Abstract
After an initial wave of coronavirus disease 2019 (COVID-19) in Haiti in summer 2020 (primarily lineage B.1), seropositivity for anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunoglobulin G (IgG) was ~40%. Variant P.1 (gamma) was introduced in February 2021, with an initially limited introduction followed by exponential local dissemination within this unvaccinated population with prior exposure to earlier SARS-CoV-2 lineages.
Haiti, the poorest country in the Western Hemisphere, is currently in the midst of a political crisis, overlaid with a humanitarian crisis compounded by spiking coronavirus disease 2019 (COVID-19) case numbers.
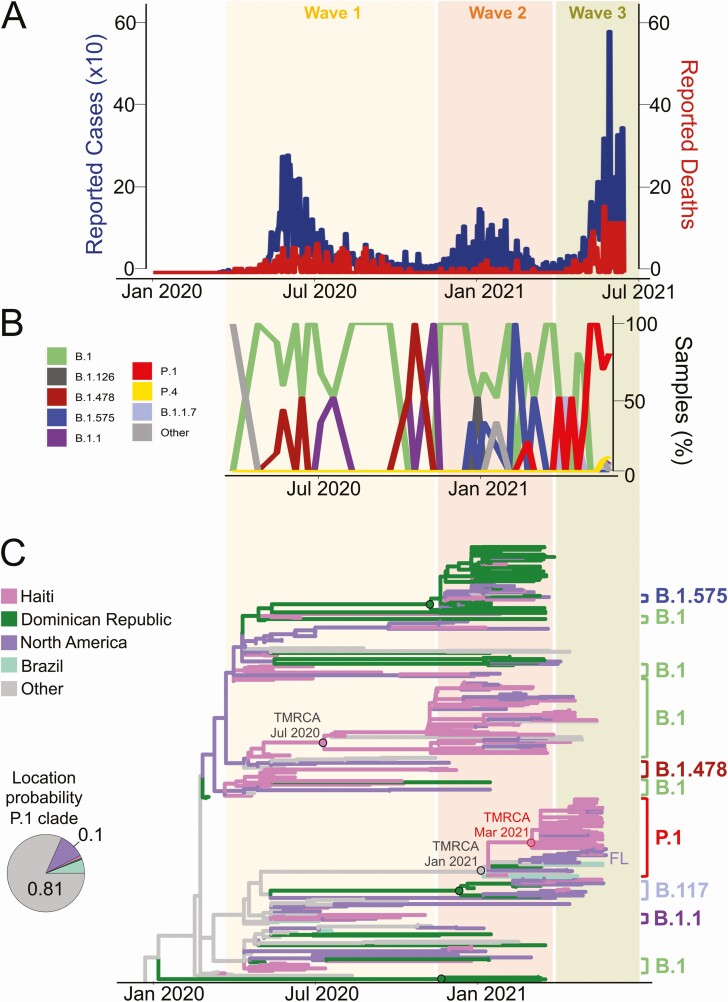
COVID-19 was first reported in Haiti on 19 March 2020. The government responded by stopping commercial passenger flights, mandating a 14-day quarantine for visitors, banning public gatherings, closing schools and factories, and asking people to wear masks. Despite these interventions, case numbers rose rapidly, peaking in early June (“first wave,” Figure 1A). In polymerase chain reaction (PCR)-based testing done by the laboratory of the GHESKIO Centers in collaboration with the Haitian Ministere de la Sante Publique et de la Population, 62% of 1814 clinical nasopharyngeal samples tested in May were positive for severe acute respiratory syndrome (SARS-CoV-2), reflecting the intensity of this initial epidemic. Based on these observations, and with an awareness of local conditions and the limitations in health infrastructure in Haiti, there were major concerns that hospitalization rates and deaths would rapidly increase, with potentially disastrous public health outcomes [1].
Figure 1.
