SUMMARY
Many diseases, including cancer, stem from aberrant activation or overexpression of oncoproteins that are associated with multiple signaling pathways. Although proteins with catalytic activity can be successfully drugged, the majority of other protein families, such as transcription factors, remain intractable due to their lack of ligandable sites. In this study, we report the development of TRAnscription Factor TArgeting Chimeras (TRAFTACs) as a generalizable strategy for targeted transcription-factor degradation. Herein, we show that TRAFTACs, which consist of a chimeric oligonucleotide that simultaneously binds to the transcription factor-of-interest (TOI) and to HaloTag-fused dCas9 protein, can induce degradation of the former via the proteasomal pathway. Application of TRAFTACs to two oncogenic TOIs, NF-κB and brachyury, suggests that TRAFTACs can be successfully employed for the targeted degradation of other DNA-binding proteinsThus, TRAFTAC technology is potentially a generalizable strategy to induce degradation of other transcription factors both in vitro and in vivo.
Keywords: PROTACs, transcription factors, undruggable, E3 ligase, proteasome, dCas9, degradation, HaloTag, Brachyury, Zebrafish
INTRODUCTION
PROteolysis TArgeting Chimeras (PROTACs) are heterobifunctional molecules that target disease-causing proteins for proteasomal degradation (Sakamoto et al., 2001, Lai and Crews, 2017). PROTAC molecules simultaneously bind to both the target protein and an E3 ligase to induce proximity-dependent ubiquitination of the former, thereby tagging the target protein for proteasome-mediated degradation (Lai and Crews, 2017, Nalepa et al., 2006). This technology was developed two decades ago and has successfully been applied to cytoplasmic, nuclear proteins, and membrane-associated proteins (BCR-Abl and the receptor tyrosine kinases EGFR and FLT3) (Zhang et al., 2020, Lai et al., 2016, Burslem et al., 2019, Burslem et al., 2018, Zhang et al., 2019). Targeted protein degradation offers several advantages over conventional small molecule-based inhibition including the potential to expand the “druggable” proteome to encompass non-catalytic, scaffolding proteins in deregulated biological systems. However, degradation of such proteins by PROTACs still requires a target ligand, and the development of ligands for proteins lacking a well-defined active site is both challenging and time-consuming (Verdine and Walensky, 2007, Lazo and Sharlow, 2016).
Transcription factors (TFs) are DNA-binding proteins that directly or indirectly regulate gene expression and upon deregulation, are among the known drivers of many pathologies. Therefore, much effort has been devoted to therapeutically target TFs implicated in human diseases (Sharifnia et al., 2019). TFs often mediate their regulatory functions through DNA and protein-protein interactions. However, unlike kinases or other druggable proteins, development of small molecule inhibitors for TFs is truly challenging due to their lack of enzymatic activity and ligandable pockets. Therefore, development of alternative strategies to target such proteins is crucial. A small number of TFs, such as the estrogen and androgen receptors that drive breast and prostate cancer, respectively, have been drugged by small molecules, although the emerging resistance to such therapies hinders their long-term effectiveness (Ali et al., 2016, Osborne et al., 2004, Rice et al., 2019, Watson et al., 2015). Recently, these ligands were incorporated into PROTACs to induce the degradation of estrogen and androgen receptors: ARV-471 and ARV-110 targeting estrogen and androgen receptors (respectively) have already progressed to Phase 1 clinical trials and demonstrated high tolerability and safety in humans (Salami et al., 2018, Hu et al., 2019, Sun et al., 2019). In addition, a selective STAT3-targeting PROTAC was recently developed (Bai et al., 2019). However, steroid receptors and STAT3 are among the minority of TFs possessing a ligand binding site that controls their activity. Because most TFs mediate their regulatory functions through interactions with DNA and/or with other proteins, they frequently lack enzymatic activity and ligandable pockets – necessary features that have been successfully exploited in developing small molecule inhibitors for more readily druggable proteins. Therefore, development of alternative, generalizable strategies to target TFs is paramount.
Since TFs mediate their transcriptional activity by interacting with DNA and other accessory proteins, a variety of protein-DNA interaction inhibitors and protein-protein interaction inhibitors (PPIs) have been developed (Yang et al., 2013). For instance, pyrrole-imidazole polyamide binds to the minor grove of DNA and inhibits binding of the pro-inflammatory transcription factor, NF-κB (Wurtz et al., 2002). The transcription factor Myc forms a heterodimer with Max to bind to enhancer box sequences to regulate gene expression (Walhout et al., 1997). Several groups have successfully developed PPIs that inhibit Myc/Max interaction, eliciting effects on gene expression (Raffeiner et al., 2014, Castell et al., 2018, Choi et al., 2017). As a complementary strategy to these PPIs, stabilizing molecules of Max homodimers have also been developed to reduce the availability of Max to form transcriptionally active Myc/Max heterodimers (Jiang et al., 2009, Struntz et al., 2019). The aforementioned approaches were directed at the TF themselves, but indirect approaches that block TF activity have also been reported. For example, specific STAT3/5 inhibitors have been developed to block nuclear STAT3 translocation by inhibiting its phosphorylation, dimerization and DNA binding (Chen et al., 2018, Dai et al., 2018). The targeting of upstream effectors, another indirect approach to inhibit TFs, has also been well-studied (Liu et al., 2017, Rios-Fuller et al., 2018): for example, multiple NF-κB pathway inhibitors do so by targeting upstream signaling components, such as SRC/Syk, PI3k/Akt and IKK (Reddy et al., 1997, Karin et al., 2004, Pande and Ramos, 2005). BET protein-targeting PROTACs that induced BRD4 degradation have also reduced downstream oncogenic c-Myc levels, leading to improved survival in mice (Raina et al., 2016, Winter et al., 2015). These previous approaches to overcome oncogenic TF overexpression and/or hyperactivity have motivated us to develop a strategy that bypasses the need for target protein ligand development, while readily and directly targeting a transcription factor-of-interest (TOI).
DESIGN
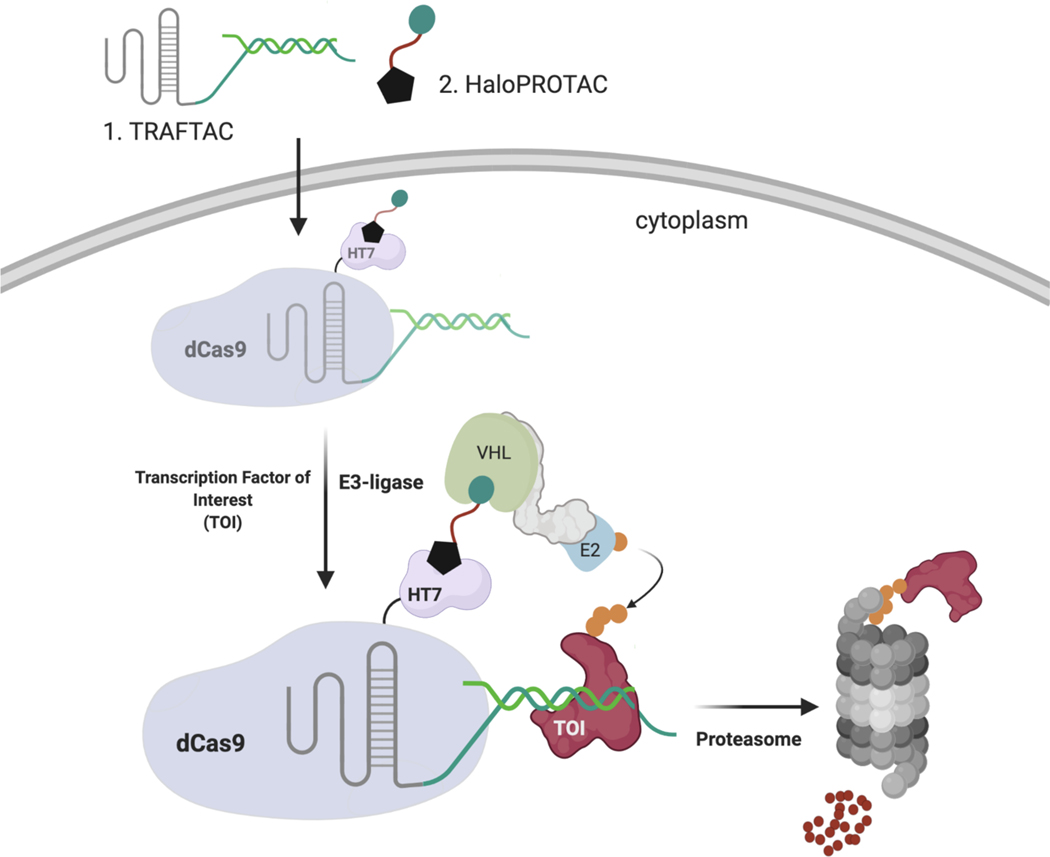
In the current study, we have developed a generalizable strategy to induce TF degradation by co-opting the cellular degradation machinery. While development of traditional PROTACs for TF degradation is challenging due to the lack of cognate small molecule ligands, TFs are proteins that elicit their function via binding to specific DNA sequences. Therefore, by taking advantage of the intrinsic TF DNA-binding ability, we developed chimeric oligos with TF-specific DNA sequences attached to an E3 ligase recruiting moiety, and we termed these chimeric molecules, TRAnscription Factor TArgeting Chimeras, or TRAFTACs. Our chimeric oligo consists of a TOI-binding double stranded DNA covalently linked to a Cas9 CRISPR-binding RNA. The dsDNA binds to TOI, while the CRISPR-RNA binds to an ectopically-expressed dCas9-Halotag7 fusion protein (dCas9HT7) fusion protein. Incubation with a haloPROTAC then recruits the VHL-E3 ligase to the vicinity of the DNA-bound TOI via the complexed fusion protein, inducing ubiquitination and proteasomal degradation of the TOI (Figure 1). In this proof-of-concept study, we demonstrate that TRAFTACs can recruit both TOI and the E3-ligase via the intermediate protein dCas9HT7, and in doing so induce degradation of the disease-relevant TFs NF-κB and brachyury in the presence of the haloPROTAC.
Figure 1. Schematic Representation of the TRAnscription Factor TArgeting Chimeras (TRAFTACs).
Heterobifunctional dsDNA/CRISPR-RNA chimera (TRAFTAC) recruits E3 ligase complex through dCas9-HT7 in the presence of haloPROTAC. Heterobifunctional TRAFTAC binds to dCas9-HT7 via its RNA moiety while dsDNA portion of the chimera binds to the transcription factor of interest (TOI). Addition of haloPROTAC recruits VHL E3 ligase complex to the vicinity of TOI. TRAFTAC-mediated proximity induced ubiquitination directs TOI for proteasomal degradation.
