory expression persists throughout quiescence and reactivation. We did not find significant differences in the period of oscillations but we did find that oscillations in quiescent cells were of better quality. This could be due to the fact that quiescent cells are less motile and interact with fewer neighbors, therefore Notch signaling in these cells may be less interrupted by intercellular Notch lateral inhibition (Kageyama et al., 2019).
With regards to HES1 levels, we did not find an increase in the HES1 level overall upon induction of quiescence or a consistent pattern of HES1 oscillatory peaks reaching higher levels or being wider compared to proliferative conditions. This is in contrast to what had been previously reported, where HES1 level was found to increase in quiescent embryonic NSCs in vitro (Sueda et al., 2019). However, it is possible that the use of different BMP ligands to establish quiescence or the use of the same ligand at a different concentration could differentially affect the level of HES1. In our cultures, we find that BMP4 addition induces expression of Id genes and Id4 is the most upregulated. All ID proteins have been reported to interact with HES1 (Jogi et al., 2002). In particular ID2 has been found to release the negative autoregulation of HES1 and increase HES1 level, but without interfering with HES1 repression on its downstream targets (Bai et al., 2007). On the other hand, ID4 can heterodimerize with ID1, ID2 and ID3 and inhibit their biological activity (Sharma et al., 2015) and in this way retain the HES1 oscillations by preventing HES1 to interact with the other IDs (Boareto et al., 2017). It is therefore possible that during establishment of quiescence in response to BMP signaling, differential expression of ID proteins can differentially affect HES1 level, with ID2 for example increasing HES1 level and ID4 inhibiting the function of ID2, thus preventing upregulation of Hes1.
To assess the role of HES1 dynamics in controlling the entry and exit of NSCs from quiescence we first generated a new HES1:mSCARLET transgenic mouse line, where both alleles of the Hes1 locus were tagged with the mSCARLET-I fluorophore, to monitor HES1 endogenous expression. By introducing exogenously a persistent HES1 input, under the control of a moderate strength UbC promoter, we found that we can override the endogenous HES1 oscillations without increasing the total HES1 level above the physiological range. Under these conditions proliferation of NSCs, as judged by Ki67 staining, was not affected. This is line with what has been previously reported where NSCs derived from HES1 mutant mice, with dampened HES1 oscillations, showed no differences in proliferation and neuronal differentiation compared to WT NSCs in vitro (Ochi et al., 2020). Accordingly, mathematical modeling of the Notch/HES regulatory module in NSCs predicted that loss of HES1 oscillations is dispensable for low proneural factor activity that is required for NSC proliferation (Boareto et al., 2017).
We then assessed how loss of HES1 oscillations affects entry into quiescence and reactivation. Upon induction of quiescence, NSCs expressing a persistent HES1 input stopped proliferating, similar to control cells; however, upon reactivation they failed to re-renter the cell cycle. We cannot conclude, though, whether this is a complete block from exiting quiescence or a delayed reactivation. Varying depths of quiescence, accounting for differences in responsiveness to reactivation, have been previously described in adult NSCs but also other systems such as muscle stem cells (MuSCs) and hematopoietic stem cells (HSCs) (Llorens-Bobadilla et al., 2015; Rodgers et al., 2014; Laurenti et al., 2015). Thus, it is possible that persistent HES1 expression alters the depth of quiescence in NSCs. But how can this be achieved? It has been previously reported that changes in the pattern of expression of a transcription factor can differentially affect expression of downstream targets (Ashall et al., 2009; Lane et al., 2017; Hao and O'Shea, 2011; Hansen and O'Shea, 2013; Chen et al., 2020) For example, it has been recently shown using elegant optogenetic tools that downstream targets of the Crz1 transcription factor in yeast have higher gene expression in response to pulsatile input, whereas others responded better to continuous inputs (Chen et al., 2020). Indeed, in our study we found that persistent HES1differentially affects expression of Ascl1 and the cell cycle inhibitor p21 even when expressed at low levels. Ascl1 was found to be downregulated in both proliferative and quiescent NSCs expressing persistent HES1 at low levels, whereas p21 was upregulated only in quiescent conditions. With regards to Ascl1, it is possible that persistent HES1 alters the dynamics of Ascl1 as well as reducing its level (from oscillatory to low sustained). It has been shown before that quiescent cells with low Ascl1 expression need to both upregulate expression of Ascl1 and resume Ascl1 oscillations in order for the cells to reactivate (Sueda et al., 2019). Hence, it is likely for persistent HES1 to block reactivation of quiescent cells by not allowing Ascl1 to increase its level or resume oscillatory expression or both. On the other hand, increased p21 expression in the presence of persistent HES1 expression may also impede reactivation of quiescent cells by not allowing them to progress through the cell cycle when they are stimulated to do so. P21 expression is also induced by p53 (Karimian et al., 2016) however we did not find p53 levels to be significantly altered. Alternatively, persistent HES1 expression may act as a ‘sponge’ of ID proteins which are overexpressed in response to BMP4. Dimerization of ID proteins with E proteins have been found to increase the half-life of IDs (Bounpheng et al., 1999; Lingbeck et al., 2005); thus, it is possible that persistent HES1 expression may ‘trap’ ID proteins and protect them from degradation and therefore retain them in the cells for longer even after removal of the BMP4 stimulus. While much remains to be done to understand the functional significance of these gene expression changes, they serve as a proof-of-principle that oscillatory HES1 expression affects downstream targets differently than persistent expression.
