Abstract
Salmonella enterica subspecies enterica serotype Dublin (S. enterica Dublin) emerged for the first time in New York, Pennsylvania, and Ohio in 1988. Since that time this host-adapted serotype has spread throughout the veal- and dairy beef-raising operations in the region; very few dairy farms have experienced clinical S. enterica Dublin infections. This study details the epidemiology of the outbreaks in cattle. During the period 1988 through 1995, nine New York and four Pennsylvania counties have been affected; 13 different locations were involved in New York, and 10 were involved in Pennsylvania. The morbidity and mortality and seasonal distribution of outbreaks, which totaled 35, is described. The antimicrobial susceptibility pattern of isolates revealed that many of the strains were resistant to a number of commonly used drugs. Clinical case details and pathology information are provided, with a caution to clinicians and microbiologists presented with suspect animals, i.e., most cases occurred in older calves, which is atypical for salmonellosis for this region (calves were 8 or more weeks old) and presented as pneumonia and septicemia rather than the primarily diarrheal syndrome that is more typically recognized for the region. The epidemiology of cases is analyzed through cluster analysis of bacterial isolates and their fatty acid methyl ester profiles; at least six clones appeared in the region during the study period. Results of the epidemiology analysis are used to support a hypothesis regarding the source of S. enterica Dublin for the region and its manner of dissemination.
Salmonella enterica subsp. enterica serotype Dublin is a host-adapted serotype predominantly found in cattle and occasionally in swine, sheep, horses, and zoological animals (5, 7, 11, 50). Salmonellosis in animals always presents a potential zoonotic threat. S. enterica Dublin in endemic areas has caused severe disease in people who drink raw milk from infected carrier cows. Many of these people are immunosuppressed individuals. Human patients present with septicemia, osteomyelitis, and meningitis, often ending in death (8, 13, 47, 52).
Today, the public and the veterinary and medical communities are aware of the issues of “emerging and reemerging infectious diseases” (49). There are perhaps many factors contributing to the increase in emerging diseases, e.g., with cattle salmonellosis, especially S. enterica Dublin, the movement of cattle on trucks, intermixing of animals at sale barns, changes in housing, and feed and management practices all contribute to this increase in prevalence (25, 36, 56, 57). Now, more than ever, a coordinated approach to disease monitoring, surveillance, and both basic and applied research is needed; this, coupled with better communication among animal industries, veterinarians, public health and government regulatory authorities, and the public will help to establish preventive and control measures for emerging and reemerging infectious diseases (19, 29).
Researchers have questioned the degree of genomic diversity in bacterial species, such as S. enterica Dublin, one of the host-adapted salmonellae, which have a very limited host range (4). S. enterica Dublin is thought to be of recent evolutionary origin based on the uniformity of its multilocus enzyme genotype and analysis of fliC flagellin DNA sequences; S. enterica Dublin is closely related to and thought to have evolved from an S. enterica Enteritidis-like ancestor (32, 40). It has been possible to differentiate clones within S. enterica Dublin by employing multilocus enzyme electrophoresis (MLEE), pulsed-field gel electrophoresis (PFGE), ribotyping, restriction fragment length polymorphism analysis, restriction enzyme fragmentation pattern (REFP) analysis, various PCR techniques, and IS200 typing (4, 9, 22, 32, 35). These studies show that it is possible to track strains of S. enterica Dublin involved in outbreaks of disease.
Salmonella infections in cattle result in the problems of accurate diagnosis and the herd-level prevention and control of infections. Salmonella strains of serogroups B, C, and E have commonly been found in cattle in the Northeastern United States (26, 36). Until 1967 S. enterica Dublin (serogroup D) was only found west of the Rocky Mountains and was traditionally considered to be a “Western” disease. Since that time it has been making a steady progression eastward as animals and their products have been extensively moved (5). While this organism has been found east of the Rockies in other host animal species as early as 1968, in January 1980 the first case of S. enterica Dublin in cattle east of the Rocky Mountains occurred in Indiana (5, 7). In 1988 S. enterica Dublin (serogroup D) appeared for the first time simultaneously in New York, Pennsylvania, and Ohio in cattle (12), and it has been spreading in the Northeastern region. While not yet considered to be endemic on Northeastern dairy farms, S. enterica Dublin has been found frequently in veal and dairy beef-raising operations.
The goals of this study are to present the descriptive epidemiology of S. enterica Dublin infections in New York and Pennsylvania, to alert clinicians and clinical microbiologists to the different clinical presentation of the serotype Dublin-infected calf, to describe the most appropriate choice of specimens for culture diagnosis, and to describe the epidemiology of S. enterica Dublin strains through the quantitative evaluation of their fatty acid methyl esters (FAME) in cluster analysis.
MATERIALS AND METHODS
Bacterial culture and identification of Salmonella isolates.
All fecal and tissue specimen manipulations were performed in a biological safety cabinet type 2A by using an aseptic technique carried out by trained microbiology staff. Standard microbiological procedures were used throughout the study (10, 41). Bacterial isolates, after being serogrouped, were biochemically identified using the Sensititre’s Automated Microbiology System’s AP80 panel (Sensititre Microbiology System Division, AccuMed International, Inc., Westlake, Ohio).
Salmonella serotyping.
Biochemically confirmed Salmonella were referred to the National Veterinary Services Laboratories, Veterinary Services, Animal Plant Health Inspection Service, U.S. Department of Agriculture Services (NVSL, VS, APHIS, USDA), Ames, Iowa, for complete serotyping by standard techniques (10). All isolates confirmed as Salmonella enterica subsp. enterica serotype Dublin were included in the study.
Bacterial isolates.
All S. enterica Dublin isolates came from outbreaks of disease in New York and Pennsylvania cattle from 1988 until 1995. Fecal or tissue specimens were obtained during the New York State Diagnostic Laboratory’s ongoing program of culture surveillance of spontaneously occurring disease in cattle. These diagnostic specimens originated from New York and Pennsylvania veterinary practitioners. A “case” was defined as a clinically ill animal having signs of pneumonia, diarrhea, or septicemia. Each case represents one animal; a single colony was taken from the culture of a case animal, and this is referred to as an isolate. In all, 114 isolates of S. enterica Dublin were studied; these represented all of the cases of diseased cattle received at the laboratory (plus a few zoological animals) during this time period. As with all diagnostic sources of data, our data were also subject to submission bias. All isolates were lyophilized and stored at −70°C and were also subcultured on Trypticase soy agar slants (BBL) and stored at room temperature for daily use.
In addition, 96 isolates of S. enterica Dublin from around the United States were obtained from the USDA APHIS NVSL in Ames, Iowa, for comparison. These isolates originally were cultured from cattle specimens during 1992 by veterinary diagnostic laboratories in 21 states and then submitted to the NVSL for serotyping.
Antimicrobial susceptibility testing.
Until March 1989, the disc diffusion method of Bauer et al. (3) was used for susceptibility testing. With the disc diffusion test, six drugs were routinely tested for their effect against gram-negative enteric bacteria: ampicillin (10 μg), chloramphenicol (30 μg), gentamicin (10 μg), neomycin (30 μg), tetracycline (30 μg), and trimethoprim (1.25 μg)-sulfadiazine (23.75 μg); chloramphenicol was used as a marker only because of the prohibition of its use in food and fiber animals. All products, including Mueller-Hinton agar, drug discs, and the disc dispenser, were from BBL.
In March 1989, the disc method was replaced by the broth dilution susceptibility test to determine the MIC for the Salmonella strains. The protocol of the National Committee for Clinical Laboratory Standards for fast-growing aerobic bacteria was followed (31). The following drugs were used in a customized MIC panel: amikacin, ampicillin, ceftiofur, cephalothin, chloramphenicol, enrofloxacin, gentamicin, neomycin, tetracycline, trimethoprim-sulfadiazine (Sensititre Microbiology System Division, AccuMed International, Inc.). Not all drugs were used in each assay since the drugs used in the custom panels changed over the years. Panels were inoculated and read with the Sensititre system.
FAME analysis. (i) Standardized growth conditions and culture medium.
The protocols for cultivating bacteria for fatty acid analysis are critical to the success and reproducibility of the analysis (see below). All bacterial isolates were grown on a single lot of medium as detailed below. Bacterial isolates were grown on Trypticase soy broth agar (TSBA) plates (BBL) at 28 ± 1°C for 23 ± 1 h. Up to 30 isolates were grown up at any one time for subsequent processing.
(ii) Harvesting bacterial cells.
