Abstract
DNA double-strand breaks may be induced by endonucleases, ionizing radiation, chemical agents, and mechanical forces or by replication of single-stranded nicked chromosomes. Repair of double-strand breaks can occur by homologous recombination or by nonhomologous end joining. A system was developed to measure the efficiency of plasmid gap repair by homologous recombination using either chromosomal or plasmid templates. Gap repair was biased toward gene conversion events unassociated with crossing over using either donor sequence. The dependence of recombinational gap repair on genes belonging to the RAD52 epistasis group was tested in this system. RAD51, RAD52, RAD57, and RAD59 were required for efficient gap repair using either chromosomal or plasmid donors. No homologous recombination products were recovered from rad52 mutants, whereas a low level of repair occurred in the absence of RAD51, RAD57, or RAD59. These results suggest a minor pathway of strand invasion that is dependent on RAD52 but not on RAD51. The residual repair events in rad51 mutants were more frequently associated with crossing over than was observed in the wild-type strain, suggesting that the mechanisms for RAD51-dependent and RAD51-independent events are different. Plasmid gap repair was reduced synergistically in rad51 rad59 double mutants, indicating an important role for RAD59 in RAD51-independent repair.
Transformation of fungi with plasmid DNA has yielded important insights into the mechanisms of recombination and double-strand break repair (DSBR). Early studies by Hinnen et al. (17) provided evidence for integration of circular nonreplicating plasmids by homologous recombination into the yeast genome. In a subsequent study, Orr-Weaver et al. (38) elaborated more thoroughly the way in which circular and linear nonreplicating DNA molecules recombine with homologous chromosomal sequences. They showed that DNA ends are highly recombinogenic and interact directly with homologous sequences. If two restriction cuts are made within a plasmid region homologous to chromosomal DNA, thereby producing a double-strand gap, the resulting deleted linear plasmids transform at a high frequency and are faithfully repaired during the integration process. Using linear replicating plasmids, Orr-Weaver and Szostak (37) reported the recovery of approximately equal numbers of integrated and nonintegrated plasmids and concluded that gene conversion by double-strand gap repair can occur either with or without crossing over. These studies formed the basis for the DSBR model (65). They also observed circularization of linear plasmid DNA, suggesting the presence of additional, recombination-independent repair pathways. Subsequent studies of plasmid gap repair in Saccharomyces cerevisiae and Ustilago maydis indicated a lower association of crossing over with gene conversion (12, 44). Studies in Drosophila and mouse cells have also shown a very low association of crossing over (<5%) during DSBR (35, 48).
When plasmids capable of autonomous replication are cut within regions that have no homology to yeast genomic sequences, repair of the break can occur by nonhomologous end joining (7). In yeast, the efficiency of this process is dependent on the types of ends produced. Cohesive ends are efficiently repaired by precise end joining in a reaction dependent on HDF1, HDF2, MRE11, RAD50, XRS2, DNL4, and LIF1 (31, 32, 51, 69). Cohesive ends generated in genomic DNA by either HO or EcoRI endonucleases are also efficiently repaired by the end-joining pathway, indicating that plasmid and chromosomal breaks are repaired by similar mechanisms (23). Repair of blunts ends is inefficient in wild-type cells (7). This contrasts with mammalian cells, which show efficient joining of a variety of DNA ends (50).
DNA repair-deficient strains have proven useful for understanding the genetic control of end joining, and the ease of recovering plasmids has allowed a molecular analysis of the type of end-joining event. Although the effects of mutations in DNA repair genes on mating-type switching (28, 60), direct repeat (25, 30), and inverted-repeat recombination (1, 45, 46) have been studied extensively, very few studies have dealt with the role of RAD genes in homology-dependent plasmid double-strand gap repair. The initial studies of plasmid gap repair demonstrated an essential role for RAD52 (38) and subsequent studies showed a requirement for RAD50, RAD53, RAD54, and RAD57, but the repair events were not analyzed in detail (14, 15, 42).
Although all of the genes of the RAD52 epistasis group are required for the repair of ionizing radiation-induced DNA damage, the mutants show considerable heterogeneity in recombination assays. RAD52 has a unique position within the group in that it is required for most spontaneous and induced mitotic recombination events, for the formation of joint molecules in the rDNA locus, and for single-strand annealing (46, 59, 73). The rad51, rad54, rad55, and rad57 mutants form a subgroup with similar phenotypes. In these mutants, joint molecules are still detected at the rDNA locus, and they are proficient at several double-strand break (DSB)-initiated and spontaneous mitotic recombination events (45, 60, 73). For example, although natural mating-type switching is lethal in rad51 mutants, repair of an HO endonuclease-induced DSB can occur under certain circumstances if the donor sequence is unsilenced and on a plasmid (60). Furthermore, a DSB introduced into one allele of the MAT locus in rad51 diploids can be efficiently repaired by strand invasion and replication primed from the invading strand to restore the chromosome arm (27). Single-strand annealing is also independent of RAD51, RAD54, RAD55, and RAD57 (19).