In summary, we provide evidence that the HES1 dynamics are required for NSCs to exit quiescence in vitro, suggesting that oscillations are needed for cells to transition from one state to another rather than just maintaining a progenitor state. This revised view on the functional significance of HES1 is consistent with previous findings showing that when Her6 (orthologue of mouse HES1 in zebrafish) oscillations are diminished due to the increase of noise, cells are unable to transition from progenitor state to differentiation in zebrafish (Soto et al., 2020), whereas the quality and occurrence of HES5 oscillations in mouse spinal cord increases as cells approach the transition to differentiation (Manning et al., 2019). The significance of HES1 oscillations in controlling the state of quiescence may also have implications in tissue regeneration and in cancer where quiescent cancer stem cells are considered to drive cancer progression and metastasis (Chen et al., 2016; Lee et al., 2020; Phan and Croucher, 2020). Overall, our findings highlight the importance of oscillatory expression in controlling cell state transitions, in contrast to the prevailing view that they are only needed to maintain a progenitor state.
Limitations of the study
In this study, we find that switching HES1 expression from oscillatory to sustained impedes reactivation of quiescent NSCs. A more accurate manipulation of the HES1 dynamics and/or levels would be beneficial in elucidating other aspects of oscillatory expression that may be relevant in controlling this transition.
Validating our findings in an in vivo experimental system would increase the relevance of the study for therapeutic applications in regeneration and cancer.
A more high-throughput exploration of how oscillatory versus sustained HES1 expression is decoded by downstream molecular targets would deepen mechanistic understanding.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Ki67 | BD Biosciences | Cat# 550609; RRID:AB_393778 |
| Rabbit polyclonal anti-SOX2 | Abcam | Cat# ab97959; RRID:AB_2341193 |
| Rabbit polyclonal anti-PAX6 | BioLegend | Cat# 901301; RRID:AB_2565003 |
| Rabbit polyclonal anti-GFP | Molecular probes | Cat# A-11122; RRID:AB_221569 |
| Goat anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11008; RRID:AB_143165 |
| Goat anti-mouse Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11004; RRID:AB_2534072 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human FGF-2 | PeproTech | Cat# 100-18B |
| Recombinant mouse EGF | PeproTech | Cat# 315-09 |
| Recombinant human BMP-4 | PeproTech | Cat# AF-120-05ET |
| Laminin | Sigma | Cat# L2020 |
| TRIzol reagent | Thermo Fisher Scientific | Cat# 15596018 |
| GlycoBlue Coprecipitant | Thermo Fisher Scientific | Cat# AM9515 |
| DNase-I | New England BioLabs | Cat# M0303 |
| EnGenSpy Cas9 NLS | New England BioLabs | Cat# M0646 |
| PFA – Pierce 16% Formaldehyde | Thermo Fisher Scientific | Cat# 28908 |
| Triton-X 100 | Sigma | Cat# T9284 |
| Western blocking reagent solution | Sigma | Cat# 11921673001 |
| ProLong Gold antifade mountant with DAPI | Thermo Fisher Scientific | Cat# P36935 |
| D-Luciferin | Promega | Cat#E1603 |
| Critical commercial assays | ||
| RNasy Mini kit | Qiagen | Cat# 74104 |
| Lipofectamine 3000 | Invitrogen | Cat# L3000001 |
| Superscript III | Invitrogen | Cat# 18080051 |
| Power SYBR green PCR master mix | Applied Biosystems | Cat# 4367659 |
| Deposited data | ||
| Code for oscillatory analysis of bioluminescence and fluorescence time series data | Zenodo | https://doi.org/10.5281/zenodo.5510278 |
| Raw imaging data and processed datasets | Figshare | https://figshare.com/projects/HES1_protein_oscillations_are_necessary_for_neural_stem_cells_to_exit_from_quiescence/122704 |
| Experimental models: Organisms/strains | ||
| Mouse: LUC2-HES1 BAC | IMSR | Cat# RBRC06013; RRID:IMSR_RBRC06013 |
| Mouse: HES1mScarlet-I/mScarlet-I | This paper | N/A |
| C57BL6/J | Envigo | N/A |
| Oligonucleotides | ||
| Primers for qPCR see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pRRL-UbC-mVenus:Hes1 | This paper | N/A |
| pLNT-UbC-NuVenus | This paper | N/A |
| Software and algorithms | ||
| Fiji | http://fiji.sc | RRID:SCR_002285 |
| Imaris | http://www.bitplane.com/imaris/imaris | RRID:SCR_007370 |
| MATLAB | http://www.mathworks.com/products/matlab/ | RRID:SCR_001622 |
| FlowJo | https://www.flowjo.com/solutions/flowjo | RRID:SCR_008520 |
| GraphPad Prism | http://www.graphpad.com/ | RRID:SCR_002798 |
| RStudio | http://www.rstudio.com/ | RRID:SCR_000432 |
| ggplot2 | https://cran.r-project.org/web/packages/ggplot2/index.html | RRID:SCR_014601 |
| PyCorrFit software | Muller et al. (2014) | N/A |
| Other | ||
| 35mm glass bottom culture dish | Greiner BIO-ONE | Cat# 627860 |
Resource availability
Lead contact
Additional information and requests for resources and reagents will be fulfilled by the lead contact, Nancy Papalopulu (Nancy.papalopulu@manchester.ac.uk)
Materials availability
The mouse line and plasmids generated in this study are available upon request from the lead contact.
