Abstract
Import of carrier proteins from the cytoplasm into the mitochondrial inner membrane of yeast is mediated by a distinct system consisting of two soluble 70-kDa protein complexes in the intermembrane space and a 300-kDa complex in the inner membrane, the TIM22 complex. The TIM22 complex contains the peripheral subunits Tim9p, Tim10p, and Tim12p and the integral membrane subunits Tim22p and Tim54p. We identify here an additional subunit, an 18-kDa integral membrane protein termed Tim18p. This protein is made as a 21.9-kDa precursor which is imported into mitochondria and processed to its mature form. When mitochondria are gently solubilized, Tim18p comigrates with the other subunits of the TIM22 complex on nondenaturing gels and is coimmunoprecipitated with Tim54p and Tim12p. Tim18p does not cofractionate with the TIM23 complex upon immunoprecipitation or nondenaturing gel electrophoresis. Deletion of Tim18p decreases the growth rate of yeast cells by a factor of two and is synthetically lethal with temperature-sensitive mutations in Tim9p or Tim10p. It also impairs the import of several precursor proteins into isolated mitochondria, and lowers the apparent mass of the TIM22 complex. We suggest that Tim18p functions in the assembly and stabilization of the TIM22 complex but does not directly participate in protein insertion into the inner membrane.
Most mitochondrial proteins are synthesized in the cytosol with a cleavable N-terminal presequence that specifies import into mitochondria via the general protein import pathway (25, 27, 32, 36). This pathway is mediated by cytosolic chaperones, a hetero-oligomeric TOM complex in the mitochondrial outer membrane, a Tim17p-Tim23p complex (referred to as the TIM23 complex) in the inner membrane, an ATP-dependent import motor associated with the matrix face of the TIM23 complex, and soluble proteins in the matrix involved in the proteolytic maturation and folding of the imported proteins (9, 11, 27–29).
Over the past few years it has become clear that mitochondria possess an additional pathway which affects import of hydrophobic inner membrane proteins (17, 22, 27). This pathway diverges from the general import pathway after the TOM channel (23). As the hydrophobic precursor exits that channel, it is met by one of the two soluble 70-kDa protein complexes that transfer it across the intermembrane space (1, 19, 21, 39). One of these 70-kDa complexes contains Tim9p and Tim10p, while the other contains Tim8p and Tim13p (20). Both complexes generally deliver hydrophobic proteins to an inner membrane complex specialized for the insertion of membrane proteins. This insertion complex, referred to as the TIM22 complex, has an apparent mass of 300 kDa and contains the membrane proteins Tim22p and Tim54p, the peripheral membrane protein Tim12p, and a small proportion of Tim9p and Tim10p (1, 18, 19, 21, 39). The 70-kDa Tim9p-Tim10p complex can also deliver some membrane proteins to the TIM23 complex and, possibly, to additional, as-yet-unknown insertion sites (24).
A typical protein imported by this novel pathway is the ADP/ATP carrier (AAC), which contains six membrane-spanning regions (7, 30). AAC lacks a cleavable N-terminal targeting sequence, carrying instead targeting information in discrete regions throughout the polypeptide chain (7, 30). Yeast has about three dozen members of the mitochondrial metabolite carrier family (26). The novel pathway probably imports all of these, as well as many other integral proteins of the mitochondrial inner membrane (4, 16, 24).
Here we describe Tim18p, a novel subunit of the TIM22 complex. We initially identified Tim18p as a protein that coimmunoprecipitated with Tim54p and then confirmed that it is indeed a subunit of the TIM22 complex. Tim18p is an integral inner membrane protein derived from a precursor protein of 21.9 kDa which is processed to its mature form within mitochondria. Its deletion decreases the growth rate of yeast by half and is synthetically lethal with temperature-sensitive (ts) mutations in Tim9p or Tim10p. Mitochondria prepared from a Tim18-less strain are defective in the import of several precursor proteins, have lowered levels of Tim23p, and contain a TIM22 complex of smaller apparent size.
MATERIALS AND METHODS
Plasmids and strains.
For in vitro transcription and translation, the DNA fragments encoding Tim23p (15) and Tim54p were subcloned into pGEM3Z (Promega), the fragment encoding Tim22p was subcloned into pSP64 (Promega, 19), the fragments encoding Tim18p and Coq2p and AAC2 were subcloned into pSP65. The plasmid carrying TIM17 was kindly provided by Nikolaus Pfanner (University of Freiburg, Freiburg, Germany).
Standard conditions were used for the growth, manipulation, and transformation of yeast strains (10, 14). The yeast strain containing a ts Tim10p (tim10-1) has been described previously (19, 21). The strain containing a ts Tim9p was constructed by the same methods as for the tim10-1 strain (19, 21). Single-step gene-replacement was used to replace the TIM18 gene with the KANR marker in the diploid yeast strain GA74 (41); after sporulation, haploid segregants carrying the replacement marker were selected. A gene encoding Tim18p C-terminally tagged with a hemagglutinin (HA) epitope was constructed by PCR with Pfu polymerase by using a primer encoding the epitope (CYPYDVPDYASL) and was then subcloned into the centromeric plasmid pRS306 (URA3 marker) (37). This recombinant plasmid was transformed in the Δtim18 strain.