Rad51 has significant homology to bacterial RecA proteins and catalyzes DNA strand exchange in vitro (54, 64). Rad54 and the Rad55-Rad57 heterodimer enhance the efficiency of the Rad51-mediated strand exchange reaction, consistent with genetic studies indicating similar phenotypes of the respective mutants (20, 43, 45, 63). Rad52 stimulates the Rad51-promoted strand exchange reaction by overcoming the inhibitory effects of replication protein A (6, 36, 55, 62). This is consistent with studies showing physical interactions between these proteins (40, 54) and genetic analysis indicating that RAD52 is epistatic to RAD51 (46). The observation that high levels of certain types of recombinational repair can occur in the absence of RAD51 suggests that alternate mechanisms for homologous pairing and strand invasion exist in S. cerevisiae. RAD59 was identified by its requirement for RAD51-independent mitotic recombination of inverted repeats (4). However, Rad59 shows 28% identity to Rad52 instead of homology to the RecA family of proteins. Although RecA-like proteins have formed the paradigm for homologous pairing and strand exchange, recent studies suggest that a different class of proteins, exemplified by bacteriophage lambda β protein, provide an alternate pathway for recombinational repair (71, 72). Rad52 shows no primary sequence homology to β protein, but both proteins form ring structures and catalyze strand annealing in vitro (22, 34, 41, 56).
Two alternative hypotheses for RAD51-independent recombination have been suggested. First that RAD51, RAD54, RAD55, and RAD57 gene products do not play a direct role in recombination but instead are required to facilitate DNA strand invasion into otherwise inaccessible sequences (60). This hypothesis was put forward to explain the occurrence of RAD51-independent DSBR when the donor for repair was expressed and plasmid borne. If the hypothesis presented by Sugawara et al. (60), is correct, we would expect rad51 mutants to be defective in all assays that involve repair from a chromosomal donor but not when the donor sequences are expressed and on a plasmid. Second, an alternative explanation to the donor accessibility model is that recombination is RAD51 independent when the event can be resolved as a crossover. Based on evidence obtained from inverted-repeat recombination experiments (45, 46), we proposed that the RAD51 pathway, including RAD54, RAD55 and RAD57, leads primarily to noncrossover recombinants and is not involved in the recombination pathway that results in crossovers. A system for intermolecular plasmid gap repair was developed to test these hypotheses.
MATERIALS AND METHODS
Plasmids.
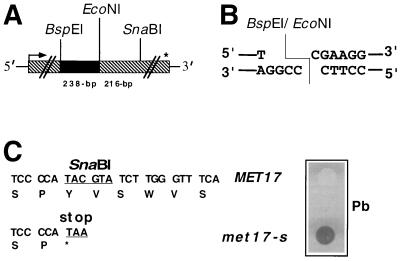
The source of the gene encoding O-acetylhomoserine-O-acetylserine sulfhydralase, MET17 (21), was plasmid pGC3 (ATTC 87440), which contains a 2.5-kb SpeI fragment of the genomic MET17 locus cloned into pRS414 (57). To obtain a restriction fragment length polymorphism nonsense mutation marker (met17-s), oligonucleotide-directed insertion mutagenesis was performed on the unique SnaBI site in MET17 of plasmid pSB99 (see Fig. 1, nucleic acid position 943) using the Quik Change site-directed mutagenesis kit (Stratagene). Plasmid pSB99 was constructed by cloning the 1.8-kb XbaI MET17 fragment from pGC3 into the XbaI site of pBluescript (Stratagene). Plasmid pSB99 was used as the template and oligonucleotides oSB2 (5′-AGAACAATCCCCATAACGTATCTTGGGTTTC-3′) and oSB1 (5′-AAACCCAAGATACGTTATGGGGATTGTTC-3′) were used to direct the sequence change. The nucleotides in the oligomers that caused the PCR-mediated mutation of the SnaBI site (TACGTA) are underlined. Plasmids derived from the mutagenesis were screened for the absence of a SnaBI site and candidates were further analyzed by DNA sequencing to confirm that a stop codon was introduced (see Fig. 1). The mutagenized plasmid pSB99 was named pSB99-1. Plasmid pSB112 was constructed by replacing the XbaI-XbaI MET17 fragment in pGC3 with the mutagenized MET17 of pSB99-1. To create plasmid pSB115, used for the two-step replacement of the chromosomal MET17 locus by met17-s, a BamHI-NotI met17-s fragment was isolated from pSB112 and ligated to the BamHI-NotI fragment of the integrating vector pRS406 (57).
FIG. 1.