Experimental model and subject details
Cell culture of primary NSCs
Primary NSCs were isolated from dissected LGEs of E13.5 embryos from LUC2-HES1 BAC (Cat# RBRC06013; RRID:IMSR_RBRC06013) reporter mice or HES1mScarlet-I/mScarlet-I mice and cultured in complete NS media as previously described (Pollard, 2013), supplemented with 10ng/ml FGF-2 (PeproTech, Cat# 100-18B), 10ng/ml EGF (PeproTech, Cat# 100-18B) and 2μg/ml laminin (Sigma, Cat# L2020) (proliferation media). LUC2-HES1 BAC mice were obtained by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan (Imayoshi et al., 2013). For induction of quiescence 22K-32K cells per square centimetre were plated in proliferating media and 24h later (or when cells reached 60–70% confluency) they were washed twice with 1xPBS while attached, and the media was replaced with complete NS media supplemented with 20ng/ml FGF-2 and 50ng/ml BMP4 (PeproTech, Cat# AF-120-05ET) (quiescence media). To reactivate the cells, following at least 3 days of culture in quiescent media, the cells were washed twice with 1xPBS while attached, and the media was replaced with proliferating media without replating the cells.
Generation of HES1mScarlet-I/mScarlet-I mice
We used the EASI-CRISPR strategy (Quadros et al., 2017) to generate a C-terminally tagged HES1:mSCARLET-I mouse. Two sgRNA targeting the STOP codon of the Hes1 gene were selected using the Sanger WTSI website http://www.sanger.ac.uk/htgt/wge/ (Hodgkins et al., 2015). sgRNA sequences (GTGGCGGAACTGAGAGCCTC-AGG and TGAGGCTCTCAGTTCCGCCA-CGG) were purchased as crRNA oligos, which were annealed with tracrRNA (both oligos supplied by Integrated DNA Technologies) in sterile, RNase free injection buffer (TrisHCl 1mM, pH 7.5, EDTA 0.1mM) by combining 2.5 μg crRNA with 5 μg tracrRNA, heating to 95°C, and slowly cooling to room temperature. To generate the long single strand DNA donor repair template, a homology flanked flexible linker-3xFLAG-mScarletI DNA sequence was cloned and used as a template in an initial PCR reaction with primers Hes1_lssDNA_F catgctcccggccgccatgggaattcggtaccaacagtgggacctcggt and Hes1 lssDNA R caagttcgtttttagtgtccgtcagaagagagaggtgggctagggactttacgggtagcagtggcctgaggctctcacttgtacagctcgtccatgcc. This amplicon appended the universal pGEM_dual_Bio_F primer sequence (catgggaattcggtac) at the 5’ end, and the lssDNA comprising 5’H(96nt)-linker-FLAG-mScarletI-3’H(88nt) was purified as described (Bennett et al., 2020). For embryo microinjection the annealed sgRNA were complexed with EnGen Cas9 protein (New England Biolabs, Cat# M0646) at room temperature for 10 minutes, before addition of long ssDNA donor (final concentrations; each sgRNA 20 ng/μl, Cas9 protein 40 ng/μl, lssDNA 10 ng/μl). The injection mix was directly microinjected into C57BL6/J (Envigo) zygote pronuclei using standard protocols. Zygotes were cultured overnight and the resulting 2 cell embryos surgically implanted into the oviduct of day 0.5 post coitum pseudopregnant mice. Potential founder mice were screened by PCR, using primers that flank the homology arms (Geno F ttgcctttctcatccccaac, Geno R gcagtgcatggtcagtcac), used in combination with internal mScarlet-I primers (mScarlet-I R GTCCTCGAAGTTCATCACGC, mScarlet-I F TCCCCTCAGTTCATGTACGG). 1/8 pups (pup #7) was PCR positive for both reactions, and Sanger sequencing confirmed perfect integration of the gene tag. Note that pup #5 demonstrated a positive PCR result for one of the two reactions, perhaps indicating an illegitimate repair event (Codner et al., 2018). Pup #7 was bred forward with a WT C57BL6/J mouse. Germline transmission was confirmed through PCR and sequencing and a colony was established. Both HET and HOM mice developed normally. All animal work was performed under UK Home Office project licences (P08B76E2B) according to the conditions of the Animals (Scientific Procedures) Act 1986.