For testing synthetic lethal interactions, a tim10-1 strain was used in which TIM10 had been replaced by HIS3, and the ts tim10-1 allele had been integrated at the leu2 locus (19, 21); this yielded strain Δtim10::HIS3 tim10-1:LEU2. This strain was mated to the strain lacking Tim18p (Δtim18::KANR), the resulting diploid was allowed to sporulate, and the asci were subjected to tetrad analysis. A tim9-3 strain was used in which TIM9 had been replaced by TRP1 and the ts tim9-3 allele had been integrated at the leu2 locus; the resulting strain was Δtim9::TRP1 tim9-3:LEU2. It was mated to the strain lacking Tim18p (Δtim18::KANR), and the diploid was subjected to tetrad analysis. Viability of dissected spores was tested at 25°C and auxotrophic markers were screened on the appropriate media.
Coimmunoprecipitation of Tim18p with Tim54p.
Protein A-Sepharose beads (Pharmacia; 600 mg) were incubated first with bovine serum albumin and then for 1.5 h with 7.5 ml of anti-Tim54p or anti-Tim44p rabbit sera made up to a final volume of 15 ml with 50 mM K+-HEPES (pH 7.4). Beads were washed and incubated with 0.04% glutaraldehyde in 0.1 M sodium phosphate (pH 8.0) for 30 min at room temperature. After further incubation with 0.2 M Tris-Cl (pH 8.5) for 1.5 h, followed by a wash, the beads were incubated for 3 min with 0.1 M glycine-HCl (pH 2.5) and then neutralized with 1 M Tris base. Mitochondria (26 mg) from the wild-type yeast strain D273-10B were suspended in 3 ml of 0.6 M sorbitol–20 mM K+-HEPES (pH 7.4) containing 3.4 mg of bovine serum albumin per ml and diluted with an equal volume of buffer A (20 mM Na+-HEPES [pH 7.0], 30 mM NaCl, 10% glycerol, 20 μM ZnSO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK]). After 15 min on ice, mitochondria were pelleted by centrifugation, resuspended at 5 mg/ml in buffer A, and preextracted by mixing with 1 volume of 0.9% n-dodecyl maltoside in buffer A. After 10 min on ice, insoluble material was pelleted by centrifugation, resuspended in buffer A to 10 mg/ml, and solubilized with 1 volume of 3% digitonin in buffer A. After 15 min on ice, insoluble matter was removed by centrifugation (128,000 × g for 15 min at 2°C), and the supernatant was loaded onto two 35-ml 15 to 25% linear glycerol gradients in buffer A and centrifuged in a Beckman TST 28.38 ultracentrifuge rotor at 90,000 × g for 16 h at 2°C. Fractions of 1.5 ml were collected, and 10 μl of each was blotted onto a nitrocellulose sheet, which was then immunoblotted with an antiserum against Tim54p. Fractions enriched for Tim54p were pooled and used for immunoprecipitation.
For immunoprecipitation, the pooled fractions were incubated first with protein A-Sepharose coupled to anti-Tim44 serum to remove nonspecifically bound proteins and then with protein A-Sepharose coupled to anti-Tim54p antiserum for 2.5 h. After incubation, the beads were pelleted and washed briefly with 0.5% digitonin in buffer A. Washed beads were eluted with 1 ml of 0.5% sodium dodecyl sulfate (SDS) in 0.1 M glycine-HCl (pH 2.5), and eluates were neutralized and precipitated with 4 volumes of methanol at −20°C overnight. The precipitated samples were resolved by electrophoresis on a 10% Tris-tricine gel (35). The gel was stained with Coomassie brilliant blue and, after destaining, the bands of interest were excised and subjected to in-gel tryptic digestion. Peptides were identified by electrospray mass spectrometry (6).
Import of radiolabeled proteins into isolated mitochondria.
Mitochondria were purified from lactate-grown yeast cells (13) and assayed for in vitro protein import as described elsewhere (31). Proteins were synthesized in a rabbit reticulocyte lysate in the presence of [35S]methionine after in vitro transcription of the corresponding gene by SP6 or T7 polymerase. The reticulocyte lysate containing the radiolabeled precursor was incubated with isolated mitochondria at the indicated temperatures in import buffer (1 mg of bovine serum albumin per ml, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, 20 mM K+-HEPES [pH 7.4]). Where indicated, the potential across the mitochondrial inner membrane was dissipated with 1 μM valinomycin. Nonimported radiolabeled proteins were removed by treatment with 100 μg of trypsin or 50 μg of proteinase K per ml for 15 to 30 min on ice. Trypsin was inhibited with 200 μg of soybean trypsin inhibitor per ml, and proteinase K was inhibited with 1 mM PMSF. To generate mitoplasts, mitochondria in import buffer were diluted with 9 volumes of 20 mM K+-HEPES (pH 7.4) and incubated at 4°C for 30 min (12). For alkali extraction, mitochondria from an import reaction were sedimented by centrifugation, resuspended to 0.1 mg/ml in 100 mM Na2CO3, and incubated for 30 min at 4°C (8). Supernatant and pellet were separated by centrifugation at 100,000 × g for 15 min.
Blue native gel electrophoresis.
