Abstract
The secreted frizzled related proteins (SFRPs) are extracellular inhibitors of WNT pathway signaling. Methyl-CpG binding domain protein 2 (MBD2) and enhancer of zeste homolog 2 (EZH2) are core members of the methylated DNA binding domain (MBD) and polycomb group (PcG) protein families for epigenetic regulation, respectively. This study aimed to ascertain the potential role of MBD2 and EZH2 proteins in colorectal cancer (CRC) and its effects on the expression of SFRP. Bioinformatics, real-time quantitative polymerase chain reaction (qPCR) and western blot analysis were used to detect the expression of MBD2, EZH2, and SFRP in CRC cell lines and tissues. The functions of MBD2 and EZH2 in regards to cell proliferation, cell cycle distribution, apoptosis and invasion were examined in CRC cell lines. Methylation-specific PCR (MSP) was used to detect the methylation status of the SFRP promoter. The results revealed that the mRNA expression levels of SFRP were significantly decreased in CRC tissues and cell lines compared to these levels in the adjacent tissues and NCM460, respectively. However, the mRNA levels of EZH2 and MBD2 genes were highly expressed in CRC cell lines. We found that reducing MBD2 and EZH2 expression together remarkably inhibited and decreased the proliferation, migration and invasion abilities of the CRC cell lines compared to reducing one of each. Flow cytometric analysis showed that knockdown of MBD2 and EZH2 together in CRC affected cell apoptosis and the cell cycle progression more effectively than knockdown of one of each. The mRNA expression of SFRP1 was reactivated by silencing of MBD2 but not EZH2 in SW480 and HCT116 cells. SFRP4 and SFRP5 mRNA expression was reactivated by silencing of EZH2 but not MBD2 only in SW480 cells. However, depletion of both MBD2 and EZH2 restored SFRP1, SFRP2, SFRP4, and SFRP5 mRNA expression more effectively in CRC cells. Interestingly, there was no significant change in the methylation status of SFRP1, SFRP2, SFRP4, and SFRP5 gene promoter between before and after interference with MBD2, EZH2, and both. In conclusion, our results suggest that silencing of MBD2 and EZH2 simultaneously was able to rescue the expression of SFRP and inhibit the proliferation of CRC cells more effectively. However, the underlying regulatory mechanism system of MBD2 and EZH2 for SFRP in CRC requires further research.
Keywords: MBD2, EZH2, SFRP, secreted frizzled related proteins, colorectal cancer, DNA methylation
Introduction
Wnt signaling pathway activation is a hallmark of cancer stem cells. Tumor cells can lose the ability to form tumors by modulating Wnt signaling activity. Abnormal activation of the Wnt pathway is associated with a variety of human tumors, such as breast cancer (1), liver cancer (2), lung cancer (3), and promotes tumor cell proliferation and migration.
Colorectal cancer (CRC) is one of the most common tumors found in the digestive system (4). Continuous activation of the Wnt signaling pathway is characteristic of colorectal tumors (5). As an extracellular antagonist of the Wnt pathway, the secreted frizzled related proteins (SFRPs) can competitively inhibit signaling and prevent overactivation of the Wnt pathway (6). However, our recent meta-analysis results indicated that hypermethylation of the SFRP promoter is inversely related to its expression in most tumors and contributes to canceration, at least in CRC (7). Furthermore, our previous study found that SFRP family gene promoter methylation occurred in the early stage of colorectal tumorigenesis, and maintained high frequency methylation in adenomas and adenocarcinomas (8). The methylation rate of SFRP1, SFRP2, SFRP4, and SFRP5 were found to be 93.1, 83.3, 36.1, and 52.8% in colorectal carcinoma (8). Removal of SFRP gene promoter methylation was found to effectively restore gene expression, and to inhibit the abnormal activation of the Wnt signaling pathway, indicating that SFRP gene silencing is associated with hypermethylation of promoter region DNA, which may be the cause of the development of CRC (8,9).
Two classes of proteins have been implicated in the interaction between DNA methylation and histone modifications, possibly related to the formation of tumor-specific epigenomes, the polycomb group (PcG) protein family and the methyl-CpG-binding domain (MBD) protein family. It is known that PcG proteins do not function alone, but are assembled into polycomb repressive complexes 1 and 2, which play important roles in embryonic development, stem cell self-renewal, and cell proliferation through epigenetic modification of target genes (10–12). As an effector component of PRC2, enhancer of zeste homolog 2 (EZH2) can recruit DNA-methyltransferase (DNMT)1, DNMT3A, and DNMT3B to the promoter region of the target gene, and bind to the gene promoter to maintain gene promoter methylation (13,14). It has been reported that EZH2 can promote cell proliferation, invasion and metastasis, and can also endanger DNA damage and repair (15). At the same time, experimental results have shown that tumor cell lines depend on polycomb repressive complex 2 (PRC2) activity, for example, knockout of EZH2 or other PRC2 core components, and the use of small-molecular substances to inhibit EZH2, can reduce the proliferation of cell lines derived from various types of cancer (16). MBD2 plays a transcriptional inhibitory role in tumors mainly by recruiting histone deacetylase complexes NuRD/Mi-2 and Sin3A to form transcriptional repressors (17,18), such as the silencing of the KAI1 gene in prostate cancer PC3 cells (19). We suspect that silencing of MBD2 and EZH2 can alter the methylation status of the SFRP gene promoter and then affect its expression to inhibit the WNT signaling pathway, which may be another target for colorectal tumor treatment.
In the present study, our aim was to explore the potential role of MBD2 and EZH2 proteins in CRC and their effects on the expression of SFRP.
Materials and methods
Analysis of GEPIA and METCH databases
GEPIA (Gene Expression Profiling Interactive Analysis) is an online application developed by Peking University that can be used to analyze the differential expression of genes in cancer and normal tissues (http://gepia.cancer-pku.cn/) (20). GEPIA is a web-based tool for analyzing normal and tumor sample RNA sequencing data based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GETx) data (tumor samples from TCGA dataset, normal samples from two datasets). In the present study, we observed the mRNA expression level of SFRP genes in the GEPIA database with the settings P≤0.01 and |log2(FC)| ≥1. Under this condition, we selected 275 tumor samples from the TCGA database and 349 normal samples from two databases in COAD and 92 tumor samples from TCGA database and 318 normal samples from two databases in READ to analyze the expression of SFRP in CRC. The relationship between SFRP gene expression and promoter methylation was analyzed by the METCH database (http://methhc.mbc.nctu.edu.tw/php/index.php) (21) [Tumor=275, Normal=349 (41 from TCGA database); READ Tumor=92, Normal=318 (10 from TCGA database)].
Cell cultures
Normal colon mucosa cell line (NCM460) and human colorectal cancer cell lines (HCT116, SW480, HT29 and DLD1) were donated from the team of Zhao Qiu, Department of Gastroenterology, Zhongnan Hospital of Wuhan University (Wuhan, China). All cells were cultured in a high glucose medium (DMEM, Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS, Hangzhou Sijiqing Biological Engineering Materials Co., Ltd.) in a humidified atmosphere at 37°C and 5% CO2.
