Abstract
Stress is the response of an organism to demands for change, yet excessive or chronic stress contributes to nearly all psychiatric disorders. The advent of high-throughput transcriptomic methods such as single cell RNA sequencing poses new opportunities to understand the neurobiology of stress, yet substantial barriers to understanding stress remain. Stress adaptation is an organismal survival mechanism conserved across all organisms, yet there is an infinity of potential stressful experiences. Unraveling shared and separate transcriptional programs for adapting to stressful experience remains a challenge, despite methodological and analytic advances. Here we review the state of the field focusing on the technologies used to study the transcriptome for the stress neurobiologist, and also attempt to identify central questions about the heterogeneity of stress for those applying transcriptomic approaches. We further explore how postmortem transcriptome studies aided by preclinical animal models are converging on common molecular pathways for adaptation to aversive experience. Finally, we discuss approaches to integrate large genomic datasets with human neuroimaging and other datasets.
Keywords: PTSD, MDD, Stress transcriptomics, Postmortem brain, Animal behavior
1. Introduction
Neuropsychiatric disorders cause considerable disability and burden economically and to the health of people around the world. Significant efforts have been made to understand these disorders; however, their underlying molecular pathology remains elusive. Preclinical and clinical studies indicate that psychiatric disorders onset is multifactorial: part environment and part genetic and arise from structural and molecular changes in corticolimbic and mesolimbic circuits (Jovanovic et al., 2010; Jovanovic and Ressler, 2010; Shin and Liberzon, 2010). Specifically, aversive psychosocial stressors are known to cause major depressive disorder (MDD) and post-traumatic stress disorder (PTSD). There is now a substantial literature describing the genetic architecture and risk for developing MDD (Wray et al., 2018) and PTSD(Gelernter et al., 2019; Nievergelt et al., 2019). However, few of the identified risk variants have been functionally annotated.
The human genetic code contains 3 billion base pairs and an its annotation (introns, exons, intergenic regions, etc.) is still an area of extensive research (Kundaje et al., 2015). The transcriptome is the “read out” of the genetic code and studying it reduces the dimensions from 3 billion base pairs to approximately 25,000 coding transcripts. Transcriptomic studies of neuropsychiatric disorders in both animal models and human postmortem tissue provide the best strategy for understanding how risk variants and stress exposure affect the molecular pathology of the central nervous system (CNS). Postmortem brain tissue provides us with a direct measurement of how risk variants may affect gene expression and perturbation of these genes in animal models provides critical functional validation. The transcriptome can be thought of a molecular phenotype of a particular trait or illness state in the same way as neurological disruptions such as plaques and tangles are phenotypic of Alzheimer's disease. This analogy is particularly relevant as there are no macro-neurological features that distinguish patients with psychiatric disorders. For example, functional genomic studies of autism have consistently identified atypical gene co-expression networks across multiple ASD cohorts (Adhya et al., 2021; Wang et al., 2018).
There are currently major efforts to understand the molecular pathology of stress disorders. Preclinical work on stress disorders has been directed at understanding how much of the disease transcriptomic changes are recapitulated by each stress model (Scarpa et al., 2020). Increased collection of human postmortem tissue from donors with psychiatric illness has contributed to advances in understanding the transcriptomic changes of MDD and PTSD and provided comprehensive genomic atlases which can be used to identify clinically relevant changes in animal model comparisons. Here we review the current state of technologies used in transcriptomic studies: from RNA-sequencing of bulk-tissue to single cell-types. We explore how animal models have contributed to our understanding of the molecular pathology of MDD and PTSD. And finally, we describe how postmortem genomics work has revolutionized our understanding of MDD and PTSD and how integrating these large, rich data sets with other human analyses such as neuroimaging are providing critical information necessary for better biomarker identification and therapeutic design.
2. Transcriptomic methods
Technical advances have made high-fidelity measurements of gene expression changes following stress at cellular resolution possible (Macosko et al., 2015), as well as enabling simultaneous large-scale studies of other cellular measurements (“multi-omics”) (Macaulay et al., 2017). This rapidly advancing methodological approach is yielding insights into fundamental building blocks of pathophysiology across biomedical fields. Here we provide a brief summary of approaches to transcriptomic measurements relevant to understanding stress in order to facilitate psychiatric neuroscience investigators who wish to adopt these approaches in the understanding of stress-related disorders.
Recent advances in our understanding of the genomic structure of the mammalian CNS have been directed by the development of high throughput sequencing approaches such as RNA-seq. RNA-seq provides three to four magnitudes more information than previous gene expression systems. Deep sequencing has been applied to many different genomic levels including genomic DNA sequencing (DNA-seq) (Yu et al., 2017), non-coding RNA profiling (smRNA-seq) (Choi et al., 2005; Xu et al., 2011; Zhou et al., 2021), DNA-protein interactions (ChiP-seq) (Barski et al., 2007) and DNA structure (e.g., open chromatin) (ATAC-seq) (Bryois et al., 2018). The actual process of RNA-seq is detailed elsewhere but we will briefly review here. RNA is extracted and ribosome depleted before cDNA synthesis. Ribosome depletion has become the gold standard for RNA sample preparation as it removes biological contaminants (rRNA and mitochondrial RNA) that may obscure other relevant molecular changes. Each cDNA fragment (usually 75–100 bp) is sequenced normally in a paired end fashion where the fragment is sequenced 3′ to 5′ and then again from 5′ to 3’. The resulting sequence reads are aligned to the genome of interest but can also be assembled without reference. This allows for unprecedented analysis of the transcriptome-not just what is expressed but also how it is expressed (i.e., alternative splicing, exonic SNP detection, and novel transcript prediction).
A major limitation of bulk-tissue RNA sequencing, particularly in the brain, lies in the intrinsic heterogeneity of cell-types within brain regions – any anatomically-based collection of RNA samples will be a weighted average of expression levels of all cells present, and potentially biased by cell-size and other confounds. In order to overcome this barrier, several solutions have been developed even prior to the development of single-cell sequencing (Macosko et al., 2015). Cell-type specific RNA sequencing can be obtained via methods that use a Cre-recombinase systems to tag translating ribosomes with GFP within specific cell-types (translating ribosome affinity purification, TRAP) (Doyle et al., 2009). Conditional transgenic animals can thus be used to isolate actively translating mRNAs from a cell-type of interest within anatomically localized tissue, as long as a transgenic Cre mouse exists for that cell type.
The development of microfluidic-based approaches such as droplet-based RNA sequencing (Macosko et al., 2015), in turn, has produced a dramatic change in neuronal gene expression studies over the past few years (Ofengeim et al., 2017). Instead of being limited to a small number of pre-specified cell-types in transgenic animals, individual cells within a brain region can be isolated and sequenced separately after embedding them onto gel droplets and adding nucleic acid barcodes which allow for later identification of transcripts by cell-type. The power of this approach lies in the ability to sequence individual cells at scale in an unbiased fashion, although the high dimensionality of single cell sequencing data presents new bioinformatic challenges (Vallejos et al., 2017). Single nucleus sequencing, in particular, permits extraction of mRNA with a high correlation to expression levels in the entire cell thus permitting extensions of droplet-based approaches to difficult tissues such as human postmortem brain tissue (Lake et al., 2016).
Extensions of droplet-based sequencing are rapidly progressing, particularly with regard to acquiring other data simultaneously on the sequenced cells. Spatial RNA sequencing, in which nucleic acid barcodes for two-dimensional position are applied before subsequent droplet-based sequencing (Rodriques et al., 2019), permit fine-scale localization of expression across anatomical axes and was named the 2020 method of the year by Nature Methods (Marx, 2021). Multiplexing of genomic information such as chromatin accessibility sequencing (ATAC-seq) or even protein expression levels within individual cell-types (Macaulay et al., 2017) may allow stress biologists to understand the impact of transcriptomic changes on cellular state.
3. Models of stress for transcriptomic studies
Hans Selye defined stress in 1936 as “the non-specific response of the body to any demand for change,“(Selye, 1998) an organismal response to a challenge. The brain serves a central role in this response, taking in sensory and physiological evidence of this demand for change and orchestrating behavioral responses. Alterations in neural circuit functions related to stress have a causal role in major psychiatric illnesses (McEwen, 2008) such as depression, anxiety, posttraumatic stress disorder, and substance abuse. Three brain regions in particular have contributed to our knowledge of the cellular and molecular mechanisms of stress in the brain. The hippocampus is perhaps the best studied structure. Stress causes dendritic shrinkage and loss of spines in the hippocampus. The prefrontal cortex suffers from debranching and shrinkage of dendrites after chronic stress (McEwen et al., 2016). Traumatic stressors have been shown to increase spine density of basolateral amygdala dendrites and induce loss of spines in the medial amygdala. These structural changes have been implicated in the development of PTSD behaviors (Mitra et al., 2005). Molecularly, stress induces transcriptomic and epigenetic changes to excitatory amino acids, glucocorticoids, and chromatin modifiers. Extracellular and intracellular mediators such as endocannabinoids and brain-derived neurotropic (BDNF) have also been shown to play a role in stress development (McEwen et al., 2016).
Similarly, the ability to induce stress in animal systems is essential to understanding the transcriptomic signatures of stress-induced psychopathology (Scharf and Schmidt, 2012). As the kind of stress that is believed to lead to psychiatric illness is not under the experimenter's control in humans, such studies will always be limited to observational or quasi-experimental approaches (James and Ice, 2006). Thus, the induction of psychological stress in animals is an essential component of understanding transcriptomic programs induced by aversive experience. While the larger project of summarizing animal models of stress lies outside the purview of our review, we choose to focus here on the key aspects of animal models of stress that have impacted our understanding of transcriptional programs induced by it.
Following McEwen and colleagues (McEwen, 2008; Mcewen, 2004), these experimental models can broadly be classified into those relating to an individual episode of stress (acute stress) or those relating to a cumulative experience of stress (allostatic load; chronic stress). The former is amenable to an experimental strategy in which tissue is collected from animals at a series of time points after an initial event and used for sequencing. The latter, in contrast, have typically been collected at a fixed delay after a more persistent stressful experience in order to create a contrast between two conditions (stressed and unstressed). Importantly the effects of acute and chronic stress are frequently different in terms of transcriptomic changes (Simmons et al., 2020). One large gene expression atlas of different kinds of stress in rodent brain meta-analyzed gene expression data from 18 projects studying the effect of stress on the transcriptome of the brain (Flati et al., 2020). They observed a consistent pattern between acute and chronic stressors and brain gene expression. GO analysis of genes modulated a short time after stress (acute) enriched for genes involved in enzymatic activities and transcription factors. At longer time points after stress (chronic) functional annotation of DEGs revealed enrichment for structural changes such as axonogenesis and synapse organization. These findings are consistent with known structural and plasticity responses to chronic and acute stress.
Acute stress offers the ability to identify direct gene expression mediators of persistent changes after stress (Floriou-Servou et al., 2021). A recent study of the ventral hippocampus after exposure to three different types of acute stressors found that gene expression patterns related to different stressors were quite divergent, but that there were some common changes across different stress types (Floriou-Servou et al., 2018). Since acute stress responses evolve over hours to days, future studies and modeling efforts may facilitate understanding the temporal evolution and key elements of stress responsivity.