RESULTS
NF-κB Engages with the dCas9HT7 Ribonucleocomplex
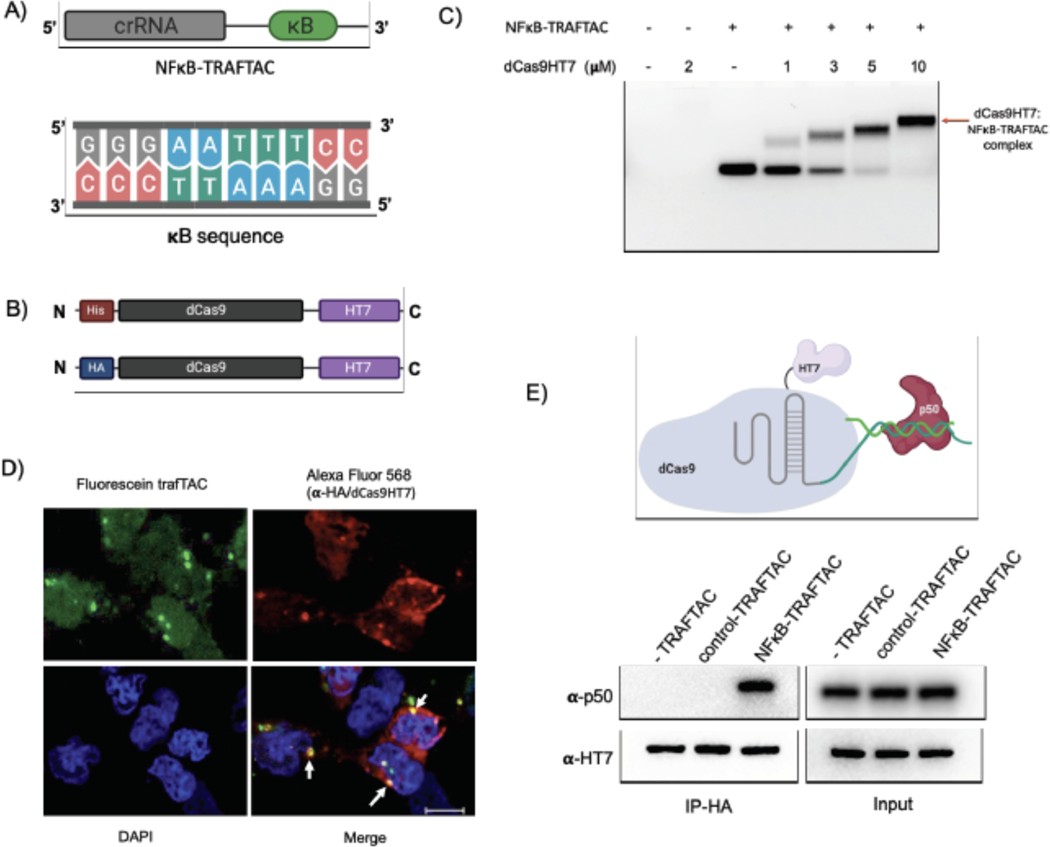
NF-κB binds to kappa B sequences upstream of its target genes (Chen et al., 1998). Therefore, we selected the kappa B sequence as the NF-κB binding element in our initial TRAFTAC design (Figure 2A). A chimeric oligonucleotide containing the kappa B sequence at the 3’ end of a CRISPR RNA (crRNA) was synthesized. The double-stranded chimeric oligo (NFκB-TRAFTAC) was generated by annealing with the reverse complementary oligo of the kappa B sequence (Figure S1A). Using an electrophoretic mobility shift assay (EMSA), we first evaluated the ability of crRNA to engage the dCas9HT7 fusion protein when the former is presented as a part of the double stranded chimeric oligo. The data indicated that NFκB-TRAFTAC engaged with purified dCas9HT7 fusion protein as the band is shifted upwards in the agarose gel (Figures 2C and S2A). Also, the observance of a single major band for NFκB-TRAFTAC indicated the stability of the chimeric molecule after the annealing reaction. Subsequently, we evaluated the interaction of the chimeric oligo with the fusion protein within cells. We transfected a fluorescein-labeled TRAFTAC into HEK293 cells that stably overexpress HA-tagged dCas9 fused at its C-terminus to HT7 (CT-dCas9HT7) (Figure 2B). First, to test whether the NFκB-TRAFTAC successfully enters the cell, we analyzed cells by confocal microscopy after the transfection of fluorescein-labeled TRAFTAC (Figure S1C). The intracellular green signal indicated that transfection of the chimeric NFκB-TRAFTAC was successful (Figure S2B). Next, we analyzed the cells by confocal microscopy to test whether NFκB-TRAFTAC can engage with dCas9HT7 in cells. After fixation, the cells were incubated with anti-HA primary antibody followed by Alexa Fluor 568-labeled secondary antibody. The co-localization of fluorescein signal with immunofluorescence signal (Figure 2D) suggested that dCas9HT7 fusion protein can engage NFκB-TRAFTAC in cells. Next, we tested for NF-κB (p50) engagement with the dCas9HT7 ribonucleocomplex (dCas9HT7:NFκB-TRAFTAC) by performing an immunoprecipitation experiment. Cell lysates were incubated with either NFκB-TRAFTAC or a control-TRAFTAC and then subjected to immunoprecipitation by HA agarose beads. The eluted samples were probed for p50 and HT7, and the results indicated that p50 binds to dCas9HT7 only in the presence of the NFκB-TRAFTAC (Figure 2E). Overall, the data indicate that NFκB-TRAFTAC engages both the dCas9HT7 fusion protein and p50.
Figure 2. Binding Experiments for NFκB-TRAFTAC, CT-dCas9HT7 and p50.
A) A diagram illustrating the architecture of NF-κB-TRAFTAC and the NF-κB binding kappaB sequence.
B) Schematic of bacterial and mammalian expression CT-dCas9HT7 constructs. After bacterial expression, protein was purified by His-tag affinity column purification. While the bacterial expression vector has a His-tag sequence, it was replaced by HA-tag for mammalian expression.
C) Different concentrations of purified CT-dCas9HT7 were incubated with NFκB-TRAFTAC and the protein:oligo complexes separated on an agarose gel. Data represent the ability of dCas9HT7 to bind to the modified-double stranded chimeric NFκB-TRAFTAC.
D) Immunofluorescence data illustrating in cellulo TRAFTAC engagement with fusion dCas9HT7. Cells were transfected with fluorescein labeled TRAFTAC for 12 h and cells fixed, permeabilized and probed with anti-HA antibody, followed by secondary antibody conjugated to Alexa Fluor 568. Images were captured using a confocal microscope. Scale bar: 25 μm.
E) Immunoprecipitation of p50 and ribonucleocomplex (dCas9HT7:NFκB-TRAFTAC). Stable cells lysates with dCas9HT7 were treated with NFκB-TRAFTAC and control-TRAFTAC. After 1 h, dCas9HT7 was immunoprecipitated using HA antibody conjugated beads and eluted samples were probed with antibodies as shown.
TRAFTACs Induce NF-κB Degradation
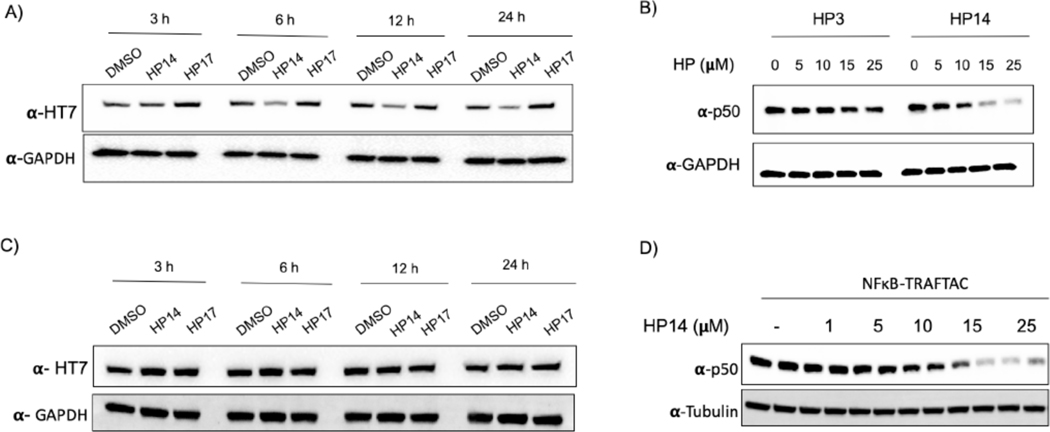
In TRAFTAC design, we adapted a RNA-binding but catalytically dead CRISPR Cas9 (dCas9) protein as an intermediate spacer protein that brings the TOI and E3 complex into close proximity (Qi et al., 2013). Specifically, in our HEK293 cell line, we stably overexpressed CT-dCas9HT7, to simultaneously bind both the von-Hippel Lindau E3 ubiquitin ligase (VHL), via a haloPROTAC, and the TOI, through a TRAFTAC (Figure 1). In previous studies, VHL-based haloPROTACs were designed to induce degradation of directly-bound HaloTag fusion proteins (Buckley et al., 2015), and their ability to do so required that the haloPROTAC not exceed an optimal length. However, in this study, our intent is to employ HaloTag7 as a ligand inducible E3-recruiting element to cause TOI degradation but not that of the dCas9HT7 fusion. Therefore, we first carried out an initial haloPROTAC screening using existing haloPROTACs and several newly synthesized haloPROTACs to select the optimal haloPROTAC that spares CT-dCas9HT7 from degradation. Our previous study suggested that haloPROTACs with 3-PEG linkers are potent degraders of directly bound HaloTag7 fusion proteins, but that ability of haloPROTACs to induce degradation of a directly bound substrate protein was decreased with increasing linker lengths (Buckley et al., 2015). Therefore, we synthesized several haloPROTACs with longer linker lengths and screened for their ability to induce CT-dCas9HT7 degradation. As anticipated, we observed that longer haloPROTACs are poor degraders of CT-dCas9HT7: more specifically, haloPROTACs HP3, HP8 and HP10 with shorter linker lengths (3 and 4 PEG units) induced CT-dCas9HT7 degradation even at 2.5 μM, whereas the comparatively longer HP13 (7 PEG units) and HP14 (9 PEG units) did not induce significant CT-dCas9HT7 degradation (Figures S2C and S2E, F). Two other, even longer haloPROTACs, HP15 and HP16 (12 PEG units) were also tested for their ability to degrade CT-dCas9HT7 and consistently, we did not observe significant CT-dCas9HT7 degradation (Figures S2 D, F). Therefore, we selected HP13, HP14, HP15 and HP16 as our haloPROTAC panel to test in TRAFTAC studies.
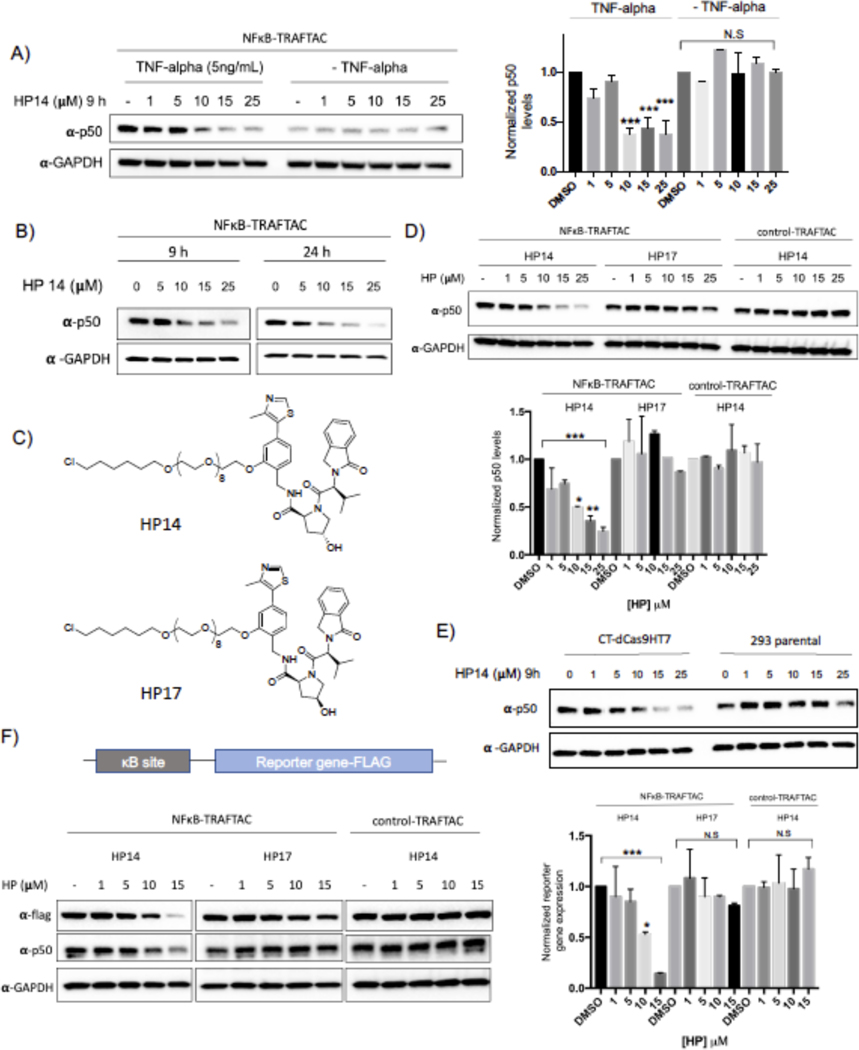
Before testing the ability of TRAFTACs to induce p50 degradation, we determined the maximum NFκB-TRAFTAC concentration that does not induce cellular cytotoxicity. The data obtained from MTS assays suggested that NFκB-TRAFTAC can be introduced to cells up to 50 nM without inducing significant cytotoxicity by lipofection (Figure S3A). To ensure lack of toxicity, we transfected a lower concentration (25 nM) of NFκB-TRAFTAC into CT-dCas9HT7 stable cells followed by the treatment of HP14 in the presence or absence of 5 ng/mL of TNF-alpha for 12 h. HP14 induced p50 degradation only in the presence of TNF-alpha (Figure 3A). Latent NF-κB stays in the cytoplasm as an inactive heterotrimeric complex until extracellular stress signal is introduced. The inhibitor of Kappa B (IκB) binds to the NF-κB, physically masking its kappa B-binding sequence. However, TNF-alpha induces the release of active NF-κB from the inhibitory complex and facilitates TRAFTAC binding and subsequent degradation by NFκB-TRAFTAC. TNF-alpha treatment also induces the proteolytic processing of the NFκB precursor, p105 into p50 (Mellits et al., 1993, Orian et al., 2000). Therefore, overall TRAFTAC accessible p50 levels are higher in the presence of TNF-alpha, compared to untreated cells (Figure 3A, right panel). We also tested our panel of haloPROTACs to determine the effect of linker length in TRAFTAC-mediated p50 degradation. Interestingly while HP13 treatment induced p50 degradation to a similar extent as in HP14 treated cells, HP15 with a 12 PEG unit linker did not induce p50 degradation (Figure S3D). Left-handed haloPROTAC (HP16) with 12 PEG unit linker also did not induce p50 degradation.
Figure 3. NF-κB Degradation by TRAFTACs.
A) HP14 induces p50 degradation in the presence of TNF alpha. Briefly, cells were transfected with NFκB-TRAFTAC and after 16 h, cells were treated with increasing HP14 concentrations. After 1h, cells were treated with or without TNF-alpha for indicated times.