For bacterial-cell harvesting, 40 mg of cell mass in the late log phase of growth was harvested from the third quadrant of the TSBA plates by using a 10-μl disposable plastic inoculating loop; cells were then weighed and placed into a teflon screw-capped test tube (13 by 100 mm).
(iii) Saponification.
The lipids of the bacterial cell were saponified with 1 ml of an NaOH-methanol solution (45 g of NaOH, 150 ml of methanol, 150 ml of distilled water) added to each tube, vortexed for 15 s, and then heated at 100°C in a water bath for 5 min. Tubes were then removed from the water bath, vortexed for an additional 15 s, and then heated again at 100°C for 25 min.
(iv) Methylation of fatty acids.
After the tubes were cooled in a water bath to room temperature, free fatty acids were methylated by adding 2 ml of a hydrochloric acid-methanol solution (325 ml of 6.00 N HCl, 275 ml of methanol) to the tube, followed by vortexing for 15 s and heating at 80 ± 1°C for 10 min in a water bath.
(v) Extraction of FAME.
After a rapid cooling in an ice bath to room temperature, the FAME were extracted from the acidic aqueous phase by the addition of 1.25 ml of a hexane–methyl-tert-butyl ether solution (1:1 [vol/vol]) to the tube followed by end-over-end rotation of the tubes for 10 min on a hematology rotator. For the base wash, the lower acidic aqueous phase was aspirated and discarded; the upper FAME extract layer was washed and neutralized by adding 3 ml of an NaOH solution (10.8 g of NaOH, 900 ml of distilled water), tightly capping the tubes, and rotating the tubes for 5 min on a hematology rotator; this step also served to remove any free fatty acids and traces of reagents. Two-thirds of the upper FAME solvent layer was then pipetted into an autosampler vial and capped for later chromatographic analysis.
Gas chromatography.
FAME were separated with a Hewlett-Packard 5890 gas-liquid chromatograph (GC) fitted with a capillary column (Ultra 2; cross-linked 5% phenylmethyl siloxane; 25 m by 0.2 mm [inner diameter] by 0.33-μm film thickness; Hewlett Packard, Avondale, Pa.) coated with phenylmethyl silicone and a flame ionization detector; hydrogen was the carrier gas. The FAME were identified by using a chromatography work station, the MIDI-Sherlock System (Microbial ID, Inc., Newark, Del.) and associated software, which was linked to the GC. The MIDI Sherlock System uses a standard calibration mixture containing FAME in 0.8 ml of hexane (straight-chain saturated nC9:0 to nC20:0, plus two and three hydroxy acids; MIDI, Inc.) to standardize the chromatography.
Quality control.
Standard quality control procedures performed for all FAME studies included controls at the growth and extraction levels. Single colonies were picked for each bacterial isolate for culture and extraction to minimize isolate variation. A single batch of TSBA medium was used for the entire study to reduce any variation due to batch to batch media differences. Bacterial cells were grown under controlled culture conditions, i.e., 23 h at 29°C in air and harvested only from the third quadrant of the agar plate (early-log-phase cells with the most stable and reproducible fatty acid levels). A total of 40 mg of cell mass (wet weight) was weighed out for each assay. Quality control also included controls at the GC system level. Only 10 test samples were injected into the injection port liner of the GC before these were replaced with clean and freshly packed liners so as not to introduce any error into the FAME analyses from clogged liners. The entire GC injection procedure was under computer control. The GC was calibrated at the beginning of each run from a commercially available calibration mix, and the GC was recalibrated automatically after every 11 injections, all of which was under computer control. The hydroxy compounds in the calibration mixture are especially sensitive to changes in pressure and temperature relationships and to contamination of the injection port liner and thus are important quality control checks.
Analysis of chromatograms.
The MIDI-Sherlock System software and the Hewlett-Packard ChemStation software run concurrently on the system, i.e., the ChemStation software stores the raw data, produces the sample chromatogram, and locates the FAME. The MIDI-Sherlock System software then identifies FAME by integrating the peak location versus named peaks from the standard calibration mix that is run as a calibration control. The MIDI software also produces a report of the microorganism identification after a comparison of the FAME composition to the FAME library entries stored in its database (2).
Cluster numerical analysis of FAME data.
To study the relatedness of S. enterica Dublin isolates, the quantitative FAME data produced by the MIDI-Sherlock System was evaluated with two programs from Microbial ID, Inc. “Cluster” is a designation of a major grouping of isolates that is readily visible from the dendrogram and two-dimensional (2D-Plot) analyses without additional numerical analysis. Clustering of strains was then further evaluated numerically by the unweighted-pair-group method of arithmetic averages (UPGMA) according to the method of Sneath and Sokal (44); this resulted in a one-dimensional dendrogram. The distance along the dendrogram scale at which isolates paired was noted as the euclidian distance (ED). Principle component analysis of FAME data according to the method of Gower (14) produced a 2D-Plot to show the distribution of groupings of the isolates. From the 2D-Plot, if one measures the ED spanned by the cluster along the x axis and then multiplies this value by the distance spanned by the cluster along the y axis, this calculation yields the ED2.
Reproducibility of the FAME method for S. enterica Dublin.
To test the reproducibility of the FAME method for the gram-negative bacterium S. enterica Dublin, a single field isolate of S. enterica Dublin was grown according to the standard culture protocol (see above) on 6 different days in replicates of five or six plates each (this represented the within-run variability). The growth from each plate was harvested and treated as a separate unknown bacterial strain. FAME data obtained from these analyses were processed by cluster analysis by using the dendrogram and 2D-Plot software programs (see below). This reproducibility study was used to determine the conventions for interpreting the results of cluster analyses, especially with regard to the status of a group of isolates as a “clone,” defined here as a divergent line within the population of S. enterica Dublin. The between-run variability was assessed by using all the data points generated from each of the six daily runs in a single dendrogram and 2D-Plot.
Descriptive epidemiology of S. enterica Dublin outbreaks.
Each suspect case of salmonellosis was accompanied by information on the location and size of the herd of origin, on the herd morbidity and case fatality rates, and on the dates of onset of the problem. In addition, the details of the clinical case presentation were supplied by the veterinarian referring the case to the laboratory for cultural confirmation of disease. The Necropsy Service of the Department of Pathology at the College of Veterinary Medicine, Cornell University, provided details of the gross pathology and histopathology of the cases.
RESULTS
National and regional frequency of S. enterica Dublin.
The national and Northeastern regional frequency distributions of S. enterica Dublin cases as reported by the NVSL, VS, APHIS, USDA, are presented in Table 1. During 1988 and 1989, the first and second years of the outbreaks in New York, Pennsylvania, and Ohio, isolates from these three states made up 26 and 12%, respectively, of the total Salmonella enterica Dublin cases reported for the United States by the NVSL. During the next 6 years, this percentage dropped and, it has remained at ca. 5% of the U.S. total. The outbreaks occurring in New York, Pennsylvania, and Ohio during the years 1988 to 1993 correlated with the national overall increase in Dublin cases reported to the USDA.
TABLE 1.
USDA Statistics for S. enterica Dublin for New York, Pennsylvania, Ohio, and New England versus the overall U.S. totalsa
| Location | No. of isolates in federal fiscal yrb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1986 | 1987 | 1988 | 1989 | 1990 | 1991 | 1992 | 1993 | 1994 | 1995 | |
| U.S. total | 169 | 227 | —c | 544 | 660 | 574 | 540 | 417 | 415 | 264 |
| New Englandd | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| New York | 0 | 0 | 0 | 66 | 8 | 11 | 3 | 0 | 2 | 3 |
| Pennsylvania | 0 | 0 | 0 | 48 | 55 | 11 | 20 | 7 | 7 | 4 |
| Ohio | 0 | 0 | 0 | 29 | 13 | 7 | 4 | 13 | 7 | 5 |
Data were compiled from USDA APHIS VS NVSL Salmonella Serotyping Laboratory records (Ames, Iowa) and from New York State Diagnostic Laboratory records.
The federal fiscal year (FY) has varied over time (month/year): FY86 = 10/85–9/86; FY87 = 10/86–9/87; data are not available for 10/87–6/88; FY89 = 7/88–6/89; FY90 = 7/89–6/90; FY91 = 7/90–6/91; FY92 = 7/91–6/92; FY93 = 7/92–6/93; FY94 = 7/93–6/94; FY95 = 7/94–6/95. The first case of S. enterica Dublin in New York occurred in October 1988, which falls in federal FY89. Numbers represent individual isolates sent to NVSL that were confirmed to be S. enterica Dublin, and not individual locations.