Method details
Cell transfection, qPCR and RNA extraction
Primary NSCs were transfected with Lipofectamine 3000 (Invitrogen, Cat# L3000001) according to manufacturers’ instructions. 22K–32K cells per square centimeter were plated in proliferating media and 24h later (or when cells reached 60–70% confluency) the cells were transfected with 80ng plasmid per square centimetre. For induction of quiescence (where applicable), the proliferation media was replaced with quiescence media the day after transfection.
To generate the pRRL-UbC-mVenus:Hes1 plasmid, the UbC-mVenus:Hes1 sequence was subcloned into a new lentiviral transfer vector previously described (Seppen et al., 2002; Gilham et al., 2010). Briefly, the UbC-mVenus:Hes1 sequence was released from the pLNT UbC-mVENUS:HES1 plasmid (Phillips et al., 2016) using Pac1 (followed by blunting with T4 DNA polymerase) and KpnI and cloned into the pRRL lenti backbone which was released with Pme1 and KpnI. To generate pLNT-UbC-NuVenus plasmid (nuclear VENUS driven by the UbC promoter) the mVenus cDNA was PCR-amplified from the pLNT-UbC-mVENUS lenti backbone plasmid (Bagnall et al., 2015) using a forward primer containing SV40 NLS and then cloned back to the same vector.
For E13.5 LUC2:HES1 NSCs, RNA was extracted from cells cultured in proliferative (1-2days), quiescent (3d) or reactivated (2d) conditions using the RNeasy kit (Qiagen, Cat# 74104) as per manufacturers’ instructions.
For WT E13.5 LGE NSCs transfected with either UbC-mVENUS or UbC-mVENUS:HES1, cells positive for mVENUS expression were sorted directly in 1ml TRIzol reagent (Thermo Fisher Scientific, Cat# 15596018) that was kept on ice. Following a gentle mix with the pipette the samples were left at RT for 5min. 200μl of chloroform were then added to each sample and samples were centrifuged at 14,000rpm for 15 min at 4°C. The clear phase of the solution was transferred into a new Eppendorf tube to which 500μl of isopropanol and 1ul of GlycoBlue (15ml/ml) (Thermo Fisher Scientific, Cat# AM9515) were added. Samples were kept at −20°C for a minimum of 30 min or overnight and then centrifuged at 14,000 rpm for 15 min at 4°C. The supernatant was removed and pellets were washed with 70% ethanol and then centrifuged at 14,000 rpm for 10 min. Pellets were reconstituted in RNAse-free water and treated with DNase-I (New England BioLabs, Cat# M0303) as per manufacturers’ instructions.
cDNA was prepared using Superscript III (Invitrogen, Cat# 18080051) as per manufacturers’ instructions and qPCR was performed with SYBR green (Applied Biosystems, Cat# 4367659) using the primers listed in Table S1. CT values were normalized to reference genes Gapdh and ActB.
Immunostaining and image analysis
Primary WT NSCs transfected with either UbC-mVENUS or UbC-mVENUS:HES1 were fixed 1–2d after transfection in proliferative conditions, 4d after transfection in quiescent conditions (kept in quiescent media for 3d), and 6d after transfection in reactivated conditions (kept in reactivation media for 2 d). Cells were fixed in 4% PFA (Thermo Fisher Scientific, Cat# 28908) for 25 min, followed by permeabilization for 5 min with 0.5% Triton-X-100 (Sigma, Cat# T9284) diluted in PBS. Serum blocking was performed for 20 min at RT in 1xWBR (Western Blocking Reagent) (Sigma, Cat# 11921673001) solution. Cells were incubated with primary antibodies at 4°C overnight and with secondary antibodies at room temperature for 40 min. Coverslips were mounted using ProLong diamond antifade mountant with DAPI (Invitrogen, Cat# P36935). Primary antibodies were mouse anit-Ki67 (1:200, BD Biosciences, Cat# 550609; RRID:AB_393778), rabbit anti-SOX2 (1:200, Abcam, Cat# ab97959; RRID:AB_2341193), rabbit anti-PAX6(1:300, BioLegend, Cat# 901301;RRID:AB_2565003) and rabbit anti-GFP (for mVENUS detection) (1:500, Molecular probes, Cat# A-11122; RRID:AB_221569). Secondary antibodies were anti-rabbit 488 (1:500, Thermo Fisher Scientific, Cat# A-11008; RRID:AB_143165) and anti-mouse 568 (1:500, Thermo Fisher Scientific, Cat# A-11004; RRID:AB_2534072). Slides with Ki67/DAPI staining were collected with a Nikon Eclipse 80i using a Plan Fluor 20x/0.5 DIC N2 objective and a 1280x1024 format, slides with SOX2/Ki67/DAPI or PAX6/Ki67/DAPI staining were collected with a Zeiss LSM880 Inverted Airyscan using a Fluar 20x/0.75 M27 and a 1024x1024 format, slides with GFP/Ki67/DAPI staining were collected either with a Zeiss LSM 880 Airy Scan Upright using a Fluar 20x/0.75 and a 1192x1192 format or with a Zeiss LSM880 Inverted Airyscan using a Fluar 20x/0.75 M27 and a 1024x1024 format. For fluorescence intensity analysis of mVENUS expression the GFP signal was excited with two different 488nm laser powers (1% and 0.2%). The high power setting was for capturing dimmer GFP positive cells and the lower power setting was for capturing bright GFP positive cells, so that there was no saturated signal. Images were processed with Fiji (RRID:SCR_002285) and positive cells were scored based on detection of expression above background of control images only treated with secondary antibodies. For fluorescence intensity analysis, single nuclei detection was performed based on segmentation of DAPI signal. Images of DAPI were smoothed and thresholded to produce individual nuclear regions. Single nuclear regions were used as masks and applied onto fluorescent channels to produce a mean intensity readout per nucleus. Positive cells were identified based on mean nuclear intensity above negative control images. Intensities were estimated as mean fluorescent units per nucleus. To account for variability between experimental conditions and the control, we reported as positive the nuclei with intensity levels above the control as well as the non-DAPI regions indicative of background intensity.