B) TRAFTACs induce p50 degradation within 9 h of HP14/TNF-alpha treatment.
C) Chemical structures of HP14/TNF-alpha and the inactive epimer control (HP17).
D) VHL and TRAFTAC-dependent p50 degradation. Cells were transfected with either with NFκB-TRAFTAC or control-TRAFTAC followed by the treatment of HP14/TNF-alpha or the HP17/TNF-alpha for 12 h. Cell lysates were probed for p50 and GAPDH.
E) TRAFTAC induced p50 degradation is dependent on dCas9HT7. Stable cells overexpressing dCas9HT7 and parental cells were transfected with chimeric oligo and the experiment performed as mentioned above.
F) NFκB-TRAFTAC mediated p50 degradation elicit an effect on downstream gene expression.
(Not significant (N.S); * P < 0.03; ** p<0.002; *** p<0.0002; **** p<0.0001; n: Independent biological replicates; n=2)
Next, we assayed for TRAFTAC-mediated p50 degradation at both 9 and 24 h post HP14/TNF-alpha treatment. The data indicated that TRAFTACs could induce significant p50 degradation after 9 h of TNF-alpha/HP14 treatment. However, the degradation levels of p50 only slightly improved over a longer 24 h treatment (Figure 3B). Moreover, TRAFTAC-mediated degradation was further intensified in the presence of protein synthesis inhibitor cycloheximide (CHX) demonstrating that TRAFTACs significantly lowered p50 half-life (Figure S3E). To confirm whether p50 degradation is dependent on VHL recruitment, we next compared TRAFTAC-induced degradation in response to HP14 to its inactive epimer control (HP17) (Figure 3C). Gratifyingly, p50 degradation was observed only in the cells treated with HP14, whereas no significant p50 degradation was observed in HP17-treated cells (Figure 3D left and center panel). Next, stable cells overexpressing CT-dCas9HT7 were transfected with NFκB-TRAFTAC or control-TRAFTAC (Figure S1B). Cells were subsequently treated with HP14 for 12 h and cell lysates probed for p50 levels. As shown in Figure 3D, HP14 induced p50 degradation in NFκB-TRAFTAC transfected cells. In contrast, cells transfected with control-TRAFTAC did not induce significant p50 loss, suggesting that TRAFTAC-mediated p50 degradation is dependent on the dsDNA portion of the TRAFTAC (Figure 3D-right panel). Furthermore, qRT-PCR data indicated that total RNA levels were not affected in response to TRAFTAC and HP14 treatment suggesting that the observed p50 degradation occurred as a posttranslational event (Figure S3F). In addition to p50 degradation, NFκB-TRAFTAC also induced the degradation of second NFκB heterodimeric subunit, RelA (p65) (Figure S3C). Furthermore, parental cells, without CT-dCas9HT7 expression, did not induce p50 degradation, confirming that the observed target degradation is dependent on dCas9HT7 as well (Figure 3E). Finally, we performed a reporter gene assay to test the effect of TRAFTAC-mediated p50 degradation on downstream gene expression. Cells stably expressing CT-dCas9HT7 were transiently transfected with a NF-κB reporter plasmid that expresses FLAG tagged reporter gene under the control of NF-kB. After TRAFTAC transfection and HP14 treatment, cell lysates were analyzed for reporter gene expression. The results demonstrated a loss in κB promoter activation that was consistent with the p50 degradation observed only in response to NFkB-TRAFTAC. Importantly, the control-TRAFTAC did not reduce reporter levels, nor did co-treatment with the inactive haloPROTAC HP17. We observed that only HP14 co-treatment repressed reporter gene expression (Figure 3F).
HaloTag7 Position Relative to dCas9 Governs Its Susceptibility to Degradation
We employed 2.5 μM haloPROTACs in the initial screening to select the best haloPROTAC for TRAFTAC investigations. While HP14 did not induce p50 degradation at that concentration, our data indicated that significant p50 degradation occurred at concentrations over 10 μM HP14. However, HP14 also induced CT-dCas9HT7 degradation within that concentration range (Figure 4A). Thus, we tested whether the observed p50 degradation was an indirect consequence of overall CT-dCas9HT7:NFκB-TRAFTAC:p50 complex degradation (Figure S4A). This was accomplished using HP3, a comparatively shorter haloPROTAC that we already had demonstrated will cause degradation of CT-dCas9HT7 (Figure S4B). We hypothesized that the shorter haloPROTAC (HP3) should be much more restrictive in terms of the steric range of protein ubiquitination it permits the recruited VHL, and therefore we tested whether or not HP3 also induces p50 degradation. To this end, cells stably overexpressing CT-dCas9HT7 were transfected with NFκB-TRAFTAC followed by the treatment of either HP3 or HP14. Although HP3 could induce significant degradation of CT-dCas9HT7, it did not induce p50 degradation. In contrast, HP14 induced the degradation of both (Figures 4B and S4B). Clearly, the data showed that ligand-mediated degradation of CT-dCas9HT7 does not mandate concomitant p50 degradation. Thus, HP14-mediated p50 degradation is not an obligatory result of CT-dCas9HT7 fusion protein degradation. Having successfully “uncoupled” degradation of the two complexed proteins, we next set about engineering the reciprocal scenario: facilitation of p50 degradation while sparing degradation of dCas9HT7.
Figure 4. Positioning of HT7 in dCas9 Governs the HT7 Fusion Protein Degradation by haloPROTACs.
A) HP14 induces degradation of CT-dCas9HT7 fusion protein. The cell line stably overexpressing CT-dCas9HT7 was treated with HP14 (20 μM) and HP17 (20 μM) and lysed at indicated time points. Lysates were probed for HT7 and GAPDH.
B) The haloPROTAC HP3 (possessing a shorter linker) did not induce p50 degradation, in contrast to HP14.
C) Cells stably expressing NT-dCas9HT7 were treated with HP14 and lysed at indicated time points.
D) NFκB-TRAFTAC/HP14/TNF-alpha treated NT-dCas9HT cells were lysed and probed for p50 and tubulin levels.
To improve the TRAFTAC system accordingly, we generated a construct with HT7 at the N-terminus of dCas9 (NT-dCas9HT7). The positioning of susceptible lysine residues and the presentation and orientation of the E3 complex are important factors for a successful PROTAC-mediated substrate degradation (Smith et al., 2019, Buckley et al., 2015, Buhimschi et al., 2018, Jaime-Figueroa et al., 2020). Therefore, we speculated that fusion of HaloTag7 at the opposite end of the dCas9 might alter the HP14-induced dCas9HT7 degradation. To investigate this, we generated cells stably overexpressing NT-dCas9HT7 protein and incubated them with increasing concentrations of HP14, followed by probing for NT-dCas9HT7 levels. Interestingly, we observed significantly less NT-dCas9HT7 degradation by HP14 compared to that of CT-dCas9HT7 (Figure 4C). Next, we evaluated the same cell line to test whether NT-dCas9HT7 can be used to induce p50 degradation. First, we transfected NFκB-TRAFTAC followed by HP14 and TNF-alpha treatment for 9 h. Then cell lysates were probed for p50 levels. The data showed that NT-dCas9HT7 can, indeed, induce p50 degradation (Figure 4D). From the Cas9 crystal structure bound to guide RNA, it is apparent that the 3’ end of the guide RNA, the N-terminus and C-terminus of Cas9 are all positioned at the same face of the protein (PDB ID:4OO8; Figure S4C) (Nishimasu et al., 2014). This serendipitous positioning of the dCas9 and crRNA offers an explanation as to how the E3 ligase recruited via both NT- and CT-HaloTag7 can access NFκB-TRAFTAC-bound proteins and induce their degradation. Overall, the data suggest that dCas9 protein is less susceptible to HP14-mediated degradation when HaloTag7 is fused at the N-terminal end and therefore provides a better tool for use in TRAFTAC design. Therefore, we employed the NT-dCas9HT7 fusion protein (which retains the N-terminal HA tag) in subsequent experiments.
TRAFTACs are Generalizable: TRAFTACs Induce Brachyury Degradation
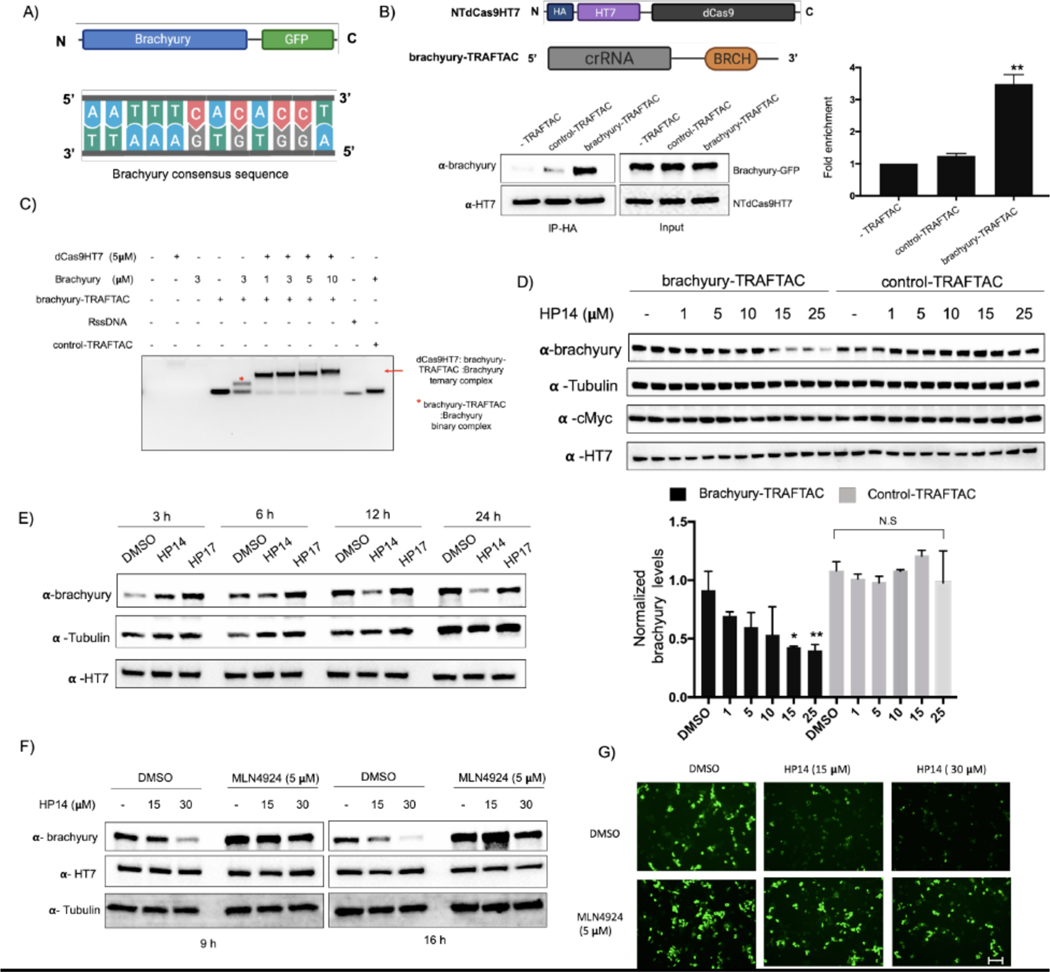
TRAFTAC technology is based on the intrinsic ability of TFs to bind to specific DNA sequences. Therefore, we hypothesized that TRAFTACs should be adaptable to target a variety of transcription factors for proteasomal degradation by simply customizing the DNA sequence specific to a TOI. To investigate the generalizability of TRAFTACs to target different TFs, we extended our approach to induce the degradation of the T-box transcription factor T (TBXT), also known as brachyury. Brachyury is a key transcription factor that regulates notochord formation in embryonic stages of development, and its downregulation leads to tail development defects (Zhu et al., 2016, Stemple et al., 1996). Recent studies have also suggested that brachyury knockdown leads to the inhibition of tumorigenesis, cell migration and invasion, suggesting a key role in cancer metastasis (Chen et al., 2020). Furthermore, genomic amplification of the locus that harbors brachyury gene and germline tandem duplication of the brachyury gene are associated with cancer such as chordoma (Bosotti et al., 2017, Presneau et al., 2011, Yang et al., 2009, Pillay et al., 2012). Given the importance of brachyury in cancer, we sought to target it for proteasomal degradation using the TRAFTAC technology. To this end, we synthesized a chimeric oligo comprised of a dsDNA portion that recognizes and binds brachyury along with the previously-used crRNA sequence that recruits NT-dCas9HT7, the E3 ligase-recruiting intermediary-fusion-protein (Figures 5A and S4F). First, we confirmed binary (dCas9HT7:brachyury-TRAFTAC) and ternary (dCas9HT7:brachyury-TRAFTAC:brachyury) complex formation by EMSA experiments. Consistent with NFκB-TRAFTAC EMSA data, we showed that brachyury-TRAFTAC engages the dCas9HT7 fusion protein (Figure S4D). Further, data suggest a successful ternary complex formation of dCas9HT7:brachyury-TRAFTAC:brachyury, whereas a control-TRAFTAC failed to do so (Figure 5C). Next, we tested whether brachyury could engage the NT-dCas9HT7 ribonucleocomplex. We generated a cell line stably overexpressing both brachyury-GFP and NT-dCas9HT7 proteins. The cell lysates were subjected to immunoprecipitation with HA antibody attached to agarose beads in the absence of a chimeric oligo or in the presence of control-TRAFTAC or brachyury-TRAFTAC. The eluates were probed for dCas9HT7 protein and brachyury-GFP using antibodies against HaloTag7 and brachyury respectively. The results indicated that brachyury selectively engages NT-dCas9HT7 protein only when brachyury-TRAFTAC is present, confirming successful brachyury recruitment by the chimeric oligo to the dCas9HT7 fusion protein (Figure 5B).