RNA interference and transfection
CRC cells were seeded in a 6-well plate (50,000 cells/ml). Transfection was initiated when the cell density reached 50–60%. Cells were transfected with siRNA using a transfection reagent (GenMute™ siRNA, cat no. SL100568; GenePharma) according to the manufacturer's protocol. Approximately 24 h later, the cells were harvested for analysis as described below. The small interfering RNA (siRNA) sequences targeting human MBD2, EZH2, and negative control (NC) were purchased from Guangzhou RiboBio Co., Ltd. The siRNA sequences used in the study are listed as follows: MBD2-001 forward, 5′-GAGGCUACAAGGACUUAGUTT−3′ and reverse, 5′-ACUAAGUCCUUGUAGCCUCTT−3′; MBD2-002 forward, 5′-GAUGAUGCCUAGUAAAUUATT−3′ and reverse, 5′-UAAUUUACUAGGCAUCAUCTT−3′; MBD2-003 forward, 5′-CCUGGGAAAUACUGUUGAUTT−3′ and reverse, 5′-AUCAACAGUAUUUCCCAGGTT−3′; EZH2-2196 forward, 5′-GCAGCUUUCUGUUCAACUUTT−3′ and reverse, 5′-AAGUUGAACAGAAAGCUGCTT−3′; EZH2-488 forward, 5′-GACUCUGAAUGCAGUUGCUTT−3′ and reverse, 5′-AGCAACUGCAUUCAGAGUCTT−3′; EZH2-1952 forward, 5′-CCUGACCUCUGUCUUACUUTT−3′ and reverse, 5′-AAGUAAGACAGAGGUCAGGTT−3′. A nonspecific control siRNA was used as a control: NC forward, 5′-UUCUCCGAACGUGUCACGUTT−3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT−3′.
RNA isolation and quantitative real-time PCR (qPCR)
CRC cells were placed in an EP tube containing 1 ml of TRIzol reagent (Thermo Fisher Scientific, Inc.). Total RNA extraction was performed according to the manufacturer's instructions. The resulting RNA was dissolved in RNase-free water and immediately stored at −80°C. The RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was then synthesized using a ReverTra Ace qPCR RT kit (Toyobo Life Science). qPCR was performed using SYBR-Green PCR master mix in a Biorad 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR primers for MBD2, EZH2, and GAPDH were purchased from QingKe. The specific primers used are as follows: MBD2 forward, 5′-GCAAGCCTCAGTTGGCAAG-3′ and reverse, 5′-ATCGTTTCGCAGTCTCTGTTT−3′; EZH2 forward, 5′-TCCTACATCCTTTTCATGCAACAC−3′ and reverse, 5′-GCTCCCTCCAAATGCTGGTA-3′; SFRP1 forward, 5′-TGGCCCGAGATGCTTAAGTG-3′ and reverse, 5′-GACACACCGTTGTGCCTTG-3′; SFRP2 forward, 5′-CGACATGCTTGAGTGCGAC-3′ and reverse, 5′-CTTTGGAGCTTCCTCGGTGG-3′; SFRP4 forward, 5′-TCACCCATCCCTCGAACTCA-3′ and reverse, 5′-CATCATCCTTGAGCGCCACT-3′; SFRP5 forward, 5′-CCTCCAGTGACCAAGATCTGC−3′ and reverse, 5′-TCCTTGATGCGCATTTTGACC−3′; GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-GCAGGAGGCATTGCTGATGAT−3′. GAPDH was used as a control. All reactions were run in triplicate, and the results were analyzed and expressed relative to threshold cycle (CT) values and then converted to fold change values (2−ΔΔCq) (22).
Western blot analysis
Cells were lysed with RIPA protein extraction reagent (Beyotime Institute of Biotechnology) supplemented with 1% phenylmethanesulfonyl fluoride (Seebio Science & Technology Co., Ltd.). Protein concentrations were measured using an enhanced BCA protein assay kit (Beyotime Institute of Biotechnology). Each lane contained equal amounts of protein (30 µg). Proteins were separated using 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were then transferred to a polyvinylidene fluoride membrane (Millipore, USA). The membranes were blocked with 5% milk for 2 h at room temperature. The membranes were then placed in a TBST solution containing anti-SFRP1 antibody (dilution 1:500; cat. no. AB_2764730; ABclonal Biotechnology Co., Ltd.), SFRP2 (dilution 1:1,000; cat. no AB_2766193; ABclonal Biotechnology Co., Ltd.), SFRP4 (dilution 1:1,000; cat. no. AB_2767011; ABclonal Biotechnology Co., Ltd.), SFRP5 (dilution 1:1,000; cat. no. AB_2772201; ABclonal Biotechnology Co., Ltd.), MBD2 (dilution 1:500; cat. no. ab38646; Abcam), and EZH2 antibodies (dilution 1:1,000; cat. no. ab186006; Abcam) overnight (~12 h) at 4°C. The following day, after washing with TBST, the blots were incubated with an HRP-labelled anti-rabbit (dilution 1:5,000, cat. no. GB233303-1, Servicebio) secondary antibody for 2 h at room temperature. The blots were visualized using a super ECL detection reagent (Solarbio, Beijing, China). ImageJ 1.8.0 (NIH) was used for densitometry. Each set was repeated at least three times.
Wound-healing assay
CRC cells were seeded in a 6-well plate (50,000 cells/ml). Twenty-four hours post-transfection, the cells had reached 70–80% confluency. A straight scratch was created using a sterile yellow pipette tip. PBS was added to the 6-well plate to remove floating cells. The original culture medium was replaced with 0.5% FBS for another 24 h as previously described (23). All tests were repeated three times. Cell migration was observed and imaged at 0, 24, and 36 h with a digital camera (×40 magnification; OLYMPUS U-RFL-T; Olympus Corp.).
Cell proliferation assay
At 24 h post-transfection, SW480 and HCT116 cells were trypsinized to provide suspensions that were then seeded in 96-well plates at densities of 2,000 cells/well. The cell proliferation rates were calculated using the CCK-8 assay at 0, 24, 48, and 72 h. Briefly, 10 µl of CCK-8 was added to each well, and the cells were allowed to incubate at 37°C in 5% CO2 for 2 h. The absorbance value of each well at 570 nm was recorded. Each experiment was repeated at least three times.
Cell invasion assay
A 24-multiwell insert plate with a small chamber (BD Biosciences) containing an 8.0-micron pore size Matrigel-coated membrane was used for the cell invasion assays. Briefly, Matrigel liquid was added to the chamber the day before the experiment and placed in a 4°C refrigerator overnight to solidify. On the next day, the cells of each group that had been transfected in a 6-well plate were digested and centrifuged. A total of 1×105 cells were seeded into the upper chambers (coated in Matrigel) in serum-free medium. The 24-well plates were filled with 600 µl of DMEM containing 20% FBS as a chemo-attractant. After the plates were incubated at 37°C for 24 h, the non-invasive cells above the chamber were gently wiped with a wet cotton swab. The cells below the chamber were fixed with 4% paraformaldehyde for 30 min, stained with 0.1% crystal violet for 20 min, and then counted under an optical microscope (40× magnification). Each set was repeated at least three times.