While chronic stress manipulations vary, the most common have either been repetitive unpredictable sensory stimuli (Girgenti et al., 2019), social defeat stress (Bagot et al., 2016), or early life stress (Peña et al., 2017). Social defeat stress, in which a mouse is exposed repeatedly to another aggressive mouse over 10 days, leads to differential expression of large numbers of genes throughout limbic circuits (Bagot et al., 2016). One advantage of this model is the ethological nature of the stress manipulation, as well the ability to distinguish resilient vs susceptible animals by their level of future social interaction. In transcriptomic studies, this has been used to identify gene networks and key gene expression programs. In one remarkable study, early life stress induced widespread and persistent changes in gene expression in the ventral tegmental area (Peña et al., 2017). Interestingly, this response was orchestrated by a brief change in expression of the transcription factor OTX2 which was not sustained into adult gene expression changes. Importantly, these studies highlight the critical need of tying gene expression changes to neuronal activity as both studies provide causal evidence for specific genes in the control of the stress response. Chronic stress has an impact on the individual cell's regulation of gene expression. One group performed single cell RNA-seq on each component of the HPA axis and identified a previously unknown subcluster of cells that were Abcb1b+ and which play an important role in the plasticity and adaptation processes of chronic stress in the adrenal cortex and highlights the need to move this work to the single cell-type level (Lopez et al., 2021).
However, neurobiological research on stress has reached a major obstacle - the very “non-specific” nature of the phenomenon has blocked further study (Simmons et al., 2020). The wide variety of ways to induce stress lead to diverse and often contradictory responses in neural circuits. Stress is typically induced in neurobiological studies by the standardized application of different aversive experiences – say, 4 h of physical restraint (Conrad et al., 1999) or ten days of a mouse being placed for 10 min per day with an aggressive mouse (Golden et al., 2011). These approaches have yielded tremendous insight into the neurobiology of stress but are difficult to compare across conditions. In the next section, we address one potential approach to this problem.
4. Exploring stress space with transcriptomics
Understanding the nature of stressful experiences requires environmental manipulations that vary across multiple dimensions: sensory variability, higher-order unpredictability, controllability, aversion, chronicity (McGonagle and Kessler, 1990; Peters et al., 1998; Seligman, 1972). The space of potentially stressful events is infinite, with contemporary human stress induced by diverse socioeconomic factors (Lantz et al., 2005) such as social isolation, threat of danger, grief, economic resource instability, diminished appetitive stimuli, loss of control and increased threat of violence. In human psychological studies, the observation that stressors of specific types can induce diverse behavioral manifestations has been an important facet of study going back to the beginning of the field. Yet in animal neurobiology research, the ability to compare across causal manipulations of distinct stressors has been limited by the difficulty of exploring an infinite space of stressors (although see recent efforts at comparison across acute stressors (Floriou-Servou et al., 2018)).
Typically, neurobiologists choose one or a few distinct types of stress, which can be thought of as points in the space of potential stressors. Yet, comparing neurobiological manifestations between two classes of stressors is equivalent to choosing two points, and does not permit inference into the nature of the structure of stressors within that space. Given that stress typically constitutes a series of aversive experiences over time, each individual incident within the stressful experience can be considered as a small directed perturbation in stress space. One quantitative way to conceptualize the transcriptomics of stress is as a mapping from the space of potential stressful (uncertainty- or aversion-inducing) (Peters et al., 2017) experiences to the space of cellular states.
In some ways, our understanding of transcriptomics of stress is thus limited by our ability to control and elicit stress in model systems. As discussed in the previous section, stress in animal models is usually either a single acute or chronic stressor with contrasts between stressed and unstressed conditions. However, this level of quantitative and individualized description has been limited by the lack of theoretical model for stress in which to localize transcriptomics data. In order to truly understand the commonalities across behavioral stress and its cell-type specific transcriptomic consequences, it may be necessary to develop improved behavioral programs and modeling approaches as well as make comparisons with human postmortem tissue of stress disorders. Work is currently underway to identify the molecular intersections between animal models of stress and postmortem tissue genomics work. One study compared a large postmortem, MDD cohort to three separate stress paradigms to identify whether one model better captured the molecular pathology of MDD than another (Scarpa et al., 2020). Interestingly, the authors found that each model captured different aspects of the depression molecular signature. The authors suggest that this is not an impediment but an opportunity to expand the types of mouse models of MDD. Because all three behaviors in this study capture parts of the MDD molecular profile, future work should be expanded into other stress disorders such as PTSD where a complete molecular profile of the animal models is still lacking. Further, future work in the depression model area should be to expand this into cell type-specific work. Despite the tremendous progress in transcriptomic methods, linking new bioinformatic methods for understanding cellular state trajectories after stress to an improved quantitative understanding of stress itself may be essential to understanding how stress alters neural circuit function.
5. Bioinformatic approaches to brain transcriptomic studies
5.1. Differential gene and isoform identification
The original, most direct and informative analyses of transcriptomic data involve identifying lists of differentially expressed genes between sample groups such as discrete brain regions, diseased tissue, or even between the two sexes. RPKM or FPKMs counts for individual transcripts are normalized based on the total number of effective counts for a particular gene in each sample. Programs such as EdgeR(Robinson et al., 2010), DESeq (Love et al., 2014) and LimmaVoom (Law et al., 2014) perform well for most data sets and are effective in ranking differentially expressed genes. EdgeR and DESeq2 both assume that no transcripts are differentially expressed. DESeq uses a geometric normalization strategy where a scaling factor for a given sample is computed as the median of the ratio for each gene and its read count over its geometric mean across all samples. EdgeR calculates a weighted mean of log ratios between sample cohorts after exclusion of the most expressed genes and genes with the largest log ratios. Limmavoom was historically used to analyze microarray data but has been updated to work with sequencing data. Limma/voom matches distributions of gene counts across samples to normalize expression (Shahjaman et al., 2020).
Alternative splicing (AS) is a critical component of gene expression regulation. Through the inclusion or exclusion of exons and intronic sequences, the transcriptome adds to the diversity of the proteome (Pan et al., 2008). AS can have a profound effect on gene function and different isoforms of the same gene can be involved in different or even opposing functions. AS can be measured using traditional RNA-seq methods. Numerous bioinformatical tools have been developed for identifying alternative splicing patterns from RNA-seq including LeafCutter (Li et al., 2018), MISO (Katz et al., 2010), rMATs (Shen et al., 2014), MAJIQ (Vaquero-Garcia et al., 2016), RSEM (Li and Dewey, 2011), Kallisto (Bray et al., 2016), and Salmon (Patro et al., 2017). While RNA-seq data is generally analyzed on the gene level these packages can be used to analyze this data on the exon level to detect and quantify novel splicing events. AS is usually reported as percent spliced, a metric of the percentage of how efficiently sequences of interest are spliced into a full transcript (Li et al., 2018). A recent study (Gandal et al., 2018b) by the Geschwind group identified that more than 25% of the human frontal cortex exhibits differential alternative splicing and that isoform-level changes captured the largest disease effects for donors with autism spectrum disorder, schizophrenia and bipolar disorder. While there is a high degree of common polygenicity in these disorders these findings suggest that alternative splicing of the transcriptome most differentiates these disorders from one another.
Traditionally, RNA-seq is performed on small fragments of RNA (75-150bp). Current detection methods for identifying AS events relies on concatenation of these small sequence fragments. The ability to sequence longer pieces of cDNA are allowing for more accurate identification of splicing events. These so-called long read sequencing assays allow sequencing of full-length transcripts or up to 10 kb or longer (Wang et al., 2016). These sequences can be compared to known AS annotations or can be used for de novo splicing identification. While this technology has not been applied extensively to brain tissue, one recent report (Estill et al., 2021) looked at isoform usage in the nucleus accumbens (NAc) of mice and found that 46% of the detected transcripts harbored novel splicing events not detected by previous RNA-seq experiments, indicating that we are missing significant amounts of AS events using normal RNA-seq for isoform detection and that long read Iso-seq technologies will be necessary for complete isoform detection.
5.2. Pathway analysis and gene set enrichment
Gene-set enrichment has emerged as one of the most popular analyses available on large genomic data sets. Functional annotation analyses seek to aid in interpretation of large transcriptomic and methylomic datasets by utilizing a statistical methodology to identify functionally related groups of genes annotated using systems such as Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for biological process and pathway identification. While very popular, recently evidence suggests significant inflation of significant findings relative to expectations in both mouse and human datasets. One study found that the probability of a GO category being reported as significant in a given dataset increases with its estimated false-positive rate (Fulcher et al., 2020). They showed that this bias is driven by co-expression patterns within GO categories and by spatial autocorrelation. It is also important to point out that many GO and KEGG pathways were organized from large cancer datasets that make interpretation using brain-specific transcriptomic profiles difficult. One mouse study characterizing the sex-specific, single cell transcriptomics of SST and Pvalb interneuron populations provides an example case (Girgenti et al., 2019). Differentially expressed genes in female SST + interneurons that had been exposed to chronic unpredictable stress enriched for GO category GO: pancreatic adenocarcinoma, a category which on the face of it would seem have little to do with stress. However, examination of the GO found it was primarily made up of growth factors including vascular endothelial growth factor (VEGF), fibroblast growth factor receptors (FGFRs) which has known roles in both angiogenesis and neurogenesis pathways with known roles in the regulation of stress and antidepressant use. Taken together, these findings point toward a critical need for development of a brain specific gene ontology organization.
5.3. Gene Co-Expression analysis and deep learning
Transcriptional regulation plays a key role in many homeostatic functions and in disease states. Traditionally, bioinformatic approaches to transcriptomic datasets have focused on the identification of genome-wide significant differentially expressed genes and is the logical extension from the traditional candidate gene studies that predominated early gene profiling studies. However, additional tools have been developed to provide far more information about transcriptional regulation and organization in tissue. One approach that has been consistently popular is gene co-expression network analysis. The underlying biological principle of this analysis is that genes do not operate alone and likely require co-regulated transcriptional control. Thus, gene levels with correlated levels of expression across cohorts are grouped into network modules and differences in module connectivity and membership provide insight into dysregulated gene assemblies. Further, these modules can also be enriched for cell-type specific markers, allowing for identification of the cell types likely responsible for the aberrant transcriptional differences.
A popular tool for this type of analysis is Weighted Gene Co-Expression Network Analysis (WGCNA) (Langfelder and Horvath, 2008). This tool is been used to study co-expression patterns in normal brain tissue (Oldham et al., 2008), major depressive disorder (Labonté et al., 2017) and PTSD (Girgenti et al., 2021). Importantly, co-expression analysis can be used in prioritizing illness-state DEGs and linking these to genetic risk. For example, the largest postmortem study of PTSD brain tissue identified a PTSD associated co-expression network with numerous down regulated molecular key drivers important in interneuron function (Girgenti et al., 2021). Using a global, step-wise approach the authors combined transcriptome-wide imputation (transcriptome-wide association (TWAS)) using eQTL data from the GTEx portal and integrated it with the largest PTSD GWAS generated by the Million Veteran Program. One of the molecular key drivers identified in their WGCNA modules, ELFN1, achieved transcriptome-wide significance for PTSD by TWAS. This was an important first step in identifying convergences between cortical co-transcriptomics and PTSD genetic signals, a pipeline that can be exploited to identify high confidence molecular targets for potential therapeutics for PTSD. TWAS is quickly becoming one of the most popular transcriptomic analysis tools in stress disorders (Stein et al., 2021,Huckins et al., 2020) and its application and development has been reviewed extensively elsewhere (Cano-Gamez and Trynka, 2020; Chatzinakos et al., 2021). Deep learning technology is also being applied to large transcriptomic screens to identify gene-gene relationships (Yuan and Bar-Joseph, 2019). This study designed a novel pipeline which encoded single cell type gene expression data and followed that up with deep neural network analysis. This framework allows for detection of a diverse number of relevant biological phenomenon including transcription factor target prediction and identification of disease-related causal, co-expressed genes.
5.4. Single cell transcriptomics in brain
With an immense number of cell types and cellular connections, the brain is arguably the most complex organ in the human body. The recent single cell sequencing revolution opens a new avenue for dissecting the complexity of the human brain at the single cell level. As we have touched on previously, in no realm is the development of bioinformatics tools more crucial or expanding more rapidly than those for single cell-type analysis. In this section, we will describe broad techniques and methods for conducting single cell analysis in brain tissue.