Figure 5. TRAFTACs Targeting Brachyury Induces its Degradation.
A) Schematic representation of the brachyury-GFP construct and the brachyury binding DNA sequence.
B) Cell lysates that overexpress brachyury-GFP were incubated with the brachyury-TRAFTAC and the inactive control-TRAFTAC before HA immunoprecipitation. Eluted samples were probed against brachyury and HT7 levels.
C) Ternary complex formation assay for the dCas9HT7:chimeric oligo and brachyury. Purified proteins were incubated with chimeric oligos and protein:oligo complexes were separated using DNA agarose gel to analyze gel shifts as depicted in the figure.
D) Cells stably expressing brachyury-GFP were transfected with brachyury-TRAFTAC or control-TRAFTAC followed by HP14 treatment. Cells were then lysed and analyzed for brachyury, tubulin, c-Myc and HT7 levels. (Not significant (N.S); * P < 0.03; ** p<0.002; n: Independent biological replicates; n=2)
E) Time dependent degradation of brachyury-GFP. Cells stably expressing brachyury-GFP were transfected with brachyury-TRAFTAC and treated with HP14 and HP17. Cells were lysed at indicated time points, subjected to western blotting and probed for brachyury, tubulin and HT7.
F) Stable cells overexpressing brachyury-GFP were treated with HP14 and MLN4924 for 9 and 16 h. Cells were lysed and probed for brachyury, HT7 and tubulin.
G) The brachyury-GFP fluorescence signal was captured after HP14 and MLN4924 co-treatment. Scale bar: 75 μm.
To test whether brachyury-targeting TRAFTACs induce target degradation, cells stably expressing brachyury-GFP were transfected with brachyury-TRAFTAC or control-TRAFTAC, followed by the treatment with increasing HP14 concentrations. Western blot analysis indicated that brachyury-TRAFTAC induces brachyury-GFP degradation at low micromolar HP14 concentrations. Further, HP14 induced brachyury-GFP degradation within 12 h and reached maximum degradation after 24 h (Figure 5E). The inactive epimer, HP17 did not induce brachyury-GFP degradation indicating that the observed degradation is dependent on VHL-E3 ligase recruitment (Figure S4G). Similarly, the control-TRAFTAC did not induce brachyury-GFP degradation in response to HP14 suggesting that the observed degradation is also dependent on brachyury-TRAFTAC double stranded DNA (Figure 5D). Additionally, HP14 co-treatment with a neddylation inhibitor, MLN4924, failed to induce brachyury-GFP degradation, further confirming that TRAFTAC induces brachyury degradation via the proteasomal pathway (Figure 5F). We also investigated brachyury-GFP levels using fluorescence microscopy and observed GFP signal reduction, further confirming the observed degradation pattern in the western blot analysis (Figure 5G). Overall, the data show that the brachyury-targeting TRAFTAC can successfully induce brachyury-GFP fusion protein degradation via the proteasome.
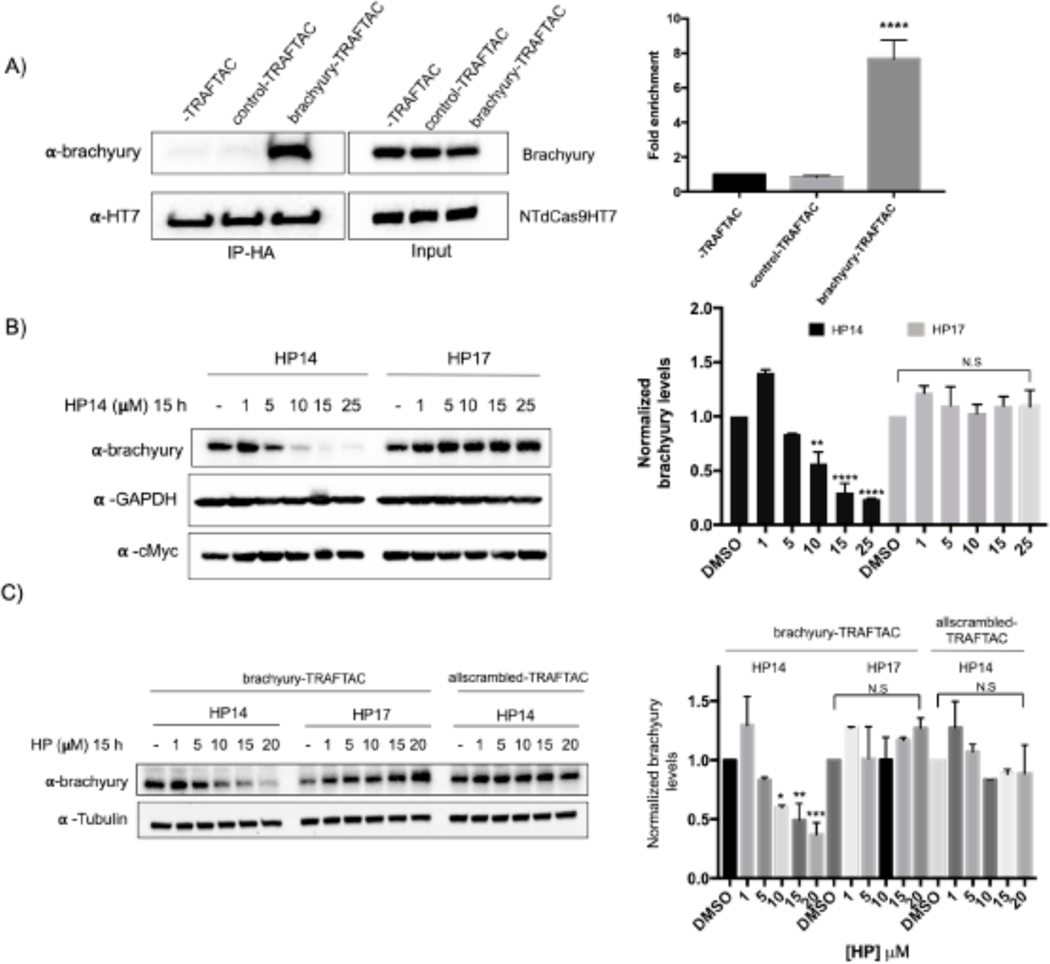
Next, we investigated the potential of TRAFTACs to induce proteasomal degradation of endogenous brachyury protein. Brachyury is predominantly expressed in early embryonic development. However, it has been shown that brachyury is also expressed in several cancer cell lines, including Hela cells. Therefore, we selected Hela cells to test TRAFTAC-mediated endogenous brachyury degradation. We first evaluated whether brachyury engages dCas9HT7 through brachyury-TRAFTAC, by performing an immunoprecipitation experiment using Hela cell line that expresses NT-dCas9HT7. The data indicate that brachyury successfully binds dCas9HT7 only in the presence of brachyury-TRAFTAC (Figure 6A). The data obtained after the transfection of brachyury-TRAFTAC and treatment with HP14 at different concentrations indicated that TRAFTACs can also successfully degrade endogenous brachyury protein at concentrations of HP14 < 10 μM (Figure 6B, left panel). We have confirmed that the observed endogenous brachyury degradation is VHL-dependent as the epimer control failed to induce brachyury degradation (Figure 6B, right panel). Furthermore, the data obtained with the control-TRAFTAC oligo confirmed that TRAFTAC-induced endogenous-brachyury degradation is DNA-sequence dependent (Figure S4E). To further confirm sequence dependency, we also tested the levels of other transcription factors. We analyzed p65 and c-Myc levels using the same experimental protocol and the data indicated that brachyury-targeting TRAFTAC did not affect either p65 or c-Myc levels (Figures 6B and S4H). This suggests that TRAFTAC-mediated degradation is selective and displays a minimal effect on other DNA binding proteins. Furthermore, we tested an all-scrambled version of the chimeric oligo, allscrambled-TRAFTAC, composed of both scrambled crRNA and scrambled dsDNA sequences (Figure S4F). The western blot analysis showed that upon transfection of allscrambled-TRAFTAC, HP14 failed to induce brachyury degradation compared to brachyury-TRAFTAC transfected cells (Figure 6C). Collectively, the data suggest that TRAFTAC-mediated degradation of endogenous brachyury is dependent on the specific dsDNA sequence, dCas9, VHL and the proteasomal pathway.
Figure 6. Endogenous Brachyury Degradation by TRAFTACs.
A) Co-immunoprecipitation of brachyury in the presence and absence of brachyury-TRAFTAC or control-TRAFTAC oligos. Cell lysates were subjected to HA immunoprecipitation and eluted samples were probed for brachyury and HT7.
B) Hela cells transiently transfected with NT-dCas9HT7 followed by a second transfection with brachyury-TRAFTAC. Then HP14 and HP17 were treated and cell lysates were prepared after 15 h and probed for brachyury, c-Myc and tubulin.
C) Hela cells stably overexpressing NT-dCas9HT7 were transfected with either with brachyury-TRAFTAC or allscrambled-TRAFTAC followed by HP14 or HP17 treatment. Cell lysates were then probed for brachyury and tubulin levels. (Not significant (N.S); * P< 0.03; ** p<0.002; *** p<0.0002; **** p<0.0001; n: Independent biological replicates; n=2)
Brachyury-TRAFTAC phenocopies mutant brachyury in zebrafish
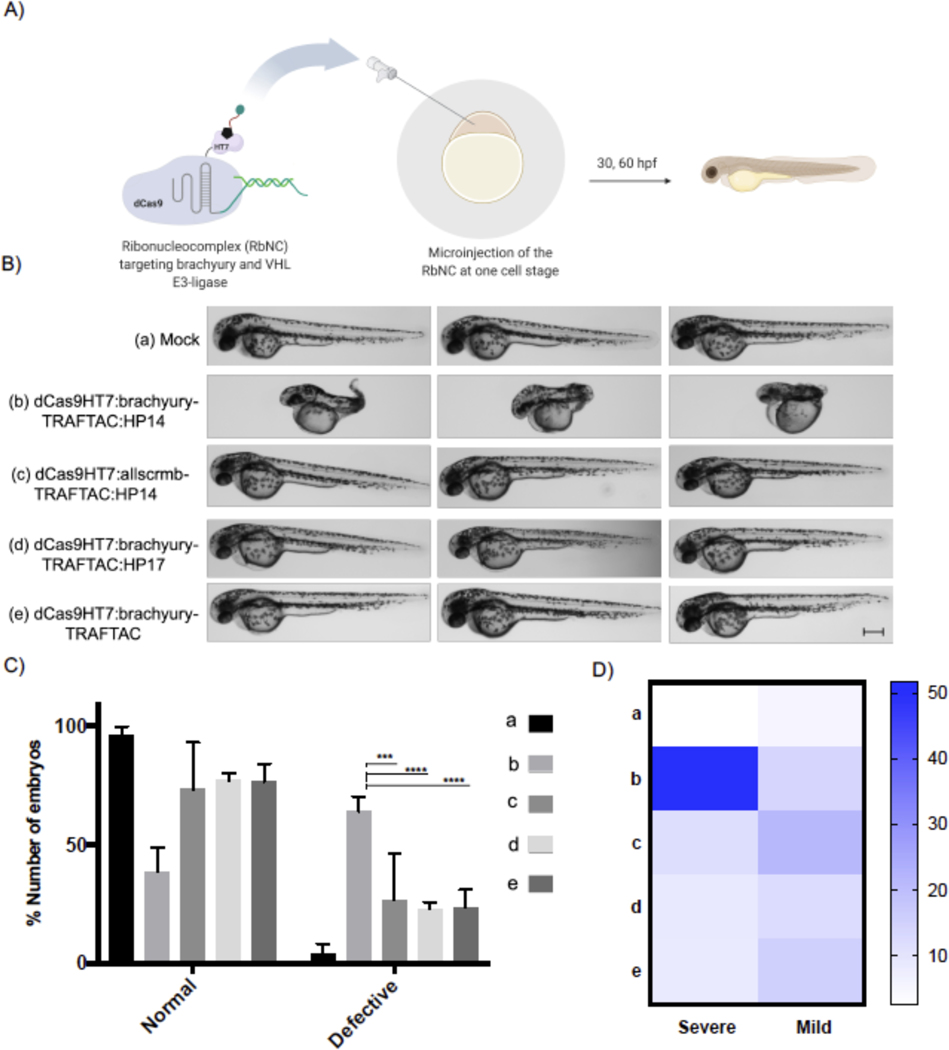
Having confirmed that brachyury-TRAFTAC can efficiently degrade both fusion and endogenous brachyury in human cells, next we sought to extend the applicability of TRAFTACs to an in vivo setting. Brachyury is an essential protein that expressed during early embryonic stages and plays a central role in the formation of the embryonic framework of vertebrates (Herrmann, 1991, Wilson et al., 1995). Zebrafish no tail a (ntla) protein is a mutant form of brachyury associated with truncated or completely abolished notochord and tail somites, highlighting the essential function of brachyury in many aspects of embryonic development including cell movements and tail outgrowth (Halpern et al., 1993, Kimelman, 2016). Therefore, we chose zebrafish as a model system to evaluate the effect of brachyury-TRAFTACs in tail formation. We noticed that fortuitously the VHL recognition motif of HIF1A and the HIF1A binding domain of VHL are conserved in zebrafish and human (Figures S5A, B) (van Rooijen et al., 2009). This suggests the direct applicability of HP14 in zebrafish as HP14 is based on human-VHL-binding HIF1A peptide sequence (Figures S5C, D). To introduce TRAFTAC system into zebrafish embryos, we first generated the ribonucleocomplexes (RbNCs) consisting of dCas9HT7; brachyury- or control- or fluorescein-TRAFTAC; and HP14 or HP17 or TAMRA-HT7 ligand (Figure S6A). We employed the same brachyury-TRAFTAC used in initial cell-based degradation experiments since human brachyury and zebrafish brachyury have been shown to bind to the similar DNA sequence (PDB: 6F58) (Müller and Herrmann, 1997, Morley et al., 2009).