Data were not available for the period 10/87 to 6/88.
Connecticut, Rhode Island, Massachusetts, Vermont, New Hampshire, and Maine.
Antimicrobial susceptibility: disc diffusion and broth dilution MIC.
From 1973 to 1989 the Kirby-Bauer method of determining antimicrobial susceptibility was used in the New York State Diagnostic Laboratory. During this time, different drugs, including ampicillin, chloramphenicol, gentamicin, neomycin, tetracycline, and trimethoprim-sulfadiazine, were used at different times against enteric bacteria. All isolates tested were resistant to ampicillin, chloramphenicol, neomycin, and tetracycline and were sensitive to gentamicin and trimethoprim-sulfadiazine.
After 1989, the laboratory switched to the broth dilution MIC method with a commercial automated testing system and a custom MIC testing panel. In general, all S. enterica Dublin strains tested, with few exceptions, were resistant to ampicillin, chloramphenicol, neomycin, and tetracycline and were sensitive to amikacin, cefoxitin, cephalothin, enrofloxacin, gentamicin, and trimethoprim-sulfadiazine at the MIC levels shown in Table 5.
TABLE 5.
Descriptive epidemiology and antibiograms of S. enterica Dublin outbreaks in New York and Pennsylvania, 1988 to 1995
| Date (mo/yr) | Locationa | Type of operationb | Herd census (n) | Herd morbidity rate (%) | Case fatality rate (%) | |
|---|---|---|---|---|---|---|
| 10/88 | N1 | Dairy beef | 225 | 22 | 60 | |
| 10/88 | P1 | Dairy beef | 303 | 18 | 2 | |
| 11/88 | N2 | Dairy beef | 300 | 4 | 17 | |
| 11/88 | P1 | Dairy beef | 249 | 80 | 18 | |
| 11/88 | N3 | Dairy beef | 450 | 2 | 12 | |
| 11/88 | N4 | Veal | 108 | 25 | 100 | |
| 11/88 | N5 | Dairy | 80 | 1 | 100 | |
| 1/89 | N3 | Dairy beef | 470 | 11 | 6 | |
| 2/89 | N2 | Dairy beef | 300 | 2 | –d | |
| 2/89 | N3 | Dairy beef | 500 | 40 | 12 | |
| 3/89 | N3 | Dairy beef | 500 | 8 | 30 | |
| 3/89 | P2 | Veal | 145 | 1 | 0 | |
| 3/89e | P3 | Veal | – | – | – | |
| 5/89 | P4 | Veal | 150 | 23 | 41 | |
| 6/89 | P2 | Veal | – | – | – | |
| 6/89 | P3 | Veal | – | – | – | |
| 8/89 | P5 | Veal | 150 | 22 | 91 | |
| 8/89 | P6 | Veal | 65 | 18 | 50 | |
| 10/89 | P3 | Veal | 58 | 7 | 0 | |
| 12/89 | N6 | Veal | – | – | – | |
| 1/90 | P7 | Dairy beef | 400 | 3 | – | |
| 5/90 | N7 | Dairy beef | 230 | 30 | 0 | |
| 7/90 | N8 | Veal | 100 | 30 | 100 | |
| 3/91 | N9 | Veal | 160 | 6 | 50 | |
| 4–5/91 | N7 | Dairy beef | 200 | 3 | 20 | |
| 9/91 | N9 | Veal | 160 | 10 | 5 | |
| 10/91 | N10 | Dairy beef | 150 | 19 | 100 | |
| 4/93 | P8 | Sheep dog | 6 Fetuses | 100 | 100 | |
| 4/93 | P9 | Veal | 400 | 19 | 33 | |
| 10/93 | N11 | Dairy beef | 150 | 7 | 70 | |
| 9/94 | N12 | Dairy | 400 | <1 | 0 | |
| 2/95 | N13 | Tiger | 2 | 100 | 0 | |
| 3/95 | N13 | Cheetah | 1 | 100 | 0 | |
| 8/95 | P10 | Heifer replacement | 80 | 13 | 40 |
| Age (wks) | Susceptibility profile |
|---|---|
| 12 | AM-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(S)c |
| 11 | AM-(R), C-(R), GM-(S), N-(I), TE-(R), TS-(S) |
| 12 | AM-(R), C-(R), GM-(S), N-(I), TE-(R), TS-(S) |
| 10 | AM-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(S) |
| 9–15 | AM-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(S) |
| 10 | AM-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(S) |
| Adult (6 yr) | AM-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(S) |
| 14 | Not tested |
| Calf | AK-(R), AM-(R), CP-(R), C-(R), GM-(S), N-(R), TE-(R), TS-(R) |
| 14 | AK-(S), AM-(R), CP-(S), C-(R), GM-(S), N-(R), TE-(R), TS-(S) |
| 15 | Not tested |
| 8–10 | Not tested |
| Calf | AM-R(>32)f CP-S(2), C-R(>32), GM-S(≤1), N-R(>16), TE-R(>16), TS-S(0.25) |
| 16 | AK-S(<8), AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(>16), TE-R(>16), TS-S(≤0.12) |
| 3–16 | AM-S(0.5), CP-S(1), C-S(≤4), GM-S(≤1), N-S(≤2), TE-S(≤2), TS-S(≤0.12) |
| Calf | AM-R(>32), CP-MS(8), C-R(>32), GM-S(≤1), N-R(16), TE-R(>16), TS-S(≤0.12) |
| 7 | AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(>16), TE-R(>16), TS-S(0.25) |
| 8–10 | AM-R(>32), CP-S(2), C-R(>32), EN-S(≤0.12), GM-S(≤1), TE-R(>16), TS-S(0.12) |
| 14 | AK-S(≤8), AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(>16), TE-R(>16), TS-S(0.12) |
| 10 | AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(16), TE-R(>16), TS-S(≤0.12) |
| 10 | AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(16), TE-R(>16), TS-S(≤0.12) |
| 16 | AM-R(>32), CP-MS(8), C-R(>32), GM-S(2), N-R(>16), TE-R(>16), TS-R(2) |
| 3 | AM-R(>32), CP-MS(16), C-R(>32), GM-S(≤1), N-R(>16), TE-R(>16), TS-S(0.12) |
| 10 | AK-S(≤4), AM-R(>32), CP-S(4), C-R(>32), GM-S(≤1), N-MS(8), TE-R(>16), TS-S(≤0.12) |
| 3 | AK-S(≤4), AM-R(>32), CP-S(2), C-R(>32), GM-S(≤1), N-R(16), TE-R(>16), TS-S(≤0.12) |
| 8 | AK-S(≤8), AM-R(>32), CF-S(≤1), CP-S(≤4), C-R(>32), EN-S(≤0.25), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| 9 | AK-S(≤8), AM-R(>32), CF-S(≤1), CP-S(≤4), C-R(>32), EN-S(≤0.25), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| Fetus | AK-S(≤8), AM-R(>32), CF-S(≤0.5), CP-S(≤4), C-R(>32), EN-S(≤0.5), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| 11 | AK-S(≤8), AM-R(>32), CF-S(≤0.5), CP-S(≤4), C-R(>32), EN-S(≤0.5), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| 12 | AK-S(≤8), AM-R(>32), CF-S(≤0.5), CP-S(≤4), C-R(>32), EN-S(≤0.5), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| Adult | AK-S(≤8), AM-S(0.5), CF-S(≤1), CP-S(≤4), C-R(32), EN-S(≤0.5), GM-S(≤0.5), TE-S(≤2), TS-S(≤0.5) |
| Adult | AK-S(≤8), AM-R(>32), CF-S(≤1), CP-S(8), C-R(32), EN-S(≤0.5), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
| Adult | Not tested |
| 8 | AK-S(≤8), AM-R(>32), CF-S(≤1), CP-S(8), C-R(32), EN-S(≤0.5), GM-S(≤0.5), TE-R(>16), TS-S(≤0.5) |
N, New York; P, Pennsylvania. For example, N1 is the first outbreak location in New York, etc.
Type of operation: dairy beef, bull calves from dairy breeds of cattle raised to between 1,000 and 1,200 pounds before going to market (usually “preconditioned” by weaning, castration, dehorning, and vaccinating before they enter the rearing facility); veal, bull calves from dairy breeds raised to the age of 12 to 14 weeks either on a milk replacer diet (hence the name “milk fed”) or on a milk diet supplemented with hay and grain; dairy, farm with calves and adult dairy breed animals that produce milk for retail sale; zoological (i.e., exotic animal, bird, or reptile collection used for exhibit and/or study); heifer replacement, contract raising of female calves from multiple farms for eventual return to original farm of origin.