Mitochondria (2.5 mg/ml) were solubilized in 20 mM K+-HEPES (pH 7.4)–50 mM NaCl–10% glycerol–2.5 mM MgCl2–1 mM EDTA, plus either 0.16% n-dodecyl maltoside or 1% digitonin (Boehringer Mannheim). After 30 min on ice, insoluble material was removed by centrifugation at 100,000 × g for 10 min, and the solubilized proteins were analyzed by blue native gel electrophoresis on a 6 to 16% linear polyacrylamide gradient (5, 21, 33, 34).
Miscellaneous.
An 86-residue C-terminal fragment of Tim54p was expressed in Escherichia coli as a glutathione S-transferase fusion protein and, following cleavage by thrombin and gel purification, was used to raise monospecific antisera in rabbits by standard procedures. Determining the subcellular localization of proteins (12) and coimmunoprecipitation assays (21) were done as described earlier. Mitochondrial membrane potential was determined by the uptake of the lipophilic cation triphenylmethylphosphonium (TPMP) (2). For this assay, 250 μg of mitochondria were incubated in import buffer lacking bovine serum albumin, with 50 nCi of [3H]TPMP per ml and 500 nM unlabeled TPMP for 3 min. They were then sedimented by centrifugation, and [3H]TPMP in the supernatant and pellet were quantified by liquid scintillation counting. The membrane potential was estimated from the accumulation ratio, after subtracting TPMP accumulation measured in a parallel incubation containing 4 μM carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP), and assuming a mitochondrial volume of 0.7 μl per mg of protein (3). Protein concentration was assayed by the bicinchoninic acid (BCA) method (Pierce) with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence of the TIM18 gene has been submitted to GenBank under accession number BankIt300568 AF200324.
RESULTS
Identification and localization of Tim18p.
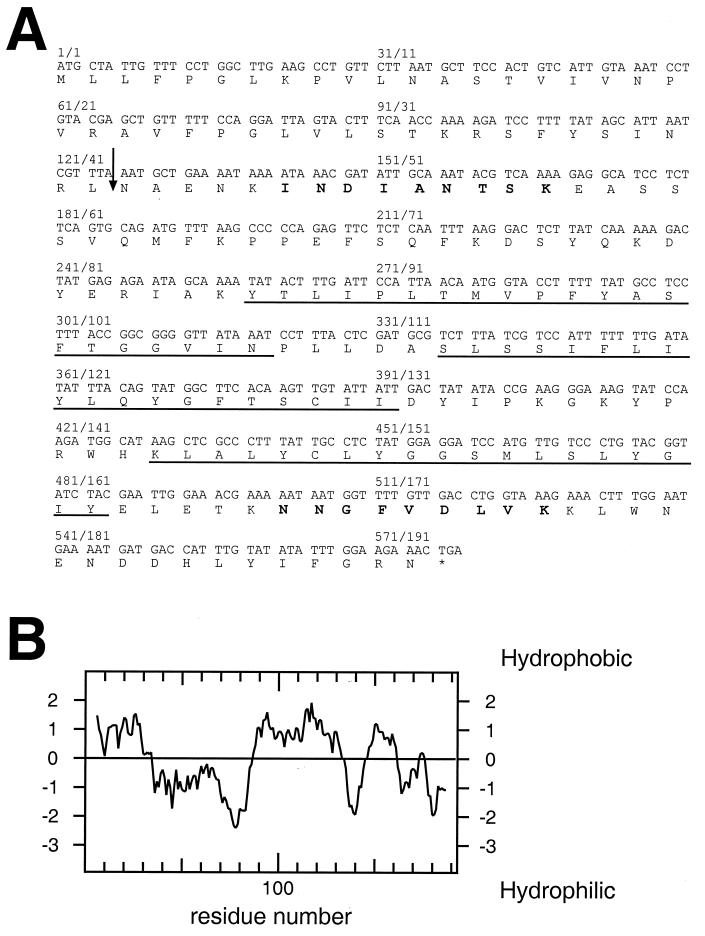
To identify novel components of the TIM22 complex, yeast mitochondria were preextracted with n-dodecyl maltoside and then solubilized with digitonin. The solubilized proteins were further separated by centrifugation through a glycerol gradient, and fractions enriched for Tim54p were immunoprecipitated with monospecific antisera against Tim54p. In addition to Tim10p and Tim22p, a band of ∼18 kDa was seen that was absent from the control precipitates with nonimmune serum (not shown). Tryptic digestion of this protein band followed by electrospray mass spectrometry identified two peptides (Fig. 1A, boldface letters), whose sequences corresponded to the open reading frame (ORF) yOR297c in the yeast genome that encodes a 21.9-kDa polypeptide of unknown function (Fig. 1A). We designated this ORF TIM18. Its predicted product, Tim18p, has a predicted cleavage site for mitochondrial processing peptidase (RXXS/A [40]) between amino acids 42 and 43 (Fig. 1A, arrow), and three putative transmembrane regions (Fig. 1B). It has significant homology to the succinate dehydrogenase membrane anchor subunit (Sdh4p) of yeast mitochondria (39% identical, 58% similar) and to the putative product of ORF yLR164w (53% identical, 74% similar).
FIG. 1.
Sequence of the TIM18 gene and its product, Tim18p. (A) The DNA sequence of TIM18 (ORF yOR297c) and the deduced amino acid sequence of Tim18p are shown. The two peptide sequences initially identified by tryptic digestion of the protein followed by electrospray mass spectrometry are in boldface. The three putative transmembrane segments are underlined, and the predicted mitochondrial peptidase processing site is indicated by an arrow. (B) Hydropathy plot of Tim18p calculated by using the Kyte-Doolittle algorithm.