5.4.1. Preprocessing and quality control (QC)
Computational modeling of scRNA-seq data is challenging due to its ultra-high dimensionality, low capture efficiency, and high level of technical noise. Therefore, a series of QC steps are vital for accurate downstream analyses. For instance, bulk RNA-seq QC tools, such as FastQC, can be employed to check the sequencing quality of scRNA-seq data. Then cells with very few genes detected, mitochondrial genes or extremely high portion of reads mapped to the spike-ins will be removed. Further, several tools have been developed to further remove doublets to avoid artifactual libraries generated from more than one cell (Bais and Kostka, 2019; Bernstein et al., 2020; McGinnis et al., 2019; Wolock et al., 2019). In addition, multi-sample scRNA-seq analysis usually suffers from severe batch effects when integrating data generated by distinct operators at different times or from multiple laboratories using disparate protocols and sequencing platforms. Recent computational methods such as MNN and KBET can be used to correct batch effects for improved performance (Büttner et al., 2019; Haghverdi et al., 2018). Single cell type transcriptomics datasets are prone to noise and circumstantially there has been advances in tools for processing these datasets particularly in the areas of cluster and drop out analysis (Bouland et al., 2021).
5.4.2. Cell level analysis
Normalization is usually carried out to correct unwanted biases (e.g., dropout, sequencing depth, and capture efficiency). Recently, a generalized linear model has been widely used to omit the need for heuristic parameterizations in normalization (e.g., pseudocount addition and log-transformation), benefiting downstream analytical tasks such as variable gene selection, dimensional reduction, and differential expression (Hafemeister and Satija, 2019). Afterward, highly variable genes are selected as informative features to perform various dimension reducing techniques. To identify distinct cell populations, we can perform either supervised clustering methods based on known marker genes or identify de novo cell types using unsupervised algorithms based on k-means, hierarchical clustering, density-based clustering and graph-based clustering. In addition, to accommodate a continuous spectrum of cellular status, cell trajectories and pseudotime can be reconstructed based on scRNA-seq using various tools, such as Monocle (Qiu et al., 2017), Waterfall (Shin et al., 2015), Wishbone (Setty et al., 2016), and CellRouter (Rocha et al., 2018).
5.4.3. Gene level analysis
Distinct from bulk level analysis, scRNA-seq data can identify differentially expressed genes (DEGs) between disease and controls within each subpopulation or group of cells. Although traditional methods designed for bulk RNA-seq DE analysis can be used, recently several methods have been specifically developed for scRNA-seq data with improved performance, such as MAST (Finak et al., 2015), SCDE (Kharchenko et al., 2014), and DEsingle (Miao et al., 2018).
5.4.4. Network level analysis
Various network inferences, such as gene co-expression network and gene regulatory network (GRN) have been widely conducted using bulk RNA-seq data. A number of existing tools such as WGCNA and SCENIC can be directly applied to scRNA-seq data although caution is warranted due to the higher level of noise in single cell experiments. Several recent tools, such as PIDC(Chan et al., 2017) and CNNC(Yuan and Bar-Joseph, 2019), have been developed specifically for GRN construction using scRNA-seq data. Additionally, inter-cellular communication is particularly critical for the development and functionality of the human brain and one obvious advantage of scRNA-seq data is the ability to systematically dissect the cellular connectome. Recently several tools have been developed to construct cell-to-cell communication networks, such as cellPhoneDB (Efremova et al., 2020), CellChat (Jin et al., 2021), and cellTalkDB (Shao et al., 2020), from single cell data.
Recently, several high profile papers have begun mapping the single cell transcriptomic contribution to the human and mouse brain (Darmanis et al., 2015; Hodge et al., 2019; Lake et al., 2018). These studies have challenged our traditional views of the number of potential neuronal subtypes found in the brain. For example, the Hodge et al. paper identified 75 distinct cell type clusters, including 24 types of excitatory neurons and 45 types of inhibitory neurons which precise laminar placement. Most of these clusters represent new, previously unknown cell subtypes. While Hodge is considered the standard by which other brain single cell type studies are being compared, one caveat of the study was the general lack of identification of novel non-neuronal cell subtypes. For example, it is likely that microglia would cluster into at least two transcriptional cell types (activated and quiescent) and there is good evidence of at least 2 separate oligodendrocyte populations in human cortex defined by myelination status (Kleijn et al., 2019). The classification of all cell types in the brain is one of the primary goals of neuroscience and will require the creation of additional bioinformatic tools to parse out the different cell types and subtypes. Further, this work could also help to identify molecules converging between different species (e.g., mice and non-human primates) as these are used extensively as models of human disease.
6. Transcriptomic insights into the molecular pathology of stress implicates interneuron and microglial dysfunction
Postmortem studies of the brains of psychiatric patients has proved essential in understanding the molecular effects of these disorders. This work has been aided by the creation of many large brain banks collecting tissue across numerous neurological and neuropsychiatric disorders. Further, this work has expanded rapidly by the establishment of several large consortia such as Common Mind (Fromer et al., 2016), BrainSeq (Consortium et al., 2018), and PsychENCODE (Wang et al., 2018) which are compiling large disparate genomics datasets and uniformly analyzing them to increase the number of brains analyzed.
Recent postmortem studies of stress disorders including major depressive disorder (MDD) (Duric et al., 2010; Kang et al., 2012; Labonté et al., 2017; Ota et al., 2014; Seney et al., 2018) and post-traumatic stress disorder (PTSD) (Girgenti et al., 2021; Licznerski et al., 2015, Logue et al., 2021; Young et al., 2015) have identified several hundred differentially expressed (DE) genes. Most postmortem studies of the MDD brain have focused on the prefrontal cortex, hippocampus and nucleus accumbens. Evidence has consistently pointed to GABAergic interneuron dysfunction, specifically the subtype that express the neuropeptide somatostatin (SST), as having a significant role in MDD pathology (Levinson et al., 2010; Luscher et al., 2011). Transcript levels of SST have been reported to be reduced in the dorsolateral prefrontal cortex (DLPFC) and the anterior cingulate cortex (ACC) across cortical layers and the amygdala (Seney et al., 2015; Tripp et al., 2011). Studies in knock out mice of SST demonstrate a causal role in several MDD behavioral models, increased corticosterone, and reduction in brain-derived neurotrophic factor (BDNF) and Gad67 transcripts (Soumier and Sibille, 2014). Reductions in interneuron marker genes including parvalbumin (PV), VIP, and SST have also been reported in schizophrenia and autism spectrum disorder (Chung et al., 2018; Gandal et al., 2018a). However, there is inconsistent evidence of reduced PV and VIP expression in MDD (Fee et al., 2017) indicating significant differences in the cell types driving the molecular pathology of these disorders.
Nearly 50% of patients diagnosed with PTSD are comorbid for MDD (Flory and Yehuda, 2015; Kessler et al., 1995; Rytwinski et al., 2013). Recently, the largest postmortem study of PTSD frontal cortex was performed using a matched MDD cohort to disentangle the differences between the two disorders (Girgenti et al., 2021). Similar to MDD, this study also discovered GABAergic signaling deficits in PTSD and identified the interneuron-specific transporter ELFN1 as significantly down regulated and associated with PTSD by transcriptomic imputation using the largest PTSD GWAS from the Million Veteran Program (Fig. 1). While this decrease in SST expression would seem to be a function of comorbidity between MDD and PTSD, formally testing the differentially expressed (DE) gene overlap revealed few DEGs in common: 111 DEGs in the DLPFC, 67 in the mOFC, 14 in the dACC, and 0 in the sgPFC with the most significant being ceruloplasmin (Cp), c-c motif chemokine ligand 2 (Ccl2), and ADAM Metallopeptidase with Thrombospondin Type 1 Motif 2 (Adamts2) (Girgenti et al., 2021). Further, the transcriptomic correlation between PTSD, MDD, and an aggregated, uniformly analyzed MDD profile from the PsychENCODE consortium (Gandal et al., 2018a) revealed no significant correlation between the transcriptomic patterns of the two disorders suggesting different molecular pathologies. However, it should be noted that there was also a lack of correlation between the MDD cohorts of both studies as well. While there are many possible reasons for this, we believe it stems from technical variance between meta-analyzed microarray data with RNA-seq data and differences in MDD diagnosis criteria. The meta-analysis by Gandal was from multiple labs and different brain banks making it likely that MDD cases varied especially for depression subtype, ancestry, trauma load, etc. Twelve DEGs were shared between cohorts and were consistent in fold change direction. Perhaps the most interesting common DEG was corticotropin releasing hormone 1 (CRH). CRH has been a fundamental neurobiological correlate of stress disorders and MDD in particular for twenty years (Holsboer, 2000; Pariante and Miller, 2001). It has been extensively studied for its role in regulating HPA axis function in the stressed brain (Menke, 2019) and the robustness of these findings are highlighted by its appearance in the two largely disparate datasets. On the other hand, a separate PTSD transcriptomic study found in this issue (Logue et al., 2021) compared expression changes between their PTSD cohort and the Girgenti PTSD cohort. 50% of this cohort overlaps with donors in the Girgenti study and they found that 17% of their nominally significant DEGs overlapped with the Girgenti cohort- significantly greater than would be expected by chance. Additionally, the Girgenti cohort is made up of tissue from two brain banks (UPMC and the VA National PTSD Brain Bank) and despite no overlap between donors in the UPMC group, the Logue cohort had significant overlap in DEGs with this group alone. Further, the Logue cohort found significant global correlation (r = 0.75, p < 2.2 × 10−16) overall between their transcriptomic signature and the Girgenti cohort. The strong correlation between two PTSD transcriptomic data sets versus weak correlation between two MDD transcriptomic datasets implies greater differences not only between MDD and PTSD as disorders but also in the way MDD is diagnosed or presents and suggests a need to better evaluate how we diagnose depression in future brain bank collections.
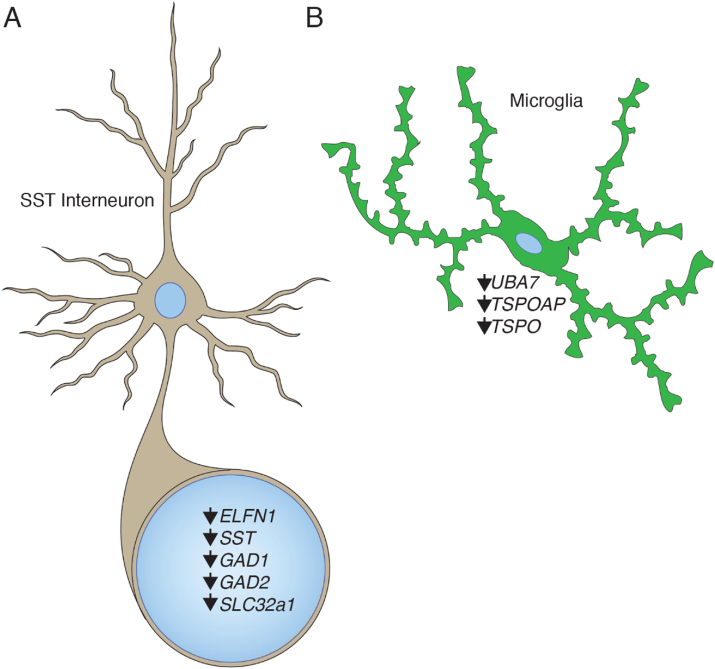
Fig. 1.
Cell type-specific transcriptomic changes in PTSD frontal cortex. A. GABA-related key drivers and transcripts exhibiting regional (DLPFC) down regulation in PTSD postmortem brain. B. Microglial marker genes are down regulated in PTSD frontal cortex.