Bioluminescence and fluorescence imaging
For bioluminescence imaging 200K-300K primary NSCs were plated on 35-mm glass bottom dishes (Greiner BIO-ONE, Cat# 627860) in media supplemented with 1mM D-luciferin (Promega, Cat#E1603) and imaged on an Olympus UK LV200 inverted bioluminescence microscope using a 40x oil objective. Cells were kept at 37°C in 5% CO2 throughout the imaging period. Images were acquired with a 10-min exposure and a 2x2 binning. Imaging was performed either in proliferating conditions (cells had been cultured in proliferation media for at least 1 d) or quiescent conditions (cells had been cultured in quiescence media for at least 3 d) or reactivation conditions (cells had been reactivated in proliferation media for at least 2 d). To perform continuous bioluminescence imaging of cells transitioning from proliferating to quiescent conditions, imaging was paused to wash the cells, while the dish was firmly attached to the stage, and replace the proliferation media with quiescent, supplemented with fresh 1mM D-luciferin.
For dual fluorescence imaging of mVENUS and mSCARLET-I fluorophores, 200K-300K primary NSCs were plated on 35-mm glass bottom dishes and imaged 1 or 2 d after transfection on a Nikon A1-R confocal microscope using a Plan Fluor 40x/0.75 objective, a 1024x1024 format and with sequential scanning to avoid any bleed through from one channel to another. Images were acquired every 15–20min with z-stacking covering a 35μm depth. Bioluminescent and fluorescent movies were analysed with the Imaris imaging software (RRID:SCR_007370) and individual cells were manually tracked using the ‘Spots’ function. Background traces (i.e., detection of bioluminescence or fluorescence signal from a cell-free area) were collected alongside from the same field of imaging for every movie that was analysed. Prior to tracking, the bioluminescent images were subjected to 3x3 median filter to remove bright spot artefacts from cosmic rays. For fluorescence intensity and oscillatory analysis saturated tracks of mVENUS expression (18% of cells) were excluded.
Detection of oscillations
For the detection of oscillators we considered only single-cell HES1 timeseries of 5.5h length (approximately double the expected period of 2-3h). The bioluminescence/fluorescence HES1 timeseries of each cell trace was normalized per experiment by background subtraction. For this, the average timeseries of 2–4 backgrounds collected per video were smoothed using a nonlinear polynomial (order 2) to account for minor fluctuations due to detection noise. The smoothed background timeseries were then subtracted from the bioluminescence/fluorescence timeseries corresponding to each cell at each available time point. This produced pre-processed timeseries of HES1 in both bioluminescence and fluorescence experiments.
To identify statistically significant HES1 oscillatory expression, we used a statistical method developed by Phillips et al. (Phillips et al., 2017). Timeseries of bioluminescent or fluorescent HES1 were first detrended to account for long-term trends. We used a squared covariance detrending approach with lengthscale 7.5h, consistent with previous guidelines (Phillips et al., 2017). The detrended timeseries were then analyzed using Gaussian processes (GP). Briefly, the GP approach uses experimental timeseries to assess whether the signal is better described by a non-oscillatory fluctuating or oscillatory covariance model. The approach contains an internal calibration of white noise detector followed by a stringent 5% false discovery rate statistical test to determine oscillatory timeseries. For the bioluminescence data we used the variance of background timeseries to measure detection noise. For the fluorescence data this is not sufficient, as nuclear signal also contains auto-fluorescence, and here we used a modified approach which infers the variance of noise from the fluorescent data by optimizing the relationship between the LLR and signal-to-noise ratio as previously described (Manning et al., 2019). GP fitting producing null parameter estimates were excluded. As an additional quality control, we only report as oscillatory timeseries with a minimum of 2 peaks per track. These additional exclusions affected less than 15% of data per experiment. The code for the analysis of bioluminescence and fluorescence time-series is available on Zenodo:https://doi.org/10.5281/zenodo.5510278
Fluorescence correlation spectroscopy
Primary HES1mScarlet-I/mScarlet-I NSCs were plated in 35-mm glass bottom dishes (22K-32K cells per square centimetre) and transfected with pRRL-UbC-mVENUS:HES1 plasmid. FCS experiments were performed 2 d after transfection using a Zeiss LSM880 Inverted Airyscan with a Fluar 40x/1.30 Oil M27 objective while the cells were kept at 37°C and 5% CO2. The fluorescent signals were collected on the same track where mVENUS fluorescence was excited with a 488nm laser and emission was collected between 508-552nm, and mSCARLET-I fluorescence was excited with 561nm laser and emission was collected between 606-677nm. Data from mVENUS/mSCARLET-I doublepositive cells or mSCARLET-I single positive cells were collected using 3x8sec or 1x15sec runs with 0.05-0.1% 488nm laser power and 0.05% 561nm laser power, which were optimized to reduce bleaching. Traces with large spikes/drops in the count rate and bleaching >20% were excluded. Also measurements from high mVENUS expressing cells with saturated levels (15% of cells) were omitted as they cannot be deemed accurate. Results were comparable between the long acquisition sequences and triplicate short sequences. FCS auto-correlations were then analyzed individually with the PyCorrFit software (Muller et al., 2014). Autocorrelation curves were fitted to one-component diffusion model with triplet state and the Levenberg-Marquardt minimization algorithm was applied with a number of neighboring data points j=3. The structural parameter (SP) was fixed at 4. The autocorrelation curves with the best model fitting were used and a counts per particle cut-off of 0.05KHz and 0.1KHz for the mVENUS and mSCARLET-I particle estimation respectively was applied.