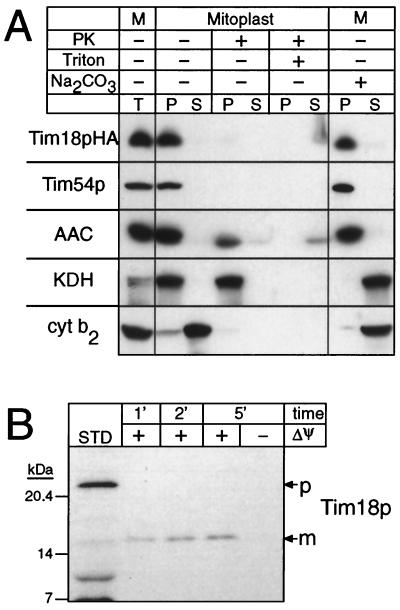
To determine the intracellular location of Tim18p, we epitope tagged the protein with an HA tag and expressed it from a single-copy plasmid in a strain (described below) whose TIM18 gene had been disrupted. The HA-tagged Tim18p is functional since its expression ameliorated the growth rate of the TIM18 deletion strain to wild-type levels. In addition import of Tim23p into mitochondria prepared from the Tim18pHA-expressing strain was as efficient as import into wild-type mitochondria in contrast to the much slower rate observed with the deletion strain (data not shown). Subfractionation of the transformants followed by immunoblotting showed that the HA-tagged Tim18p is present in the mitochondria together with the mitochondrial proteins Tim54p, AAC, α-ketoglutarate dehydrogenase (KDH), and cytochrome b2 (Cyt b2) (Fig. 2A). When the mitochondrial outer membrane was ruptured by osmotic shock, the tagged Tim18p remained associated with the mitoplasts, but its C-terminal HA tag became susceptible to proteolytic digestion (Fig. 2A). Like AAC and Tim54p, the tagged Tim18p was not extracted by alkali (Na2CO2), indicating that it is an integral membrane protein (Fig. 2A). To confirm that Tim18p is a mitochondrial protein, radiolabeled Tim18p was incubated with energized mitochondria. Tim18p was imported rapidly and processed to its mature form (Fig. 2B). We conclude that Tim18p is an integral protein of the mitochondrial inner membrane whose C terminus is exposed to the intermembrane space.
FIG. 2.
Tim18p is a mitochondrial inner membrane protein. (A) Mitochondria were isolated from a strain that carries a disrupted TIM18 gene and expresses Tim18pHA on a low-copy plasmid. Mitoplasts were prepared by incubation in 20 mM K+-HEPES (pH 7.4) at 4°C for 30 min in the presence or absence of proteinase K (100 μg/ml) or Triton X-100 (0.1%) as indicated. Pellet (P) and supernatant (S) were then separated by centrifugation. Mitochondria (M) were also extracted with 0.1 M Na2CO3 for 30 min on ice, followed by centrifugation at 100,000 × g for 15 min to separate the pellet and supernatant. Equivalent amounts of protein were analyzed by SDS-PAGE and immunoblotting with rabbit antisera monospecific for Tim54p, ADP/ATP carrier (AAC), KDH, and Cyt b2. Tim18pHA was probed with a mouse monoclonal antibody against the HA tag, followed by rabbit anti-mouse immunoglobulin G (IgG). All blots were decorated with 125I-labeled protein A and autoradiographed. T, untreated mitochondria. (B) Radiolabeled Tim18p precursor was synthesized in vitro and incubated with mitochondria for the indicated times in the absence or presence of a membrane potential (ΔΨ). After import, samples were treated with proteinase K to remove nonimported precursor and further processed as described previously (31). Samples were then analyzed by SDS-PAGE and fluorography. STD, percentage of the total precursor added to the import assay.
Tim18p is a subunit of the TIM22 complex.
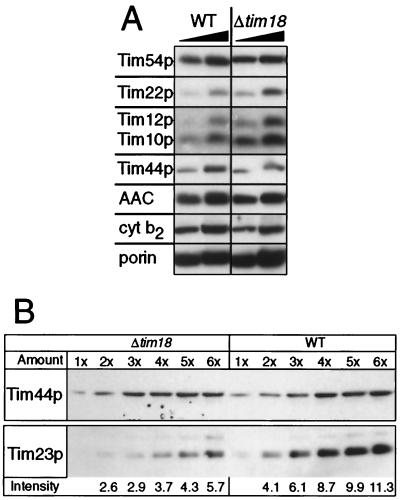
To investigate the role of Tim18p in mitochondrial function, we disrupted the TIM18 gene. The resulting Δtim18 strain was viable, but its growth rate on nonfermentable carbon sources was about half that of the corresponding wild-type strain (not shown). The Δtim18 strain had only half as much Tim23p (Fig. 3B) but twice as much Tim12p and Tim10p as the wild-type strain (Fig. 3A). The levels of Tim54p, Tim22p, Tim44p, AAC, Cyt b2 and porin were not significantly different from those of the wild-type strain (Fig. 3A).
FIG. 3.