Different combinations of RbNCs were microinjected (~300pL from 1 mg/mL) into zebrafish embryos at single cell stage and the injected embryos were transferred to a 29˚C incubator (Figure 7A). Interestingly, the embryos that were injected with dCas9HT7:brachyury-TRAFTAC:HP14 had significantly higher percentage of severe defects in their tail development compared to the embryos injected with mock solution; scrambled-TRAFTAC- or inactive epimer (HP17)-containing RbNCs; or RbNC without haloPROTAC (Figures 7B, C). While mock injected embryos showcased no defects in tail development, the embryos injected with other control RbNCs had tail developmental defects to a lesser, limited degree. These low degrees of defects can be possibly attributed to double stranded DNA-mediated decoy effects. We have noticed that dCas9HT7:brachyury-TRAFTAC:HP14 injected embryos had significantly higher percentage of severe defects (Figure 7D). TRAFTAC mediated tail deformation was evident even at 30 hpf (Figure S6C). Moreover, we found that injected RbNC is stable even after 18 hpf in vivo as a stable fluorescence signal was seen in the embryos injected with fluorescently labelled RbNC with fluorescein and TAMRA-HT7 ligand (Figure S6B). Overall, the data demonstrated that TRAFTAC technology can be successfully implemented into in vivo settings to achieve desired phenotype associated with targeted transcription factors. Many of existing methods to control embryonic notochord and tail development have been carried out at the genetic level by either siRNA or CRISPR-mediated knock down or knockout approaches. While these technologies provided invaluable insights to how brachyury and other transcriptional regulator control embryonic development, they lack spatiotemporal activity to understand how these proteins regulate different stages of embryonic development. With desired optimizations to TRAFTACs, this approach will open an avenue to explore spatiotemporal regulation of embryonic development by DNA binding proteins including TFs.
Figure 7. Brachyruy-TRAFTAC Induces tail Formation Defects in Zebrafish.
A) Schematic illustration of the experimental work flow.
B) Buffer (mock, a), dCas9HT7:brachyury-TRAFTAC:HP14 (b), dCas9HT7:allscrmb-TRAFTAC:HP14 (c), dCas9HT7:brachyury-TRAFTAC:HP17 (d) and dCas9HT7:brachyury-TRAFTAC (e) were introduced to zebrafish via microinjection and embryos were analyzed at 30- and 60-hours post fertilization (hpf). Embryos injected with dCas9HT7:brachyury-TRAFTAC:HP14 displayed significant, severe tail deformation compared to the embryos injected with mock and other controls RbNCs. Scale bar: 250 μm.
C) Percentages of number of normal and defective embryos in each group treated with different RbNCs (A-E). (n: Independent biological replicates; n=4, *** p<0.0002; **** p<0.0001 in Two-Way ANOVA; Test: Tukey; confidence level: 95%, number of embryos per group = 30–50)
D) A heat map showing the percentages of severely and mildly defective embryos in each study group. Percentages were calculated compare to the total number of embryos within the corresponding group. (n: Independent biological replicates; n=4, *** p<0.0002; **** p<0.0001, 30–50 embryos per group)
DISCUSSION
Despite the advances in chemotherapeutics, many cancer-driving proteins have remained intractable to current therapeutic modalities. Small molecule inhibitors are collectively one of the most accepted and clinically approved therapeutic strategies to treat many diseases including cancer. However, development of small molecule inhibitors, beyond those that target a few proteins classes (e.g. kinases and GPCRs) is challenging due to the lack of ligandable sites. Transcription factors are one such protein class that is difficult to drug. TFs regulate their downstream gene expression through a collective network of protein-protein and protein-DNA interactions, and they lack enzymatic activity. Therefore, direct targeting of TFs is challenging. In this study, we have co-opted the DNA binding ability of TFs to develop TRAFTACs -- chimeric oligos that can simultaneously bind to a TOI and E3 ligase via an intermediary dCas9HT7 fusion protein. By employing TRAFTACs we have demonstrated that DNA-binding proteins such as TFs can be degraded via the proteasomal pathway. We have successfully validated the proof-of-concept application of TRAFTAC technology in degrading transcription factors NF-κB and brachyury.
TNF-alpha induces IκB phosphorylation and its subsequent degradation by the proteasome (independent of TRAFTAC). The dissociation of NF-κB from IκB inhibitory complex exposes the DNA binding region of NF-κB and facilitates its binding to the double-stranded DNA portion of the TRAFTAC chimeric oligo (Hayden and Ghosh, 2014). Upon haloPROTAC administration, VHL E3 ligase recruitment induces subsequent proteasomal degradation of NF-κB. This feature of TRAFTACs to target “active” TFs offers an additional layer of selectivity to NF-κB proteasomal degradation. Hence, NF-κB-TRAFTACs could display a selectivity towards diseased cells with hyperactive NF-κB signaling, while sparing healthy cells with basal levels of NF-κB signaling (Xia et al., 2018). In designing TRAFTAC technology, we have adopted a dCas9-HaloTag7 fusion protein as an intermediary protein: the dCas9HT7 fusion protein binds simultaneously to the TOI through the chimeric TRAFTAC and to the VHL-E3 complex through HP14. HaloPROTACs can be efficient degraders of HaloTag fusion proteins, but herein we have determined that the positioning of the HaloTag protein (N- or C-terminus) relative to dCas9HT7 significantly affected the resulting fusion protein’s susceptibility to HP14-mediated degradation. While C-terminal HaloTag fusion (CTdCas9HT7) is more prone to undergo HP14-mediated degradation at micromolar concentrations, the N-terminal HaloTag fusion is less susceptible. Our data also suggested that a potential use of C-terminal-HaloTag fusion Cas systems in CRISPR-mediated gene editing: to achieve less off-target effects by inducing the degradation of engineered Cas proteins after the desired genome editing via administration of a haloPROTAC (specifically, the shorter HP3) (Cho et al., 2014, Gangopadhyay et al., 2019, Tu et al., 2017).
Previous brachyury knockdown studies have demonstrated its critical roles in notochord fate during embryonic development. Although transgenic animal models such as knockout zebrafish models are available to study the biology of a given protein during embryonic development, the lack of spatiotemporal control of such systems hinders studying differential biology during different stages of embryonic development. In this study we have demonstrated successful application of TRAFTACs in zebrafish by introducing TRAFTACs into embryos in the form of a ribonuclear complex. Injection of RbNCs induced significantly higher percentage of embryos with severe tail developmental defects, suggesting an efficient and on-target effect in vivo by brachyury-TRAFTAC. TRAFTAC technology can be readily adapted to target other transcription factors by replacing the double-stranded DNA portion of the chimeric oligo. Thus, TRAFTAC degradation platform provides the flexibility to target many DNA-binding proteins for proteasomal degradation both in vitro and in vivo.
Although many indirect strategies have been developed, only a few strategies have been demonstrated to directly inhibit transcription factor activity. Development of decoy elements that transiently inhibit TFs by interfering with its DNA binding ability has been successfully demonstrated (Morishita et al., 2004, De Stefano, 2011, Egashira et al., 2008). Further, decoys that directly bind at the minor grove of dsDNA have also been developed for many TFs including STAT3 and NF-κB (Wurtz et al., 2002, Sen et al., 2012, Lee et al., 2018, Imbaby et al., 2020). However, these strategies function via occupancy-driven pharmacology, meaning these strategies provide only a transient blockade of the targeted transcription factors for so long as the DNA binding is sterically blocked. Thus, to elicit robust inhibitory effects, these elements should display a sustained interaction with their target sites.
Significantly, like PROTACs, TRAFTACs can exhibit an event-driven pharmacology that requires only transient interaction of TOI with the chimeric oligo to induce TF degradation. Furthermore, also similar to PROTACs, TRAFTACs also could be catalytic since the ternary complex of chimeric oligo:dCas9HT7:E3-ligase can bind to another molecule of the TOI and induce its degradation after having completed the first ubiquitination cycle of a previously-bound TOI. Therefore, transient interaction is enough to induce TOI degradation, in contrast current DNA-protein interaction inhibitors that possess limited inhibitory effects. Moreover, TRAFTACs are generalizable to many transcription factors with a known DNA binding sequence. We anticipate that most transcription factors could be successfully targeted for proteasomal degradation by changing the dsDNA portion of the chimeric oligo. Therefore, this method provides a straightforward strategy to target a broad range of DNA-binding proteins with minor changes to the system, thus offering an efficient approach to investigate unknown biology of known DNA-binding proteins in a rapid and robust way. Also, since TRAFTACs can be introduced as a ribonucleocomlplex, the current strategy would provide increased in vivo stability compared to short oligonucleotide derived-decoy elements.
As a future extension of this study, we anticipate modifying the current strategy to induce degradation of promoter-bound transcription factors in a gRNA-dependent and locus-dependent manner. This strategy would also provide an avenue to repress a single, targeted gene by inducing the degradation of its promoter-bound transcription factor without altering the expression of other genes that are controlled by the same transcription factor. Collectively, the TRAFTAC technology holds potential, both as a chemical biology tool and a potential therapeutic strategy for several reasons: first, many oncogenic transcription factors have already been identified as potential drug targets; second, synthesis of the chimeric oligo is simple and straightforward; third, chimeric oligos are readily adaptable to target different TFs; fourth, structural information of the transcription factor is not necessary; fifth, laborious small molecule ligand discovery campaigns are not required, and finally; sixth, TRAFTACs only require transient interaction with the target transcription factor to induce TF degradation relative to decoy or antisense oligonucleotide approaches, which require persistent engagement with target proteins to achieve the desired inhibitory effects. In fact, TRAFTAC could be a potential therapeutic strategy to treat diseases in which a transcription factor activity is dysregulated. It is also important to highlight that the current version of TRAFTACs is required to deliver as a dCas9HT7:TRAFTAC complex into cells. Therefore, the efficiency of this system might be hindered by its limited bio-availability. However, once coupled with an efficient delivery strategy, TRAFTAC technology has the potential to target hard-to-drug transcription factors and other DNA-binding proteins for proteasomal degradation in patients in the future.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
requests or information regarding resources and reagent should be addressed to Dr. Craig M. Crews (craig.crews@yale.edu).