There is considerable evidence for the role of microglia in many neural processes including the stress response (Wohleb et al., 2016). Chronic stress induced overactivation of microglia has been implicated in reduced neurogenesis in the hippocampus and in synaptic protein reduction in prefrontal cortex of subjects with MDD (Wohleb, 2016). A role for microglial dysfunction in PTSD has been suspected based on blood transcriptomic work identifying inflammatory and immune related gene dysregulation (Passos et al., 2015). Recent work has identified dysregulation of microglia in the CNS PTSD transcriptome. The immune gene UBA-7 was identified by TWAS as associating with PTSD and is a significant transcriptomic key driver in females with PTSD (Girgenti et al., 2021)(Fig. 1). Another study using this same dataset stratified their cohorts (PTSD and neurotypical controls) by normal and high BMI for each sex (Stone et al., 2020). They identified numerous DEGs across comparisons and identified the cytokine IL-1B as a putative upstream regulator of transcription in PTSD males with high BMI. Previous work in the periphery of PTSD subjects has identified regulation of IL-1B (Passos et al., 2015). However, high BMI alone has not been shown to regulate IL-1B levels in human prefrontal cortex (Lauridsen et al., 2017), suggesting a possible molecular intersection between PTSD and BMI in human brain and may imply further functional implications as genetic variation in IL-1B has been linked to risk of PTSD in males (Hovhannisyan et al., 2017). Taken together, these transcriptomic findings point to vulnerabilities in neuroimmune function as promoting behavioral and neurobiological consequences of stress disorders.
7. Cell-type specificity of stress induced transcriptomic changes
One of the goals of gene regulation biology is to understand the contribution individual cell types have in regulating the transcriptome. The human brain is made of many diverse cell types each contributing its own unique molecular signature to a “bulk-tissue” RNA-seq experiment (Newman et al., 2019). One possible confound is that shifts in cell type proportions may accompany a particular neuropsychiatric disorder and that may affect our ability to translate a particular gene expression difference. There are currently several methods for cell type deconvolution of gene expression from bulk-tissue RNA-seq data including CIBERSORTx (Newman et al., 2019), Braininablender (Hagenauer et al., 2018), and Bisque(Jew et al., 2020). These programs quantify relative population proportions from bulk tissue transcriptomics by using cell type-specific expression profiles derived from single -cell/nuclei sequencing data from specific regions as a background dataset. CIBERSORTx was used in a study examining the transcriptome of the PTSD frontal cortex (Girgenti et al., 2021). The study identified regional changes in cell type proportions, most notably a significant increase in excitatory neurons and a decrease in microglia.
Bioinformatic inference of cell type proportions and gene expression from bulk-tissue RNA-seq is limited, however. There are subtypes of interneurons and layer specific excitatory neurons that are generally missed and undoubtedly reflect important information on how gene regulation is altered in a given disorder. To overcome this hurdle several laser cell-capture microscopy techniques (LCM) have been developed. A recent LCM study of gene expression isolated granule cells from the dentate gyrus in a large cohort of schizophrenia, bipolar disorder, and major depressive postmortem tissue (Jaffe et al., 2020). Few MDD DEGs (7) were identified. The authors then compared a subset of their MDD cohort treated with an SSRI to those that were not. Interestingly, none of the DEGs identified were involved in adult neurogenesis, a process where there is considerable evidence of SSRI's function in treating depression (Eisch and Petrik, 2012). This finding and the relatively few genes identified across all diagnostic groups, highlights the limits of this technology. In addition, LCM suffers from antibody specificity issues and throughput- while it is relatively easy to isolate granule cells in the hippocampus it is much more difficult to adopt the described LCM methods to a more cell-diffuse region such as the prefrontal cortex.
To more robustly identify cell type-specific changes, isolation of individual cells or their nuclei is necessary for transcriptomic interrogation. Single-nuclei transcriptomics (snRNA-seq) is an emerging technology that has been used to study postmortem brains from several neuropsychiatric and neurological disorders however, there are currently few studies that have used this on postmortem tissue of stress disorders. The first study using postmortem dlPFC of donors with MDD used snRNA-seq on 80,000 nuclei from 17 MDD donors and 17 normal controls (Nagy et al., 2020). The study used 10X Genomics Chromium version 2 and isolated mRNA from approximately 3000 nuclei per sample. They identified 26 clusters (transcriptional cell types) with most (60%) having significant numbers of DEGs. They identified 96 DEGs almost 50% of which occurred in the excitatory neuron and oligodendrocyte precursor cells (OPC) clusters. Three DEGs had been identified in previous MDD postmortem studies (FADS2, CKB, and KAZN) and 26 have been previously linked to mental illness including the synaptic genes GRIN2A and Synapsin 1. The authors point out that cell-type proportions are difficult to estimate from snRNA-seq datasets in large part because of the inconsistency in dissections. However, deconvolution of a bulk-tissue RNA-seq dataset from a separate, large MDD cohort did find reductions in the oligodendrocyte and OPC populations of the dlPFC(Girgenti et al., 2021). This study (Nagy et al., 2020) was the first to use single cell type gene expression profiling and sets the groundwork necessary for future studies focused on disentangling the role of each cell type in the diseased and neurotypical brain.
8. Integration of neuroimaging and transcriptomics of stress disorders
Human neuroimaging using functional magnetic resonance imaging (MRI) and positron-emission tomography (PET) scanning have proved indispensable to understanding how brain regions are connected and formed and the role that particular molecules play in disease state. This interest has led to the creation of large consortia, such as ENIGMA whose goal is correlating genetic variation with image-derived phenotypes (Thompson et al., 2020). The ENIGMA consortium has made substantial progress in linking neuroimaging derived structural changes with genetic research through over 200 studies across a wide-array of neuropsychiatric disorders.
There is currently much interest in integrating neuroimaging with gene expression states rather than allelic variation (Arnatkevic̆;iūtė et al., 2019; Fornito et al., 2018). Multiple factors outside of genetic architecture can affect a genes activity and as a result, the mechanisms through which variants influence how a phenotype manifests are unclear. This work correlates gene expression levels with variation in one or more imaging-derived phenotypes. Modern advances in large gene expression assays have allowed these studies to move from single gene correlates to genome-wide level. Gene expression assays provide a direct measure of genes activity and combined with neuroimaging can help resolve how spatial variation on the molecular level manifests on the structural level. Large multi-region transcriptomic atlases such as the Allen Human Brain Atlas which is comprised of measures for more than 20,000 genes from over 3702 spatially distinct brain tissue samples offers the greatest range of coverage for healthy brain gene expression. There is now unprecedented capacity to link gene function to structural brain organization particularly as relates to canonical resting-state networks; fiber connections between discrete regions; and perhaps most importantly for this review pathological changes in brain disorders.
Recent studies linking stress disorders by integrating human imaging with postmortem brain transcriptomics has been quite fruitful in advancing our understanding of the neurobiology of major depression and PTSD. Indeed, work in this area has shown that there is significantly higher cortical mGluR5 availability in PTSD in vivo using a radioligand that binds to mGluR5 protein (Holmes et al., 2017). Dysfunction of the glutamate system has been implicated in trauma and stress psychopathology. Up regulation of the transcript SHANK1, a mGluR5 cell membrane anchor was identified in a concurrent postmortem transcriptomic study. Another recent study using PET imaging of the microglial marker TSPO found prefrontal-limbic availability was lower in patients with PTSD (Bhatt et al., 2020). The TSPO and microglial-associated genes TSPOAP1 (an upstream regulator of TSPO) and TNFRSF14 were found to be down regulated in postmortem PTSD cortex (Fig. 1). These findings suggest that PTSD is associated with suppression of the immune response (through microglia) and not through neuroimmune activation as previously thought. Taken together, these studies highlight how combining neuroimaging with gene expression studies can link structural findings to a molecular mechanisms.
9. Conclusion
Transcriptomic studies have provided critical information on the pathophysiology of stress disorders. While genetic studies have illuminated the inherited genetic risks for these disorders, transcriptomics has provided a window into understanding the functional output of these risks and of the illness itself. Our current understanding of the molecular pathology of MDD suggests disruptions in GABAergic signaling, specifically in the SST interneuron subtype (Bajbouj et al., 2006; Levinson et al., 2010; Sanacora et al., 1999). SST expression is reduced in the frontal cortex and other regions and its reduction appears to correlate with symptom severity (Seney et al., 2015; Tripp et al., 2011). SST expression is also reduced in the frontal cortex of PTSD subjects and suggests a molecular intersection between these two highly comorbid disorders. Further, neuroimmune dysfunction has been reported in both MDD and PTSD though neuroimaging of live subjects and suggests that this occurs in opposing directions, with MDD being characterized with increases in inflammatory signaling and PTSD with immune suppression and reduced microglial signaling (Bhatt et al., 2020). It should be noted that there are currently no strongly identified genetic risk loci associated with GABAergic signaling or microglia, highlighting the need to move genetic work to functional genomic studies.
Future work in the transcriptomics of stress disorders should be focused in three directions: single cell RNA-sequencing, multi -omics integration and comparative overlap of animal models with postmortem findings. The brain is comprised of a myriad of cell types and it is critical that we understand how these individual cells contribute to the mechanistic function of the cell types involved, as some gene pathways may have effects in only single cell types or opposite effects in different cell types. Bulk-tissue transcriptomics would miss the roles of such genes and pathways. Multi-omic approaches should be explored as well. Currently, most genetic variants fall within non-coding regions and their impact on the transcriptome and by extension cell biology is obscured. snATAC-seq and HiC assays will be crucial for understanding how the genomic and nuclear structure of the cell regulates the transcriptome. Spatial transcriptomics is emerging as one of the most promising genomic technologies. The spatial organization of the brain defines many of its functions with different regions exhibiting differing patterns of cell morphology and physiology. Outside approaches such 3-dimensional organoids and exosomes will also be critical as will identifying the animal models with the closest molecular changes to those measured in human (postmortem) brain. There is much more to be gained through transcriptomic work in the field of stress-disorders and basic research in this area is needed to identify promising therapeutic targets.
CRediT authorship contribution statement
Jing Zhang: Writing – review & editing. Alfred P. Kaye: Writing – original draft. Jiawei Wang: Writing – original draft. Matthew J. Girgenti: Conceptualization, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors report no financial or biomedical COIs.
Acknowledgements
This work was supported by the Department of Veterans Affairs, Veteran Health Administration, VISN1 Career Development Award (M.J.G). The Glenn H. Greenberg Fund for Research on Stress and Resilience, 10x Genomics Pilot Grant (A.P.K.). The National Center for PTSD (A.P.K. and M.J.G), and Brain and Behavior Research Foundation Young Investigator Awards (A.P.K. and M.J.G.), and NIMH (MH123896 to J.Z.).The views expressed here are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs (VA) or the U.S. government. The data used for the analyses described in this manuscript were obtained from Girgenti et al. (Girgenti et al., 2021).The authors declare no biomedical financial interests or potential conflicts of interest.