Analysis of cell apoptosis and cell cycle distribution
The cells were seeded in 6-well plates at 5×105 cells/well. After transfection with siRNA for 24 h, SW480 and HCT116 cells were harvested. Cycle and apoptosis kits (MultiSciences, China) were used to detect apoptosis and cell cycle distribution of the SW480 and HCT116 cells according to the manufacturers' instructions. The cell cycle distribution and apoptosis rate were analyzed using a flow cytometer (Beckman CytoFLEX FCM). Specific methods were carried out according to our previous experiments (24).
Methylation specific PCR (MSP)
DNA was extracted using TIANamp Genomic DNA Kit (Tiangen Biotech Co.). Sodium bisulfite treatment was performed using an EZ DNA Methylation Kit (Zymo Research). The regions of the methylated SFRP1, SFRP2, SFRP4, and SFRP5 promoters by MSP analysis were chromosome 8 (41309333-41309459, length: 126 bp), chromosome 4 (153788916-153789054, length: 138 bp), chromosome 7 (37916853-37916965, length: 112 bp) and chromosome 10 (97772023-97772159, length: 136 bp), respectively. The sequence information of the SFRP primers used in the MSP experiments are shown in Table I.
Table I.
SFRP primer sequence information for MSP experiments.
| Gene name | Primer sequence |
|---|---|
| SFRP1(U) | F: 5′-GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT-3′ |
| R: 5′-CCTACGATCGAAAACGACGCGAACG-3′ | |
| SFRP2(U) | F: 5′-TTTTGGGTTGGAGTTTTTTGGAGTTGTGT-3′ |
| R: 5′-AACCCACTCTCTTCACTAAATACAACTCA-3′ | |
| SFRP4(U) | F: 5′-GGGGGTGATGTTATTGTTTTTGTATTGAT-3′ |
| R: 5′-CACCTCCCCTAACATAAACTCAAAACA-3′ | |
| SFRP5(U) | F: 5′-GTAAGATTTGGTGTTGGGTGGGATGTTT-3′ |
| R: 5′-AAAACTCCAACCCAAACCTCACCATACA-3′ | |
| SFRP1(M) | F: 5′-TGTAGTTTTCGGAGTTAGTGTCGCGC-3′ |
| R: 5′-CCTACGATCGAAAACGACGCGAACG-3′ | |
| SFRP2(M) | F: 5′-GGGTCGGAGTTTTTCGGAGTTGCGC-3′ |
| R: 5′-CCGCTCTCTTCGCTAAATACGACTCG-3′ | |
| SFRP4(M) | F: 5′-GGGTGATGTTATCGTTTTTGTATCGAC-3′ |
| R: 5′-CCTCCCCTAACGTAAACTCGAAACG-3′ | |
| SFRP5(M) | F: 5′-AAGATTTGGCGTTGGGCGGGACGTTC-3′ |
| R: 5′-ACTCCAACCCGAACCTCGCCGTACG-3′ |
MSP, methylation specific PCR; U, unmethylated; M, methylated; SFRP, secreted frizzled related protein.
Statistical analysis
The correlation between SFRP expression and its methylation were confirmed by Pearson correlation coefficients. The data presented in the text and figures were analyzed using one-way analysis of variance (ANOVA) with a Bonferroni correction. Data are presented as the mean standard ± deviation. Statistical analyses were performed using SPSS 22.0 (IBM Corp). A P-value of <0.05 was considered as indicative of statistical significance.
Results
Expression of SFRP, MBD2, and EZH2 in colorectal tumor tissues and cells
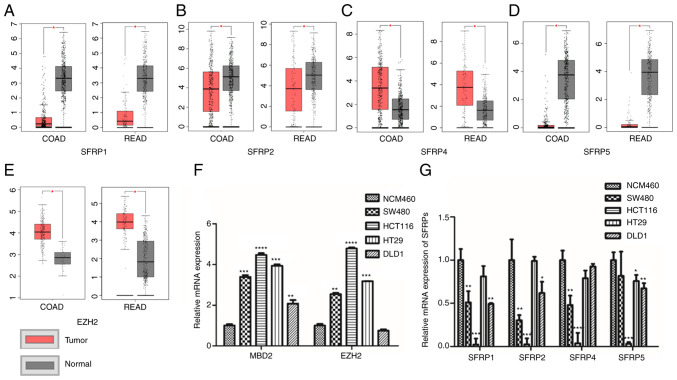
The TCGA database (including 275 colorectal tumor tissues and 349 normal tissues) demonstrated that the expression of SFRP1, SFRP2, and SFRP5 genes were lower in CRC tissues compared (COAD, colon adenocarcinoma; READ, rectum adenocarcinoma) to that in the corresponding paratumorous normal tissue (Fig. 1A, B and D), whereas EZH2 and SFRP4 expression levels were higher in the tumor tissue than levels in the matching normal colorectal tissues (Fig. 1C and E). Furthermore, the expression of SFRP1, SFRP2, SFRP4, and SFRP5 genes were all lower in the CRC cell lines than that noted in the normal colorectal mucosal cells (NCM460) (Fig. 1G). However, the expression levels of MBD2 and EZH2 were much higher in the CRC cells rather than in normal colorectal mucosal epithelial cells (NCM460) (Fig. 1F).
Figure 1.
Expression levels of SFRPs, MBD2 and EZH2 in colorectal tumor tissues and cells. The expression of SFRP1, (A) SFRP2, (B) SFRP4, (C) SFRP5 (D) and EZH2 (E) in TCGA and GTEx database [COAD: Tumor=275, Normal=349 (41 from TCGA database); READ Tumor=92, Normal=318 (10 from TCGA database)]. The mRNA expression of MBD2, EZH2 (F) and SFRPs (G) in normal colorectal epithelial cells (NCM460) and colorectal tumor cell lines (SW480, HCT116, HT29 and DLD1) was detected by qPCR. COAD, colon adenocarcinoma; READ, rectum adenocarcinoma; COAD num(T)=275; num(N)=349; READ num(T)=92; num(N)=318. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. normal colonic mucosal tissues or normal colon mucosa cell line (NCM460). SFRPs, secreted frizzled related proteins; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
Correlation between SFRP gene expression and promoter methylation in colorectal tumors
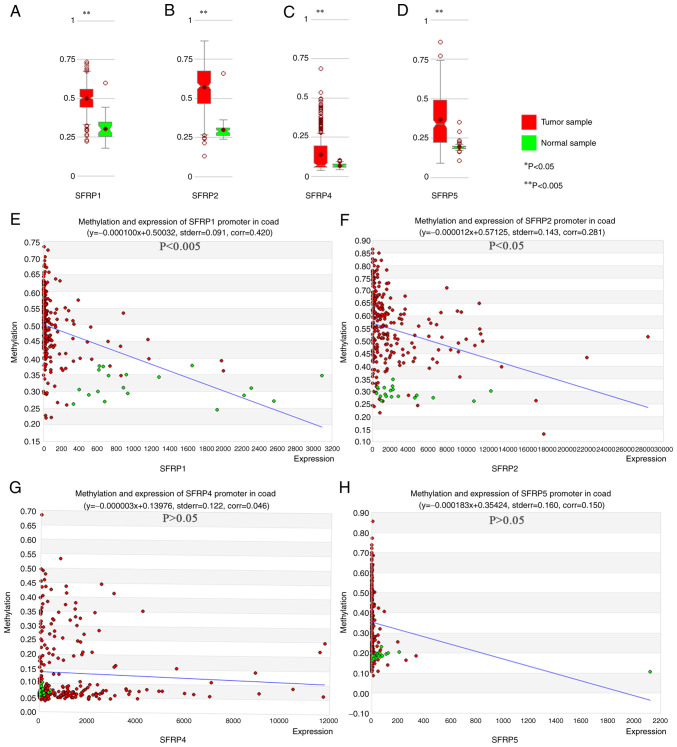
MethHC online database confirmed the greater degree of SFRP1, SFRP2, SFRP4, and SFRP5 promoter methylation in tumor tissue compared with that noted in the paired normal samples (Fig. 2A-D). In colorectal tumors, the expression of SFRP1 and SFRP2 genes were inversely proportional to the degree of promoter methylation (Fig. 2E and F, P<0.05). However, the expression of SFRP4 and SFRP5 genes exhibited no obvious relationship with the degree of promoter methylation (Fig. 2G and H, P>0.05).