Kirby-Bauer Susceptibility profile. Abbreviations: AK, amikacin; AM, ampicillin; CF, ceftiofur; CP, cephalothin; C, chloramphenicol; EN, enrofloxacin; GM, gentamicin; N, neomycin; TE, tetracycline; TS, trimethoprim-sulfadiazine; (R), resistant; (S), sensitive; (I), intermediate; (MS), moderately sensitive.
–, No data were available.
Until March 1989, the disc diffusion (Kirby-Bauer) method of susceptibility testing was utilized. After this time the broth dilution MIC method was used.
Broth dilution MIC profile. “R(32)” indicates by the letter before the parentheses (i.e., R, S, MS, or I) the MIC value for that drug (i.e., resistant, sensitive, moderately sensitive, or intermediate; the number in parentheses indicates the MIC value for the drug in micrograms per milliliter.
Epidemiology of S. enterica Dublin. (i) reproducibility of FAME method for S. enterica Dublin.
A single isolate of S. enterica Dublin grown on 5 different days in replicate platings on five or six TSBA plates produced linkages of replicates at the following levels in dendrogram analyses (day 1, 2.8 ED; day 2, 1.8 ED; day 3, 2.9 ED; day 4, 2.6 ED; day 5, 3.0 ED); these same replicates showed clustering of replicates at the following levels when analyzed in the 2D-Plot: day 1, 8.0 ED2; day 2, 2.4 ED2; day 3, 9.0 ED2; day 4, 4.0 ED2; and day 5, 4.0 ED2. (These values represented the within-run variability.) The use of all of the data from the six daily runs (between-run variability) in a single dendrogram analysis resulted in a linkage of strains at 3.3 ED and in a 2D-Plot analysis clustering at 12.0 ED2.
(ii) Conventions for the interpretation of dendrogram and 2D-Plot analyses.
Reproducibility studies (see above) with the gram-negative bacterium S. enterica Dublin support the following interpretive criteria: in the dendrogram, isolates that link with a ≤3 ED are likely to be clones; in the 2D-Plot, isolates with a clustering of ≤9 ED2 are likely to be clones. Other studies of gram-positive and gram-negative bacteria have stated that isolates linking in the dendrogram at ≤2 ED and in the 2D-Plot at ≤25 ED2 are likely to be clones (39, 42).
(iii) Cluster analysis of the FAME data.
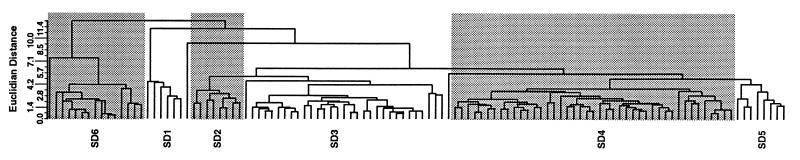
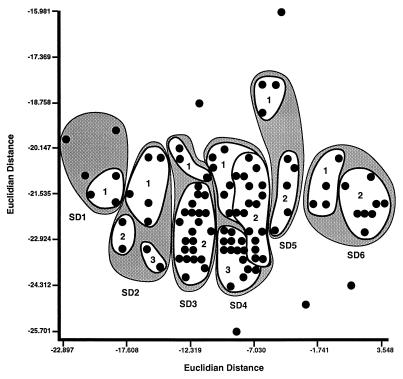
Within the first six months of the initial outbreaks of disease in New York and Pennsylvania, 35% of the cattle operations of the study were already infected with S. enterica Dublin. That number rose to 52% within the first year. The FAME profiles of all isolates of S. enterica Dublin when analyzed by clustering techniques produced a dendrogram and a 2D-Plot containing six readily identifiable clusters of isolates. Within these clusters, however, there were a number of possible clones that were apparent when the definitions of cluster analysis were used (Table 2, Fig. 1 and 2).
TABLE 2.
Cluster analysis of S. enterica Dublin isolates from New York and Pennsylvania
| Cluster and clonea | No. of isolates in:
|
EDb | ED2 c | |
|---|---|---|---|---|
| Cluster | Clone | |||
| All isolates | 114 | 12.1 | 257.0 | |
| SD1 | 6 | 4.3 | 22.2 | |
| 1 | 3 | ≤2.0 | 1.8 | |
| SD2 | 9 | 3.6 | 11.5 | |
| 1 | 5 | ≤2.0 | 6.2 | |
| 2 | 2 | ≤2.0 | 0.2 | |
| 3 | 2 | ≤2.0 | 0.7 | |
| SD3 | 30 | 4.3 | 12.3 | |
| 1 | 3 | ≤2.0 | 2.0 | |
| 2 | 27 | ≤2.0 | 8.0 | |
| SD4 | 43 | 3.6 | 21.0 | |
| 1 | 7 | ≤2.0 | 3.4 | |
| 2 | 20 | ≤2.0 | 17.5 | |
| 3 | 16 | ≤2.0 | 5.3 | |
| SD5 | 8 | 4.3 | 3.9 | |
| 1 | 3 | ≤2.0 | 1.2 | |
| 2 | 5 | ≤2.0 | 3.9 | |
| SD6 | 13 | 4.3 | 14.0 | |
| 1 | 4 | ≤2.0 | 2.9 | |
| 2 | 9 | ≤2.0 | 5.5 | |
| Total | 109d | 109 | ||
A cluster is a major grouping of isolates that is readily visible from the dendrogram (Fig. 1) and the 2D-Plot analyses (Fig. 2), e.g., “SD1” stands for the first cluster of S. enterica Dublin. A clone is defined as strains grouping at an ED of ≤3 as determined by dendrogram analysis (UPGMA) (Fig. 1) and/or grouping at ≤9 ED2 by 2D-Plot analysis (principal component analysis) (Fig. 2), e.g., “SD1, 1” signifies that within the first cluster of S. enterica Dublin “SD1” there is a clone indicated as clone 1.
ED is a measure of the literal distance between two objects when they are viewed as points in the two-dimensional space formed by their attributes. This value is read directly from the dendrogram scale in Fig. 1 and is the point on the scale where a group of bacterial isolates join up or pair together.
ED2 is the ED squared. In the 2D-Plot, the ED spanning from the left edge of the cluster of bacterial isolates along the x axis to the right edge of the cluster, multiplied by the distance spanned by the bottom edge of the cluster along the y axis to the top edge of the cluster, gives the ED2. These values are read directly from the x and y axes of Fig. 2.
The totals for the groups add only to 109 and not to 114 because five outlier isolates that did not cluster with the other strains were not included in the Table.
FIG. 1.
Clustering of S. enterica Dublin isolates as shown in a dendrogram produced by the MIDI-Sherlock program. This dendrogram shows the relationship of 114 S. enterica Dublin isolates and their FAME analyses obtained by UPGMA cluster analysis. The designations SD1, SD2, etc., stand for Salmonella Dublin 1, Salmonella Dublin 2, etc., and represent the clusters of isolates referred to in Tables 3 and 4 and in Fig. 2.
FIG. 2.
Clustering of S. enterica Dublin isolates as shown in a 2D-Plot adapted from the plot produced by the MIDI-Sherlock program. This figure shows the relationship of 114 S. enterica Dublin isolates and their FAME analyses obtained by principal component cluster analysis. The designations SD1, SD2, etc., are shown as shaded areas and represent the clusters of Fig. 1 and Tables 3 and 4. The numbered white areas within the shaded clusters correspond to the clones described in the text and referred to in Tables 3 and 4. Points not held within circles are outliers.
Table 3 presents a geographical and temporal distribution of S. enterica Dublin clusters. The clusters consisted of isolates occurring in 3 to 12 different locations per cluster. Within a cluster of isolates, one or more clones were found at multiple locations. For example, five individual locations were infected within the first cluster of S. enterica Dublin isolates (designated SD1), i.e., N6, P7, N7, N8, and N9, and of these a clone designated clone 1 was found which contained isolates from three locations: N6, P7, and N9. Three locations were contained in the second cluster, SD2, which contained three clones (clones 1, 2, and 3), etc.
TABLE 3.