Distribution of Haitian SARS-CoV-2 lineages, reported cases and deaths over time, and phylogeography of SARS-CoV-2 in Haiti. A, Counts of reported cases (×10) and deaths by WHO (https://covid19.who.int/) are shown in blue and red, respectively. B, Distribution of SARS-CoV-2 pango-lineages circulating in Haiti through time and colored according to the legend. Other lineages are defined as present within < 2% of the total sample population. C, Maximum likelihood ancestral state reconstruction of the origin of the COVID-19 epidemic in Haiti performed with TreeTime using genomes from Haiti and other countries linked to Haiti sequences via genetic similarity. Branch lengths are scaled in time by molecular clock calibration and colored according to origin as in the legend. Dots at nodes are colored based on the ancestry; dots circled in black or red indicate ≥ 90% or ≥ 70% (only the case of P.1 Haitian clade) statistical bootstrap support for the clade. B.1 and P.1 Haitian clades are shown. COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WHO, World Health Organization.
The expected major impact on public health did not occur, with the number of reported cases dropping in July 2020, and new cases and deaths declining to very low levels through the end of 2020 [2]. Nonpharmacologic interventions that had been put in place at the beginning of the outbreak were lifted or generally ignored. Although peak case numbers reported in June 2020 were still lower than might be expected with respect to the country’s population (approximately 11 million), testing capabilities for SARS-CoV-2 were known to be extremely limited, leading to an assumption that actual case numbers were substantially higher than reflected by official case counts; it was also believed that high levels of seropositivity in the general population were playing a role in keeping the disease in check. To obtain a more accurate estimate of infection rates in this time period, GHESKIO conducted serologic studies among groups of asymptomatic individuals between 5 August 2020 and 12 January 2021; these and subsequent studies were approved, as appropriate, by ethical committees in Haiti and at the University of Florida. Studies were conducted by using the COVID-Presto lateral flow immunochromatographic assay manufactured by AAZ (Boulogne-Billancourt, France). The manufacturer reports 100% sensitivity and specificity for IgG (by 16 days after onset of infection) for this assay in its package insert; in independent testing of IgG sensitivity and specificity were both reported to be 92% [3]. Of 4550 persons tested by GHESKIO, 1783 (39%) had IgG directed against SARS-CoV-2. The group tested included 236 persons who had a PCR-confirmed history of infection with SARS-CoV-2 dating back 3–6 months; only 132 (56%) of this group were seropositive when the serologic testing was done.
In January 2021, there was again an increase in reported case numbers (“second wave,” Figure 1A) followed by a decline in February, with periods in March and April 2021 when no new cases were officially reported. Case numbers started rising in the latter part of April, with major peaks (“third wave,” Figure 1A) occurring in June and July, accompanied by anecdotal reports of increasing hospitalizations and deaths, particularly among persons with higher socioeconomic status.
To assess factors driving the changing patterns of infections in Haiti, we analyzed 116 nasopharyngeal samples collected by GHESKIO between May 2020 and May 2021 and identified by reverse transcriptase PCR as being SARS-CoV-2 positive; samples were collected from persons with clinical syndromes ranging from mildly symptomatic to severe, including 5 patients who died. From these samples, we successfully obtained nearly full genome viral sequence data for 97 strains (details in Supplementary Material). A data set of all 140 SARS-CoV-2 genomes currently available from Haitian patients (97 generated in this study plus 43 currently listed on GISAID) was assembled and analyzed with Phylogenetic Assignment of Named Global Outbreak Lineages to assess viral variants. Twelve lineages were identified (Table 1; Figure 1B), with the B.1 variant appearing to be the predominant one during the first 2 epidemic waves between March 2020 and February 2021. The increase in case numbers in January 2021 (second wave) also corresponded with the appearance of lineage B.1.575, which has subsequently disappeared, and the variant of concern (VOC) alpha (B.1.1.7). Viral demographic dynamics of the first 2 waves mirror the virus Bayesian skyline plots (Figure S1) inferred from phylodynamic analysis of full genome sequences (see Supplementary Material), showing an initial increase in virus diversity followed by a plateau (first and second wave), or eventual decline (second wave). In February 2021, the first VOC gamma (P.1) sequence was identified from a Haitian patient, with gamma strains responsible for virtually all infections within the third, currently ongoing, epidemic wave. The genealogy of gamma strains from Haiti clearly shows a star-like topology, typical of an epidemic outburst, also confirmed by the strong level of star-like signal in the sequence data (Figure S1C). Haitian gamma strains are characterized by low genetic diversity (2.3e-4, Standard Error = 2.8e-5 with no evidence of protein mutations specific to the Haitian P.1 strains) and appear to have emerged in a short timeframe that is compatible with sudden exponential growth, possibly because of limited introduction followed by 1 or more super spreading events.