Deletion of Tim18p decreases the steady-state levels of Tim23p. (A) The mitochondrial levels of Tim54p, Tim22p, Tim12p, Tim10p, Tim44p, AAC, Cyt b2, and porin are not decreased by deleting Tim18p. Mitochondria (50 and 100 μg of protein, left to right) from wild-type (WT) yeast or yeast lacking Tim18p (Δtim18) were analyzed by SDS-PAGE and immunoblotting with monospecific rabbit antisera against the proteins indicated on the left. Blots were decorated with 125I-labeled protein A and autoradiographed. (B) The mitochondrial levels of Tim23p are decreased by deleting Tim18p. Mitochondria (1× = 30 μg protein) from wild-type (WT) yeast or yeast lacking Tim18p (Δtim18) were analyzed as in panel A with rabbit antisera monospecific for Tim23p or Tim44p. The BCA assay confirmed that equal amounts of wild-type and Δtim18 mitochondria had been loaded. Intensity: β-particle emissions (103) for Tim23p were quantified by using a β-imager. The intensities for Tim44p (used as a control) were identical for wild-type and Δtim18 strains.
As a further test for the function of Tim18p, we looked for genetic interactions between Tim18p and either Tim10p or Tim9p. Deletion of Tim18p was synthetically lethal with either the ts tim10-1 mutation (19) or the ts tim9-3 mutation at 25°C. Synthetic lethality was shown by the absence of viable haploid Δtim18 tim10-1 or Δtim18 tim9-3 meiotic segregants from the appropriate crosses (Table 1). The genetic interaction between Tim18p and either Tim9p or Tim10p suggests that Tim18p functions in the mitochondrial import of inner membrane proteins.
TABLE 1.
Deletion of TIM18 is synthetically lethal with the ts tim9-3 and tim10-1 mutations
| Genotype of haploid progeny | Segregation analysis
|
|
|---|---|---|
| Viabilityc | Frequencyd | |
| Δtim18 tim9-3a | ||
| TIM9 leu2 TIM18 | + | 0.125 |
| TIM9 tim9-3:LEU2 TIM18 | + | 0.138 |
| TIM9 leu2 Δtim18::KANR | + | 0.129 |
| Δtim9::TRP1 leu2 TIM18 | − | 0.108 |
| TIM9 tim9-3:LEU2 Δtim18::KANR | + | 0.112 |
| Δtim9::TRP1 leu2 Δtim18::KANR | − | 0.134 |
| Δtim9::TRP1 tim9-3:LEU2 TIM18 | + | 0.125 |
| Δtim9::TRP1 tim9-3:LEU2 Δtim18::KANRe | − | 0.129 |
| Δtim18 tim10-1b | ||
| TIM10 leu2 TIM18 | + | 0.122 |
| TIM10 tim10-1:LEU2 TIM18 | + | 0.132 |
| TIM10 leu2 Δtim18::KANR | + | 0.112 |
| Δtim10::HIS3 leu2 TIM18 | − | 0.138 |
| TIM10 tim10-1:LEU2 Δtim18::KANR | + | 0.143 |
| Δtim10::HIS3 leu2 Δtim18::KANR | − | 0.128 |
| Δtim10::HIS3 tim10-1:LEU2 TIM18 | + | 0.107 |
| Δtim10::HIS3 tim10-1:LEU2 Δtim18::KANRe | − | 0.117 |
The diploid strain TIM9/Δtim9::TRP1 TIM18/Δtim18::KANR leu2/tim9-3:LEU2 was sporulated, and the haploid progeny from 58 tetrads was analyzed for viability and segregation of LEU2, TRP1, and KANR markers. The genotype of the inviable spores was deduced assuming a 2:2 segregation of auxotrophic markers.
The diploid strain TIM10/Δtim10::HIS3 TIM18/Δtim18::KANR leu2/tim10-1:LEU2 was sporulated, and the haploid progeny from 49 tetrads was analyzed for viability and segregation of LEU2, HIS3, and KANR markers. The genotype of the inviable spores was deduced assuming a 2:2 segregation of auxotrophic markers.
Growth on rich glucose medium at room temperature: +, viable; −, lethal.
Expected frequency of independent segregation of each marker is 0.125, P < 0.01 (χ2 test).
Progeny with this genotype contain only the tim9-3 or tim10-1 allele and the KANR allele (replacing the TIM18 gene) and were inviable, indicating that disruption of TIM18 is synthetically lethal with the tim9-3 and tim10-1 ts alleles.