Temporal and geographical distribution of S. enterica Dublin clusters and clones in New York and Pennsylvania
| Cluster and clonea | Geographic distribution by date (yr, mo) and locationb
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1988
|
1989
|
1990
|
1991
|
1993
|
1994
|
1995
|
||||||||||||||||
| Oct. | Nov. | Jan. | Feb. | Mar. | June | Aug. | Oct. | Dec. | Jan. | May | July | Mar. | Apr.-May | Sep. | Oct. | Apr. | Oct. | Sep. | Feb. | Mar. | Aug. | |
| SD1 | N6 | P7 | N7 | N8 | N9 | N7 | ||||||||||||||||
| 1 | N6 | P7 | N7 | |||||||||||||||||||
| SD2 | ||||||||||||||||||||||
| 1 | N3 | N3 | N3 | |||||||||||||||||||
| P1 | ||||||||||||||||||||||
| 2 | P1 | P2 | ||||||||||||||||||||
| 3 | N3 | |||||||||||||||||||||
| SD3 | ||||||||||||||||||||||
| 1 | N3 | N9 | ||||||||||||||||||||
| 2 | P1 | P1 | N3 | N3 | N3 | P3 | ||||||||||||||||
| N4 | P3 | |||||||||||||||||||||
| N2 | N2 | |||||||||||||||||||||
| SD4 | ||||||||||||||||||||||
| 1 | N3 | N3 | P2 | P3 | N10 | |||||||||||||||||
| 2 | N3 | N3 | N3 | P3 | P3 | P3 | N8 | N7 | N9 | |||||||||||||
| N3 | P2 | |||||||||||||||||||||
| 3 | N3 | N3 | N3 | P3 | P4 | P6 | ||||||||||||||||
| N2 | N2 | N3 | ||||||||||||||||||||
| SD5 | ||||||||||||||||||||||
| 1 | P3 | N6 | N7 | |||||||||||||||||||
| 2 | N6 | P3 | N8 | N9 | ||||||||||||||||||
| N3 | ||||||||||||||||||||||
| SD6 | ||||||||||||||||||||||
| 1 | N1 | N8 | N9 | |||||||||||||||||||
| 2 | N5 | N7 | P9 | N11 | N12 | N13 | N13 | P10 | ||||||||||||||
See Table 2, footnote a, for an explanation of the designations cluster and clone.
The notation “N6” means the sixth outbreak in New York; the notation “P1” means the first outbreak in Pennsylvania, etc.
After the first year of the outbreaks in which 12 locations were affected, we did not see recurring infections at these first farm locations but instead saw new locations affected (Table 3). Cluster SD2 was only found during the first year of the outbreaks, whereas SD1 was seen only in the second year. Clusters SD3, SD4, and SD5, though found mostly in the first year, were seen sporadically at new locations in the second and third years as well. Cluster SD6 was seen throughout the study period but appeared, for the most part, in many new locations in the later years of the study. Some clusters appeared on the same farm sporadically over a 6- to 9-month period but usually not for longer than this.
(iv) Cluster analysis of New York and Pennsylvania isolates with isolates from other areas of the United States.
By using dendrogram analysis of FAME, 73 S. enterica Dublin isolates from the NVSL were compared with the 114 isolates from New York and Pennsylvania. At least 19 of these isolates could be paired with isolates from 17 other states at levels which would define them as clones, i.e., ≤3 ED (Table 4). Isolates from each of the clusters already noted from New York and Pennsylvania, i.e., SD1 to SD6, could be grouped with U.S. isolates at the clonal level. There were 17 clusters noted for the combined U.S.–New York-Pennsylvania sample, which contained 22 clones. The clusters and clones identified in the analysis of the New York-Pennsylvania isolates were conserved when the isolates of the combined data set were analyzed, thus illustrating further the reliability of the clusters and clones identified.
TABLE 4.
Cluster analysis of New York and Pennsylvania S. enterica Dublin isolates with other U.S. isolates
| New York-Pennsylvania clustera | Locationb and isolation date (mo/yr) | States with isolates grouped with the New York-Pennsylvania isolates at an ED of ≤3 in the dendrogramc |
|---|---|---|
| SD1 | N7 (5/90) | AL, CO, IA, IL, WI |
| SD1 | N7 (5/91) | ID, MN, ND |
| SD1 | P7 (1/90) | ID, MN, MO, ND |
| SD1 | N6 (12/89), N8 (7/90), N9 (3/91) | ND |
| SD2 | N3 (1/89) | AL, TN |
| SD2 | N3 (11/88), N3 (2/89) | AZ, CA, MO, WA |
| SD3 | N3 (1/89), N9 (3/91) | SD |
| SD4 | P3 (10/89) | AZ, ID |
| SD4 | N2 (11/88), N3 (3/89) | UT |
| SD5 | N3 (2/89), N8 (7/90) | TX |
| SD6 | N1 (10/88), N8 (7/90), N9 (3/91) | GA |
Cluster corresponds to the clusters of Tables 2 and 3. (See Table 2 for the definition of cluster.)
N, New York; P, Pennsylvania. “N7” refers to the seventh outbreak in New York. Only isolates grouping at the level of clone are listed, i.e., with a value of ≤3 ED on the dendrogram.
AL, Alabama; AZ, Arizona; CA, California; CO, Colorado; GA, Georgia; IA, Iowa; ID, Idaho; IL, Illinois; MO, Missouri; MN, Minnesota; ND, North Dakota; SD, South Dakota; TN, Tennessee; TX, Texas; UT, Utah; WA, Washington; WI, Wisconsin.
Descriptive epidemiology of S. enterica Dublin outbreaks in New York and Pennsylvania. (i) Geographic distribution.
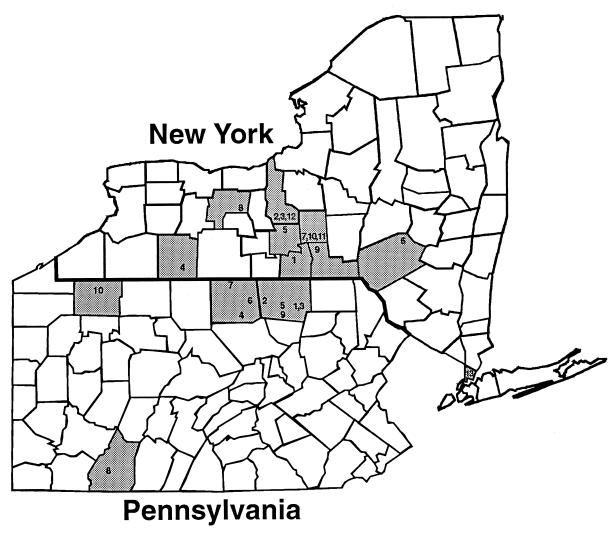
From October 1988 to 1998, nine New York State counties had S. enterica Dublin clinical cases; a total of 13 different locations were involved. All cases were found in cattle-based operations except for three zoological cases found in tigers and cheetahs. During the same time period, four Pennsylvania counties were affected, involving a total of 10 different locations. All of the Pennsylvania cases came from cattle operations, except for one case of aborted pups in a sheep dog. In both states a total of 35 outbreaks have occurred (Table 5 and Fig. 3).
FIG. 3.
Geographical distribution of the New York and Pennsylvania S. enterica Dublin outbreaks from 1988 to 1995. Numbers within each shaded county area correspond to the location designation given in all of the Tables (e.g., “1” on the New York map corresponds to NY1; “3” on the Pennsylvania map corresponds to PA3).
(ii) Types of cattle operations affected.
During the early months of the outbreaks in New York and Pennsylvania, the new dairy beef industry operations were affected by S. enterica Dublin. Later cases appeared in numerous veal operations throughout the region. Only two clinical cases in adult cattle were detected on dairy farms during the study. Case details and census information were supplied by the referring veterinarian and animal owner (Table 5). The dairy beef operations tended to have a higher animal census than the veal-raising operations. The herd morbidity rates varied greatly in all types of cattle operations, as did the case fatality rates. The herd morbidity rate was calculated by dividing the total number of ill animals by the herd census and multiplying by 100 to get a percentage. The case fatality rate was calculated by taking the number of clinically ill animals that died, dividing this value by the total number of clinically ill animals, and multiplying the total 100 to get a percentage. The average case fatality rate for dairy beef animals was 29%, for veal calves it was 47%, and for adult dairy cows it was 50%.
(iii) Temporal distribution.
Outbreaks in cattle occurred throughout the year without obvious seasonality (Table 5).
(iv) Clinical presentation and pathology.