References
- Adhya D., Swarup V., Nagy R., Dutan L., Shum C., Valencia-Alarcón E.P., Jozwik K.M., Mendez M.A., Horder J., Loth E., Nowosiad P., Lee I., Skuse D., Flinter F.A., Murphy D., McAlonan G., Geschwind D.H., Price J., Carroll J., Srivastava D.P., Baron-Cohen S. Atypical neurogenesis in induced pluripotent stem cells from autistic individuals. Biol. Psychiatr. 2021;89:486–496. doi: 10.1016/j.biopsych.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnatkevic̆iūtė, A. Fulcher B.D., Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage. 2019;189:353–367. doi: 10.1016/j.neuroimage.2019.01.011. [DOI] [PubMed] [Google Scholar]
- Bagot R.C., Cates H.M., Purushothaman I., Lorsch Z.S., Walker D.M., Wang J., Huang X., Schlüter O.M., Maze I., Peña C.J., Heller E.A., Issler O., Wang M., Song W., Stein JasonL., Liu X., Doyle M.A., Scobie K.N., Sun H.S., Neve R.L., Geschwind D., Dong Y., Shen L., Zhang B., Nestler E.J. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais A.S., Kostka D. Scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2019;36:1150–1158. doi: 10.1093/bioinformatics/btz698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajbouj M., Lisanby S.H., Lang U.E., Danker-Hopfe H., Heuser I., Neu P. Evidence for impaired cortical inhibition in patients with unipolar major depression. Biol. Psychiatr. 2006;59:395–400. doi: 10.1016/j.biopsych.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein N.J., Fong N.L., Lam I., Roy M.A., Hendrickson D.G., Kelley D.R. Solo: doublet identification in single-cell RNA-seq via semi-supervised deep learning. Cell Syst. 2020;11:95–101. doi: 10.1016/j.cels.2020.05.010. e5. [DOI] [PubMed] [Google Scholar]
- Bhatt S., Hillmer A.T., Girgenti M.J., Rusowicz A., Kapinos M., Nabulsi N., Huang Y., Matuskey D., Angarita G.A., Esterlis I., Davis M.T., Southwick S.M., Friedman M.J., Girgenti M.J., Friedman M.J., Duman R.S., Krystal J.H., Duman R.S., Carson R.E., Krystal J.H., Pietrzak R.H., Cosgrove K.P. PTSD is associated with neuroimmune suppression: evidence from PET imaging and postmortem transcriptomic studies. Nat. Commun. 2020;11:2360. doi: 10.1038/s41467-020-15930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouland G.A., Mahfouz A., Reinders M.J.T. Differential dropout analysis captures biological variation in single-cell RNA sequencing data. Biorxiv. 2021 doi: 10.1101/2021.02.01.429187. 2021, 02.01.429187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Bryois J., Garrett M.E., Song L., Safi A., Giusti-Rodriguez P., Johnson G.D., Shieh A.W., Buil A., Fullard J.F., Roussos P., Sklar P., Akbarian S., Haroutunian V., Stockmeier C.A., Wray G.A., White K.P., Liu C., Reddy T.E., Ashley-Koch A., Sullivan P.F., Crawford G.E. Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat. Commun. 2018;9:3121. doi: 10.1038/s41467-018-05379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M., Miao Z., Wolf F.A., Teichmann S.A., Theis F.J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods. 2019;16:43–49. doi: 10.1038/s41592-018-0254-1. [DOI] [PubMed] [Google Scholar]
- Cano-Gamez E., Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.E., Stumpf M.P.H., Babtie A.C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 2017;5:251–267. doi: 10.1016/j.cels.2017.08.014. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzinakos C., Georgiadis F., Daskalakis N.P. GWAS meets transcriptomics: from genetic letters to transcriptomic words of neuropsychiatric risk. Neuropsychopharmacology. 2021;46:255–256. doi: 10.1038/s41386-020-00835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.K., Yu U., Yoo O.J., Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- Chung D.W., Chung Y., Bazmi H.H., Lewis D.A. Altered ErbB4 splicing and cortical parvalbumin interneuron dysfunction in schizophrenia and mood disorders. Neuropsychopharmacology. 2018;43:2478–2486. doi: 10.1038/s41386-018-0169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C.D., Magariños A.M., LeDoux J.E., McEwen B.S. Repeated restraint stress facilitates fear conditioning independently of causing hippocampal CA3 dendritic atrophy. Behav. Neurosci. 1999;113:902–913. doi: 10.1037/0735-7044.113.5.902. [DOI] [PubMed] [Google Scholar]
- Consortium T.B., Jaffe A.E., Straub R.E., Shin J.H., Tao R., Gao Y., Collado-Torres L., Kam-Thong T., Xi H.S., Quan J., Chen Q., Colantuoni C., Ulrich W.S., Maher B.J., Deep-Soboslay A., Cross A.J., Brandon N.J., Leek J.T., Hyde T.M., Kleinman J.E., Weinberger D.R. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 2018;21:1117–1125. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Gephart M.G.H., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G., Bupp S., Shrestha P., Shah R.D., Doughty M.L., Gong S., Greengard P., Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2009;139:1022. doi: 10.1016/j.cell.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V., Banasr M., Licznerski P., Schmidt H.D., Stockmeier C.A., Simen A.A., Newton S.S., Duman R.S. A negative regulator of MAP kinase causes depressive behavior. Nat. Med. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Eisch A.J., Petrik D. Depression and hippocampal neurogenesis: a road to remission? Science. 2012;338:72–75. doi: 10.1126/science.1222941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estill M., Ribeiro E., Francoeur N.J., Smith M.L., Sebra R., Yeh S.-Y., Cunningham A.M., Nestler E.J., Shen L. Long read, isoform aware sequencing of mouse nucleus accumbens after chronic cocaine treatment. Sci Rep-uk. 2021;11:6729. doi: 10.1038/s41598-021-86068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee C., Banasr M., Sibille E. Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: cortical microcircuit and therapeutic perspectives. Biol. Psychiatr. 2017;82:549–559. doi: 10.1016/j.biopsych.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., Linsley P.S., Gottardo R. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flati T., Gioiosa S., Chillemi G., Mele A., Oliverio A., Mannironi C., Rinaldi A., Castrignanò T. A gene expression atlas for different kinds of stress in the mouse brain. Sci Data. 2020;7:437. doi: 10.1038/s41597-020-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floriou-Servou A., Ziegler L. von, Stalder L., Sturman O., Privitera M., Rassi A., Cremonesi A., Thöny B., Bohacek J. Distinct proteomic, transcriptomic, and epigenetic stress responses in dorsal and ventral Hippocampus. Biol. Psychiatr. 2018;84:531–541. doi: 10.1016/j.biopsych.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Floriou-Servou A., Ziegler L. von, Waag R., Schläppi C., Germain P.-L., Bohacek J. The acute stress response in the multi-omic era. Biol. Psychiatr. 2021 doi: 10.1016/j.biopsych.2020.12.031. [DOI] [PubMed] [Google Scholar]
- Flory J.D., Yehuda R. Comorbidity between post-traumatic stress disorder and major depressive disorder: alternative explanations and treatment considerations. Dialogues Clin. Neurosci. 2015;17:141–150. doi: 10.31887/DCNS.2015.17.2/jflory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A., Arnatkevičiūtė A., Fulcher B.D. Bridging the gap between connectome and transcriptome. Trends Cognit. Sci. 2018;23:34–50. doi: 10.1016/j.tics.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R., Klei L.L., Kramer R., Pinto D., Gümüş Z.H., Cicek A.E., Dang K.K., Browne A., Lu C., Xie L., Readhead B., Stahl E.A., Xiao J., Parvizi M., Hamamsy T., Fullard J.F., Wang Y.-C., Mahajan M.C., Derry J.M.J., Dudley J.T., Hemby S.E., Logsdon B.A., Talbot K., Raj T., Bennett D.A., Jager P.L.D., Zhu J., Zhang B., Sullivan P.F., Chess A., Purcell S.M., Shinobu L.A., Mangravite L.M., Toyoshiba H., Gur R.E., Hahn C.-G., Lewis D.A., Haroutunian V., Peters M.A., Lipska B.K., Buxbaum J.D., Schadt E.E., Hirai K., Roeder K., Brennand K.J., Katsanis N., Domenici E., Devlin B., Sklar P. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher B.D., Arnatkevičiūtė A., Fornito A. Overcoming bias in gene-set enrichment analyses of brain-wide transcriptomic data. Biorxiv. 2020 doi: 10.1101/2020.04.24.058958. 2020, 04.24. [DOI] [Google Scholar]
- Gandal M.J., Haney J.R., Parikshak N.N., Leppa V., Ramaswami G., Hartl C., Schork A.J., Appadurai V., Buil A., Werge T.M., Liu C., White K.P., Consortium C., Consortium P., Group, iPSYCH-B.W. Horvath S., Geschwind D.H. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal M.J., Zhang P., Hadjimichael E., Walker R.L., Chen C., Liu S., Won H., Bakel H. van, Varghese M., Wang Y., Shieh A.W., Haney J., Parhami S., Belmont J., Kim M., Losada P.M., Khan Z., Mleczko J., Xia Y., Dai R., Wang D., Yang Y.T., Xu M., Fish K., Hof P.R., Warrell J., Fitzgerald D., White K., Jaffe A.E., Consortium P., Peters M.A., Gerstein M., Liu C., Iakoucheva L.M., Pinto D., Geschwind D.H. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362 doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J., Sun N., Polimanti R., Pietrzak R., Levey D.F., Bryois J., Lu Q., Hu Y., Li B., Radhakrishnan K., Aslan M., Cheung K.-H., Li Y., Rajeevan N., Sayward F., Harrington K., Chen Q., Cho K., Pyarajan S., Sullivan P.F., Quaden R., Shi Y., Hunter-Zinck H., Gaziano J.M., Concato J., Zhao H., Stein M.B., Program, D. of V.A.C.S.P. (#575B) and M.V. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat. Neurosci. 2019;22:1394–1401. doi: 10.1038/s41593-019-0447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti M.J., Wang J., Ji D., Cruz D.A., Alvarez V.E., Benedek D., Brady C., Davis D.A., Holtzheimer P.E., Keane T.M., Kowell N., Logue M.W., McKee A., Marx B., Mash D., Miller M.W., Scott W.K., Stein T., Ursano R., Wolf E.J., Stein M.B., Gelernter J., Young K.A., Huber B.R., Williamson D.E., Friedman M.J., Krystal J.H., Zhao H., Duman R.S. Transcriptomic organization of the human brain in post-traumatic stress disorder. Nat. Neurosci. 2021;24:24–33. doi: 10.1038/s41593-020-00748-7. [DOI] [PubMed] [Google Scholar]