Physical map of MET17, the molecular structure of the gap, and the phenotype conferred to cells by the met17-s mutation. (A) The hatched box indicates the 1.5-kb ORF (arrow, start codon; ∗, stop codon). MET17 plasmids to be used as substrates in the gap repair assay were digested with BspEI and EcoNI to produce a 238-bp gap, indicated by a black box. The plasmid and chromosomal DNA donor sequences contain a nonsense mutation that destroyed a SnaBI site (met17-s) 216 bp downstream of the EcoNI site. (B) The gap produced by BspEI-EcoNI digests consists of noncomplementary 5′ overhangs that overlap in one nucleotide (C · C) and is expected to provide a poor substrate for ligation. However, degradation or melting of the EcoNI end could provide microhomologies (C · G and/or GG or CC) for annealing to the overhang produced by BspEI digestion. (C) The first sequence shows the SnaBI site in the MET17 allele; the second line of sequence indicates the A insertion at the SnaBI site, which creates a stop codon and disrupts the recognition site for SnaBI. The met17-s mutation confers to cells a dark brown phenotype when grown on medium supplemented with lead (Pb).
Centromeric and nonreplicating gap repair plasmids pSB103 and pSB101 were constructed by ligation of the BamHI-EagI MET17 fragment isolated from pGC3 into the pRS416 (CEN6 ARSH4 URA3) and pRS406 (URA3) vectors, respectively (57). For the construction of the autonomously replicating gap repair plasmid pSB110, ARSH4 (8) was first PCR amplified from the centromeric ARS plasmid pRS414. Primers oSB11 (5′-AGACTCTAGGGGGACGTCGATCGCCAACAA-3′) and oSB12 (5′-TTTCTTAGGACGGACGTCGATCGCTTGCCTG-3′) were designed in a way that AatII sites (underlined) flanked the PCR-amplified ARS element. The PCR product was digested with AatII and cloned into the AatII site of pSB101. To obtain pSB118, the plasmid DNA donor in the plasmid by plasmid gap repair experiment, the BamHI-EagI MET17 fragment from pGC3 was first cloned into pRS414 (CEN6 ARSH4 TRP1) to yield pSB104. The MET17 in pSB104 was then replaced by the met17-s allele to generate pSB118.
All plasmids were amplified in Escherichia coli strain DH5αF′ [F′ endA1 hsdR17 (rK− mK+) supE44 thi-1 recA gyrA96 relA1 Δ(argF-lacZYA)U169, P80 lacZΔM15).
Media and strains.
All media were prepared as described previously (53) with minor modifications. Synthetic complete (SC) medium lacked cysteine, since this amino acid can be converted into methionine via MET17-independent pathways (10). Selective media lacking 1 amino acid (aa) are designated SC−aa, e.g., selection for LEU2 disrupted genes was performed in SC−Leu which is SC with all the amino acids W303 needs for growth except leucine. Lead (Pb2+) plates were prepared by dissolving 3 g of peptone, 5 g of yeast extract, 200 mg of ammonium sulfate, and 40 g of glucose and suspending 20 g of agar in 1 liter of water. The suspension was autoclaved, the agar solution was cooled to 55°C, 2 ml of lead nitrate, Pb(NO3)2 (0.5 mg/ml of water), was added, and the solution was mixed vigorously before the plates were poured (10). Standard genetic techniques were used to manipulate yeast strains (53).