Material availability
Except for the TBXTA-c005 plasmid, which was obtained under MTA from Dr. Opher Gileadi from University of Oxford, other reagents or plasmids are available from CMC on request. Chemical compounds were synthesized using commercially available materials. Cell lines, other reagents and antibodies were purchased from commercial vendors described in the key resource table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ECL rabbit IgG, HRP-linked whole Ab | GE Health care | Cat#NA934; RRID: AB_772206 |
| ECL mouse IgG, HRP-linked whole Ab | GE Health care | Cat# NA931; RRID: AB_772210 |
| GAPDH (14C10) | Cell Signaling | Cat# 2118S; RRID: AB_561053 |
| alpha tubulin: Alexa Fluor 488 | Invitrogen | Cat#53-4502-82;RRID:AB_1210525 |
| NFkB1 p105/p50 | Cell Signaling | Cat# 3035S; RRID: AB_330564 |
| NFkB p65 | Cell Signaling | Cat# 8242S; RRID: AB_10859369 |
| brachyury | Cell Signaling | Cat# 81694S; RRID: AB_2799983 |
| c-Myc (9E10) | Santa Cruz | Cat# sc-40; RRID: AB_627268 |
| HaloTag | Promega | Cat# G921A; RRID: AB_2688011 |
| HA-tag | Cell Signaling | Cat# 3724S; RRID: AB_1549585 |
| DYKDDDDK Tag | Cell Signaling | Cat# 14793S; RRID: AB_2572291 |
| Bacterial and virus strains | ||
| E. coli BL21-RIPL codon plus | Agilent Technologies | Cat# 230280 |
| NEB 5-alpha | New England BioLabs | Cat# C2987H |
| Rosetta 2 DE3 | Sigma | Cat# 71397 |
| Chemicals, peptides, and recombinant proteins | ||
| Tumor Necrosis Factor-α human recombinant protein | Sigma | Cat# T0157–10UG |
| dCas9 recombinant protein | Crews Lab | N/A |
| Ni-NTA agarose | QIAGEN | Cat# 30250 |
| Cycloheximide | Sigma | Cat# C104450 |
| RNAiMAX transfection reagent | Thermo Fisher Sci | Cat# 13778–150 |
| MLN4924 | selleckchem | Cat# S7109 |
| Janelia Fluor 646 (JF646) HaloTag Ligand | Promega | Cat# ga1120 |
| Critical commercial assays | ||
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystem | Cat# 4368814 |
| KAPA SYBR FAST qPCR Master Mix Kit | Kapa Biosystems | Cat# KK4600 |
| Experimental models: cell lines | ||
| HEK293T | ATCC | Cat# CRL-3216 |
| Flp-In T-Rex 293 | Thermo Fisher Sci | Cat# R78007 |
| HeLa | ATCC | Cat# CCL-2 |
| Experimental models: organisms/strains | ||
| Zebrafish (Danio rerio) wildtype strain TL | Scott Holley Lab, Yale University | N/A |
| Oligonucleotides | ||
| F_CTdCas9:ACCTGACTATGCTGGAGTG GATAAGAAATACUCAATAGGCTTAGCT ATCGGC | Yale School of Medicine | N/A |
| R_CTdCas9:ATCAGCGGGTTTAACCGGA AATCUCCAGAGTAGACAGCC | Yale School of Medicine | N/A |
| F_CTPCDNA5:AGATTTCCGGTTAAACC CGCTGAUCAGCCTCGAC | Yale School of Medicine | N/A |
| R_CTPCDNA5:AGTATTTCTTATCCACTC CAGCATAGTCAGGUACGTCATAAGGG | Yale School of Medicine | N/A |
| RNA:DNA chimeric oligos | This manuscript | Figure S1 |
| pCDNA5-HA-NTdCas9HT7 | This manuscript | Table S1 |
| pCDNA5-HA-NTdCas9HT7-NLS | This manuscript | Table S1 |
| Recombinant DNA | ||
| pET302-6His-dCas9-Halo | Deng et al Proc Natl Acad. 2015 | Addgene, Cat# 72269 |
| pCDNA5-HA-CTdCas9HT7 | This manuscript | N/A |
| pCDNA5-HA-NTdCas9HT7-NLS | This manuscript | N/A |
| pNL-Flag-Nluc-GFP | This manuscript | N/A |
| TBXTA-c005 | Opher Gileadi SGC expression vectors | Addgene Cat# 139754 |
| Software and algorithms | ||
| Image Lab 6.0 | BioRad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| Graphpad | Prism | https://www.graphpad.com/scientific-software/prism/ |
Data and code availability
DNA sequences for the plasmids used to overexpress dCas9HT7 fusion proteins are included at the end of the supplemental information.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293T, Flp-In T-Rex 293 and HeLa cells (derived from a female adult, Source: ATCC) were cultured as described in the Method Details.
Microbe strains
NEB5-alpha and E. coli Rosetta™ 2 DE3 strains were purchased from New England Biolab and Sigma, respectively. BL21-RIPL codon plus strain was purchased from Agilent Technologies.
Animal models
Zebrafish (Danio rerio; both female and male) raised and cared according to standard protocols under the supervision of the Yale University Institutional Animal Care and Use Committee.
METHOD DETAILS
Cloning
pCDNA5-CT-dCas9HT7 and pCDNA5-NT-dCas9HT7 constructs were generated by USER cloning strategy. For pCDNA5-CT-dCas9HT7, CT-dCas9HT7 ORF sequence was directly amplified using pET302-CT-dcas9HT7 plasmid as the template (Forward primer: 5’ACCTGACTATGCTGGAGTGGATAAGAAATACUCAATAGGCTTAGCTATCGGC3’; Reverse primer: 5’ATCAGCGGGTTTAACCGGAAATCUCCAGAGTAGACAGCC3’) and the pCDNA5 vector was amplified using pCDNA5 empty vector (Forward primer: 5’ AGATTTCCGGTTAAACCCGCTGAUCAGCCTCGAC3’; Reverse primer: 5’ AGTATTTCTTATCCACTCCAGCATAGTCAGGUACGTCATAAGGG3’). Amplified insert and vector PCR products were gen purified and subjected to USER cloning reaction using USER enzyme (NEB). Reaction mixture was then transformed into NEB5 cells and single colonies were sequenced to identify successful cloning products. To generate pCDNA5-NT-dCas9HT7 construct, a 2 step USER cloning strategy was employed. First, HaloTag7 sequence was amplified without the stop codon using pET302-CT-dcas9HT7 (Forward primer:5’ACGTACCTGACTATGCTGGAGCAGAAAUCGGTACTGGCTTTCCATTCG3’; Reverse primer: 5’ATCAGCGGGTACCGGAAATCUCCAGAGTAGACAGC3’) as the template and subjected to USER cloning reaction together with amplified pCDNA5 vector (Forward primer:5’AGATTTCCGGTACCCGCTGAUCAGCCTCG3’; Reverse primer: 5’ ATTTCTGCTCCAGCATAGTCAGGTACGUCATAAGGG3’). In the second step, (Forward primer:5’ATTTCCGGTGGTGGCTCCAGAUCTGTGGATAAGAAATACTCAA TAGGCTTAGCTATCGGC3’; Revisers primer: 5’ AGCGGGTTTAGTCACCTCCTAGCU GACTCAAATCAATGC3’) dCas9 sequence was amplified with a stop codon and subjected to USER cloning reaction with the amplified vector obtained from the clone generated in the first step (Forward primer: 5’ AGCTAGGAGGTGACTAAACCCGCUGAT CAGCCTCG3’; Reverse primer: 5’ ATCTGGAGCCACCACCGGAAAUCTCCAGAGTAG ACAGC 3’). N terminal HA tag was introduced by the USER primers in both cases.
Protein purification
C-terminal HaloTag7-containing dCas9 fusion protein was expressed in E. coli BL21-RIPL codon plus bacterial cells. BL21 cells were transformed with 50 ng of plasmid DNA encoding dCas9HT7 and transformed cells were plated on carbenicillin-containing agar plates. On the following day, a single colony was selected and inoculated in 5 mL of LB and incubated overnight at 37 degrees. After 16 h, bacterial cells were diluted in 1 L of LB and shook at 37 degrees until OD600 reaches 0.8. Cells were kept on ice and induced with 0.5 mM IPTG and incubated 20 h at 18 degrees in a shaker. Cells were subjected to lysis (20 mM HEPES pH 8, 1mM MgCl2, 10% glycerol, 300 mM NaCl, 1 mM ß-ME and 1X protease inhibitor cocktail (Roche)) by exposing cells to 4 cycles of 30 seconds pulses and 1-minute rest on ice. Then clarified lysate was incubated with pre-washed Ni-NTA agarose beads (Agilent Technologies) for 1 h, at 4 degrees. The Ni-NTA beads were washed twice with wash buffer A and wash buffer B (Wash buffer A:20 mM HEPES pH 8, 1 mM MgCl2, 10% glycerol, 300 mM NaCl, 5 mM imidazole; Wash buffer B:20 mM HEPES pH 8, 1 mM MgCl2, 10% glycerol, 300 mM NaCl, 35 mM imidazole). Enriched dCas9HT7 protein was eluted with the elution buffer (20 mM HEPES pH 8, 1mM MgCl2, 10% glycerol, 300 mM NaCl, 300 mM imidazole). Eluted protein was further purified by gel filtration chromatography using a Superdex 200 column (GE Healthcare) in the storage buffer (20 mM HEPES pH 8, 1mM MgCl2, 300 mM NaCl). Purified protein was dialyzed against the storage buffer containing 10% glycerol. Purity of the dCas9HT7 protein was assessed by Coomassie staining.
TBXTA-c005 was expressed in E. coli Rosetta™ 2 DE3 Competent bacterial cells. Competent cells were transformed with 100 ng of plasmid DNA encoding TBXTA-c005 and transformed cells were plated on kanamycin containing LB plates. The following day, cells were inoculated into a 100 mL LB starter culture and incubated at 37 degree until OD600 reached 0.6. The culture was spun down and pellet resuspended in 2 L of TB media under selection of kanamycin (selection for TBXT-c005) and chloramphenicol (selection for Rosetta™ 2 pRARE-2 plasmid). Culture was grown at 37 degrees in a shaker until OD600 reached 1.2. The culture was induced with 0.3 mM IPTG and incubated overnight at 18 degrees in a shaker. Cells were subject to lysis in buffer A (50mM HEPES pH 7.5, 500mM NaCl, 10mM imidazole, 5% glycerol, 1mM ß-ME) by exposing cells to 10 minutes of sonication using 50% Dutt Cycle and 35% output control on the Branson Sonifier 450. Clarified lysate was applied to 5 mL Ni column on the AktaPure™ and eluted over 15 column volumes using buffer B (50mM HEPES pH 7.5, 500mM NaCl, 300mM imidazole, 5% glycerol, 1mM ß-ME). Fractions were resolved by SDS-PAGE. Pooled fractions were subject to dialysis against 4 L Dialysis Buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, and 1 mM ß-ME) at 4 degrees overnight. Protein was aliquoted and flash frozen at 50 μM by liquid nitrogen. TBXTA-c005 was a gift from Opher Gileadi (Addgene plasmid # 139754; http://n2t.net/addgene:139754; RRID:Addgene139754)
Oligonucleotide annealing
Single stranded TRAFTACs and reverse oligonucleotides were dissolved in ultra-pure, RNAase free water. All the steps in this protocol were carried out in a clean, RNAase/DNAase-free environment. All the equipment and plasticware in this protocol were treated with RNAase Away prior to their use. Single stranded TRAFTACs and single stranded reverse oligonucleotides were mixed (final concentrations of TRAFTACs were set to 25 μM) in 1X annealing buffer (10 mM Tris, pH 7.5, 50 mM NaCl and 1 mM EDTA) and incubated for 5 minutes in a water bath at 95 degrees. Then, the hot-plate was turned off and the samples left to cool down to room temperature over 1–2 h. Double stranded TRAFTACs were mixed well, aliquoted and stored at −80 for the future use.
EMSA
Increasing concentrations of purified dCas9HT7 protein were incubated with or without 500 nM of NFκB-TRAFTAC or brachyury-TRAFTAC for 30 min at RT. Then the mixture was separated in a 1% agarose gel for 30 minutes at constant 120 mV and images were captured using ChemiDoc system (BioRad). For ternary complex formation assay, 3 μM of dCas9HT7 and increasing concentrations of purified brachyury was incubated with or without brachyury-TRAFTAC or control-TRAFTAC for 30 minutes at RT. Gel shifts were analyzed as described above.
Cell culture
Human embryonic kidney cells HEK293 cells and Hela cells were grown in Dulbecco’s Modified Eagles Medium (DMEM) containing 10% heat inactivated fetal bovine serum (FBS), 5 ug/mL streptomycin and 5 U/mL penicillin. All the cell culture experiments and maintenance were carried out in a humidified incubator at 37 degrees and 5% CO2 supplementation. One day prior to the transfection of chimeric oligos, cells (3.5×106) were split into 6 cm cell culture dishes in complete growth medium. On the day of transfection cell culture medium was replaced with 3mL of transfection medium (DMEM 10%FBS in DMEM). Chimeric oligo transfection was performed using RNAi-Max according to the manufacturer’s protocol. Briefly, 25 nM of chimeric oligo and 15 μl of RNAi-Max were mixed in 125 μl of OPTI-MEM medium in two separate Eppendorf tubes. Mix two tubes together after 5 minutes and incubated for another 5 minutes prior to the addition to cells. After 6 h of transfection, the cells were split into 6-well plates and the transfection medium containing chimeric oligo was evenly divided into each well. Then cells were incubated for another 24 h and replaced with 1 mL of fresh transfection medium containing different concentrations of HaloPROTACs and incubated for 1 h prior to the addition of 0.25 mL of 5 ng/mL of TNF-alpha. Cells were then incubated for desired time at 37 degrees followed by cell lysis. Cell lysates were prepared by scraping off the cells using lysis buffer (25 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% NP40, 5% glycerol and 1X protease inhibitor cocktail from Roche) and lysates were centrifuged at high speed (14 000 rpm) for 10 minutes and clear supernatant was collected for further experiments.