- Girgenti M.J., Wohleb E.S., Mehta S., Ghosal S., Fogaca M.V., Duman R.S. Prefrontal cortex interneurons display dynamic sex-specific stress-induced transcriptomes. Transl. Psychiatry. 2019;9:292. doi: 10.1038/s41398-019-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.A., Covington H.E., Berton O., Russo S.J. A standardized protocol for repeated social defeat stress in mice. Nat. Protoc. 2011;6:1183–1191. doi: 10.1038/nprot.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenauer M.H., Schulmann A., Li J.Z., Vawter M.P., Walsh D.M., Thompson R.C., Turner C.A., Bunney W.E., Myers R.M., Barchas J.D., Schatzberg A.F., Watson S.J., Akil H. Inference of cell type content from human brain transcriptomic datasets illuminates the effects of age, manner of death, dissection, and psychiatric diagnosis. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B.P., Shehata S.I., Aevermann B., Beller A., Bertagnolli D., Brouner K., Casper T., Cobbs C., Dalley R., Dee N., Ding S.-L., Ellenbogen R.G., Fong O., Garren E., Goldy J., Gwinn R.P., Hirschstein D., Keene C.D., Keshk M., Ko A.L., Lathia K., Mahfouz A., Maltzer Z., McGraw M., Nguyen T.N., Nyhus J., Ojemann J.G., Oldre A., Parry S., Reynolds S., Rimorin C., Shapovalova N.V., Somasundaram S., Szafer A., Thomsen E.R., Tieu M., Quon G., Scheuermann R.H., Yuste R., Sunkin S.M., Lelieveldt B., Feng D., Ng L., Bernard A., Hawrylycz M., Phillips J.W., Tasic B., Zeng H., Jones A.R., Koch C., Lein E.S. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S.E., Girgenti M.J., Davis M.T., Pietrzak R.H., DellaGioia N., Nabulsi N., Matuskey D., Southwick S., Duman R.S., Carson R.E., Krystal J.H., Esterlis I., Group, the T.S.B.S. Altered metabotropic glutamate receptor 5 markers in PTSD: in vivo and postmortem evidence. Proc. Natl. Acad. Sci. U.S.A. 2017;114(31):8390–8395. doi: 10.1038/nprot.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/s0893-133x(00)00159-7. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan L., Stepanyan A., Arakelyan A. Genetic variability of interleukin-1 beta as prospective factor from developing post-traumatic stress disorder. Immunogenetics. 2017;69:703–708. doi: 10.1007/s00251-017-1016-4. [DOI] [PubMed] [Google Scholar]
- Huckins L.M., Chatzinakos C., Breen M.S., Hartmann J., Klengel T., Almeida A.C. da S., Dobbyn A., Girdhar K., Hoffman G.E., Klengel C., Logue M.W., Lori A., Maihofer A.X., Morrison F.G., Nguyen H.T., Park Y., Ruderfer D., Sloofman L.G., Rooij S.J.H. van, Consortium, P.W.G. of P.G. Baker D.G., Chen C.-Y., Cox N., Duncan L.E., Geyer M.A., Glatt S.J., Im H.K., Risbrough V.B., Smoller J.W., Stein D.J., Yehuda R., Liberzon I., Koenen K.C., Jovanovic T., Kellis M., Miller M.W., Bacanu S.-A., Nievergelt C.M., Buxbaum J.D., Sklar P., Ressler K.J., Stahl E.A., Daskalakis N.P. Analysis of genetically regulated gene expression identifies a prefrontal PTSD gene, SNRNP35, specific to military cohorts. Cell Rep. 2020;31:107716. doi: 10.1016/j.celrep.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E., Hoeppner D.J., Saito T., Blanpain L., Ukaigwe J., Burke E.E., Collado-Torres L., Tao R., Tajinda K., Maynard K.R., Tran M.N., Martinowich K., Deep-Soboslay A., Shin J.H., Kleinman J.E., Weinberger D.R., Matsumoto M., Hyde T.M. Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat. Neurosci. 2020;23:510–519. doi: 10.1038/s41593-020-0604-z. [DOI] [PubMed] [Google Scholar]
- James G.D., Ice G.H. 2006. Measuring stress in humans; pp. 246–265. [DOI] [Google Scholar]
- Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11:1971. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T., Norrholm S.D., Blanding N.Q., Davis M., Duncan E., Bradley B., Ressler K.J. Impaired fear inhibition is a biomarker of PTSD but not depression. Depress. Anxiety. 2010;27:244–251. doi: 10.1002/da.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T., Ressler K.J. How the neurocircuitry and genetics of fear inhibition may inform our understanding of PTSD. Am. J. Psychiatr. 2010;167:648–662. doi: 10.1176/appi.ajp.2009.09071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.J., Voleti B., Hajszan T., Rajkowska G., Stockmeier C.A., Licznerski P., Lepack A., Majik M.S., Jeong L.S., Banasr M., Son H., Duman R.S. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat. Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R.C., Sonnega A., Bromet E., Hughes M., Nelson C.B. Posttraumatic stress disorder in the national comorbidity survey. Arch. Gen. Psychiatr. 1995;52:1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijn K.M.A.D., Zuure W.A., Peijnenborg J., Heuvelmans J.M., Martens G.J.M. Reappraisal of human HOG and MO3.13 cell lines as a model to study oligodendrocyte functioning. Cells. 2019;8:1096. doi: 10.3390/cells8091096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Amin V., Whitaker J.W., Schultz M.D., Ward L.D., Sarkar A., Quon G., Sandstrom R.S., Eaton M.L., Wu Y.-C., Pfenning A., Wang X., Liu M.C., Coarfa C., Harris R.A., Shoresh N., Epstein C.B., Gjoneska E., Leung D., Xie W., Hawkins R.D., Lister R., Hong C., Gascard P., Mungall A.J., Moore R., Chuah E., Tam A., Canfield T.K., Hansen R.S., Kaul R., Sabo P.J., Bansal M.S., Carles A., Dixon J.R., Farh K.-H., Feizi S., Karlic R., Kim A.-R., Kulkarni A., Li D., Lowdon R., Elliott G., Mercer T.R., Neph S.J., Onuchic V., Polak P., Rajagopal N., Ray P., Sallari R.C., Siebenthall K.T., Sinnott-Armstrong N.A., Stevens M., Thurman R.E., Wu J., Zhang B., Zhou X., Abdennur N., Adli M., Akerman M., Barrera L., Antosiewicz-Bourget J., Ballinger T., Barnes M.J., Bates D., Bell R.J.A., Bennett D.A., Bianco K., Bock C., Boyle P., Brinchmann J., Caballero-Campo P., Camahort R., Carrasco-Alfonso M.J., Charnecki T., Chen H., Chen Z., Cheng J.B., Cho S., Chu A., Chung W.-Y., Cowan C., Deng Q.A., Deshpande V., Diegel M., Ding B., Durham T., Echipare L., Edsall L., Flowers D., Genbacev-Krtolica O., Gifford C., Gillespie S., Giste E., Glass I.A., Gnirke A., Gormley M., Gu H., Gu J., Hafler D.A., Hangauer M.J., Hariharan M., Hatan M., Haugen E., He Y., Heimfeld S., Herlofsen S., Hou Z., Humbert R., Issner R., Jackson A.R., Jia H., Jiang P., Johnson A.K., Kadlecek T., Kamoh B., Kapidzic M., Kent J., Kim A., Kleinewietfeld M., Klugman S., Krishnan J., Kuan S., Kutyavin T., Lee A.-Y., Lee K., Li J., Li N., Li Y., Ligon K.L., Lin S., Lin Y., Liu J., Liu Yuxuan, Luckey C.J., Ma Y.P., Maire C., Marson A., Mattick J.S., Mayo M., McMaster M., Metsky H., Mikkelsen T., Miller D., Miri M., Mukame E., Nagarajan R.P., Neri F., Nery J., Nguyen T., O'Geen H., Paithankar S., Papayannopoulou T., Pelizzola M., Plettner P., Propson N.E., Raghuraman S., Raney B.J., Raubitschek A., Reynolds A.P., Richards H., Riehle K., Rinaudo P., Robinson J.F., Rockweiler N.B., Rosen E., Rynes E., Schein J., Sears R., Sejnowski T., Shafer A., Shen L., Shoemaker R., Sigaroudinia M., Slukvin I., Stehling-Sun S., Stewart R., Subramanian S.L., Suknuntha K., Swanson S., Tian S., Tilden H., Tsai L., Urich M., Vaughn I., Vierstra J., Vong S., Wagner U., Wang H., Wang Tao, Wang Y., Weiss A., Whitton H., Wildberg A., Witt H., Won K.-J., Xie M., Xing X., Xu I., Xuan Z., Ye Z., Yen C., Yu P., Zhang Xian, Zhang Xiaolan, Zhao J., Zhou Y., Zhu J., Zhu Y., Ziegler S., Beaudet A.E., Boyer L.A., Jager P.L.D., Farnham P.J., Fisher S.J., Haussler D., Jones S.J.M., Li W., Marra M.A., McManus M.T., Sunyaev S., Thomson J.A., Tlsty T.D., Tsai L.-H., Wang W., Waterland R.A., Zhang M.Q., Chadwick L.H., Bernstein B.E., Costello J.F., Ecker J.R., Hirst M., Meissner A., Milosavljevic A., Ren B., Stamatoyannopoulos J.A., Wang Ting, Kellis M., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Amin V., Whitaker J.W., Schultz M.D., Ward L.D., Sarkar A., Quon G., Sandstrom R.S., Eaton M.L., Wu Y.-C., Pfenning A.R., Wang X., Claussnitzer M., Liu Yaping, Coarfa C., Harris R.A., Shoresh N., Epstein C.B., Gjoneska E., Leung D., Xie W., Hawkins R.D., Lister R., Hong C., Gascard P., Mungall A.J., Moore R., Chuah E., Tam A., Canfield T.K., Hansen R.S., Kaul R., Sabo P.J., Bansal M.S., Carles A., Dixon J.R., Farh K.-H., Feizi S., Karlic R., Kim A.-R., Kulkarni A., Li D., Lowdon R., Elliott G., Mercer T.R., Neph S.J., Onuchic V., Polak P., Rajagopal N., Ray P., Sallari R.C., Siebenthall K.T., Sinnott-Armstrong N.A., Stevens M., Thurman R.E., Wu J., Zhang B., Zhou X., Beaudet A.E., Boyer L.A., Jager P.L.D., Farnham P.J., Fisher S.J., Haussler D., Jones S.J.M., Li W., Marra M.A., McManus M.T., Sunyaev S., Thomson J.A., Tlsty T.D., Tsai L.-H., Wang W., Waterland R.A., Zhang M.Q., Chadwick L.H., Bernstein B.E., Costello J.F., Ecker J.R., Hirst M., Meissner A., Milosavljevic A., Ren B., Stamatoyannopoulos J.A., Wang Ting, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B., Engmann O., Purushothaman I., Menard C., Wang J., Tan C., Scarpa J.R., Moy G., Loh Y.-H.E., Cahill M., Lorsch Z.S., Hamilton P.J., Calipari E.S., Hodes G.E., Issler O., Kronman H., Pfau M., Obradovic A.L.J., Dong Y., Neve R.L., Russo S., Kasarskis A., Tamminga C., Mechawar N., Turecki G., Zhang B., Shen L., Nestler E.J. Sex-specific transcriptional signatures in human depression. Nat. Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B.B., Ai R., Kaeser G.E., Salathia N.S., Yung Y.C., Liu R., Wildberg A., Gao D., Fung H.-L., Chen S., Vijayaraghavan R., Wong J., Chen A., Sheng X., Kaper F., Shen R., Ronaghi M., Fan J.-B., Wang W., Chun J., Zhang K. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., Duong T.E., Gao D., Chun J., Kharchenko P.V., Zhang K. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 2018;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz P.M., House J.S., Mero R.P., Williams D.R. Stress, life events, and socioeconomic disparities in health: results from the Americans' changing lives study. J. Health Soc. Behav. 2005;46:274–288. doi: 10.1177/002214650504600305. [DOI] [PubMed] [Google Scholar]