The clinical presentation of calf cases were those of depressed, pneumonic animals (labored breathing, nasal discharge) with or without concomitant diarrhea. Gross pathologic and histopathologic evaluation of these cases confirmed the involvement of the lungs and the intestinal tract and usually indicated that the calves had septicemic salmonellosis. These cases often had swollen, inflamed joints containing fibrin tags. The intestinal tract lesions included ileitis and mesenteric lymphadenitis, while the lung lesions ranged from acute, suppurative pneumonia to chronic bronchopneumonia. Many cases had histopathological lesions of cholecystitis and hepatitis. Calves with brain involvement had severe meningoencephalitis. The ages of affected calf cases ranged from 7 to 16 weeks of age; this is an older calf than was usually seen in the region for cases of salmonellosis (Table 5). Of the two adult dairy cattle cases, one was affected 2 days postpartum and died acutely. Its pathologic lesions included chronic, resolving mastitis, but otherwise no lesions were noted in the gastrointestinal tract. The other dairy cow was noted to have fever and diarrhea. Since it survived this clinical episode, no pathology was performed, and it was lost to follow-up.
DISCUSSION
Today bacterial population genetics and the molecular epidemiology of infectious diseases are two topics with renewed importance (30). Moreover, the tools with which we can now fingerprint the isolates of bacteria from cases of disease in both animals and humans are readily available; some of these techniques are PFGE (16), ribotyping (51), and FAME analysis (43, 46). Many disease outbreaks are caused by distinct bacterial clones (30). This is true for the salmonellae (24, 28, 33, 48) and in particular for S. enterica Dublin (9, 32, 35). S. enterica Dublin strains are thought to be very closely related and clonal; the fact that strains exhibit little diversity may reflect their adaptation to the bovine animal as a host-adapted serovar (4, 32, 40). On one level, i.e., serotyping, a laboratory may identify a Salmonella isolate as S. enterica Dublin. S. enterica Dublin may be further phage typed, but most isolates fall into just one of 11 possible types, making this tool of limited value in outbreak investigation (11, 15). MLEE has been able to subdivide S. enterica Dublin into four electrophoretic types, Du1, Du2, Du3, and Du4, but, unfortunately, most U.S. isolates belong to the Du1 type, limiting the usefulness of MLEE in epidemiology studies (4, 40). IS200 typing did not subdivide S. enterica Dublin strains at all (9, 32). Ribotyping of a group of English S. enterica Dublin isolates produced just two groups considered to be clones (9). Other studies with ribotyping have demonstrated as many as eight ribotypes from a larger multinational collection of S. enterica Dublin isolates (32), thus showing a potential for the use of this molecular technique in epidemiological studies of S. enterica Dublin. Also, REFP analysis of whole-cell bacterial DNA produced two major types of S. enterica Dublin and six further subtypes (35). Recently, investigators have used up to five concurrent typing methods to enhance the discrimination and tracking of S. enterica Dublin isolates. The combination of ribotyping with arbitrarily primed PCR was especially useful (22).
The use of FAME analysis for the identification of microorganisms and also for the study of the epidemiology of bacterial diseases in plants and animals has been well documented, e.g., for Borrelia (23) and Bacillus (42) spp. An excellent review by Smith and Siegel (43) discusses the concepts of FAME analysis, the MIDI-Sherlock System, the use of FAME for strain tracking, the reproducibility of the FAME, and the fatty acids of gram-positive and gram-negative bacteria. FAME analyses of bacteria are highly reproducible and relatively simple and inexpensive to perform.
Relatively few reports have been made of the FAME of Salmonella spp., e.g., a report from 1996 showed the closely related biovars S. enterica Gallinarum and Pullorum could readily be distinguished by FAME analysis (38), and another report in 1995 showed that FAME analysis was a useful technique for distinguishing vaccine strains of both S. enterica Typhimurium and Dublin from field strains of these bacteria (28). In the present study we were able to show clustering of S. enterica Dublin in outbreaks of disease in New York and Pennsylvania. In dendrogram and 2D-Plot analyses of the New York and Pennsylvania isolates of S. enterica Dublin, we noted that six clusters and a number of clones were evident (Fig. 1 and 2 and Tables 3 and 4). Since the New York, Pennsylvania, and Ohio areas were free of S. enterica Dublin prior to 1988, since serotype Dublin is found in a limited host range (cattle for the most part), and since we knew the source(s) of calves for the dairy beef and veal locations were local farms that were free of S. enterica Dublin infection, we have proposed the hypothesis that a transport system (manure-laden trucks) carried S. enterica Dublin from areas of the country where it is endemic to our own area; during 1988 there were outbreaks of S. enterica Dublin in the nearby western states of Illinois and Indiana, as well as in Kentucky (12). Once S. enterica Dublin was present in the sale yard environment due to cross-contamination from animals removed from trucks, it readily spread via calves congregated there to the cattle-raising farm locations. The fact that the same clone was widely disseminated to multiple locations in New York and Pennsylvania simultaneously was interpreted to mean that S. enterica Dublin most likely spread from initial sources such as transport trucks or sale yards on to the many veal or dairy beef operations in the area. Also, the intermittent presence of the same clone at one location for up to 9 months was interpreted as being due to the long-term survival of S. enterica Dublin in the environment of the veal or dairy beef operations. The clustering of cases that seems apparent in Fig. 3 is probably due to a combination of factors. The first is the cattle distribution, since this is the major veal and dairy beef region of the two respective states. The second factor is submission bias, since these areas have ready access to the veterinary college at Cornell University for the cultural confirmation of a field differential diagnosis. However, since efficient commercial courier service is available and owners are willing to drive animals to Cornell, this submission bias is probably not a major factor. Third, these sites also get calves from common sale barns so that many types of infections, including enteric bacterial diseases, are readily spread and shared at the cattle-growing locations.
We compared 96 isolates of S. enterica Dublin available to us from areas where serotype Dublin is endemic in the United States. These isolates were obtained in 1992, the time closest to the start of the outbreaks in late 1988 and early 1989. Using cluster analysis of the FAME profiles (dendrograms), we found that 19 of our local isolates could be linked with strains from 17 other states at levels that would define them as clones (Table 4). Thus, there is plausible support of our hypothesis that contaminated transport trucks brought S. enterica Dublin into our heretofore nonaffected area. The many potential source states for S. enterica Dublin may also account for the variety of clusters (i.e., SD1 to SD6) that we were able to detect in our local area.
There are five isolates of S. enterica Dublin that did not cluster in the FAME analysis and are thus labeled as outliers. These outliers are readily seen in the 2D-Plot (Fig. 2) but are not as easily seen in the dendrogram (Fig. 1) due to the scale of the figure. The significance of the outliers is not known at this time. However, it is plausible that they may represent as-yet-unrecognized clones which may become apparent in future studies with additional isolates of S. enterica Dublin.
Infection in the region with S. enterica Dublin has been confined to the relatively transient populations of veal, which are kept for about 14 weeks before going to market and the relatively new industry called dairy beef in which dairy animals are raised as beef for up to 1.5 years before going to market. These two industries need large numbers of assembled calves and require frequent restocking. Thus, animals in these industries are exposed to significant infection risk factors such as multiple transport, comingling at sales sites, and ample opportunity for fecal-oral cycles of infection (25, 53, 55, 56, 57). Because our data are based on spontaneous disease submissions from referring veterinary clientele, these data are undoubtedly biased. Nevertheless, we feel that if S. enterica Dublin were endemic in New York and Pennsylvania dairy farms, we would have seen more than the two isolated cases of disease in adult milking cows. The two dairy cow cases, NY5 from November 1988 and NY12 from September 1994, were likely to have been acute disease cases resulting in death of the animal on at least one of the farms. Death of the animal precluded the development of a carrier state in one case and thus there should be no long-term problem for the host farms; the second cow case was lost to follow-up. In contrast, in areas of the world where this organism is endemic, carrier cattle serve as a continuing focus of infection for the herd and especially for young stock (36, 53, 57).
A danger to the Northeastern region, however, is the potential residual environmental contamination present in veal and dairy beef operations and also the sale yards and trucks that handled the infected animals. S. enterica Dublin may be spread to other farms via direct or indirect means by the normal course of farm operations and the traffic (both human and animal) that this entails; the organism may also be spread via birds, such as the abundant gulls of the area’s waterways.