Immunoprecipitation
Cells that overexpress dCas9HT7 together with either p50 or brachyury-GFP was lysed lysis buffer (25 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% NP40, 5% glycerol and 1X protease inhibitor cocktail from Roche) or IP buffer (25 mM Tris pH 7.4, 150 mM NaCl, 0.4% NP40, 5% glycerol and 1X protease inhibitor cocktail from Roche) respectively. Lysates (1 mg for each sample) were incubated with 100 nM of NFκB-TRAFTAC, brachyury-TRAFTAC or 3’controlcrRNA for 1 h, at RT. Then cell lysates mixture was incubated with 25 ul of pre-washed HA-agarose beads (Sigma) and top up to 500 ul with 1X TBS. Then tubes were incubated at 4 degrees overnight in a rotator. Tubes were centrifuged at 2000 rpm for 2 minutes at 4 degrees and beads were washed three times with 1 mL of lysis buffer or IP buffer for 3 times with 5 minutes incubation in a rotisserie. Beads were then eluted with 2X loading buffer containing 10% ß-ME and eluted samples were analyzed by western blotting by probing with desired primary antibodies. Chemiluminescence signal was captured using ChemiDoc system by BioRad.
Immunofluorescence
Cells were split into 8-well imaging slides one day prior to the transfection. On the day of transfection, cell culture medium was replaced with transfection medium and transfection of fluorescein-labelled chimeric oligo was carried out as described above. After 12 h of transfection, cells were fixed with 4% paraformaldehyde for 10 minutes followed by three washes with 1X PBS. Then cells were permeabilized with 0.1% Triton-X-100 in PBS for another 10 minutes. Cells were blocked for 1 h, at RT prior to the overnight incubation of anti-HA antibody. Cells then washed three times with PBS and secondary antibody conjugated to Alexa Fluor-567 was incubated for 1h at RT. After 3 washings with PBS cells were analyzed by confocal microscopy (Zeiss LSM 880 Microscope). For brachyury-GFP and fluorescently labeled zebrafish embryo (injected with RbNC labeled with TAMRA-HT7 ligand and fluorescein-TRAFTAC), fluorescence images were directly captured using fluorescence microscope (EVOS M5000 IMAGING SYSTEM) without prior fixation or permeabilization of the cells and embryos.
RNA isolation and qRT-PCR
Stable cells that overexpress CTdCas9HT7 were transfected with NFκB-TRAFTAC and control-TRAFTAC followed by HP14 or HP17 and TNF-α treatment. Cells were harvested after 9 h and RNA was isolated using RNAeasy Mini Kit from QUIGEN. Approximately 1 μg of isolated RNA was subjected to reverse transcription using a commercially available High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem). Relative RNA levels for p50 and GAPDH were analyzed using ddCt method after performing quantitative PCR in triplicates using KAPA SYBR FAST qPCR Master Mix Kit from Kapa Biosystems.
Ribonucleocomplex formation
First, purified NTdCas9HT7 protein was subjected to covalent labeling with either HP14 or HP17. Purified dCas9HT7 protein (200 μL, 2 nmoles) was incubated with HP14 in a ratio of 1:6 (12 nmoles) and 1 nmole of dCas9HT7 with 6 nmoles of HP17 at room temperature for 1 h separately. After 1 h incubation, samples were transferred to centrifugal device (30 kDa, 0.5 mL Amicon ultrs-0.5 filter units) and top up to 400 μL with dCas9HT7 storage buffer. Then filter units were centrifuged at high speed for 15 minutes until the samples were concentrated to 50 μL to remove excess HP14 and HP17. Samples containing dCas9HT7:HP14 was diluted back to 100 μL and split into two Eppendorf tubes for riboncleocomplex formation. Then dCas9HT7:HP14, dCas9HT7:HP17 and dCas9HT7 were incubated with allscrmb-TRAFTAC or brachyury-TRAFTAC in a molar ratio of 1:1 and incubated for 1 h at RT. Ribonucleocomplexes were aliquoted and stored in −80 freezer until use. To generate fluorescently labeled ribonucleocomplex, dCas9HT7 reacted with TAMRA-HT7 ligand and subsequently incubated with fluorescein labeled-TRAFTAC as described above.
Zebrafish Husbandry and Microinjection:
Zebrafish (Danio rerio) are raised and cared for according to standard protocols as approved by the Yale University Institutional Animal Care and Use Committee.
Briefly, adults from the wildtype strain TL were placed pairwise, but separate, in mating tanks the night before the microinjections. The following morning the pairs were joined and eggs from the ensuing natural spawning collected in embryo media. Approximately 350 picoliters of 1 mg/mL of RbNCs plus 0.125% phenol red were injected into the blastomere of one-cell stage embryos. In all assays these embryos were raised at 28.6°C and phenotypically scored at 24 hours post-fertilization. For fluorescent microscopy analysis, embryos were injected with RbNC without phenol red. Sex-specific data were not collected as zebrafish do not have a strictly genetic sex determination mechanism, and sex is determined after the developmental timepoints.
QUANTIFICATION AND STATISTICAL ANALYSIS
Western blots quantitation was performed using Image Lab 6.0 software from BioRad. GraphPad Prism 7.0a was used to calculate statistical significances. Statistical significances were calculated using Two-Way ANOVA (Tukey-test at 95% confidence level) using duplicate or quadruplicates values and p-values of <0.03 indicates statistical significance. Number of replicates and p-values are presented in the respective figure legend.
Supplemental Table (Table S1)
DNA sequences of the dCas9HT7 constructs used in this study. Related to the STAR methods.
Supplementary Material
SIGNIFICANCE.
Transcription factors (TFs) are conventionally considered hard to drug proteins due to their mode of action. TFs bind DNA or other accessory proteins to maintain the desired gene expression pattern that required by the cell. Unlike small molecule inhibitors, targeted protein degraders only require a simple binding element to such hard-to-drug proteins to induce their degradation and abrogate biological functions. In this study, we have developed a generalizable strategy to target transcription factors for proteasomal degradation by utilizing a double-stranded DNA as the TF-recruiting element and a conjugated oligo sequence that binds to an E3 ligase-recruiting dCas9HT7 fusion protein. Chimeric oligos, or TRAFTACs, induce the recruitment of TF and E3 ligase to proximity and facilitate the ubiquitination of the former in the presence of HaloPROTAC. We have successfully shown the application of this strategy to degrade two oncogenic TFs, NF-κB and brachyury. Our data also demonstrated that targeting the zebrafish brachyury by TRAFTACs can induce no tail phenotype, suggesting, TRAFTACs as an in vivo compatible and potential therapeutic strategy to degrade disease-relevant TFs. Furthermore, current TRAFTAC technology can be readily exploited to target promoter bound TFs using a gRNA that binds to a proximal DNA sequence. Thus, targeting promoter-bound TF will allow us to selectively repress a single gene while circumventing off target gene expression.
ACKNOWLEDGEMENT
We thank Dr. John Hines for the critical reading of the manuscript, insightful comments and critiques. We also thank Dr. Miriam A. Genuth and Dr. Dörthe Jülich for valuable inputs and discussions. We are thankful to Dr. Brian M. Linhares for the support during brachyury protein purification and Dr. Todd Douglas for providing the plasmid for NT-dCas9HT7 purification. Funding provided by NIH (R01GM127876) to S.A.H. C.M.C. is funded by the NIH (R35CA197589) and is supported by an American Cancer Research Professorship. (Figures were created with BioRender.com)
Footnotes
DECLARATION OF INTEREST
C.M.C is founder, shareholder, and consultant to Arvinas, Inc. and Halda, LLC, which support research in his laboratory.
REFERENCE
- ALI S, RASOOL M, CHAOUDHRY H, P NP, JHA P, HAFIZ A, MAHFOOZ M, ABDUS SAMI G, AZHAR KAMAL M, BASHIR S, ALI A. & SARWAR JAMAL M. 2016. Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation, 12, 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAI L, ZHOU H, XU R, ZHAO Y, CHINNASWAMY K, MCEACHERN D, CHEN J, YANG C-Y, LIU Z, WANG M, LIU L, JIANG H, WEN B, KUMAR P, MEAGHER JL, SUN D, STUCKEY JA & WANG S. 2019. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell, 36, 498–511.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSOTTI R, MAGNAGHI P, DI BELLA S, COZZI L, CUSI C, BOZZI F, BELTRAMI N, CARAPEZZA G, BALLINARI D, AMBOLDI N, LUPI R, SOMASCHINI A, RADDRIZZANI L, SALOM B, GALVANI A, STACCHIOTTI S, TAMBORINI E. & ISACCHI A. 2017. Establishment and genomic characterization of the new chordoma cell line Chor-IN-1. Scientific reports, 7, 9226–9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCKLEY DL, RAINA K, DARRICARRERE N, HINES J, GUSTAFSON JL, SMITH IE, MIAH AH, HARLING JD & CREWS CM 2015. HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins. ACS chemical biology, 10, 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUHIMSCHI AD, ARMSTRONG HA, TOURE M, JAIME-FIGUEROA S, CHEN TL, LEHMAN AM, WOYACH JA, JOHNSON AJ, BYRD JC & CREWS CM 2018. Targeting the C481S Ibrutinib-Resistance Mutation in Bruton’s Tyrosine Kinase Using PROTAC-Mediated Degradation. Biochemistry, 57, 3564–3575. [DOI] [PubMed] [Google Scholar]
- BURSLEM GM, SCHULTZ AR, BONDESON DP, EIDE CA, SAVAGE STEVENS SL, DRUKER BJ & CREWS CM 2019. Targeting BCR-ABL1 in Chronic Myeloid Leukemia by PROTAC-Mediated Targeted Protein Degradation. Cancer Res, 79, 4744–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURSLEM GM, SONG J, CHEN X, HINES J. & CREWS CM 2018. Enhancing Antiproliferative Activity and Selectivity of a FLT-3 Inhibitor by Proteolysis Targeting Chimera Conversion. J Am Chem Soc, 140, 16428–16432. [DOI] [PubMed] [Google Scholar]
- CASTELL A, YAN Q, FAWKNER K, HYDBRING P, ZHANG F, VERSCHUT V, FRANCO M, ZAKARIA SM, BAZZAR W, GOODWIN J, ZINZALLA G. & LARSSON L-G 2018. A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Scientific Reports, 8, 10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN FE, HUANG DB, CHEN YQ & GHOSH G. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature, 391, 410–3. [DOI] [PubMed] [Google Scholar]
- CHEN M, WU Y, ZHANG H, LI S, ZHOU J. & SHEN J. 2020. The Roles of Embryonic Transcription Factor BRACHYURY in Tumorigenesis and Progression. Frontiers in Oncology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN Y, JI M, ZHANG S, XUE N, XU H, LIN S. & CHEN X. 2018. Bt354 as a new STAT3 signaling pathway inhibitor against triple negative breast cancer. J Drug Target, 26, 920–930. [DOI] [PubMed] [Google Scholar]
- CHO SW, KIM S, KIM Y, KWEON J, KIM HS, BAE S. & KIM J-S 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research, 24, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI SH, MAHANKALI M, LEE SJ, HULL M, PETRASSI HM, CHATTERJEE AK, SCHULTZ PG, JONES KA & SHEN W. 2017. Targeted Disruption of Myc– Max Oncoprotein Complex by a Small Molecule. ACS Chemical Biology, 12, 2715–2719. [DOI] [PubMed] [Google Scholar]
- DAI X, YIN C, ZHANG Y, GUO G, ZHAO C, WANG O, XIANG Y, ZHANG X. & LIANG G. 2018. Osthole inhibits triple negative breast cancer cells by suppressing STAT3. J Exp Clin Cancer Res, 37, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE STEFANO D. 2011. Oligonucleotides decoy to NF-kappaB: becoming a reality? Discov Med, 12, 97–105. [PubMed] [Google Scholar]
- EGASHIRA K, SUZUKI J, ITO H, AOKI M, ISOBE M. & MORISHITA R. 2008. Long-term follow up of initial clinical cases with NF-kappaB decoy oligodeoxynucleotide transfection at the site of coronary stenting. J Gene Med, 10, 805–9. [DOI] [PubMed] [Google Scholar]
- GANGOPADHYAY SA, COX KJ, MANNA D, LIM D, MAJI B, ZHOU Q. & CHOUDHARY A. 2019. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry, 58, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALPERN ME, HO RK, WALKER C. & KIMMEL CB 1993. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell, 75, 99–111. [PubMed] [Google Scholar]
- HAYDEN MS & GHOSH S. 2014. Regulation of NF-κB by TNF family cytokines. Seminars in immunology, 26, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERRMANN BG 1991. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development, 113, 913. [DOI] [PubMed] [Google Scholar]