All strains are derivatives of W303-1A and W303-1B (66) with the corrected RAD5 allele (11) and are listed in Table 1. The met17-s mutant LSY693 was generated by two-step replacement (70) of MET17 in strain YKH10a using the URA3-integrating plasmid pSB115. First, plasmid pSB115 was cut with BspEI to target integration at MET17. Then Ura+ transformants were selected for 5-fluoroorotic acid (5-FOA) resistance and screened for the met17-s-derived brown phenotype when grown on Pb2+ plates (see Fig. 1). To obtain the met17::ADE2 (PMET17-ADE2) mutant strain LSY694, the complete MET17 open reading frame (ORF) in W303-1A was replaced by the ADE2 ORF using a microhomology-mediated one-step gene replacement method (5). For that experiment, two PCR primers were designed: a 60-mer oligonucleotide, in which the 5′ end was complementary to the 40-nucleotide proximal region of the MET17 ORF and the 3′ portion was complementary to the first 20 nucleotides of the ADE2 ORF, and a 61-mer oligonucleotide, in which the 3′ end was complementary to the distal 21 nucleotides of the ADE2 ORF and the 5′ end was complementary to the sequence that extends 40 nucleotides from the MET17 ORF. The ADE2 ORF delineated by MET17 sequences was PCR amplified from plasmid pL909, gel purified and transformed into strain W303-1A. Ade+ transformants containing the correct disruption were identified by phenotype and confirmed by Southern blot analysis.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATa | 67 |
| W303-1B | MATα | 67 |
| YKH10a | MATa ADE2 | K. Huang |
| W1588-4C | MATa RAD5 | R. Rothstein |
| W1588-4A | MATα RAD5 | R. Rothstein |
| YHK595-1C | MATα rad51::LEU2 RAD5 | H. Klein |
| YHK598-8B | MATα rad57::LEU2 RAD5med | H. Klein |
| LSY563 | MATa rad59::LEU2 | Y. Bai |
| B404-4D | MATα rad51::HIS3 rad59::LEU2 lys2 | Y. Bai |
| Y301 | MATa rad53-21 | S. Elledge |
| LSY693 | MATa ADE2 met17-s | This study |
| LSY694 | MATa met17::ADE2 | This study |
| LSY695-2B | MATα ADE2 met17-s RAD5 | This study |
| LSY695-7D | MATa ADE2 met17-s RAD5 | This study |
| LSY696-1C | MATα met17::ADE2 RAD5 | This study |
| LSY696-7A | MATa met17::ADE2 RAD5 | This study |
| LSY715 | MATα rad52::LEU2 | This study |
| LSY826 | MATa rad51::LEU2 met17-s ADE2 RAD5 | This study |
| LSY827 | MATa rad51::LEU2 met17::ADE2 RAD5 | This study |
| LSY718 | MATa rad52::LEU2 met17-s ADE2 RAD5 | This study |
| LSY723 | MATa rad52::LEU2 met17::ADE2 RAD5 | This study |
| LSY842 | MATa rad53-21 met17-s ADE2 RAD5 | This study |
| LSY843 | MATa rad53-21 met17::ADE2 RAD5 | This study |
| LSY838 | MATa rad57::LEU2 met17-s ADE2 RAD5 | This study |
| LSY839 | MATa rad57::LEU2 met17::ADE2 RAD5 | This study |
| LSY837 | MATa rad59::LEU2 met17-s ADE2 RAD5 | This study |
| LSY836 | MATa rad59::LEU2 met17::ADE2 RAD5 | This study |
| LSY841 | MATa rad51::HIS3 rad59::LEU2 met17-s ADE2 RAD5 lys2 | This study |
| LSY840 | MATa rad51::HIS3 rad59::LEU2 met17:: ADE2 RAD5 lys2 | This study |
All strains are derivatives of W303 (leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 MET17 rad5-535). Only differences in the genotype from the parental strains are indicated, except for B404-4D, which is also ade2::hisG:URA3:hisG his3::ade2-5′Δ:TRP1:ade2-n.
Strains W1588-4A and W1588-4C (Table 1) are RAD5 derivatives of W303 (a gift of R. Rothstein) that were crossed to LSY693 and LSY694 to create RAD5 strains containing the met17-s (LSY697) and met17::ADE2 (LSY698) alleles, respectively. The rad52::LEU2-disrupted strain LSY715 (MET17 rad52::LEU2 rad5-535) was constructed by transformation of W303-1B with the BamHI-digested disruption plasmid pSM20 (52). Leu+ transformants containing the rad52::LEU2 allele were identified by sensitivity to γ irradiation and confirmed by Southern blot analysis. Strains LSY697 and LSY698 were then crossed to W303 derivatives containing the appropriate rad gene mutation to create the strains listed in Table 1. Colony PCR was used to monitor the rad5-535 missense allele in genetic crosses (29).
Plasmid DNA gap repair assays.
Plasmids to be used as substrates in the gap repair assay were digested with BspEI and EcoNI (Fig. 1), and the linear DNA was gel purified. Transformation was performed by the lithium acetate transformation method (53) with 100 ng of gapped plasmid (gap repair substrate) or 100 ng of uncut plasmid (transformation efficiency control) in the presence of 50 μg of denatured salmon sperm DNA as carrier DNA. The transformed cells were diluted and plated onto SC−Ura−Met and SC−Ura media. The colonies on both plates were counted after incubation at 30°C for 3 days. The amounts of DNA used were determined to be in the linear range for uptake of DNA. The specific gap repair frequency was calculated as the number of Met+ Ura+ recombinants per microgram of transformed linearized DNA divided by the total number of Ura+ Met+ transformants per microgram of appropriate circular transformation control DNA. As a control for undigested plasmid DNA contamination of the gapped substrate, a second yeast strain containing a complete deletion of the MET17 ORF was used as a host for transformation. The rare Met+ transformants arising from this strain were considered to be due to contamination of the gapped DNA with uncut plasmid, and this number was deducted from the number of Met+ transformants obtained in the experimental strain. The gap repair experiments were repeated at least twice for each substrate and strain, and the mean gap repair frequencies are presented.