Figure 2.
MethHC online database shows the degree of DNA methylation of SFRP promoter and its relationship with gene expression in colorectal tumor and normal tissues. The degree of methylation of SFRP1, (A) SFRP2 (B), SFRP4 (C) and SFRP5 (D) promoters in colorectal tumor and normal tissues. Correlations between methylation of SFRP1 (E, corr=0.420, P<0.005), SFRP2 (F, corr=0.281, P<0.05), SFRP4 (G, corr=0.046, P>0.05) and SFRP5 (H, corr=0.150, P>0.05) promoters and gene expression in colorectal tumor and normal tissues. **P<0.005 vs. normal sample. SFRP, secreted frizzled related protein.
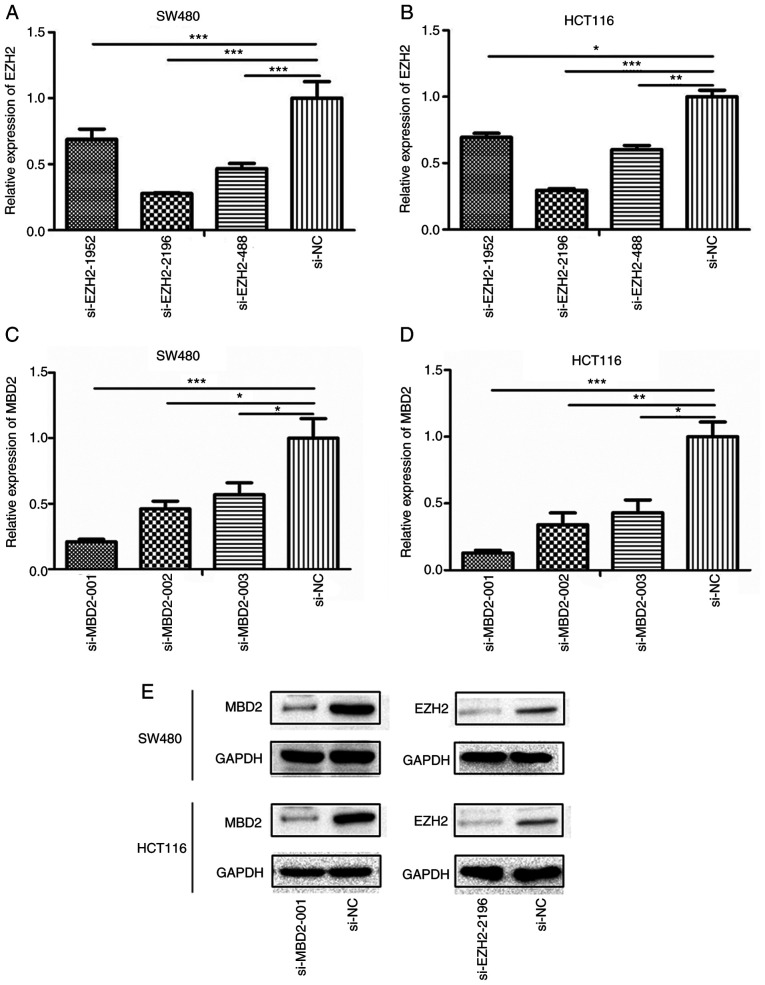
Determination of the transfection efficiency of si-MBD2 and si-EZH2
Twenty-four hours after transfection, the interference efficiency of siRNA-EZH2-1952, siRNA-EZH2-2196, siRNA-EZH2-488 and siRNA-MBD2-001, siRNA-MBD2-002, siRNA-MBD2-003 was detected by qPCR. The results showed that siRNA-EZH2-2196 and siRNA-MBD2-001 performed the best in both SW480 and HCT116 cell lines (Fig. 3A-D). Moreover, it was further verified that siRNA-EZH2-2196 and siRNA-MBD2-001 had the best interference efficiency at the protein level (Fig. 3E). Therefore, we selected these siRNAs for subsequent experiments.
Figure 3.