Antimicrobial resistance in a pathogen like S. enterica Dublin is important in cattle as a clinical treatment variable and as a risk factor in zoonotic disease (21, 47, 52). As for most other Salmonella spp., many surveys have been published over the years detailing trends in drug resistance for S. enterica Dublin (11, 20, 45, 54). As in our study, at least four other North American studies have found a considerable degree of resistance in S. enterica Dublin (6, 11, 34, 37). While most of these studies provided no information on antimicrobial usage in the host animals from which S. enterica Dublin was isolated, at least one study of California dairies indicated that up to 10% of the S. enterica Dublin isolates were chloramphenicol resistant; this study determined that such resistance was caused by use of chloramphenicol on the farm within a year prior to sampling for the study (34). The results of the present study (Table 5) will provide baseline data for evaluating future trends in antimicrobial resistance in the Northeastern region. The resistance to ampicillin, chloramphenicol, tetracycline, and neomycin noted here reflects either the use of these drugs in the cattle operations of the region and/or their use in other states from which the S. enterica Dublin originated. Neomycin and tetracycline have been commonly used as feed additives, and resistance to these drugs is common in many other enteric bacterial species, such as Escherichia coli, recovered from calves. Resistance to the fluoroquinolone drugs was not encountered in this study, nor have we seen any resistance in other salmonella serotypes from our case load. Fluoroquinolone drugs are not currently licensed for use in calves.
Salmonella serotypes in serogroups B and C generally cause more severe disease than those from serogroup E. However, there is much variation in the clinical presentation in the bovine animal depending on the host’s age, its immune status and intercurrent disease, the infecting Salmonella serotype, its dose and inherent virulence, and other environmental stress factors, such as the availability of adequate food and water and various weather conditions (1, 25, 36). S. enterica Typhimurium (serogroup B) has been the most common serotype in the Northeastern region, where calves from 2 to 8 weeks of age typically present with enteric salmonellosis, i.e., fever and bloody diarrhea with intestinal casts. This is also the typical presentation in adult cattle. In contrast, in areas where it is not endemic, most cases of S. enterica Dublin occur in older calves (8 or more weeks old) that present with septicemic salmonellosis, i.e., with fever, pneumonia, swollen joints, and sometimes diarrhea, rather than a primarily diarrheal syndrome. S. enterica Dublin infections in adult cattle may vary in presentation from fever, bloody diarrhea, and abortions in newly exposed herds to milder forms of diarrhea in endemic herds (1, 50, 55). In areas where serotype Dublin is endemic, the syndrome in calves may present with milder forms of diarrhea only (53). S. enterica Typhimurium is a non-host-adapted serotype, is not known to produce a carrier state, and is known for primarily enteric disease states. S. enterica Dublin, a host-adapted serotype, produces carriers and causes primarily septicemia (6, 7). Because of the differences in pathogenesis and in the clinical presentation in S. enterica Typhimurium- versus serotype Dublin-infected herds, appropriate specimens for the antemortem culture diagnosis of disease may differ, i.e., for Typhimurium, diagnosis would be by fecal culture, whereas for Dublin, diagnosis would be by blood cultures, tracheal washes, and fecal cultures. Serology is also more important in the diagnosis of S. enterica Dublin infections in adult cattle (where carrier states are common) than in Typhimurium disease (17, 18).
S. enterica Dublin may cause severe problems for the Northeastern dairy industry should it become endemic on dairy farms. Though infection appears not to have spread to dairies in the region, a surveillance system that relies on spontaneous disease submissions is likely to miss some cases of salmonellosis unless severe disease occurs in the herds. The present work provides basic information regarding the descriptive epidemiology and microbiology for ongoing evaluation of disease events in the field. New York State is implementing a cattle quality assurance program that will address the problems of salmonellosis prevention and control. Also, additional evaluation of isolates by PFGE and ribotyping is planned in the future. The basis for antimicrobial resistance will be evaluated by plasmid analysis as well.
ACKNOWLEDGMENTS
The MIDI-Sherlock System for FAME analysis was provided, in large part, by funds from the Unrestricted Alumni Grants program of the College of Veterinary Medicine at Cornell University to P.L.M. and also by funds from the New York State Department of Agriculture and Markets.
We also gratefully acknowledge the S. enterica Dublin isolates, helpful discussions, and data provided by David Miller and Kathy Ferris of the Salmonella Serotyping Laboratory of the NVSL, VS, APHIS, USDA, Ames, Iowa.
REFERENCES
- 1.Anderson M, Blanchard P. The clinical syndromes caused by Salmonella infection. Vet Med. 1989;84:816–819. [Google Scholar]
- 2.Anonymous. p. F-1 to F-24. In R. Paisley (ed.), Training manual: MIS whole cell fatty acid analysis by gas chromatography. MIDI, Inc., Newark, Del. 1995. Sample reports. [Google Scholar]
- 3.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Beltran P, Musser J M, Helmuth R, Farmer III J J, Frerichs W M, Wachsmuth I K, Ferris K, McWhorter A C, Wells J G, Cravioto A, Selander R K. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci USA. 1988;85:7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn B O, Sutch K, Harrington R., Jr . Proceedings of the 84th Annual Meeting of the United States Animal Health Association, Louisville, Ky. Richmond, Va: U.S. Animal Health Association; 1980. The changing distribution of Salmonella dublin in the United States; pp. 445–451. [PubMed] [Google Scholar]
- 6.Brackelsberg C A, Nolan L K, Brown J. Characterization of Salmonella dublin and Salmonella typhimurium (Copenhagen) isolates from cattle. Vet Res Commun. 1997;21:409–420. doi: 10.1023/a:1005803301827. [DOI] [PubMed] [Google Scholar]
- 7.Bulgin M S. Salmonella dublin: what veterinarians should know. J Am Vet Med Assoc. 1983;182:116–118. [PubMed] [Google Scholar]
- 8.Celum C L, Chiasson R E, Rutherford G W, Barnhart J L, Echenberg D F. Incidence of salmonellosis in patients with AIDS. J Infect Dis. 1987;156:998–1002. doi: 10.1093/infdis/156.6.998. [DOI] [PubMed] [Google Scholar]
- 9.Chowdry N, Threlfall E J, Rowe B, Stanley J. Genotype analysis of faecal and blood isolates of Salmonella dublin from humans in England and Wales. Epidemiol Infect. 1993;110:217–225. doi: 10.1017/s0950268800068138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewing W H. Identification of Enterobacteriaceae. 3rd ed. Minneapolis, Minn: Burgess Publishing Co.; 1972. The genus Salmonella; pp. 146–256. [Google Scholar]
- 11.Ferris K E, Andrews R E, Jr, Thoen C O, Blackburn B O. Plasmid profile analysis, phage typing, and antibiotic sensitivity of Salmonella dublin from clinical isolates in the United States. Vet Microbiol. 1992;32:51–62. doi: 10.1016/0378-1135(92)90006-f. [DOI] [PubMed] [Google Scholar]
- 12.Ferris K E, Miller D A. Proceedings of the 93rd Annual Meeting of the United States Animal Health Association, Las Vegas, Nev. Richmond, Va: U.S. Animal Health Association; 1989. Salmonella serotypes from animals and related sources reported during July 1988–June 1989; pp. 521–538. [Google Scholar]
- 13.Fierer J. Invasive Salmonella dublin infections associated with drinking raw milk. West J Med. 1983;138:665–669. [PMC free article] [PubMed] [Google Scholar]
- 14.Gower J C. A general coefficient of similarity and some of its properties. Biometrics. 1971;27:857–874. [Google Scholar]
- 15.Guinee P A M, vanLeeuwen W J. Phage typing of Salmonella. In: Bergan T, Norris J R, editors. Methods in microbiology. II. New York, N.Y: Academic Press; 1978. pp. 157–191. [Google Scholar]
- 16.Hampton M D, Ward L R, Rowe B, Threlfall E J. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg Infect Dis. 1998;4:317–320. doi: 10.3201/eid0402.980223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.House J K, Smith B P, Dilling G W, Roden L D. Enzyme-linked immunosorbent assay for serological detection of Salmonella dublin carriers on a large dairy. Am J Vet Res. 1993;54:1391–1399. [PubMed] [Google Scholar]