Fluorescence-activated cell sorting (FACS)
WT E13.5 LGE NSCs were transfected with UbC-mVENUS or UbC-mVENUS:HES1 plasmid as previously described. Cells were collected for sorting at either 2d after transfection while kept in proliferative conditions or 4d after transfection and after kept in quiescent conditions for 3d. Samples were sequentially gated based on forward scatter (FSC) vs side scatter (SSC) followed by depletion of doublets based on FSC vs Trigger Pulse Width. mVENUS expression was detected based on ∗488/530/40 (mVENUS) vs ∗488/710/60 (autofluorescence) plotting. Samples not transfected with the plasmids but cultured in the same conditions (i.e., proliferative or quiescent) were used as negative controls to inform the gating strategy for determining mVENUS negative and positive cells. Positive mVENUS cells were further sub-divided into low and high expressors based on the mVENUS expression distribution profile, with higher level expressing cells clustering more tightly together. The same mVENUS fluorescence intensity cut-off, separating the low from the high expressors, was applied to all sorts. Overall, from each sample, we sorted 3 populations corresponding to mVENUS negative expression, mVENUS low and mVENUS high. Cells were sorted on a BD Influx and data were analysed with FlowJo 10.8.0 (RRID:SCR_008520)
Quantification and statistical analysis
Statistical analysis was performed in GraphPad Prism 8.4.1 (RRID:SCR_002798). Data were tested for normality with a Shapiro-Wilk test to inform the choice of a parametric or non-parametric statistical test. The lower and upper hinges of the boxplots correspond to the first and third quartiles. No outliers were removed. The statistical tests, sample sizes, number of biological replicates, and p-values are reported in each figure legend.
Acknowledgments
The authors wish to thank Prof. Steve Pollard and Prof. Fiona Doetsch for advice and discussions, Dr Hayley Bennett and Maj Simonsen Jackson for their work on generating the HES1mScarlet-I/mScarlet-I mouse transgenic line, Dr Cerys Manning and Dr James Bagnall for help with FCS analysis, MMath Joshua Burton for assistance with the statistical analysis, Dr Michael Lie-A-Ling for his help with subcloning the pRRL-UbC-mVenus:Hes1 construct, Dr Ximena Soto for technical assistance and advice, MSci Joshua Hawley for help with the graphical abstract and all members of the Papalopulu lab for comments on the work. The authors would also like to thank the Biological Services Facility, the Bioimaging Facility (especially Peter March and David Spiller for their help with imaging), the Genome Editing Unit, the Flow Cytometry facilities (and especially Gareth Howell) and the Genomic Technologies Core Facility of the University of Manchester for technical support. The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome Trust and the University of Manchester Strategic Fund. This work was supported by a Sir Henry Wellcome Trust Fellowship to E.M (201380/Z/16/Z) and a Wellcome Trust Senior Research Fellowship to NP (090868/Z/09/Z)
Author contributions
Conceptualization, E.M. and N.P.; Methodology, E.M. and N.P.; Investigation, E.M., V.B., N.S., A.M. and J.D.; Resources, A.D.A.; Writing – Original draft, E.M.; Writing – Review & Editing, E.M. and N.P.; Funding Acquisition, E.M. and N.P.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as living with a disability.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103198.
Contributor Information
Elli Marinopoulou, Email: elli.marinopoulou@manchester.ac.uk.
Nancy Papalopulu, Email: nancy.papalopulu@manchester.ac.uk.
Supplemental information
Data and code availability
-
•
Raw imaging data and processed datasets have been deposited at Figshare and are publicly available as of the date of publication. The link is listed in the key resources table. Any data reported in this paper that are not present in the public repository will be shared by the lead contact upon request.