- Lauridsen J.K., Olesen R.H., Vendelbo J., Hyde T.M., Kleinman J.E., Bibby B.M., Brock B., Rungby J., Larsen A. High BMI levels associate with reduced mRNA expression of IL10 and increased mRNA expression of iNOS (NOS2) in human frontal cortex. Transl. Psychiatry. 2017;7 doi: 10.1038/tp.2016.259. e1044–e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson A.J., Fitzgerald P.B., Favalli G., Blumberger D.M., Daigle M., Daskalakis Z.J. Evidence of cortical inhibitory deficits in major depressive disorder. Biol. Psychiatr. 2010;67:458–464. doi: 10.1016/j.biopsych.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznerski P., Duric V., Banasr M., Alavian K.N., Ota K.T., Kang H.J., Jonas E.A., Ursano R., Krystal J.H., Duman R.S., Group T.S.B.S. Decreased SGK1 expression and function contributes to behavioral deficits induced by traumatic stress. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue M.W., Zhou Z., Morrison F.G., Wolf E.J., Daskalakis N.P., Chatzinakos C., Georgiadis F., Labadorf A.T., Girgenti M.J., Young K.A., Williamson D.E., Zhao X., Grenier J.G., Group, the T.S.B.R. Huber B.R., Miller M.W. Gene expression in the dorsolateral and ventromedial prefrontal cortices implicates immune-related gene networks in PTSD. Neurobiology Stress. 2021;15:100398. doi: 10.1016/j.ynstr.2021.100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.P., Brivio E., Santambrogio A., Donno C.D., Kos A., Peters M., Rost N., Czamara D., Brückl T.M., Roeh S., Pöhlmann M.L., Engelhardt C., Ressle A., Stoffel R., Tontsch A., Villamizar J.M., Reincke M., Riester A., Sbiera S., Fassnacht M., Mayberg H.S., Craighead W.E., Dunlop B.W., Nemeroff C.B., Schmidt M.V., Binder E.B., Theis F.J., Beuschlein F., Andoniadou C.L., Chen A. Single-cell molecular profiling of all three components of the HPA axis reveals adrenal ABCB1 as a regulator of stress adaptation. Sci Adv. 2021;7 doi: 10.1126/sciadv.abe4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B., Shen Q., Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol. Psychiatr. 2011;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay I.C., Ponting C.P., Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33:155–168. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., Trombetta J.J., Weitz D.A., Sanes J.R., Shalek A.K., Regev A., McCarroll S.A. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Method of the Year: spatially resolved transcriptomics. Nat. Methods. 2021;18:9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- Mcewen B.S. Protection and damage from acute and chronic stress: allostasis and allostatic overload and relevance to the pathophysiology of psychiatric disorders. Ann Ny Acad Sci. 2004;1032:1–7. doi: 10.1196/annals.1314.001. [DOI] [PubMed] [Google Scholar]
- McEwen B.S. Central effects of stress hormones in health and disease: understanding the protective and damaging effects of stress and stress mediators. Eur. J. Pharmacol. 2008;583:174–185. doi: 10.1016/j.ejphar.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S., Nasca C., Gray J.D. Stress effects on neuronal structure: Hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology. 2016;41:3–23. doi: 10.1038/npp.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337. doi: 10.1016/j.cels.2019.03.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonagle K.A., Kessler R.C. Chronic stress, acute stress, and depressive symptoms. Am. J. Community Psychol. 1990;18:681–706. doi: 10.1007/bf00931237. [DOI] [PubMed] [Google Scholar]
- Menke A. Is the HPA Axis as target for depression outdated, or is there a new hope? Front. Psychiatr. 2019;10:101. doi: 10.3389/fpsyt.2019.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Deng K., Wang X., Zhang X. DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics. 2018;34:3223–3224. doi: 10.1093/bioinformatics/bty332. [DOI] [PubMed] [Google Scholar]
- Mitra R., Jadhav S., McEwen B.S., Vyas A., Chattarji S. Stress duration modulates the spatiotemporal patterns of spine formation in the basolateral amygdala. P Natl Acad Sci Usa. 2005;102:9371–9376. doi: 10.1073/pnas.0504011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C., Maitra M., Tanti A., Suderman M., Théroux J.-F., Davoli M.A., Perlman K., Yerko V., Wang Y.C., Tripathy S.J., Pavlidis P., Mechawar N., Ragoussis J., Turecki G. Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat. Neurosci. 2020;23:771–781. doi: 10.1038/s41593-020-0621-y. [DOI] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., Diehn M., Alizadeh A.A. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt C.M., Maihofer A.X., Klengel T., Atkinson E.G., Chen C.-Y., Choi K.W., Coleman J.R.I., Dalvie S., Duncan L.E., Gelernter J., Levey D.F., Logue M.W., Polimanti R., Provost A.C., Ratanatharathorn A., Stein M.B., Torres K., Aiello A.E., Almli L.M., Amstadter A.B., Andersen S.B., Andreassen O.A., Arbisi P.A., Ashley-Koch A.E., Austin S.B., Avdibegovic E., Babić D., Bækvad-Hansen M., Baker D.G., Beckham J.C., Bierut L.J., Bisson J.I., Boks M.P., Bolger E.A., Børglum A.D., Bradley B., Brashear M., Breen G., Bryant R.A., Bustamante A.C., Bybjerg-Grauholm J., Calabrese J.R., Almeida J.M.C.- de-, Dale A.M., Daly M.J., Daskalakis N.P., Deckert J., Delahanty D.L., Dennis M.F., Disner S.G., Domschke K., Dzubur-Kulenovic A., Erbes C.R., Evans A., Farrer L.A., Feeny N.C., Flory J.D., Forbes D., Franz C.E., Galea S., Garrett M.E., Gelaye B., Geuze E., Gillespie C., Uka A.G., Gordon S.D., Guffanti G., Hammamieh R., Harnal S., Hauser M.A., Heath A.C., Hemmings S.M.J., Hougaard D.M., Jakovljevic M., Jett M., Johnson E.O., Jones I., Jovanovic T., Qin X.-J., Junglen A.G., Karstoft K.-I., Kaufman M.L., Kessler R.C., Khan A., Kimbrel N.A., King A.P., Koen N., Kranzler H.R., Kremen W.S., Lawford B.R., Lebois L.A.M., Lewis C.E., Linnstaedt S.D., Lori A., Lugonja B., Luykx J.J., Lyons M.J., Maples-Keller J., Marmar C., Martin A.R., Martin N.G., Maurer D., Mavissakalian M.R., McFarlane A., McGlinchey R.E., McLaughlin K.A., McLean S.A., McLeay S., Mehta D., Milberg W.P., Miller M.W., Morey R.A., Morris C.P., Mors O., Mortensen P.B., Neale B.M., Nelson E.C., Nordentoft M., Norman S.B., O'Donnell M., Orcutt H.K., Panizzon M.S., Peters E.S., Peterson A.L., Peverill M., Pietrzak R.H., Polusny M.A., Rice J.P., Ripke S., Risbrough V.B., Roberts A.L., Rothbaum A.O., Rothbaum B.O., Roy-Byrne P., Ruggiero K., Rung A., Rutten B.P.F., Saccone N.L., Sanchez S.E., Schijven D., Seedat S., Seligowski A.V., Seng J.S., Sheerin C.M., Silove D., Smith A.K., Smoller J.W., Sponheim S.R., Stein D.J., Stevens J.S., Sumner J.A., Teicher M.H., Thompson W.K., Trapido E., Uddin M., Ursano R.J., Heuvel L.L. van den, Hooff M.V., Vermetten E., Vinkers C.H., Voisey J., Wang Y., Wang Z., Werge T., Williams M.A., Williamson D.E., Winternitz S., Wolf C., Wolf E.J., Wolff J.D., Yehuda R., Young R.McD., Young K.A., Zhao H., Zoellner L.A., Liberzon I., Ressler K.J., Haas M., Koenen K.C. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 2019;10:4558. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengeim D., Giagtzoglou N., Huh D., Zou C., Yuan J. Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol. Med. 2017;23:563–576. doi: 10.1016/j.molmed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M.C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., Geschwind D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K.T., Liu R.-J., Voleti B., Maldonado-Aviles J.G., Duric V., Iwata M., Dutheil S., Duman C., Boikess S., Lewis D.A., Stockmeier C.A., DiLeone R.J., Rex C., Aghajanian G.K., Duman R.S. REDD1 is essential for stress-induced synaptic loss and depressive behavior. Nat. Med. 2014;20:531–535. doi: 10.1038/nm.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pariante C.M., Miller A.H. Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol. Psychiatr. 2001;49:391–404. doi: 10.1016/s0006-3223(00)01088-x. [DOI] [PubMed] [Google Scholar]
- Passos I.C., Vasconcelos-Moreno M.P., Costa L.G., Kunz M., Brietzke E., Quevedo J., Salum G., Magalhães P.V., Kapczinski F., Kauer-Sant’Anna M. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry. 2015;2:1002–1012. doi: 10.1016/s2215-0366(15)00309-0. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C.J., Kronman H.G., Walker D.M., Cates H.M., Bagot R.C., Purushothaman I., Issler O., Loh Y.-H.E., Leong T., Kiraly D.D., Goodman E., Neve R.L., Shen L., Nestler E.J. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., McEwen B.S., Friston K. Uncertainty and stress: why it causes diseases and how it is mastered by the brain. Prog. Neurobiol. 2017;156:164–188. doi: 10.1016/j.pneurobio.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Peters M.L., Godaert G.L.R., Ballieux R.E., Vliet M. van, Willemsen J.J., Sweep F.C.G.J., Heijnen C.J. Cardiovascular and endocrine responses to experimental stress: effects of mental effort and controllability. Psychoneuro. 1998;23:1–17. doi: 10.1016/s0306-4530(97)00082-6. [DOI] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E.L. da, Rowe R.G., Lundin V., Malleshaiah M., Jha D.K., Rambo C.R., Li H., North T.E., Collins J.J., Daley G.Q. Reconstruction of complex single-cell trajectories using CellRouter. Nat. Commun. 2018;9:892. doi: 10.1038/s41467-018-03214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytwinski N.K., Scur M.D., Feeny N.C., Youngstrom E.A. The Co‐occurrence of major depressive disorder among individuals with posttraumatic stress disorder: a meta‐analysis. J. Trauma Stress. 2013;26:299–309. doi: 10.1002/jts.21814. [DOI] [PubMed] [Google Scholar]
- Sanacora G., Mason G.F., Rothman D.L., Behar K.L., Hyder F., Petroff O.A., Berman R.M., Charney D.S., Krystal J.H. Reduced cortical gamma-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Arch. Gen. Psychiatr. 1999;56:1043–1047. doi: 10.1001/archpsyc.56.11.1043. [DOI] [PubMed] [Google Scholar]
- Scarpa J., Fatma M., Loh Y.-H.E., Traore S.R., Stefan T., Chen T.H., Nestler E., Labonte B. Shared transcriptional signatures in major depressive disorder and mouse chronic stress models. Biol. Psychiatr. 2020;87:S222. doi: 10.1016/j.biopsych.2020.02.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S.H., Schmidt M.V. Animal models of stress vulnerability and resilience in translational research. Curr. Psychiatr. Rep. 2012;14:159–165. doi: 10.1007/s11920-012-0256-0. [DOI] [PubMed] [Google Scholar]
- Seligman M.E.P. Learned helplessness. Annu. Rev. Med. 1972;23:407–412. doi: 10.1146/annurev.me.23.020172.002203. [DOI] [PubMed] [Google Scholar]
- Selye H. A syndrome produced by diverse nocuous agents. J. Neuropsychiatry Clin. Neurosci. 1998;10:230a–2231. doi: 10.1176/jnp.10.2.230a. [DOI] [PubMed] [Google Scholar]
- Seney M.L., Huo Z., Cahill K., French L., Puralewski R., Zhang J., Logan R.W., Tseng G., Lewis D.A., Sibille E. Opposite molecular signatures of depression in men and women. Biol. Psychiatr. 2018;84:18–27. doi: 10.1016/j.biopsych.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seney M.L., Tripp A., McCune S., Lewis D.A., Sibille E. Laminar and cellular analyses of reduced somatostatin gene expression in the subgenual anterior cingulate cortex in major depression. Neurobiol. Dis. 2015;73:213–219. doi: 10.1016/j.nbd.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., Pe’er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahjaman Md, Mollah MdM.H., Rahman MdR., Islam S.M.S., Mollah MdN.H. Robust identification of differentially expressed genes from RNA-seq data. Genomics. 2020;112:2000–2010. doi: 10.1016/j.ygeno.2019.11.012. [DOI] [PubMed] [Google Scholar]