To test for the mitotic stability of the URA3 and the MET17 markers, all the resulting Ura+ transformants were picked from one region of each transformation plate, transferred into water-filled 96-microtiter plate wells, and spotted onto SC−Ura plates. The cells were grown at 30°C for 3 days to confirm the Ura+ phenotype. The spots were replica plated onto SC− Met plates to score for the Met+ phenotype and, in parallel, onto YPD (plasmid by chromosome assay) or onto SC−Trp (plasmid by TRP1 plasmid assay). Cells were grown under conditions nonselective for Ura+ or Met+ for 2 to 3 days at 30°C and subsequently replica plated onto 5-FOA and Pb2+ plates, respectively, to assess the mitotic stability phenotype. Confluent growth on 5-FOA indicated that the Ura+ phenotype was mitotically unstable. Growth on SC−Met plates (diagnostic of Met+) but then a Met− phenotype (dark brown) when grown on Pb2+ indicated that the Met+ phenotype was unstable.
Molecular analysis of gap repair recombinants.
The mode of recombination during gap repair was determined by Southern blot analysis. Total DNA was extracted from 5-ml cultures of individual transformants (53) and digested with BamHI-SnaBI, and fragments were separated by electrophoresis through 0.7% agarose. DNA was transferred to a nylon membrane (Hybond-N; Amersham) and hybridized with a 1.5-kb BamHI-SnaBI MET17 fragment isolated from plasmid pGC3 that was 32P labeled for use as the probe.
RESULTS
Rationale for the gap repair system.
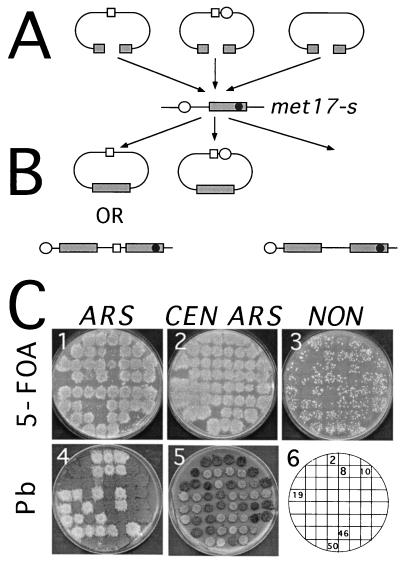
A set of recombination reporter substrates was constructed by subcloning the MET17 gene into plasmids of the pRS400 series containing the URA3 marker (57). These plasmids were gapped within the MET17 gene and used as recipients for gap repair during transformation into host strains. The gap was made in the plasmid by deleting a 238-bp fragment from the MET17 ORF with the restriction enzymes BspEI and EcoNI (Fig. 1). The restriction enzymes produce overlapping but noncomplementary ends that should be poor substrates for end-joining ligation reactions. The donor sequences for DNA repair contained a nonsense mutation at the SnaBI site of the chromosomal MET17 locus (met17-s). Since the met17-s mutation lies downstream of the region that covers the gap in the plasmid, Met+ transformants can arise by repair of the gap using chromosomal information (Fig. 2). Plasmid-by-plasmid gap repair was studied with the same rationale except that the met17-s donor sequence was located on a circular centromeric TRP1 plasmid that was introduced into a host strain in which the complete chromosomal MET17 ORF was replaced by the ADE2 ORF. Selection for the TRP1 plasmid was maintained throughout the plasmid by plasmid gap repair assay. The use of both chromosomal and plasmid donors was to test the hypothesis that RAD51 and RAD57 are not required if the donor sequence for gap repair is expressed and plasmid borne.
FIG. 2.
Rationale of the gap repair assay. (A) The double-strand gap in MET17 on either ARS (open square), CEN ARS (open square and open circle), or integrating (NON) plasmid is repaired from homologous chromosomal or plasmid met17-s sequences. (B) Repair of gapped ARS plasmids without a crossover produces a repaired MET17 plasmid and an unchanged donor sequence (chromosome or plasmid); repair associated with a crossover results in an integrated ARS plasmid. Repair of a gapped CEN ARS plasmid has to occur by a noncrossover mechanism because integration results in a dicentric chromosome or plasmid, which is inviable. Repair of the gapped plasmid that contains no ARS element has to occur by integration to yield a stable transformant. The products were drawn based on the assumption that the gap in the plasmid is not extended by nucleases over the MET17 SnaBI site. (C) The products expected from gap repair unassociated and associated with crossover were distinguished by monitoring the selective and colony color phenotypes conferred by the URA3 and MET17 markers to the recombinants and by the mitotic stability phenotype of these markers. For the identification of single patches by numbers, a grid is included in C6. Patches on independent plates are shown; note that plates 4 and 5 are not from the same master plate. Confluent growth on 5-FOA medium indicated that the Ura+ phenotype was mitotically unstable (i.e., C1 no. 2, and C2, no. 2). Secondary pop-out recombination between duplicated MET17 alleles delineating URA3 leads to the formation of papillae on 5-FOA due to the excision and loss of URA3 (i.e., C1, no. 8 and C3, no. 2). Cells displaying a dark brown phenotype when grown on lead (Pb) plates indicated that the Met+ phenotype was unstable in cells transformed with ARS plasmid (i.e., C4, no. 10) or absent (i.e., C5, no. 2). During further incubation of plate C5 for 4 to 5 days at room temperature, patches that were previously white turned a beige color and showed dark brown papillae, indicating the progressive loss of CEN ARS plasmids (i.e., C5, no. 8). White cells were diagnostic for a stable Met+ phenotype (i.e., C4 no. 2, and C5, no. 46 and 50).