-
•
All original code reported in this study has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ashall L., Horton C.A., Nelson D.E., Paszek P., Harper C.V., Sillitoe K., Ryan S., Spiller D.G., Unitt J.F., Broomhead D.S. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J.H., Hatakeyama J., Sakamoto S., Ohtsuka T., Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Bagnall J., Boddington C., Boyd J., Brignall R., Rowe W., Jones N.A., Schmidt L., Spiller D.G., White M.R., Paszek P. Quantitative dynamic imaging of immune cell signalling using lentiviral gene transfer. Integr. Biol. (Camb) 2015;7:713–725. doi: 10.1039/c5ib00067j. [DOI] [PubMed] [Google Scholar]
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev.Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bennett H., Aguilar-Martinez E., Adamson A.D. CRISPR-mediated knock-in in the mouse embryo using long single stranded DNA donors synthesised by biotinylated PCR. Methods. 2020;191:3–14. doi: 10.1016/j.ymeth.2020.10.012. [DOI] [PubMed] [Google Scholar]
- Boareto M., Iber D., Taylor V. Differential interactions between Notch and ID factors control neurogenesis by modulating Hes factor autoregulation. Development. 2017;144:3465–3474. doi: 10.1242/dev.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounpheng M.A., Dimas J.J., Dodds S.G., Christy B.A. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- Castella P., Sawai S., Nakao K., Wagner J.A., Caudy M. HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression. Mol.Cell.Biol. 2000;20:6170–6183. doi: 10.1128/mcb.20.16.6170-6183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E., Casarosa S., Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Chen H., Thiagalingam A., Chopra H., Borges M.W., Feder J.N., Nelkin B.D., Baylin S.B., Ball D.W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci.U S A. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.Y., Osimiri L.C., Chevalier M., Bugaj L.J., Nguyen T.H., Greenstein R.A., Ng A.H., Stewart-Ornstein J., Neves L.T., El-Samad H. Optogenetic control reveals differential promoter interpretation of transcription factor nuclear translocation dynamics. Cell Syst. 2020;11:336–353 e24. doi: 10.1016/j.cels.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Dong J., Haiech J., Kilhoffer M.C., Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int. 2016;2016:1740936. doi: 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codner G.F., Mianne J., Caulder A., Loeffler J., Fell R., King R., Allan A.J., Mackenzie M., Pike F.J., McCabe C.V. Application of long single-stranded DNA donors in genome editing: generation and validation of mouse mutants. BMC Biol. 2018;16:70. doi: 10.1186/s12915-018-0530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F. The glial identity of neural stem cells. Nat. Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- Doetsch F., Caille I., Lim D.A., Garcia-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Elson E.L. Fluorescence correlation spectroscopy: past, present, future. Biophys.J. 2011;101:2855–2870. doi: 10.1016/j.bpj.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y., Gotoh Y. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 2015;18:657–665. doi: 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- Gilham D.E., Lie A.L.M., Taylor N., Hawkins R.E. Cytokine stimulation and the choice of promoter are critical factors for the efficient transduction of mouse T cells with HIV-1 vectors. J. Gene Med. 2010;12:129–136. doi: 10.1002/jgm.1421. [DOI] [PubMed] [Google Scholar]
- Hansen A.S., O'Shea E.K. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol. Syst. Biol. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N., O'Shea E.K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 2011;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hodgkins A., Farne A., Perera S., Grego T., Parry-Smith D.J., Skarnes W.C., Iyer V. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31:3078–3080. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Isomura A., Kageyama R. Ultradian oscillations and pulses: coordinating cellular responses and cell fate decisions. Development. 2014;141:3627–3636. doi: 10.1242/dev.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogi A., Persson P., Grynfeld A., Pahlman S., Axelson H. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J. Biol. Chem. 2002;277:9118–9126. doi: 10.1074/jbc.M107713200. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Shimojo H., Ohtsuka T. Dynamic control of neural stem cells by bHLH factors. Neurosci. Res. 2019;138:12–18. doi: 10.1016/j.neures.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A.J., Elowitz M.B., Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Lane K., Van Valen D., DeFelice M.M., Macklin D.N., Kudo T., Jaimovich A., Carr A., Meyer T., Pe'er D., Boutet S.C., Covert M.W. Measuring signaling and RNA-seq in the same cell links gene expression to dynamic patterns of NF-kappaB activation. Cell Syst. 2017;4:458–469 e5. doi: 10.1016/j.cels.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Frelin C., Xie S., Ferrari R., Dunant C.F., Zandi S., Neumann A., Plumb I., Doulatov S., Chen J. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015;16:302–313. doi: 10.1016/j.stem.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Reed-Newman T., Anant S., Ramasamy T.S. Regulatory role of quiescence in the biological function of cancer stem cells. Stem Cell Rev. Rep. 2020;16:1185–1207. doi: 10.1007/s12015-020-10031-8. [DOI] [PubMed] [Google Scholar]