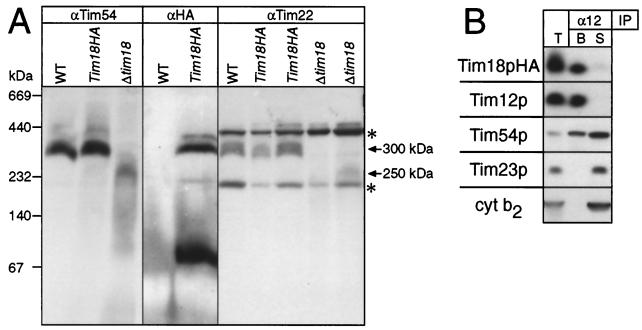
To determine whether Tim18p is a subunit of the TIM22 complex, we resolved solubilized mitochondrial proteins by nondenaturing blue native gel electrophoresis (33, 34) and probed the gels with monospecific antibodies recognizing Tim54p, Tim22p, or the HA tag of epitope-tagged Tim18p (Fig. 4A). In agreement with previous results (19), Tim 54p and Tim22p comigrated with the 300-kDa TIM22 complex when mitochondria from wild-type cells were analyzed (WT in Fig. 4A). With mitochondria from the strain expressing HA-tagged Tim18p, the tagged Tim18p comigrated with Tim54p and Tim22p (Tim18pHA in Fig. 4A). Part of the HA-tagged Tim18p also migrated as a band of ∼70 kDa, suggesting that Tim18p may also be part of a smaller complex (Fig. 4A). With mitochondria from a strain deleted for Tim18p, Tim54p and Tim22p comigrated as a diffuse complex of only ∼250 kDa (Δtim18 in Fig. 4A). The apparent size and stability of three other high-molecular-weight complexes, including F1-ATPase, the TOM complex, and cytochrome oxidase, remained unaltered in these mitochondria (data not shown). This result suggests that Tim18p is a subunit of the TIM22 complex and that its deletion specifically impairs the assembly or the stability of this complex.
FIG. 4.
Tim18p is associated with Tim22p, Tim54p, and Tim12p. (A) Blue native gels (6 to 16%) of mitochondrial proteins from wild type (WT), the strain expressing Tim18pHA, or the Δtim18 strain were blotted onto polyvinylidene difluoride membranes and immunoblotted with antisera against Tim54p (αTim54), Tim22p (αTim22), or a mouse monoclonal against the HA tag (αHA). In the αTim22 panel, 1× or 2× amounts of proteins were loaded in the two adjacent Tim18pHA and Δtim18 lanes, respectively. The bands at ∼440 and ∼200 kDa (asterisks) in the αTim22p panel are nonspecific cross-reactions (unpublished data). Arrows denote the apparent sizes of the TIM22 complex in different strains. (B) Mitochondria isolated from the Δtim18 strain expressing Tim18pHA on a low-copy-number plasmid were solubilized with 0.16% n-dodecyl maltoside and incubated with protein A-Sepharose beads containing immobilized rabbit IgG monospecific for Tim12p (α12). After a washing, bound proteins (B) were eluted with SDS-containing sample buffer, and proteins remaining in the supernatant (S) were acid precipitated and dissolved in sample buffer. Both samples, along with solubilized mitochondria (T), were analyzed by SDS-PAGE and immunoblotting with rabbit antisera monospecific for Tim12p, Tim54p, Tim23p, or Cyt b2 or with a mouse monoclonal antibody against the HA tag (followed by rabbit anti-mouse IgG). Blots were decorated with 125I-labeled protein A and autoradiographed. Tim18p was not coimmunoprecipitated by antisera to Cyt b2 or Tim23p (not shown).
To obtain additional evidence that the Tim18p is a subunit of the TIM22 complex, mitochondria containing HA-tagged Tim18p were solubilized and subjected to immunoprecipitation with antisera monospecific for Tim12p (Fig. 4B). The immunoprecipitate was then analyzed for Tim12p, Tim54p, Tim23p, Cyt b2, and the HA tag of Tim18p by SDS-polyacrylamide gel-electrophoresis (PAGE) and immunoblotting. HA-tagged Tim18p coimmunoprecipitated with Tim12p, with all of Tim22p, and with a portion of Tim54p (Fig. 4B; not shown) but not with the inner membrane protein Tim23p or the intermembrane space protein Cyt b2 (Fig. 4B). Conversely, antisera against Tim23p or Cyt b2 immunoprecipitated neither the HA-tagged Tim18p nor Tim12p (not shown). Therefore, Tim18p is not part of the TIM23 complex. We conclude that Tim18p is a subunit of the TIM22 complex in the mitochondrial inner membrane and that it interacts functionally with Tim9p and Tim10p.
Deletion of Tim18p slows import of several mitochondrial precursor proteins.
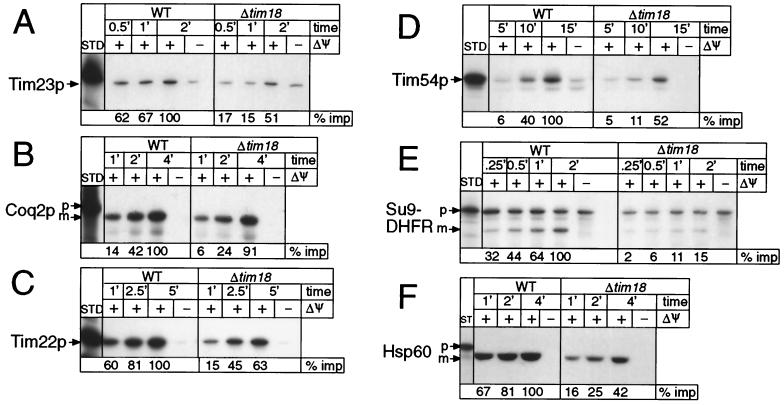
As Tim18p is a subunit of the TIM22 complex, its deletion should affect the import of proteins that require the TIM22 complex for insertion into the mitochondrial inner membrane. We therefore compared the rates of import of several radiolabeled precursor proteins into isolated mitochondria from wild-type cells and from a strain deleted for Tim18p (Fig. 5). As expected, Tim18p-less mitochondria imported the inner membrane proteins Tim23p, Tim22p, Tim54p, and Coq2p two- to threefold more slowly than the corresponding wild-type mitochondria (Fig. 5A to D). To our surprise, however, they imported Tim17p and AAC at close to normal rates (not shown) and imported and processed the matrix-targeted precursors Hsp60 and Su9-DHFR more slowly than did wild-type mitochondria (Fig. 5E and F).