- 18.House J K, Smith B P, Dilling G, Roden L D, Spier S, Picanso J. Proceedings of the Symposium of the Diagnosis and Control of Salmonella, San Diego, Calif. Richmond, Va: U.S. Animal Health Association; 1991. Serologic detection of Salmonella dublin infection in cattle; pp. 130–135. [Google Scholar]
- 19.Hughes J M, LaMontague J R. The challenges posed by emerging infectious diseases: emerging infections present opportunities for interagency coordination and collaboration. ASM News. 1994;60:248–250. [Google Scholar]
- 20.Jorgensen S T. Prevalence and molecular epidemiology of antibiotic-resistant Salmonella typhimurium and Salmonella dublin in Danish cattle. Acta Pathol Microbiol Immunol Scand Sect B. 1983;91:163–168. doi: 10.1111/j.1699-0463.1983.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 21.Kayser F H, Devaud M, Largiader F, Binswanger V. Acquisition of multiple antibiotic resistance by Salmonella dublin from the gram negative hospital flora, in a kidney allograft recipient. Zentbl Bakteriol Orig A. 1978;241:308–318. [PubMed] [Google Scholar]
- 22.Kerouanton A, Brisabois A, Grout J, Picard B. Molecular epidemiological tools for Salmonella Dublin typing. FEMS Immunol Med Microbiol. 1996;14:25–29. doi: 10.1111/j.1574-695X.1996.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 23.Livesley M A, Thompson I P, Gern L, Nuttall P A. Analysis of intra-specific variation in the fatty acid profiles of Borrelia burgdorferi. J Gen Microbiol. 1993;139:2197–2201. doi: 10.1099/00221287-139-9-2197. [DOI] [PubMed] [Google Scholar]
- 24.McDale J E, Anderson B E. Molecular epidemiology: application of nucleic acid amplification and sequence analysis. Epidemiol Rev. 1996;18:90–97. doi: 10.1093/oxfordjournals.epirev.a017919. [DOI] [PubMed] [Google Scholar]
- 25.McDonough P L. Salmonellosis: diagnostic approach to disease control and epidemiology in the bovine animal. 27th Annual Convention of the American Association of Bovine Practitioners, September 1994, Pittsburgh, Pa. Bovine Proc. 1995;27:61–68. [Google Scholar]
- 26.McDonough P L, Shin S J, Timoney J F. Salmonella serotypes from animals in New York State, 1978–1983. Cornell Vet. 1986;76:30–37. [PubMed] [Google Scholar]
- 27.McDonough P L, Timoney J F, Jacobson R H, Khakhria R. Clonal groups of Salmonella typhimurium in New York State. J Clin Microbiol. 1989;27:622–627. doi: 10.1128/jcm.27.4.622-627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsching M, Selbitz H J. Die identifizierung von Salmonella-impfstammen auf der basis der ganzzellfetsauremusteranalyse. Tieraerztl Umsch. 1995;50:413–417. [Google Scholar]
- 29.Morse S S, Hughes J M. Developing an integrated approach to emerging infectious diseases. Epidemiol Rev. 1996;18:1–3. doi: 10.1093/oxfordjournals.epirev.a017912. [DOI] [PubMed] [Google Scholar]
- 30.Musser J M. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg Infect Dis. 1996;2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard. Document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 32.Olsen J E, Skov M. Genomic lineage of Salmonella enterica serovar Dublin. Vet Microbiol. 1994;40:271–282. doi: 10.1016/0378-1135(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 33.Orskov F, Orskov I. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the Enterobacteriaceae and other bacteria. J Infect Dis. 1983;148:346–347. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- 34.Pacer R E, Spika J S, Thurmond M C, Hargrett-Bean N, Potter M E. Prevalence of Salmonella and multiple antimicrobial-resistant Salmonella in California dairies. J Am Vet Med Assoc. 1989;195:59–63. [PubMed] [Google Scholar]
- 35.Platt D J, Browning L M, Candish D. Molecular analysis of Salmonella enterica serotype Dublin: building bridges between population genetic and molecular epidemiological studies. Electrophoresis. 1995;17:667–671. doi: 10.1002/elps.1150170407. [DOI] [PubMed] [Google Scholar]
- 36.Rebhun W C. Disease of dairy cattle. Baltimore, Md: The William & Wilkins Co.; 1995. Infections of the gastrointestinal tract; pp. 169–173. [Google Scholar]
- 37.Rice D H, Besser T E, Hancock D D. Epidemiology and virulence assessment of Salmonella dublin. Vet Microbiol. 1997;56:111–124. doi: 10.1016/S0378-1135(96)01352-1. [DOI] [PubMed] [Google Scholar]
- 38.Ryll M, Bisgaard M, Christensen J P, Hinz K H. Differentiation of Salmonella Gallinarum and Salmonella Pullorum by their whole-cell fatty acid methyl ester profiles. J Vet Med Ser B. 1996;43:357–363. doi: 10.1111/j.1439-0450.1996.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 39.Sasser, M. 1992. Personal communication.
- 40.Selander R K, Smith N H, Li J, Li P, Beltran P, Ferris K E, Kopecko D J, Rubin F. Molecular evolutionary genetics of the cattle-adapted serovar Salmonella dublin. J Bacteriol. 1992;174:3587–3592. doi: 10.1128/jb.174.11.3587-3592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin S J, McDonough P L. Practical diagnostic procedures for bacterial diseases. In: Kirk R, editor. Current veterinary therapy VIII. W. B. Philadelphia, Pa: Saunders Co.; 1982. pp. 1148–1156. [Google Scholar]
- 42.Siegel J P, Smith A R, Novak R J. Comparison of the cellular fatty acid composition of a bacterium isolated from a human and alleged to be Bacillus sphaericus with that of Bacillus sphaericus isolated from a mosquito larvicide. Appl Environ Microbiol. 1997;63:1006–1010. doi: 10.1128/aem.63.3.1006-1010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A R, Siegel J P. Cellular fatty acid analysis for the classification and identification of bacteria. In: Olson W P, editor. Automated microbial identification and quantitation. Buffalo Grove, Ill: Interpharm Press, Inc.; 1996. pp. 179–222. [Google Scholar]
- 44.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman and Co.; 1973. [Google Scholar]
- 45.Sojka W J, Wray C, McLaren I. A survey of drug resistance in salmonellae isolated from animals in England and Wales in 1982 and 1983. Br Vet J. 1986;142:371–380. doi: 10.1016/0007-1935(86)90033-3. [DOI] [PubMed] [Google Scholar]
- 46.Steele M, McNab W B, Read S, Poppe C, Harris L, Lammerding A M, Odumeru J A. Analysis of whole-cell fatty acid profiles of verotoxigenic Escherichia coli and Salmonella enteritidis with the Microbial Identification System. Appl Environ Microbiol. 1997;63:757–760. doi: 10.1128/aem.63.2.757-760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor D N, Bied J M, Munro J S, Feldman R A. Salmonella dublin infections in the United States, 1979–1980. J Infect Dis. 1982;146:322–327. doi: 10.1093/infdis/146.3.322. [DOI] [PubMed] [Google Scholar]
- 48.Threlfall E J, Ward L R, Rowe B. Spread of multiresistant strains of Salmonella typhimurium phage types 204 and 193 in Britain. Br Med J. 1978;2:997. doi: 10.1136/bmj.2.6143.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidaver A K. Emerging and reemerging infectious diseases: perspectives on plants, animals, and humans. ASM News. 1996;62:583–585. [Google Scholar]
- 50.Walker R. Salmonella dublin infections in cattle in California. Bovine Proc. 1995;27:8–9. [Google Scholar]
- 51.Weidmann M, Bruce J L, Keating C, Johnson A E, McDonough P L, Batt C A. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner S B, Humphrey G L, Kamei I. Association between raw milk and human Salmonella dublin infection. Br Med J. 1979;2:238–241. doi: 10.1136/bmj.2.6184.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wray C. Proceedings of the International Symposium on Salmonella, American Association of Avian Pathologists. Kennett Square, Pa: University of Pennsylvania; 1985. Salmonella dublin infection of cattle in England and Wales: its epidemiology and control; pp. 173–181. [Google Scholar]
- 54.Wray C, McLaren I M, Beedell Y E. Bacterial resistance monitoring of salmonellas isolated from animals, national experience of surveillance schemes in the United Kingdom. Vet Microbiol. 1993;35:313–319. doi: 10.1016/0378-1135(93)90156-2. [DOI] [PubMed] [Google Scholar]
- 55.Wray C, Sojka W J. Reviews of the progress in dairy science: bovine salmonellosis. J Dairy Res. 1977;44:383–425. [PubMed] [Google Scholar]
- 56.Wray C, Todd N, McLaren I, Beedell Y, Rowe B. The epidemiology of salmonella infection in calves: the role of dealers. Epidemiol Infect. 1990;105:295–305. doi: 10.1017/s0950268800047890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wray C, Wadsworth Q C, Richards D W, Morgan J H. A three-year study of Salmonella dublin infection in a closed dairy herd. Vet Rec. 1989;124:532–535. doi: 10.1136/vr.124.20.532. [DOI] [PubMed] [Google Scholar]