Transfection efficiency of MBD2 and EZH2 in colorectal tumor cells (SW480 and HCT116). qPCR screening of the most efficient EZH2 interference sequence in SW480 (A) and HCT116 (B) cells; qPCR was used to screen the most efficient MBD2 interference sequence in SW480 (C) and HCT116 (D) cells. (E) Western blot analysis was used to identify the efficiency of si-MBD2-001 and si-EZH2-2196 in SW480 and HCT116. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
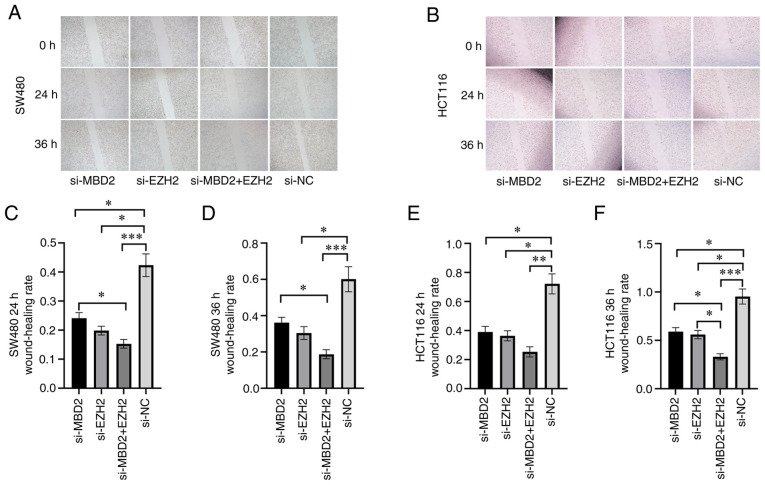
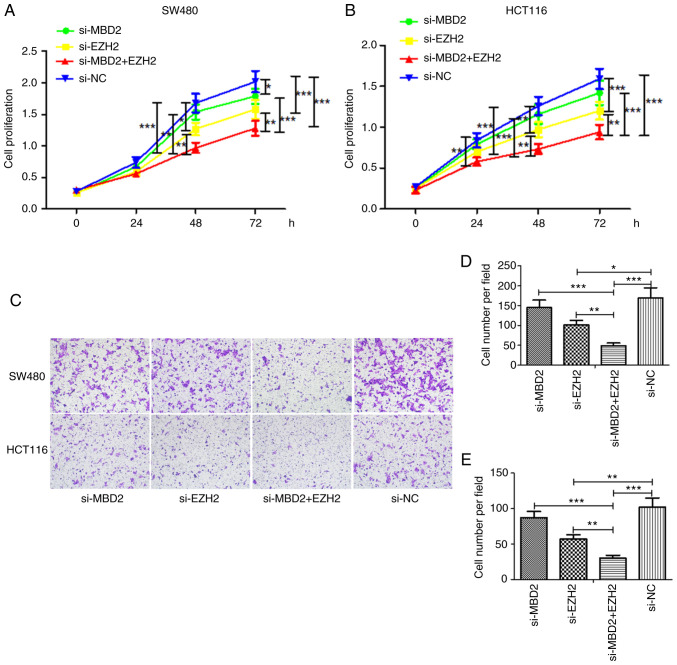
Effects of MBD2 and EZH2 knockdown on CRC cell migration
In order to detect the effect of MBD2 and EZH2 on the migratory ability of CRC cells, a scratch wound assay was used to detect the wound healing of SW480 and HCT116 cells in which MBD2 and EZH2 knockdown was conducted separately or simultaneously. Following 24 h of wounding in the SW480 cells, knockdown of either MBD2 or EZH2 significantly inhibited cell migration when compared with the negative control (si-NC) group (Fig. 4A and C). Furthermore, knockdown of both MBD2 and EZH2 simultaneously inhibited cell migration more effectively, which was statistically significant compared with the knockdown of MBD2 and EZH2, respectively (Fig. 4A and C). Compared to the si-NC group, knockdown of EZH2 or MBD2 decreased cell migration following 36 h after wounding in the SW480 cells (Fig. 4A and D); while knockdown of MBD2 and EZH2 together significantly inhibited cell migration, which was statistically significant compared with the knockdown of MBD2 alone (Fig. 4A and D). The migration of HCT116 cells following the knockdown of MBD2 and EZH2 separately or simultaneously was also decreased compared with the si-NC group 24 h after wounding (Fig. 4B and E). Following 36 h after wounding in HCT116 cells, compared with the si-NC group, knockdown of MBD2 and EZH2 separately or simultaneously inhibited cell migration; however, the group with simultaneous knockdown of MBD2 and EZH2 exhibited inhibition of cell migration to a greater degree (Fig. 4B and F). Therefore, wound healing assays showed that knockdown of MBD2 and EZH2 inhibited the migration of SW480 and HCT116 cells compared to the control group, and depletion of MBD2 and EZH2 plays a greater role in inhibiting the migration of CRC cells.
Figure 4.
Effects of MBD2 and EZH2 knockdown separately or simultaneously on CRC cell migration. Wound healing assay was used to detect knockdown of MBD2 and EZH2 separately or simultaneously on (A) SW480 and (B) HCT116 cell migration. The effects of knockdown of MBD2 and EZH2 separately or simultaneously on migration of SW480 cells were statistically analyzed following wounding at 24 h (C) and 36 h (D) The effects of knockdown of MBD2 and EZH2 separately or simultaneously on migration of HCT116 cells were statistically analyzed following wounding at 24 h (E) and 36 h. (F) *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. CRC, colorectal cancer; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
Effects of MBD2 and EZH2 on the proliferation and invasion of CRC cell lines
CCK-8 assay was used to detect the effects of MBD2 and EZH2 on the proliferation of CRC cells. In the SW480 cell line (Fig. 5A), the cell proliferation in the EZH2-knockdown group was slower than that noted in the si-NC group at 48 h; yet, knockdown of MBD2 had little effect on cell proliferation. However, knockdown of both MBD2 and EZH2 significantly inhibited the proliferation of cells, which was significantly different following knockdown of MBD2 or EZH2 alone. At 72 h, the proliferation of the cells was inhibited by knockdown of MBD2 or EZH2, and also was significantly inhibited by interference with both MBD2 and EZH2, which was significantly different following knockdown of MBD2 or EZH2 alone.
Figure 5.
Effects of MBD2 and EZH2 on proliferation and invasion of CRC cells. CCK-8 assay was used to detect the effects of MBD2 and EZH2 on the proliferation of SW480 (A) and HCT116 (B) cells. (C) The effects of MBD2 and EZH2 on the invasion of SW480 and HCT116 were detected by Transwell assay. Statistical analysis of knockdown of MBD2 and EZH2 on the invasiveness of SW480 (D) and HCT116 (E) cells. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. CRC, colorectal cancer; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
In HCT116 cells (Fig. 5B), knockdown of MBD2 or EZH2 at 24 h was statistically significant at inhibiting proliferation compared with the si-NC group, and knockdown of EZH2 at 48 and 72 h inhibited the growth of CRC cells, while knockdown of MBD2 resulted in basically the same result as in the si-NC group. Furthermore, knockdown of MBD2 and EZH2 together significantly inhibited cell proliferation, which was significantly different following knockdown of MBD2 or EZH2 alone.
The impact of MBD2 and EZH2 on CRC cell invasiveness was investigated using Matrigel invasion chambers. As shown in Fig. 5C-E, the same results were obtained in both SW480 and HCT116 cells. That is, compared with the si-NC group, knockdown of EZH2 weakened the invasiveness of CRC cells, while knockdown of MBD2 had no significant effect on the invasiveness of CRC cells. Yet, knockdown of MBD2 and EZH2 simultaneously inhibited the invasiveness of the CRC cells, which was significantly different following the knockdown of MBD2 or EZH2 alone. The results were consistent with the results of the cell proliferation.
Effects of MBD2 and EZH2 silencing on the apoptosis of CRC cells
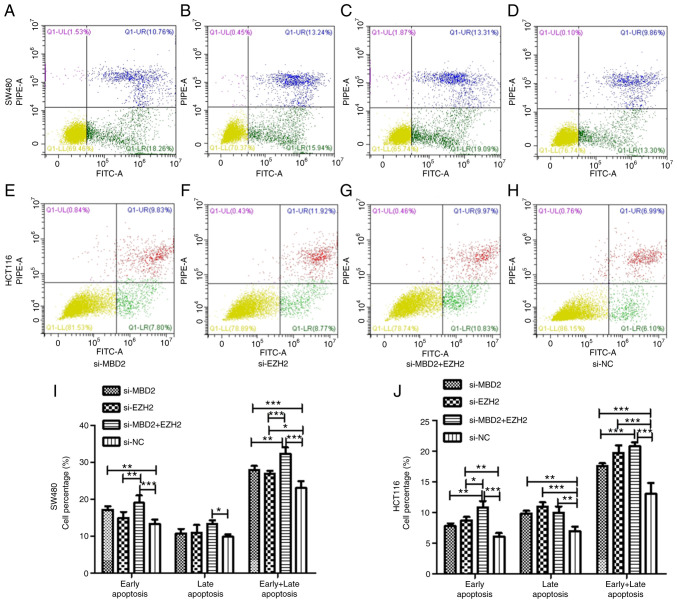
To verify the effect of MBD2 and EZH2 on cell apoptosis, we knocked down MBD2 and EZH2 separately or simultaneously in SW480 and HCT116 cells and then examined apoptosis via flow cytometry.