Table 1.
Phylogenetic Assignment of Named Global Outbreak Lineages Lineages of Haitian Strains
| Lineage | No. of Haitian Isolates |
|---|---|
| B.1 | 70 |
| B.1.1 | 6 |
| B.1.1.7 | 4 |
| B.1.126 | 3 |
| B.1.2 | 1 |
| B.1.220 | 1 |
| B.1.429 | 1 |
| B.1.478 | 10 |
| B.1.575 | 6 |
| P.1 | 34 |
| P.1.1 | 1 |
| P.4 | 3 |
The single/limited introduction scenario in Haiti of gamma strains was also confirmed by phylogenetic analysis, in which Haitian sequences were combined with the international closest genomes identified by BLAST from the same database as in Rife-Magalis et al [4] (see Supplementary Material for methods). As expected, based on the presence of multiple variants, phylogeographic inference corroborated a parapyletic ancestry of the Haitian epidemic with at least 9 independent introductions from several sources of B.1, B.1.1, B.1.478, B.1.575, alpha (B.1.1.7), and finally gamma (P.1) variants (Figure 1C, Figure S2). Within the larger monophyletic gamma clade, the majority of the isolates from Haiti formed a supported (70% bootstrap) monophyletic subclade with a most recent common ancestor in March 2021. At the base of this clade, the closest paraphyletic isolates were gamma strains from Florida and California, which would suggest initial introduction from the United States. However, alternative routes cannot be excluded, given that Brazilian, European, and the Dominican Republic isolates were also found to cluster with Haiti isolates (Haiti2-39 and Haiti3-20) collected in February and May 2021 (Figure 1C, Figure S3). Overall, although phylogeny inference uncertainty precludes firm conclusions about the origin of gamma strains’ introduction in Haiti, support for the monophyletic clade including most of the Haitian sequences clearly indicate an initially limited introduction followed by local dissemination.
COMMENT
The P.1 lineage originated in the Amazonas region of Brazil in late 2020 and was responsible for a major outbreak in Manaus [5]. Before the outbreak, a modeling study based on data from blood donors suggested that 76% of the population in Manaus had been infected with SARS-CoV-2 by October 2020, consistent with development of “herd immunity” [6]. The factors driving the subsequent occurrence of the Manaus outbreak in this setting remain controversial: although the gamma variant is a VOC known to have substantially increased infectivity, the original estimates of population seropositivity may have been inappropriately high, and there is known to have been a decrease in implementation of and adherence to nonpharmacologic interventions (including social distancing) preceding the epidemic spike [5]. However, there are also recent data from Brazil indicating that plasma from individuals previously infected with SARS-CoV-2 had an 8.6 times lower viral neutralizing capacity against a P.1 isolate than against a lineage B isolate, suggesting that the P.1 variant can escape neutralization by antibodies generated in response to polyclonal stimulation against previously circulating lineages of SARS-CoV-2 [7].