All plasmids used for gap repair contain the URA3 and MET17 genes but differ in their ability to replicate in yeast (Fig. 2). The linearized (gapped) ARS plasmid can integrate into the chromosome but can also be maintained extrachromosomally upon repair of the plasmid gap. Integration of a gapped CEN ARS plasmid creates a dicentric chromosome that cannot be maintained in cells and leads to cell death, whereas gap repair without integration results in a viable product. Linearized plasmids with no origin of replication cannot be maintained in cells unless these sequences integrate into the genome. These events can be distinguished by monitoring the selective and mitotic stability phenotypes conferred to the recombinants by the URA3 and MET17 markers. The use of different replicons for gap repair substrates was to test the hypothesis derived from studies of RAD51-independent recombination of inverted repeats that crossing over is less dependent on RAD51 function than is conversion. Based on this, we expected few recombinants when using the CEN ARS plasmid in a rad51 strain, but recombinants were expected at close to wild-type frequency when using the nonreplicative gapped plasmid.
The MET17 gene provides the advantages of another widely used colony color marker, ADE2 (47, 49) and excludes the two main disadvantages of using ADE2 in recombination studies. First, in contrast to ade2 mutants in which cellular growth is inhibited as a consequence of the toxicity of the colored by-product, the pigmentation of cells with PbS does not appear to have a deleterious effect on viability (10). The color phenotype of a strain with the met17-s allele is shown in Fig. 1. Second, the ADE2 gene harbors an autonomous replication sequence (ARS) upstream of the ADE2 ORF that might interfere with replication and DNA repair (58). For MET17, no nearby ARS is known.
Classification of gap repair products.
Genetic and physical tests were used to determine the distribution of gene conversion events associated or unassociated with a crossover. The gap repair products were grouped into categories based on the selective and mitotic stability phenotypes conferred to the recombinants by the marker genes. The Met+ Ura+ clones from the gap repair assay could have resulted from two different classes of recombination. (i) Repair of the gapped ARS plasmid by gene conversion without a crossover leads to an unstable Ura+ Met+ phenotype (Ura+u Met+u) due to loss of the plasmid under nonselective growth conditions. (ii) Repair of the plasmid followed by integration at the chromosomal met17-s locus results in a stable Ura+ Met+ phenotype (Ura+s Met+s). Transformed gapped CEN ARS and nonreplicating plasmids are constrained to remain episomal or to integrate into the genome during gap repair. Therefore, the majority of Met+ transformants from the CEN ARS plasmid were expected to be Ura+u Met+u whereas only Ura+s Met+s should be detected in assays with nonreplicating plasmids. This separation of phenotypes allowed us to monitor exclusively either gene conversion events unassociated with crossing over or conversion associated with crossing over (integration). The classification of products based on stability of the markers was confirmed by Southern blot analysis. The nonsense mutation in the met17-s allele, used as the DNA donor, has destroyed a SnaBI restriction enzyme site in MET17 and can be used to distinguish between plasmid and chromosomal alleles.
Gap repair proficiency of rad51, rad52, rad53, rad57, rad59, and rad51 rad59 mutants.
Initially, the gapped ARS plasmid was used to examine the role of plasmid versus chromosomal sequences as donors in gap repair and the dependence of plasmid-by-plasmid and plasmid-by-chromosome gap repair on RAD genes. In Rad+ cells, the efficiency of gap repair using the plasmid and chromosomal donors was comparable (Table 2). The gap repair frequencies were substantially reduced in hosts containing rad51, rad52, rad57, and rad59 mutations compared to wild-type cells, independent of the origin of the donor DNA (Table 2). The rad52 mutant showed the greatest decrease in gap repair (>500-fold), the rad51 and rad57 mutants showed an intermediate decrease (67- to 110-fold), and the rad59 strain showed a 21- to 57-fold decrease. Although not analyzed in detail, a rad55 strain showed a similar reduction in gap repair efficiency to the rad51 and rad57 strains. rad55 and rad57 mutants showed more severe DNA repair defects at low temperatures; however, the defect in gap repair was apparent even at 30°C, the temperature used for the plasmid gap repair assay (16, 20, 26). In the rad53-21 mutant, a moderate two- to fourfold reduction was observed, indicating a possible role of this DNA damage checkpoint gene in the regulation of gap repair. The epistatic relationship between RAD51 and RAD59 for gap repair was also assessed. The gap repair frequency in rad51 rad59 double mutants was synergistically reduced compared to that observed in rad51 or rad59 single mutants. The transformation frequency of the rad51 rad59 double mutants only slightly deviated from that observed in either single mutant alone. The highest reduction in the efficiency of transformation was observed in rad52 mutants (0.16 × 105 CFU/μg of DNA), as reported previously (38).