As is shown in Fig. 6, in SW480 cells, knockdown of MBD2 was able to increase the percentage of early apoptosis of the cells compared with the si-NC group, but there was no significant difference between knockdown of EZH2 and the si-NC group. Simultaneously, knockdown of MBD2 and EZH2 together increased the percentage of early apoptosis of the SW480 cells more significantly. Compared with the si-NC group, knockdown of MBD2 and EZH2 together also increased the late apoptosis of CRC cells, but knockdown of MBD2 or EZH2 did not affect late apoptosis. Knockdown of MBD2 and EZH2 separately and simultaneously increased the percentage of total apoptosis of CRC cells, and the percentage of total apoptosis of CRC cells by knockdown of MBD2 and EZH2 together was significantly higher following knockdown of MBD2 or EZH2 alone (Fig. 6I).
Figure 6.
Effect of the knockdown of MBD2 and EZH2 on the apoptosis of CRC SW480 and HCT116 cells. Flow cytometry was used to detect the effect of the knockdown of MBD2, (A) EZH2, (B) MBD2 and EZH2 (C) and control group si-NC (D) on apoptosis of SW480 cells. Flow cytometry was used to detect the effect of the knockdown od MBD2, (E) EZH2, (F) MBD2 and EZH2 (G) and control group si-NC (H) on apoptosis of HCT116 cells. Statistical analysis of the knockdown of MBD2 and EZH2 on the percentage of apoptotic cells of SW480 (I) and HCT116. (J) *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. CRC, colorectal cancer; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
In HCT116 cells, the results indicated that knockdown of EZH2 increased the percentage of early apoptosis of CRC cells, while knockdown of MBD2 had no significant effect on it. Knockdown of both MBD2 and EZH2 increased the percentage of early apoptosis of CRC cells, and there was a statistical difference compared with the knockdown of either MBD2 or EZH2 alone. Knockdown of MBD2 and EZH2 separately and simultaneously increased the percentage of late apoptosis of CRC cells compared with the si-NC group. Moreover, knockdown of MBD2 and EZH2 simultaneously increased the percentage of total apoptosis of the CRC cells, and the effect was more significant than the group with knockdown of EZH2, while there was no significant difference compared with the MBD2-knockdown group (Fig. 6J).
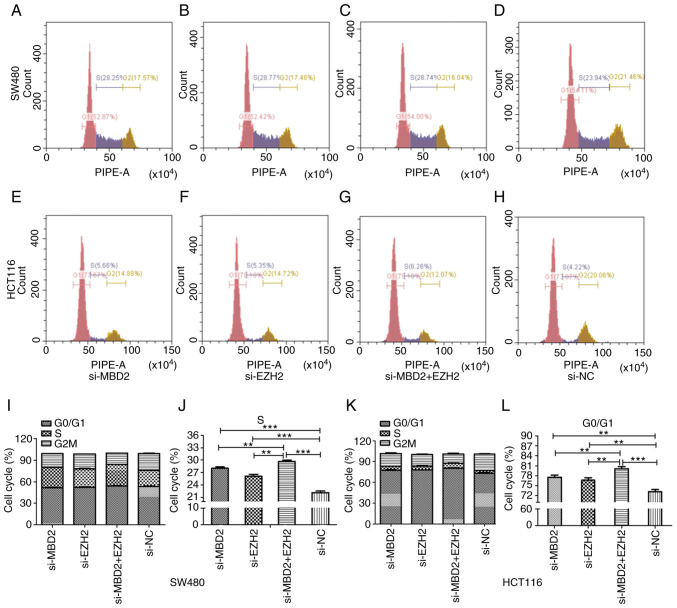
Effects of MBD2 and EZH2 silencing on the cell cycle distribution of CRC cells
As shown in Fig. 7, in SW480 and HCT116 cells, knockdown of MBD2 and EZH2 separately or simultaneously affected the cell cycle distribution compared to the si-NC group. More cells were arrested at S phase of the cell cycle compared with the si-NC group in the SW480 cell line, and there was a significant difference between the group with simultaneous knockdown of MBD2 and EZH2 and the groups with knockdown of MBD2 or EZH2 alone (Fig. 7I and J). While in HCT116 cells, there was a higher percentage of cells arrested at the G0/G1 phase of the cell cycle, and the percentage of cells at G0/G1 phase in the group with simultaneous knockdown of MBD2 and EZH2 was higher than that in the groups with knockdown of MBD2 or EZH2 alone (Fig. 7K and L).
Figure 7.
Effects of MBD2 and EZH2 on cell cycle distribution of CRC SW480 and HCT116 cells. Flow cytometry was used to detect the cell cycle distribution following knockdown of MBD2, (A) EZH2, (B) MBD2 and EZH2 (C) and control group si-NC (D) in SW480 cells. Flow cytometry was used to detect the cell cycle distribution following knockdown of MBD2, (E) EZH2, (F) MBD2 and EZH2 (G) and control group si-NC (H) in HCT116 cells. Histogram showing the effects of the knockdown of MBD2 and EZH2 on the cell cycle distribution of SW480 (I) and HCT116 (K) cells. Statistical analysis was performed to analyze the effect of the knockdown of MBD2 and EZH2 separately and simultaneously on the S phase of the cell cycle in SW480 (J) and on the G0/G1 phase of the cell cycle in HCT116 (L) cells. **P<0.01, ***P<0.001 vs. si-NC. CRC, colorectal cancer; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2.