- HU J, HU B, WANG M, XU F, MIAO B, YANG CY, WANG M, LIU Z, HAYES DF, CHINNASWAMY K, DELPROPOSTO J, STUCKEY J. & WANG S. 2019. Discovery of ERD-308 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Estrogen Receptor (ER). J Med Chem, 62, 1420–1442. [DOI] [PubMed] [Google Scholar]
- IMBABY S, MATSUDA N, TOMITA K, HATTORI K, PALIKHE S, YOKOO H. & HATTORI Y. 2020. Beneficial effect of STAT3 decoy oligodeoxynucleotide transfection on organ injury and mortality in mice with cecal ligation and puncture-induced sepsis. Scientific Reports, 10, 15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAIME-FIGUEROA S, BUHIMSCHI AD, TOURE M, HINES J. & CREWS CM 2020. Design, synthesis and biological evaluation of Proteolysis Targeting Chimeras (PROTACs) as a BTK degraders with improved pharmacokinetic properties. Bioorganic & Medicinal Chemistry Letters, 30, 126877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG H, BOWER KE, BEUSCHER AET, ZHOU B, BOBKOV AA, OLSON AJ & VOGT PK 2009. Stabilizers of the Max homodimer identified in virtual ligand screening inhibit Myc function. Molecular pharmacology, 76, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARIN M, YAMAMOTO Y. & WANG QM 2004. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov, 3, 17–26. [DOI] [PubMed] [Google Scholar]
- KIMELMAN D. 2016. A novel cold-sensitive mutant of ntla reveals temporal roles of brachyury in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists, 245, 874–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI AC & CREWS CM 2017. Induced protein degradation: an emerging drug discovery paradigm. Nature Reviews Drug Discovery, 16, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI AC, TOURE M, HELLERSCHMIED D, SALAMI J, JAIME-FIGUEROA S, KO E, HINES J. & CREWS CM 2016. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angewandte Chemie (International ed. in English), 55, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAZO JS & SHARLOW ER 2016. Drugging Undruggable Molecular Cancer Targets. Annual Review of Pharmacology and Toxicology, 56, 23–40. [DOI] [PubMed] [Google Scholar]
- LEE DS, O’KEEFE RA, HA PK, GRANDIS JR & JOHNSON DE 2018. Biochemical Properties of a Decoy Oligodeoxynucleotide Inhibitor of STAT3 Transcription Factor. International journal of molecular sciences, 19, 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU CY, HUANG TT, CHU PY, HUANG CT, LEE CH, WANG WL, LAU KY, TSAI WC, CHAO TI, SU JC, CHEN MH, SHIAU CW, TSENG LM & CHEN KF 2017. The tyrosine kinase inhibitor nintedanib activates SHP-1 and induces apoptosis in triple-negative breast cancer cells. Exp Mol Med, 49, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELLITS KH, HAY RT & GOODBOURN S. 1993. Proteolytic degradation of MAD3 (I kappa B alpha) and enhanced processing of the NF-kappa B precursor p105 are obligatory steps in the activation of NF-kappa B. Nucleic Acids Res, 21, 5059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORISHITA R, TOMITA N, KANEDA Y. & OGIHARA T. 2004. Molecular therapy to inhibit NFkappaB activation by transcription factor decoy oligonucleotides. Curr Opin Pharmacol, 4, 139–46. [DOI] [PubMed] [Google Scholar]
- MORLEY RH, LACHANI K, KEEFE D, GILCHRIST MJ, FLICEK P, SMITH JC & WARDLE FC 2009. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proceedings of the National Academy of Sciences, 106, 3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MÜLLER CW & HERRMANN BG 1997. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature, 389, 884–8. [DOI] [PubMed] [Google Scholar]
- NALEPA G, ROLFE M. & HARPER JW 2006. Drug discovery in the ubiquitin–proteasome system. Nature Reviews Drug Discovery, 5, 596–613. [DOI] [PubMed] [Google Scholar]
- NISHIMASU H, RAN FA, HSU, PATRICK D, KONERMANN S, SHEHATA, SORAYA I, DOHMAE N, ISHITANI R, ZHANG F. & NUREKI O. 2014. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell, 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORIAN A, GONEN H, BERCOVICH B, FAJERMAN I, EYTAN E, ISRAËL A, MERCURIO F, IWAI K, SCHWARTZ AL & CIECHANOVER A. 2000. SCF(beta)(-TrCP) ubiquitin ligase-mediated processing of NF-kappaB p105 requires phosphorylation of its C-terminus by IkappaB kinase. Embo j, 19, 2580–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSBORNE CK, WAKELING A. & NICHOLSON RI 2004. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br J Cancer, 90 Suppl 1, S2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANDE V. & RAMOS MJ 2005. NF-kappaB in human disease: current inhibitors and prospects for de novo structure based design of inhibitors. Curr Med Chem, 12, 357–74. [DOI] [PubMed] [Google Scholar]
- PILLAY N, PLAGNOL V, TARPEY PS, LOBO SB, PRESNEAU N, SZUHAI K, HALAI D, BERISHA F, CANNON SR, MEAD S, KASPERAVICIUTE D, PALMEN J, TALMUD PJ, KINDBLOM LG, AMARY MF, TIRABOSCO R. & FLANAGAN AM 2012. A common single-nucleotide variant in T is strongly associated with chordoma. Nat Genet, 44, 1185–7. [DOI] [PubMed] [Google Scholar]
- PRESNEAU N, SHALABY A, YE H, PILLAY N, HALAI D, IDOWU B, TIRABOSCO R, WHITWELL D, JACQUES TS, KINDBLOM L-G, BRÜDERLEIN S, MÖLLER P, LEITHNER A, LIEGL B, AMARY FM, ATHANASOU NN, HOGENDOORN PC, MERTENS F, SZUHAI K. & FLANAGAN AM 2011. Role of the transcription factor T (brachyury) in the pathogenesis of sporadic chordoma: a genetic and functional-based study. The Journal of Pathology, 223, 327–335. [DOI] [PubMed] [Google Scholar]
- QI LS, LARSON MH, GILBERT LA, DOUDNA JA, WEISSMAN JS, ARKIN AP & LIM WA 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAFFEINER P, RÖCK R, SCHRAFFL A, HARTL M, HART JR, JANDA KD, VOGT PK, STEFAN E. & BISTER K. 2014. In vivo quantification and perturbation of Myc-Max interactions and the impact on oncogenic potential. Oncotarget, 5, 8869–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAINA K, LU J, QIAN Y, ALTIERI M, GORDON D, ROSSI AMK, WANG J, CHEN X, DONG H, SIU K, WINKLER JD, CREW AP, CREWS CM & COLEMAN KG 2016. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proceedings of the National Academy of Sciences, 113, 7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REDDY SA, HUANG JH & LIAO WS 1997. Phosphatidylinositol 3-kinase in interleukin 1 signaling. Physical interaction with the interleukin 1 receptor and requirement in NFkappaB and AP-1 activation. J Biol Chem, 272, 29167–73. [DOI] [PubMed] [Google Scholar]
- RICE MA, MALHOTRA SV & STOYANOVA T. 2019. Second-Generation Antiandrogens: From Discovery to Standard of Care in Castration Resistant Prostate Cancer. Frontiers in oncology, 9, 801–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIOS-FULLER TJ, ORTIZ-SOTO G, LACOURT-VENTURA M, MALDONADO-MARTINEZ G, CUBANO LA, SCHNEIDER RJ & MARTINEZ-MONTEMAYOR MM 2018. Ganoderma lucidum extract (GLE) impairs breast cancer stem cells by targeting the STAT3 pathway. Oncotarget, 9, 35907–35921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKAMOTO KM, KIM KB, KUMAGAI A, MERCURIO F, CREWS CM & DESHAIES RJ 2001. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences, 98, 8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALAMI J, ALABI S, WILLARD RR, VITALE NJ, WANG J, DONG H, JIN M, MCDONNELL DP, CREW AP, NEKLESA TK & CREWS CM 2018. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Communications Biology, 1, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEN M, THOMAS SM, KIM S, YEH JI, FERRIS RL, JOHNSON JT, DUVVURI U, LEE J, SAHU N, JOYCE S, FREILINO ML, SHI H, LI C, LY D, RAPIREDDY S, ETTER JP, LI P-K, WANG L, CHIOSEA S, SEETHALA RR, GOODING WE, CHEN X, KAMINSKI N, PANDIT K, JOHNSON DE & GRANDIS JR 2012. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer discovery, 2, 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARIFNIA T, WAWER MJ, CHEN T, HUANG Q-Y, WEIR BA, SIZEMORE A, LAWLOR MA, GOODALE A, COWLEY GS, VAZQUEZ F, OTT CJ, FRANCIS JM, SASSI S, COGSWELL P, SHEPPARD HE, ZHANG T, GRAY NS, CLARKE PA, BLAGG J, WORKMAN P, SOMMER J, HORNICEK F, ROOT DE, HAHN WC, BRADNER JE, WONG KK, CLEMONS PA, LIN CY, KOTZ JD & SCHREIBER SL 2019. Small-molecule targeting of brachyury transcription factor addiction in chordoma. Nature Medicine, 25, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH BE, WANG SL, JAIME-FIGUEROA S, HARBIN A, WANG J, HAMMAN BD & CREWS CM 2019. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nature Communications, 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEMPLE DL, SOLNICA-KREZEL L, ZWARTKRUIS F, NEUHAUSS SC, SCHIER AF, MALICKI J, STAINIER DY, ABDELILAH S, RANGINI Z, MOUNTCASTLE-SHAH E. & DRIEVER W. 1996. Mutations affecting development of the notochord in zebrafish. Development, 123, 117. [DOI] [PubMed] [Google Scholar]
- STRUNTZ NB, CHEN A, DEUTZMANN A, WILSON RM, STEFAN E, EVANS HL, RAMIREZ MA, LIANG T, CABALLERO F, WILDSCHUT MHE, NEEL DV, FREEMAN DB, POP MS, MCCONKEY M, MULLER S, CURTIN BH, TSENG H, FROMBACH KR, BUTTY VL, LEVINE SS, FEAU C, ELMILIGY S, HONG JA, LEWIS TA, VETERE A, CLEMONS PA, MALSTROM SE, EBERT BL, LIN CY, FELSHER DW & KOEHLER AN 2019. Stabilization of the Max Homodimer with a Small Molecule Attenuates Myc-Driven Transcription. Cell Chem Biol, 26, 711–723.e14. [DOI] [PubMed] [Google Scholar]
- SUN X, GAO H, YANG Y, HE M, WU Y, SONG Y, TONG Y. & RAO Y. 2019. PROTACs: great opportunities for academia and industry. Signal Transduction and Targeted Therapy, 4, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TU Z, YANG W, YAN S, YIN A, GAO J, LIU X, ZHENG Y, ZHENG J, LI Z, YANG S, LI S, GUO X. & LI XJ 2017. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci Rep, 7, 42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN ROOIJEN E, VOEST EE, LOGISTER I, KORVING J, SCHWERTE T, SCHULTE-MERKER S, GILES RH & VAN EEDEN FJ 2009. Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood, 113, 6449–6460. [DOI] [PubMed] [Google Scholar]
- VERDINE GL & WALENSKY LD 2007. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res, 13, 7264–70. [DOI] [PubMed] [Google Scholar]
- WALHOUT AJ, GUBBELS JM, BERNARDS R, VAN DER VLIET PC & TIMMERS HT 1997. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic acids research, 25, 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON PA, ARORA VK & SAWYERS CL 2015. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nature Reviews Cancer, 15, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON V, MANSON L, SKARNES WC & BEDDINGTON RS 1995. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development, 121, 877. [DOI] [PubMed] [Google Scholar]
- WINTER GE, BUCKLEY DL, PAULK J, ROBERTS JM, SOUZA A, DHE-PAGANON S. & BRADNER JE 2015. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science, aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WURTZ NR, POMERANTZ JL, BALTIMORE D. & DERVAN PB 2002. Inhibition of DNA Binding by NF-κB with Pyrrole-Imidazole Polyamides. Biochemistry, 41, 7604–7609. [DOI] [PubMed] [Google Scholar]
- XIA L, TAN S, ZHOU Y, LIN J, WANG H, OYANG L, TIAN Y, LIU L, SU M, WANG H, CAO D. & LIAO Q. 2018. Role of the NFκB-signaling pathway in cancer. OncoTargets and therapy, 11, 2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG F, NICKOLS NG, LI BC, MARINOV GK, SAID JW & DERVAN PB 2013. Antitumor activity of a pyrrole-imidazole polyamide. Proceedings of the National Academy of Sciences, 110, 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG XR, NG D, ALCORTA DA, LIEBSCH NJ, SHERIDAN E, LI S, GOLDSTEIN AM, PARRY DM & KELLEY MJ 2009. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet, 41, 1176–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG H, ZHAO H-Y, XI X-X, LIU Y-J, XIN M, MAO S, ZHANG J-J, LU AX & ZHANG S-Q 2020. Discovery of potent epidermal growth factor receptor (EGFR) degraders by proteolysis targeting chimera (PROTAC). European Journal of Medicinal Chemistry, 189, 112061. [DOI] [PubMed] [Google Scholar]
- ZHANG X, CROWLEY VM, WUCHERPFENNIG TG, DIX MM & CRAVATT BF 2019. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nature Chemical Biology, 15, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU J, KWAN KM & MACKEM S. 2016. Putative oncogene <em>Brachyury</em> (<em>T</em>) is essential to specify cell fate but dispensable for notochord progenitor proliferation and EMT. Proceedings of the National Academy of Sciences, 113, 3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences for the plasmids used to overexpress dCas9HT7 fusion proteins are included at the end of the supplemental information.