TABLE 2.
ARS plasmid gap repair frequenciesa with chromosomal and plasmid donors
| Relevant genotype | Transformation efficiency (105 CFU/μg of DNA) | Frequency (10−2) with chromosomal donor | Fold decrease | Frequency (10−2) with plasmid donor | Fold decrease |
|---|---|---|---|---|---|
| RAD | 1.1 | 31 | 1 | 23 | 1 |
| rad51 | 0.31 | 0.32 | 98 | 0.20 | 110 |
| rad52b | 0.16 | 0.06 | 550 | 0.02 | 1,000 |
| rad53-21 | 1.1 | 12 | 3 | 11 | 2 |
| rad57 | 0.63 | 0.39 | 80 | 0.34 | 67 |
| rad59 | 0.42 | 1.5 | 21 | 0.40 | 57 |
| rad51 rad59b | 0.26 | 0.07 | 460 | <0.03 | >750 |
Gap repair frequencies were determined from the number of Ura+ Met+ transformants obtained from the gapped plasmid divided by the number of Ura+ Met+ transformants from the uncut plasmid (see Materials and Methods).
Gap repair frequencies for the rad52 strain were estimated from the accumulated data from 12 independent transformations; only 3 transformations of the rad51 rad59 strain were performed.
Gap repair events are biased toward noncrossover products.
In the previous experiment, Met+ Ura+ transformants were selected to determine the frequency of gap repair to restore the MET17 gene. To determine all possible modes of plasmid repair, Ura+ transformants were selected and then analyzed for the Met phenotype as well as mitotic stability. For example, Ura+s Met+s (integration) recombinants were distinguished from Ura+u Met+u (nonintegration) by growth on nonselective medium followed by replica plating either to medium supplemented with 5-FOA or to Pb plates (see Materials and Methods). As shown in Fig. 2, Ura+u Met+u cells formed dense patches on 5-FOA plates and cells grown on Pb-plates showed a dark brown colony color phenotype diagnostic for the loss of the MET17 gene. Secondary pop-out recombination events in Ura+s Met+s cells would be indicated by the formation of papillae on 5-FOA plates due to the excision and loss of the integrated plasmid URA3 marker by recombination of flanking homologous sequences.
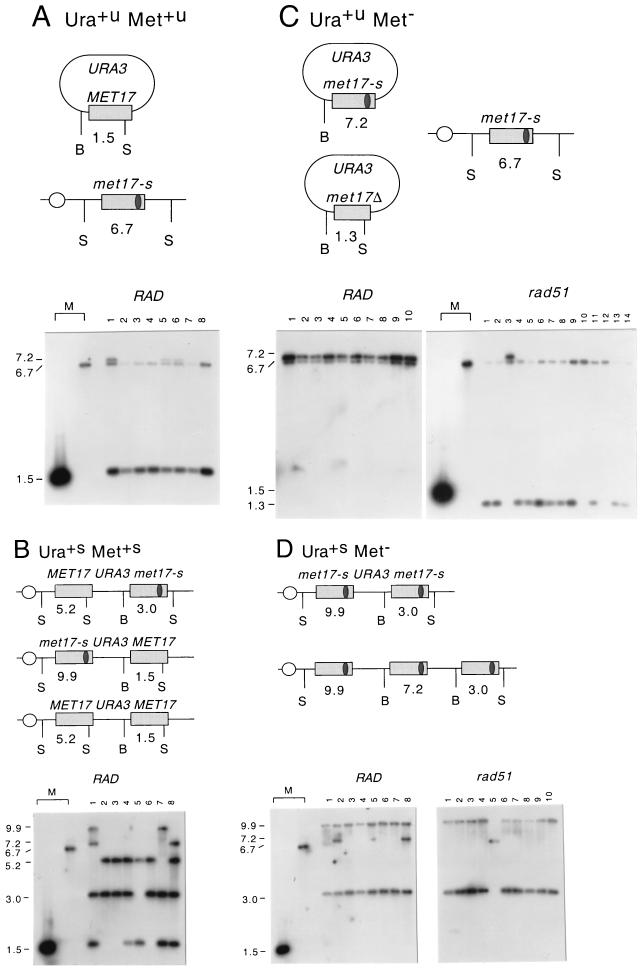
In Rad+ cells, 49% of the Ura+ transformants were Met+ (Table 3). Of the Ura+ Met+ transformants, 77% were unstable, indicative of gap repair unassociated with crossing over, and 16% were stable, indicative of gap repair associated with crossing over (integration). This distribution was confirmed by Southern blot analysis of DNA from these classes of transformants. The BamHI-SnaBI digests of total DNA isolated from 18 Ura+u Met+u transformants produced a 6.7-kb SnaBI-SnaBI chromosomal fragment and a 1.5-kb BamHI-SnaBI plasmid fragment (Fig. 3A, 8 transformants shown). In all transformants tested, the plasmid contained the wild-type SnaBI site while the chromosome retained the mutation. This pattern is diagnostic for gap repair events not associated with a crossover. In three transformants, an additional band of 7.2 kb was observed. This is most probably due to more than one plasmid entering the cell and repair to form a mixture of Met+ and Met− products (see below). Analysis of eight stable Ura+s Met+s transformants showed more complex rearrangements (Fig. 3B). Three of the eight analyzed contained the expected fragments of 5.2 and 3.0 kb, indicating the presence of two MET17 heteroalleles within the chromosomes separated by plasmid ARS URA3 sequences. The upstream allele contained a functional SnaBI site, whereas the downstream allele retained the met17-s mutation. Four transformants contained multiple, tandemly integrated copies of the plasmids. One transformant had a restriction pattern consistent with a simple integration but with two MET17 alleles.