FIG. 5.
Import of several mitochondrial precursor proteins is slowed in mitochondria deficient for Tim18p. Radiolabeled protein precursors were synthesized in vitro and incubated with either wild-type (WT) or Δtim18 mitochondria for the indicated times in the absence or presence of a membrane potential (ΔΨ). After import, samples were treated with trypsin or proteinase K to remove nonimported precursor, followed by treatment with soybean trypsin inhibitor or PMSF. Mitochondria that had been allowed to import Su9-DHFR were not treated with protease. Samples were processed and analyzed by SDS-PAGE and fluorography. Fluorographs were scanned by laser densitometry, and the longest time point for import into wild-type mitochondria was set to 100% and used to calculate the relative import (% imp). These values are given below each lane. p, intact precursor; m, mature (i.e., processed) protein; STD, percentage of the total precursor added to each import assay; A, Tim23p was imported at 15°C, STD = 10%; B, Coq2p was imported at 25°C, STD = 20%; C, Tim22p was imported at 25°C, STD = 10%; D, Tim54p was imported at 25°C, STD = 5%; E, Su9-DHFR was imported at 15°C, STD = 20%; and F, hsp60 was imported at 20°C, STD = 20%.
Since mitochondrial protein import requires a membrane potential, decreased protein import into Tim18p-less mitochondria could reflect an indirect effect resulting from decreased mitochondrial energization. We tested this possibility by measuring the mitochondrial membrane potential; it was not affected by deletion of Tim18p (wild type, 97.4 ± 10 mV; Δtim18, 109.7 ± 12.9 mV; n = 4 to 6). The lowered import rates of precursor proteins into mitochondria from the Tim18p-less strain are thus not caused by an indirect effect of a lowered potential across the mitochondrial inner membrane.
DISCUSSION
We have identified Tim18p as a new subunit of the TIM22 complex that mediates insertion of hydrophobic proteins into the mitochondrial inner membrane. This identification rests on three observations. First, Tim18p is coimmunoprecipitated with Tim22p, Tim54p, and Tim12p in the presence of nondenaturing detergents. Second, it comigrates with Tim22p and Tim54p as a 300-kDa complex upon blue native gel electrophoresis. Third, its deletion affects the apparent size of the TIM22 complex. Furthermore, Tim18p is not associated with other components of the mitochondrial protein import machinery such as the TIM23 complex.
This brings the number of known subunits of the TIM22 complex to six: Tim22p (38), Tim12p and Tim10p (19, 39), Tim54p (18), Tim9p (1, 20), and Tim18p (this study).
Tim18p not only is physically associated with the TIM22 complex but also is required for the optimal function of that complex. Deletion of TIM18 is synthetically lethal with ts mutations in either TIM9 or TIM10, whose protein products are components of both the TIM22 complex and the 70-kDa complex in the soluble intermembrane space (1, 19, 21, 39). Deletion of Tim18p also slows import of several precursor proteins into isolated mitochondria.
Deletion of Tim18p impairs import of several inner membrane proteins, including Tim22p, Tim54p, and Coq2p by two- to threefold, yet it does not lower the steady-state levels of these proteins. The slower import of Tim23p into Tim18p-less mitochondria agrees with the findings that this import is mediated by the TIM22 complex (4, 16) and that Tim18p-less mitochondria have only half as much Tim23p as do wild-type mitochondria (Fig. 3B). As Tim23p mediates import of proteins into the matrix, the decreased import and processing of matrix-targeted precursors into Tim18p-less mitochondria is thus probably a secondary consequence of the decreased abundance of Tim23p. No such defect has been reported for mutations in other components of the TIM22 complex. However, it is also possible that deletion of Tim18p affects the activity of the TIM23 complex by a more direct mechanism. It is puzzling that deletion of Tim18p affects neither import rates nor steady-state levels of AAC, even though AAC import requires the TIM22 complex. Perhaps Tim18p is not essential for the ability of the TIM22 complex to insert AAC into the mitochondrial inner membrane.
The precise role of Tim18p in the functioning of the TIM22 complex remains to be established. Tim18p probably does not bind the imported precursors directly, since we were unable to cross-link radiolabeled Tim23p to Tim18p in isolated mitochondria (not shown). The fact that Tim18p is not essential for viability, even though its deletion decreases the apparent size of the TIM22 complex, suggests that Tim18p may be involved in the assembly or the stabilization of the TIM22 complex rather than in its function. Further analysis will provide insights into how Tim18p cooperates with the known subunits of the TIM22 complex, with perhaps additional, as-yet-unknown subunits and with the two soluble 70-kDa complexes in the intermembrane space to insert hydrophobic proteins into the mitochondrial inner membrane.
ACKNOWLEDGMENTS
C. M. Koehler and M. P. Murphy contributed equally to this paper.
We are grateful to Nikolaus Pfanner for his generous gift of the plasmid carrying the TIM17 gene and to Paul Jeno for assistance with the electrospray mass spectrometry and peptide sequencing.