- Lev Bar-Or R., Maya R., Segel L.A., Alon U., Levine A.J., Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc. Natl. Acad. Sci.U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingbeck J.M., Trausch-Azar J.S., Ciechanover A., Schwartz A.L. E12 and E47 modulate cellular localization and proteasome-mediated degradation of MyoD and Id1. Oncogene. 2005;24:6376–6384. doi: 10.1038/sj.onc.1208789. [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Magnusson J.P., Frisen J. Stars from the darkest night: unlocking the neurogenic potential of astrocytes in different brain regions. Development. 2016;143:1075–1086. doi: 10.1242/dev.133975. [DOI] [PubMed] [Google Scholar]
- Manning C.S., Biga V., Boyd J., Kursawe J., Ymisson B., Spiller D.G., Sanderson C.M., Galla T., Rattray M., Papalopulu N. Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis. Nat. Commun. 2019;10:2835. doi: 10.1038/s41467-019-10734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marescal O., Cheeseman I.M. Cellular mechanisms and regulation of quiescence. Dev.Cell. 2020;55:259–271. doi: 10.1016/j.devcel.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B., Mateo J.L., Zhou B., Andersen J., Achimastou A., Urban N., van den Berg D., Georgopoulou D., Hadjur S., Wittbrodt J. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Schwille P., Weidemann T. PyCorrFit-generic data evaluation for fluorescence correlation spectroscopy. Bioinformatics. 2014;30:2532–2533. doi: 10.1093/bioinformatics/btu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K., Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146:dev156059. doi: 10.1242/dev.156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi S., Imaizumi Y., Shimojo H., Miyachi H., Kageyama R. Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development. 2020;147:dev182204. doi: 10.1242/dev.182204. [DOI] [PubMed] [Google Scholar]
- Pattyn A., Guillemot F., Brunet J.F. Delays in neuronal differentiation in Mash1/Ascl1 mutants. Dev. Biol. 2006;295:67–75. doi: 10.1016/j.ydbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Phan T.G., Croucher P.I. The dormant cancer cell life cycle. Nat. Rev. Cancer. 2020;20:398–411. doi: 10.1038/s41568-020-0263-0. [DOI] [PubMed] [Google Scholar]
- Phillips N.E., Manning C., Papalopulu N., Rattray M. Identifying stochastic oscillations in single-cell live imaging time series using Gaussian processes. PLoS Comput.Biol. 2017;13:e1005479. doi: 10.1371/journal.pcbi.1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N.E., Manning C.S., Pettini T., Biga V., Marinopoulou E., Stanley P., Boyd J., Bagnall J., Paszek P., Spiller D.G. Stochasticity in the miR-9/Hes1 oscillatory network can account for clonal heterogeneity in the timing of differentiation. Elife. 2016;5:e16118. doi: 10.7554/eLife.16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard S.M. In vitro expansion of fetal neural progenitors as adherent cell lines. Methods Mol. Biol. 2013;1059:13–24. doi: 10.1007/978-1-62703-574-3_2. [DOI] [PubMed] [Google Scholar]
- Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros R.M., Miura H., Harms D.W., Akatsuka H., Sato T., Aida T., Redder R., Richardson G.P., Inagaki Y., Sakai D. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18:92. doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.R. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S.D., Verveer P.J., Bastiaens P.I. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat.Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Seppen J., Rijnberg M., Cooreman M.P., Oude Elferink R.P. Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J. Hepatol. 2002;36:459–465. doi: 10.1016/s0168-8278(01)00308-7. [DOI] [PubMed] [Google Scholar]
- Seri B., Garcia-Verdugo J.M., McEwen B.S., Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J. Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Chinaranagari S., Chaudhary J. Inhibitor of differentiation 4 (ID4) acts as an inhibitor of ID-1, -2 and -3 and promotes basic helix loop helix (bHLH) E47 DNA binding and transcriptional activity. Biochimie. 2015;112:139–150. doi: 10.1016/j.biochi.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H., Isomura A., Ohtsuka T., Kori H., Miyachi H., Kageyama R. Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 2016;30:102–116. doi: 10.1101/gad.270785.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H., Ohtsuka T., Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Soto X., Biga V., Kursawe J., Lea R., Doostdar P., Thomas R., Papalopulu N. Dynamic properties of noise and Her6 levels are optimized by miR-9, allowing the decoding of the Her6 oscillator. EMBO J. 2020;39:e103558. doi: 10.15252/embj.2019103558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueda R., Imayoshi I., Harima Y., Kageyama R. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 2019;33:511–523. doi: 10.1101/gad.323196.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban N., Blomfield I.M., Guillemot F. Quiescence of adult mammalian neural stem cells: ahighly regulated rest. Neuron. 2019;104:834–848. doi: 10.1016/j.neuron.2019.09.026. [DOI] [PubMed] [Google Scholar]
- van Velthoven C.T.J., Rando T.A. Stem cell quiescence: dynamism, restraint, and cellular idling. Cell Stem Cell. 2019;24:213–225. doi: 10.1016/j.stem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.M., Fogarty M., Kessaris N., Richardson W.D. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 2007;27:8286–8296. doi: 10.1523/JNEUROSCI.0476-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Alder J.K., Chun J.H., Friedman A.D., Heimfeld S., Cheng L., Civin C.I. HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells. 2006;24:876–888. doi: 10.1634/stemcells.2005-0598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw imaging data and processed datasets have been deposited at Figshare and are publicly available as of the date of publication. The link is listed in the key resources table. Any data reported in this paper that are not present in the public repository will be shared by the lead contact upon request.
-
•
All original code reported in this study has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.