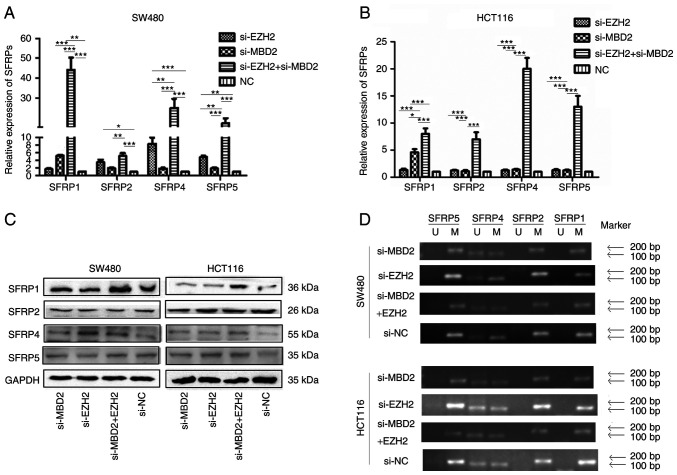
Effect of MBD2 and EZH2 silencing on SFRP gene expression in CRC cells
qPCR was used to detect the mRNA expression of SFRP before and after knockdown of MBD2 and EZH2. As shown in Fig. 8A, in the SW480 cells, the expression of SFRP1 was restored by knockdown of MBD2 (P<0.05), while the expression levels of SFRP2, SFRP4 and SFRP5 could not be restored by knockdown of MBD2 (P>0.05). The expression levels of SFRP2, SFRP4 and SFRP5 could be restored by knockdown of EZH2, while the expression of SFRP1 could not be restored by knockdown of EZH2 (P>0.05), and the expression of SFRP1, SFRP2, SFRP4 and SFRP5 could be significantly restored by simultaneous knockdown of MBD2 and EZH2 (P<0.05), which was significantly different from the groups with knockdown of MBD2 and EZH2 separately. In HCT116 cells (Fig. 8B), the expression levels of SFRP1, SFRP2, SFRP4, and SFRP5 were not restored by knockdown of EZH2. Knockdown of MBD2 restored the expression of the SFRP1 gene (P<0.05), but had no significant effect on the expression levels of SFRP2, SFRP4, and SFRP5 (P>0.05). However, knockdown of MBD2 and EZH2 markedly restored the expression of SFRP1, SFRP2, SFRP4, and SFRP5 (P<0.05), which was significantly different from the group with knockdown of MBD2 or EZH2 alone (P<0.05). The results indicated that the expression levels of SFRP1, SFRP2, SFRP4, and SFRP5 could be more effectively restored by knockdown of MBD2 and EZH2 together in CRC cells.
Figure 8.
Effects of MBD2 and EZH2 on expression and promoter methylation of SFRP genes in CRC cells. (A and B) Effects of the knockdown of MBD2 and EZH2 on SFRP mRNA in SW480 (A) and HCT116 (B) cells as detected using qPCR. (C) Effects of the knockdown of MBD2 and EZH2 on the protein expression of SFRP proteins in SW480 and HCT116 as detected using western blot analysis. (D) Effects of MBD2 and EZH2 on promoter methylation of SFRP genes in SW480 and HCT116 cells. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC. CRC, colorectal cancer; MBD2, methyl-CpG binding domain protein 2; EZH2, enhancer of zeste homolog 2; SFRP, secreted frizzled related protein.
Western blot analysis was used to investigate the effect of MBD2 and EZH2 silencing on the expression of SFRP protein in CRC cells. As shown in Fig. 8C, in SW480 and HCT116, there was no significant difference in the expression of SFRP1, SFRP2, SFRP4, and SFRP5 proteins by knockdown of MBD2 or EZH2 alone compared to the control group, while the protein levels of SFRP1 and SFRP4 were significantly restored by knockdown of MBD2 and EZH2 together.
Effect of MBD2 and EZH2 on promoter methylation of SFRP gene in colorectal cancer cells. As shown in Fig. 8D, methylation-specific PCR results showed that following either knockdown of MBD2 and EZH2 simultaneously or separately, the SFRP1, SFRP2, and SFRP5 promoters were all methylated except for partial methylation of the SFRP4 promoter in SW480 and HCT116 cells compared with control group. There was no significant change in the methylation status of SFRP1, SFRP2, SFRP4, and SFRP5 gene promoter between before and after interfering with MBD2, EZH2, and both.
Discussion
As a negative regulator of Wnt signaling, the secreted frizzled related proteins (SFRPs) can directly block the transmission of the Wnt signaling pathway and are downregulated in many types of tumors due to hypermethylation of the promoter (25–28). In the present study, we found that SFRP genes were downregulated in colorectal tumors by GEPIA and MethHC online database, and this was inversely correlated with hypermethylation of the promoter, indicating that hypermethylation of the promoter is an important reason for downregulation of SFRP genes.
Methyl-CpG binding domain protein 2 (MBD2) and enhancer of zeste homolog 2 (EZH2) are important members of the methylated DNA binding domain (MBD) and polycomb group (PcG) protein family, respectively, and play important roles in DNA methylation and histone modification. It was found that MBD2 recognizes methylated promoters and forms transcriptional repressors by recruiting histone deacetylase complexes NuRD/Mi-2 and Sin3A to inhibit gene expression (17,18,29). EZH2 as the main effector component of the PcG protein family, can recruit DNMT1, DNMT3A and DNMT3B to the promoter region of the target gene and binds to the gene promoter to maintain the stability of the gene promoter methylation, causing chromatin contraction and RNA polymerase II function pause (13). Therefore, MBD2 and EZH2 may be important molecules regulating the expression of SFRP genes in colorectal tumors.
Studies have found that MBD2 has been linked to disease such as immune system function and tumorigenesis (17,30,31), while EZH2 is closely related to tumor migration (32), proliferation (15), and invasion (33), and may be an important target for tumor treatment (34,35). The Comet team summed up the role of EZH2 in tumors, and found that EZH2 has the characteristics of promoting and suppressing cancer, indicating that the relationship between EZH2 and tumors is highly controversial (36). In our study, downregulating EZH2 was able to inhibit tumor cell proliferation and invasion, but downregulation of MBD2 did not affect it, and simultaneous knockdown of MBD2 and EZH2 significantly inhibited the migration, proliferation and cell cycle progression of colorectal tumor cells and increased apoptosis, indicating that MBD2 can enhance the biological function of EZH2 in CRC cells. Further studies have shown that blocking MBD2 in colorectal tumor cells can restore SFRP1 gene expression, but cannot restore SFRP2, SFRP4, and SFRP5 expression, indicating that MBD2 may have different regulatory mechanisms for different member genes of the same gene family. In addition, the expression of SFRP2, SFRP4, and SFRP5 in SW480 cells could be restored by blocking EZH2, while the expression of SFRP2, SFRP4 and SFRP5 in HCT116 cells could not be restored, indicating that EZH2 has different regulatory mechanisms for SFRP gene expression in different stages of tumor, and may have cell specificity. Furthermore, knockdown of both MBD2 and EZH2 could remarkably restore the expression of SFRP1, SFRP2, SFRP4, and SFRP5, indicating that MBD2 and EZH2 have synergistic effects in regulating SFRP gene expression. The results of methylation-specific PCR showed that the promoter methylation status of SFRP did not change before and after the knockdown of MBD2 and EZH2 separately or simultaneously. Taken together, MBD2 recognizes and binds to methylated CG in the MBD domain of the SFRP gene promoter, recruits histone deacetylase complexes NuRD/Mi-2 and Sin3A to form transcriptional repressors, and promotes local chromatin condensation, and to exert the function of transcriptional inhibition (18). Therefore, this effect can be enhanced by EZH2, which also has chromatin remodeling. Silencing of MBD2 and EZH2 together can block the action of histone deacetylase, alleviate the inhibition of chromatin, and help restore gene expression. This phenomenon can explain the synergistic effect of blocking MBD2 and EZH2 in restoring SFRP gene expression and the biological function of colorectal tumors.