We thank the following agencies for their support: the Swiss National Science Foundation (to G.S.), the Human Frontier Science Program Organization (to G.S.), the European Economic Union (to G.S.), the F. Louis Jeantet Foundation (to G.S.), the Damon Runyon-Walter Winchell Cancer Research Foundation (to C.M.K.), the U.S. National Science Foundation (to C.M.K.), and the European Molecular Biology Organisation (to E.O.).
REFERENCES
- 1.Adam A, Endres M, Sirrenberg C, Lottspeich F, Neupert W, Brunner M. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 1999;18:313–319. doi: 10.1093/emboj/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand M D. Measurement of mitochondrial proton motive force. In: Brown G C, Cooper C E, editors. Bioenergetics—a practical approach. Oxford, England: IRL Press; 1995. pp. 39–62. [Google Scholar]
- 3.Castrejon V, Parra C, Moreno R, Pena A, Uribe S. Potassium collapses the Δp in yeast mitochondria while the rate of ATP synthesis is inhibited only partially. Arch Biochem Biophys. 1997;346:37–44. doi: 10.1006/abbi.1997.0273. [DOI] [PubMed] [Google Scholar]
- 4.Davis A J, Ryan K R, Jensen R E. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol Biol Cell. 1998;9:2577–2593. doi: 10.1091/mbc.9.9.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker P J, Muller H, Rassow J, Pfanner N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol Chem. 1996;377:535–538. [PubMed] [Google Scholar]
- 6.Dubaquie Y, Looser R, Funfschilling U, Jeno P, Rospert S. Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping but non-identical requirement for hsp60 and hsp10. EMBO J. 1998;17:5868–5876. doi: 10.1093/emboj/17.20.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endres M, Neupert W, Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22:54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO J. 1998;17:6497–6507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Glick B, Schatz G. Import of proteins into mitochondria. Annu Rev Genet. 1991;25:21–44. doi: 10.1146/annurev.ge.25.120191.000321. [DOI] [PubMed] [Google Scholar]
- 12.Glick B S, Brandt A, Cunningham K, Muller S, Hallberg R L, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- 13.Glick B S, Pon L. Isolation of highly purified mitochondria from S. cerevisiae. Methods Enzymol. 1995;260:213–233. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 15.Haucke V, Schatz G. Reconstitution of the protein insertion machinery of the mitochondrial inner membrane. EMBO J. 1997;16:4560–4567. doi: 10.1093/emboj/16.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaldi K, Bauer M F, Sirrenberg C, Neupert W, Brunner M. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 1998;17:1569–1576. doi: 10.1093/emboj/17.6.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaldi K, Neupert W. Protein translocation into mitochondria. Biofactors. 1998;8:221–224. doi: 10.1002/biof.5520080308. [DOI] [PubMed] [Google Scholar]
- 18.Kerscher O, Holder J, Srinivasan M, Leung R S, Jensen R E. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 20.Koehler C M, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G. Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci USA. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koehler C M, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koehler C M, Merchant S, Schatz G. How membrane proteins cross the mitochondrial intermembrane space. Trends Biochem Sci. 1999;24:428–432. doi: 10.1016/s0968-0004(99)01462-0. [DOI] [PubMed] [Google Scholar]
- 23.Kubrich M, Rassow J, Voos W, Pfanner N, Honlinger A. The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane. J Biol Chem. 1998;273:16374–16381. doi: 10.1074/jbc.273.26.16374. [DOI] [PubMed] [Google Scholar]
- 24.Leuenberger D, Bally N, Schatz G, Koehler C M. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 1999;18:4816–4822. doi: 10.1093/emboj/18.17.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 26.Palmieri F, Bisaccia F, Capobianco L, Dolce V, Fiermonte G, Iacobazzi V, Indiveri C, Palmieri L. Mitochondrial metabolite transporters. Biochim Biophys Acta. 1996;1275:127–132. doi: 10.1016/0005-2728(96)00062-x. [DOI] [PubMed] [Google Scholar]
- 27.Pfanner N. Mitochondrial import: crossing the aqueous intermembrane space. Curr Biol. 1998;8:R262–R265. doi: 10.1016/s0960-9822(98)70168-x. [DOI] [PubMed] [Google Scholar]
- 28.Pfanner N, Douglas M G, Endo T, Hoogenraad N J, Jensen R E, Meijer M, Neupert W, Schatz G, Schmitz U K, Shore G C. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- 29.Pfanner N, Meijer M. The Tom and Tim machine. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- 30.Pfanner N, Neupert W. Distinct steps in the import of ADP/ATP carrier into mitochondria. J Biol Chem. 1987;262:7528–7536. [PubMed] [Google Scholar]
- 31.Rospert S, Schatz G. Protein translocation into mitochondria, P. 277–285. In: Celis J E, editor. Cell biology: a laboratory handbook. 2nd ed. Vol. 2. San Diego, Calif: Academic Press, Inc.; 1998. [Google Scholar]
- 32.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 33.Schägger H, Cramer W A, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 34.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 35.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation, of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirrenberg C, Bauer M F, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 39.Sirrenberg C, Endres M, Folsch H, Stuart R A, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 40.Von Heijne G. Cleavage-site motifs in protein targeting sequences. Genet Eng. 1992;14:1–11. doi: 10.1007/978-1-4615-3424-2_1. [DOI] [PubMed] [Google Scholar]
- 41.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]