In Haiti, we again observed a relatively high “baseline” seropositivity rate (presumably associated with infection by B.1 strains in the first wave of illness) before introduction of the gamma variant. We also found that close to one-half of people with reverse transcriptase PCR-documented SARS-CoV-2 infections no longer had detectable IgG directed against the virus when screened 3–6 months later, consistent with a waning antibody response over time. Although the Haitian government had imposed a variety of nonpharmacologic interventions at the time the virus was first recognized, these interventions had been lifted and/or were generally ignored by the fall of 2020 and thereafter, making it unlikely that changes in these interventions influenced the appearance of wave 3 of illness triggered by the gamma variant. Susceptibility would also have been influenced by the lack of vaccination within the population. The Haitian Ministere de la Sante Publique et de la Population has held discussions with COVID-19 Vaccines Global Access and World Health Organization/Pan American Health Organization regarding acquisition of COVID vaccine, and the first vaccine shipment (500 000 doses of the Moderna vaccine) arrived in mid-July 2021. However, mass vaccination campaign roll out has been slow because of underlying political instability and ongoing gang warfare, combined with vaccine hesitancy in the population. Our data highlight the potential for rapid spread of highly infectious VOCs such as gamma within unvaccinated populations, despite substantial prior exposure to earlier SARS-CoV-2 lineages. In Haiti in particular, the data also underscore the urgent need for aggressive education and vaccination campaigns to try to contain the current epidemic spike in cases driven by gamma strains.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Conception/design of work: K. Z., J. A. L., M. M. D., B. L., V. R., D. W. F., J. W. P., J. G. M., and M. S. Acquisition/analysis/interpretation of data: M. S. T., K. Z., M. N. C., B. R. M., A. R., J. W. P., J. G. M., and M. S. Drafting/revision of manuscript: M. S. T., C. M., M. N. C., J. A. L., V. R., D. W. F., J. W. P., J. G. M., and M. S.
Financial support. This work was supported in part by funds awarded to M. S. by the University of Florida Office of Research and the UF Clinical and Translational Science Institute; and by funding from the National Institute of Allergy and Infectious Diseases awarded to D. W. F. (AI111143-06S1) and to J. G. M. (AI164007).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Massimiliano S Tagliamonte, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
Carla Mavian, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
Kayvan Zainabadi, Center for Global Health, Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Melanie N Cash, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
John A Lednicky, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Environmental and Global Health, College of Public Health and Health Professions, University of Florida, Gainesville, Florida, USA.
Brittany Rife Magalis, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
Alberto Riva, Bioinformatics Core, Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, Florida, USA.
Marie Marcelle Deschamps, Les Centres GHESKIO, Port-au-Prince, Haiti.
Bernard Liautaud, Les Centres GHESKIO, Port-au-Prince, Haiti.
Vanessa Rouzier, Center for Global Health, Department of Medicine, Weill Cornell Medical College, New York, New York, USA; Les Centres GHESKIO, Port-au-Prince, Haiti.
Daniel W Fitzgerald, Center for Global Health, Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Jean William Pape, Center for Global Health, Department of Medicine, Weill Cornell Medical College, New York, New York, USA; Les Centres GHESKIO, Port-au-Prince, Haiti.
J Glenn Morris, Jr, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
Marco Salemi, Emerging Pathogens Institute, University of Florida, Gainesville, Florida, USA; Department of Pathology, Immunology, and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, Florida, USA.
References
- 1. Rouzier V, Liautaud B, Deschamps MM. Facing the monster in Haiti. N Engl J Med 2020; 383:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bazell R. Looking for the light in Haiti. Science 2020; 369:755–6. [DOI] [PubMed] [Google Scholar]
- 3. Charpentier C, Ichou H, Damond F, et al. . Performance evaluation of two SARS-CoV-2 IgG/IgM rapid tests (Covid-Presto and NG-Test) and one IgG automated immunoassay (Abbott). J Clin Virol 2020; 132:104618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rife Magalis B, Mavian C, Tagliamonte M, et al. . SARS-CoV-2 infection of BNT162b2(mRNA)-vaccinated individuals is not restricted to variants of concern or high-risk exposure environments. medRxiv 2021. [Preprint]. doi: 10.1101/2021.05.19.21257237. [Google Scholar]
- 5. Naveca FG, Nascimento V, de Souza VC, et al. . COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med 2021; 27:1230–8. [DOI] [PubMed] [Google Scholar]
- 6. Buss LF, Prete CA Jr, Abrahim CMM, et al. . Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 2021; 371:288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Souza WM, Amorim MR, Sesti-Costa R, et al. . Neutralization of SARS-coV-2 lineage P.1 by antibodies elicited through natural SARS-CoV-2 infection or vaccination with an inactivated SARS-CoV-2 vaccine: an immunological study. Lancet 2021. Available at: https://doi.org/10.1016/S2666-5247(21)00129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.