However, our previous studies have shown that histone deacetylation is not the main mechanism of SFRP gene silencing. Inhibition of histone deacetylase in colorectal cancer cell HCT116 did not restore SFRP expression (8). This mechanism cannot fully explain the role of MBD2 in restoring SFRP expression. In combination with the latest findings of transcription factor (37), we suspect that there is another possibility that transcription factors can still function in some way in the state of DNA methylation. MBD2 binds to methylated DNA and blocks the reaction between the transcription factor and methylated DNA. As shown in Fig. S1, in normal colorectal cells, the SFRP gene promoter is not methylated, the transcription factor that binds to unmethylated DNA can bind to the promoter, and SFRP can be normally transcribed; in colorectal tumor cells, the SFRP gene promoter is methylated, blocking the role of transcription factors that bind to unmethylated DNA. At the same time, MBD2, which has more advantages in binding to methylated DNA, recognizes and binds to the methylated region of the SFRP promoter, preventing transcription factor binding to methylated DNA was cut off, leading to complete inactivation of the SFRP gene in colorectal tumors; when MBD2 is blocked in colorectal tumors, the transcription factor that bound methylated DNA played its role at this time, and the SFRP genes can resume transcription.
This study preliminarily demonstrated the synergistic regulation of MBD2 and EZH2 on SFRP gene family expression and biological function in colorectal tumor cells. Subsequent research will further explore the transcription factors involved in the regulation of SFRP gene expression and clarify the specific mechanism of DNA methylation regulation of SFRP expression in colorectal tumors, and provide a new theoretical basis for the purpose of treating colorectal tumors.
Supplementary Material
Acknowledgements
The abstract was presented at the 27th United European Gastroenterology (UEG) Week, October 19–23, 2019 in Barcelona, Spain.
Funding Statement
This study was supported by the Applied Basic Research Programs of the Wuhan Science and Technology Department (2015061701011642). It was also funded by the Hubei Provincial Health and Health Commission Joint Fund Key Project (WJ2019H056).
Funding
This study was supported by the Applied Basic Research Programs of the Wuhan Science and Technology Department (2015061701011642). It was also funded by the Hubei Provincial Health and Health Commission Joint Fund Key Project (WJ2019H056).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YX designed the study, performed the experiments and drafted the manuscript. FW, JZ, JY and YL prepared the material used in the experiments and were involved in the literature search. JQ evaluated all the data and revised the manuscript. ML and JZ were involved in the statistical evaluation and in amending the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, Song Y, Meng Q, Yuan S, Luan L, et al. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8:1036. doi: 10.1038/s41467-017-01059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell JO, Monga SP. Wnt/β-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol. 2018;13:351–378. doi: 10.1146/annurev-pathol-020117-044010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft RA, et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545:355–359. doi: 10.1038/nature22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 6.Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA. 1997;94:2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y, Wang F, Qi J. Association between SFRP promoter hypermethylation and different types of cancer: A systematic review and meta-analysis. Oncol Lett. 2019;18:3481–3492. doi: 10.3892/ol.2019.10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7117. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi J, Zhu YQ. Targeting the most upstream site of Wnt signaling pathway provides a strategic advantage for therapy in colorectal cancer. Curr Drug Targets. 2008;9:548–557. doi: 10.2174/138945008784911769. [DOI] [PubMed] [Google Scholar]
- 10.Santanach A, Blanco E, Jiang H, Molloy KR, Sansó M, LaCava J, Morey L, Di Croce L. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat Commun. 2017;8:1235. doi: 10.1038/s41467-017-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracken AP, Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 12.Kerppola TK. Polycomb group complexes-many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rush M, Appanah R, Lee S, Lam LL, Goyal P, Lorincz MC. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4:404–414. doi: 10.4161/epi.4.6.9392. [DOI] [PubMed] [Google Scholar]
- 14.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 15.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takawa M, Masuda K, Kunizaki M, Daigo Y, Takagi K, Iwai Y, Cho HS, Toyokawa G, Yamane Y, Maejima K, et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 2011;102:1298–1305. doi: 10.1111/j.1349-7006.2011.01958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai AY, Wade PA. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai MA, Webb HD, Sinanan LM, Scarsdale JN, Walavalkar NM, Ginder GD, Williams DC., Jr An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015;43:3100–3113. doi: 10.1093/nar/gkv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Lee MS, Jeoung DI, Kim YM, Lee H. Promoter CpG-site methylation of the KAI1 metastasis suppressor gene contributes to its epigenetic repression in prostate cancer. Prostate. 2017;77:350–360. doi: 10.1002/pros.23274. [DOI] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL, Huang HD. MethHC: A database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43((Database Issue)):D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Zhou FF, Xie W, Chen SQ, Wang XK, Liu Q, Pan XK, Su F, Feng MH. SLC38A1 promotes proliferation and migration of human colorectal cancer cells. J Huazhong Univ Sci Technolog Med Sci. 2017;37:30–36. doi: 10.1007/s11596-017-1690-3. [DOI] [PubMed] [Google Scholar]
- 24.Xie Y, Yu J, Wang F, Li M, Qiu X, Liu Y, Qi J. ERCC6L promotes cell growth and invasion in human colorectal cancer. Oncol Lett. 2019;18:237–246. doi: 10.3892/ol.2019.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Wang J, Han X, Zhou J, Linder S. Up-regulation of human secreted frizzled homolog in apoptosis and its down-regulation in breast tumors. Int J Cancer. 1998;78:95–99. doi: 10.1002/(SICI)1097-0215(19980925)78:1<95::AID-IJC15>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 27.Wong SC, Lo SF, Lee KC, Yam JW, Chan JK, Wendy Hsiao WL. Expression of frizzled-related protein and Wnt-signalling molecules in invasive human breast tumours. J Pathol. 2002;196:145–153. doi: 10.1002/path.1035. [DOI] [PubMed] [Google Scholar]
- 28.Vincan E, Barker N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin Exp Metastasis. 2008;25:657–663. doi: 10.1007/s10585-008-9156-4. [DOI] [PubMed] [Google Scholar]
- 29.Liu K, Xu C, Lei M, Yang A, Loppnau P, Hughes TR, Min J. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J Biol Chem. 2018;293:7344–7354. doi: 10.1074/jbc.RA118.001785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Álvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol. 2015;15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 31.Wood KH, Johnson BS, Welsh SA, Lee JY, Cui Y, Krizman E, Brodkin ES, Blendy JA, Robinson MB, Bartolomei MS, Zhou Z. Tagging methyl-CpG-binding domain proteins reveals different spatiotemporal expression and supports distinct functions. Epigenomics. 2016;8:455–473. doi: 10.2217/epi-2015-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH, Li LY, La Win MK, Yau YH, Guo T, et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol. 2015;16:505–516. doi: 10.1038/ni.3125. [DOI] [PubMed] [Google Scholar]
- 33.Xia L, Zhu X, Zhang L, Xu Y, Chen G, Luo J. EZH2 enhances expression of CCL5 to promote recruitment of macrophages and invasion in lung cancer. Biotechnol Appl Biochem. 2020;67:1011–1019. doi: 10.1002/bab.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.First EZH2 inhibitor approved-for rare sarcoma. Cancer Discov. 2020;10:333–334. doi: 10.1158/2159-8290.CD-NB2020-006. [DOI] [PubMed] [Google Scholar]
- 35.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 36.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 37.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.