- Shao X., Liao J., Li C., Lu X., Cheng J., Fan X. CellTalkDB: a manually curated database of ligand–receptor interactions in humans and mice. Briefings Bioinf. 2020;22 doi: 10.1093/bib/bbaa269. [DOI] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G., Song H. Single-cell RNA-seq with Waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin L.M., Liberzon I. The neurocircuitry of fear, stress, and anxiety disorders. Neuropsychopharmacology. 2010;35:169–191. doi: 10.1038/npp.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons J.M., Winsky L., Zehr J.L., Gordon J.A. Priorities in stress research: a view from the U.S. National institute of mental health. Ann Ny Acad Sci. 2020:1–7. doi: 10.1080/10253890.2020.1781084. [DOI] [PubMed] [Google Scholar]
- Soumier A., Sibille E. Opposing effects of acute versus chronic blockade of frontal cortex somatostatin-positive inhibitory neurons on behavioral emotionality in mice. Neuropsychopharmacology. 2014;39:2252–2262. doi: 10.1038/npp.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M.B., Levey D.F., Cheng Z., Wendt F.R., Harrington K., Pathak G.A., Cho K., Quaden R., Radhakrishnan K., Girgenti M.J., Ho Y.-L.A., Posner D., Aslan M., Duman R.S., Zhao H., Program V.C.S., Program V.M.V., Polimanti R., Concato J., Gelernter J. Genomic characterization of posttraumatic stress disorder in a large US military veteran sample. Nat. Genet. 2021;2:174–184. doi: 10.1101/764001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone L.A., Girgenti M.J., Wang J., Ji D., Zhao H., Krystal J.H., Group T.S.B.R., Duman R.S. Cortical transcriptomic alterations in association with appetitive neuropeptides and body mass index in posttraumatic stress disorder. Int. J. Neuropsychopharmacol. 2020;24 doi: 10.1093/ijnp/pyaa072. pyaa072- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M., Jahanshad N., Ching C.R.K., Salminen L.E., Thomopoulos S.I., Bright J., Baune B.T., Bertolín S., Bralten J., Bruin W.B., Bülow R., Chen J., Chye Y., Dannlowski U., Kovel C.G.F. de, Donohoe G., Eyler L.T., Faraone S.V., Favre P., Filippi C.A., Frodl T., Garijo D., Gil Y., Grabe H.J., Grasby K.L., Hajek T., Han L.K.M., Hatton S.N., Hilbert K., Ho T.C., Holleran L., Homuth G., Hosten N., Houenou J., Ivanov I., Jia T., Kelly S., Klein M., Kwon J.S., Laansma M.A., Leerssen J., Lueken U., Nunes A., Neill J.O., Opel N., Piras Fabrizio, Piras Federica, Postema M.C., Pozzi E., Shatokhina N., Soriano-Mas C., Spalletta G., Sun D., Teumer A., Tilot A.K., Tozzi L., Merwe C. van der, Someren E.J.W.V., Wingen G.A. van, Völzke H., Walton E., Wang L., Winkler A.M., Wittfeld K., Wright M.J., Yun J.-Y., Zhang G., Zhang-James Y., Adhikari B.M., Agartz I., Aghajani M., Aleman A., Althoff R.R., Altmann A., Andreassen O.A., Baron D.A., Bartnik-Olson B.L., Bas-Hoogendam J.M., Baskin-Sommers A.R., Bearden C.E., Berner L.A., Boedhoe P.S.W., Brouwer R.M., Buitelaar J.K., Caeyenberghs K., Cecil C.A.M., Cohen R.A., Cole J.H., Conrod P.J., Brito S.A.D., Zwarte S.M.C. de, Dennis E.L., Desrivieres S., Dima D., Ehrlich S., Esopenko C., Fairchild G., Fisher S.E., Fouche J.-P., Francks C., Frangou S., Franke B., Garavan H.P., Glahn D.C., Groenewold N.A., Gurholt T.P., Gutman B.A., Hahn T., Harding I.H., Hernaus D., Hibar D.P., Hillary F.G., Hoogman M., Pol H.E.H., Jalbrzikowski M., Karkashadze G.A., Klapwijk E.T., Knickmeyer R.C., Kochunov P., Koerte I.K., Kong X.-Z., Liew S.-L., Lin A.P., Logue M.W., Luders E., Macciardi F., Mackey S., Mayer A.R., McDonald C.R., McMahon A.B., Medland S.E., Modinos G., Morey R.A., Mueller S.C., Mukherjee P., Namazova-Baranova L., Nir T.M., Olsen A., Paschou P., Pine D.S., Pizzagalli F., Rentería M.E., Rohrer J.D., Sämann P.G., Schmaal L., Schumann G., Shiroishi M.S., Sisodiya S.M., Smit D.J.A., Sønderby I.E., Stein D.J., Stein J.L., Tahmasian M., Tate D.F., Turner J.A., Heuvel O.A. van den, Wee N.J.A. van der, Werf Y.D. van der, Erp T.G.M. van, Haren N.E.M. van, Rooij D. van, Velzen L.S. van, Veer I.M., Veltman D.J., Villalon-Reina J.E., Walter H., Whelan C.D., Wilde E.A., Zarei M., Zelman V., Consortium E. ENIGMA and global neuroscience: a decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry. 2020;10:100. doi: 10.1038/s41398-020-0705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp A., Kota R.S., Lewis D.A., Sibille E. Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiol. Dis. 2011;42:116–124. doi: 10.1016/j.nbd.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos C.A., Risso D., Scialdone A., Dudoit S., Marioni J.C. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat. Methods. 2017;14:565–571. doi: 10.1038/nmeth.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5 doi: 10.7554/elife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Tseng E., Regulski M., Clark T.A., Hon T., Jiao Y., Lu Z., Olson A., Stein J.C., Ware D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016;7:11708. doi: 10.1038/ncomms11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu S., Warrell J., Won H., Shi X., Navarro F.C.P., Clarke D., Gu M., Emani P., Yang Y.T., Xu M., Gandal M.J., Lou S., Zhang J., Park J.J., Yan C., Rhie S.K., Manakongtreecheep K., Zhou H., Nathan A., Peters M., Mattei E., Fitzgerald D., Brunetti T., Moore J., Jiang Y., Girdhar K., Hoffman G.E., Kalayci S., Gümüş Z.H., Crawford G.E., Consortium P., Roussos P., Akbarian S., Jaffe A.E., White K.P., Weng Z., Sestan N., Geschwind D.H., Knowles J.A., Gerstein M.B. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362 doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S. Neuron–microglia interactions in mental health disorders: “for better, and for worse. Front. Immunol. 2016;7:544. doi: 10.3389/fimmu.2016.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S., Franklin T., Iwata M., Duman R.S. Integrating neuroimmune systems in the neurobiology of depression. Nat. Rev. Neurosci. 2016;17:497–511. doi: 10.1038/nrn.2016.69. [DOI] [PubMed] [Google Scholar]
- Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291. doi: 10.1016/j.cels.2018.11.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F., Bacanu S.-A., Bækvad-Hansen M., Beekman A.F.T., Bigdeli T.B., Binder E.B., Blackwood D.R.H., Bryois J., Buttenschøn H.N., Bybjerg-Grauholm J., Cai N., Castelao E., Christensen J.H., Clarke T.-K., Coleman J.I.R., Colodro-Conde L., Couvy-Duchesne B., Craddock N., Crawford G.E., Crowley C.A., Dashti H.S., Davies G., Deary I.J., Degenhardt F., Derks E.M., Direk N., Dolan C.V., Dunn E.C., Eley T.C., Eriksson N., Escott-Price V., Kiadeh F.H.F., Finucane H.K., Forstner A.J., Frank J., Gaspar H.A., Gill M., Giusti-Rodríguez P., Goes F.S., Gordon S.D., Grove J., Hall L.S., Hannon E., Hansen C.S., Hansen T.F., Herms S., Hickie I.B., Hoffmann P., Homuth G., Horn C., Hottenga J.-J., Hougaard D.M., Hu M., Hyde C.L., Ising M., Jansen R., Jin F., Jorgenson E., Knowles J.A., Kohane I.S., Kraft J., Kretzschmar W.W., Krogh J., Kutalik Z., Lane J.M., Li Yihan, Li Yun, Lind P.A., Liu X., Lu L., MacIntyre D.J., MacKinnon D.F., Maier R.M., Maier W., Marchini J., Mbarek H., McGrath P., McGuffin P., Medland S.E., Mehta D., Middeldorp C.M., Mihailov E., Milaneschi Y., Milani L., Mill J., Mondimore F.M., Montgomery G.W., Mostafavi S., Mullins N., Nauck M., Ng B., Nivard M.G., Nyholt D.R., O'Reilly P.F., Oskarsson H., Owen M.J., Painter J.N., Pedersen C.B., Pedersen M.G., Peterson R.E., Pettersson E., Peyrot W.J., Pistis G., Posthuma D., Purcell S.M., Quiroz J.A., Qvist P., Rice J.P., Riley B.P., Rivera M., Mirza S.S., Saxena R., Schoevers R., Schulte E.C., Shen L., Shi J., Shyn S.I., Sigurdsson E., Sinnamon G.B.C., Smit J.H., Smith D.J., Stefansson H., Steinberg S., Stockmeier C.A., Streit F., Strohmaier J., Tansey K.E., Teismann H., Teumer A., Thompson W., Thomson P.A., Thorgeirsson T.E., Tian C., Traylor M., Treutlein J., Trubetskoy V., Uitterlinden A.G., Umbricht D., Auwera S.V. der, Hemert A.M. van, Viktorin A., Visscher P.M., Wang Y., Webb B.T., Weinsheimer S.M., Wellmann J., Willemsen G., Witt S.H., Wu Y., Xi H.S., Yang J., Zhang F., eQTLGen 23andMe, Arolt V., Baune B.T., Berger K., Boomsma D.I., Cichon S., Dannlowski U., Geus E.C.J. de, DePaulo J.R., Domenici E., Domschke K., Esko T., Grabe H.J., Hamilton S.P., Hayward C., Heath A.C., Hinds D.A., Kendler K.S., Kloiber S., Lewis G., Li Q.S., Lucae S., Madden P.F.A., Magnusson P.K., Martin N.G., McIntosh A.M., Metspalu A., Mors O., Mortensen P.B., Müller-Myhsok B., Nordentoft M., Nöthen M.M., O'Donovan M.C., Paciga S.A., Pedersen N.L., Penninx B.W.J.H., Perlis R.H., Porteous D.J., Potash J.B., Preisig M., Rietschel M., Schaefer C., Schulze T.G., Smoller J.W., Stefansson K., Tiemeier H., Uher R., Völzke H., Weissman M.M., Werge T., Winslow A.R., Lewis C.M., Levinson D.F., Breen G., Børglum A.D., Sullivan P.F., Consortium, M.D.D.W.G. of the P.G. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li C.-X., Li Y.-S., Lv J.-Y., Ma Y., Shao T.-T., Xu L.-D., Wang Y.-Y., Du L., Zhang Y.-P., Jiang W., Li C.-Q., Xiao Y., Li X. MiRNA–miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. 2011;39:825–836. doi: 10.1093/nar/gkq832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.A., Thompson P.M., Cruz D.A., Williamson D.E., Selemon L.D. BA11 FKBP5 expression levels correlate with dendritic spine density in postmortem PTSD and controls. Neurobiology Stress. 2015;2:67–72. doi: 10.1016/j.ynstr.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Baune B.T., Licinio J., Wong M.-L. A novel strategy for clustering major depression individuals using whole-genome sequencing variant data. Sci Rep-uk. 2017;7:44389. doi: 10.1038/srep44389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Bar-Joseph Z. Deep learning for inferring gene relationships from single-cell expression data. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:27151–27158. doi: 10.1073/pnas.1911536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhu Y., Chen W., Tang Y. Emerging role of microRNAs in major depressive disorder and its implication on diagnosis and therapeutic response. J. Affect. Disord. 2021;286:80–86. doi: 10.1016/j.jad.2021.02.063. [DOI] [PubMed] [Google Scholar]