TABLE 3.
Phenotypes of Ura+ transformants derived from recombination between the gapped ARS plasmid and chromosomal met17-s donor
| Relevant genotype | No. of Ura+ transformantsa | % of Ura+ transformantsa that were:
|
|||||
|---|---|---|---|---|---|---|---|
| Ura+u Met+u | Ura+u Met− | Ura+s Met+s | Ura+s Met− | Othersb | Met+c | ||
| RAD | 616 | 38 | 38 | 8 | 13 | 3 | 49 |
| rad51 | 161 | 14 | 56 | 4 | 26 | 0 | 18 |
| rad52d | 34 | 0 | 88 | 0 | 12 | 0 | 0 |
| rad53-21 | 91 | 37 | 44 | 8 | 9 | 2 | 47 |
| rad57 | 224 | 20 | 50 | 8 | 20 | 2 | 30 |
| rad59 | 288 | 38 | 38 | 11 | 11 | 2 | 51 |
| rad51 rad59d | 18 | 5 | 78 | 5 | 11 | 0 | 11 |
Number of Ura+ transformants analyzed; the other numbers are given as percentages of the total Ura+.
Percentage of transformants classified as Ura+s Met+u and Ura+u Met+s.
Percentage of transformants that was Met+.
Low gap repair efficiency resulting in few Ura+ transformants to analyze.
FIG. 3.
Structural analysis of Ura+ transformants. Yeast DNA isolated from Ura+ transformants was digested with BamHI (B) and SnaBI (S), and Southern blot analysis was performed with a MET17 DNA fragment as the hybridization probe. (A) Schematic representation (not drawn to scale) of the expected DNA repair product to generate an unstable (u) Ura+ Met+ phenotype; also shown is the chromosomal met17-s allele. The sizes of DNA fragments that hybridize to the probe are shown. The lower panel shows a Southern blot of this class of events. (B) Schematic representation of integration events to produce a stable Ura+s Met+s phenotype. The three simplest classes are shown, although multiple integration events to produce fragments of 1.5 and/or 7.2 kb also occur. The lower panel shows a representative Southern blot of this class of events. (C) Schematic representation of events to produce a Ura+u Met− phenotype. Conversion of the plasmid MET17 to met17-s is monitored by the appearance of a 7.2-kb fragment. A 1.3-kb BamHI-SnaBI fragment that hybridizes to the probe is diagnostic for a nonhomologous end joining of the gapped plasmid substrate. For both classes, the 6.7-kb met17-s allele is unchanged. Events from a wild-type strain (RAD) are shown on the left Southern blot, and those from a rad51 strain are shown on the right. (D) Schematic representation of an integration event to produce a stable Ura+s Met− phenotype. The 9.9- and 3.0-kb fragments are diagnostic of two copies of met17-s; multiple integration results in an additional fragment of 7.2 kb. Southern blots of DNA from RAD and rad51 strains are shown below the schematic. Included as size markers (M) are SnaBI-BamHI-digested plasmid pSB110 (MET17) and genomic DNA, isolated from an untransformed tester strain (met17-s), which produce signals of 1.5 and 6.7 kb, respectively. Fragment sizes are given in kilobase pairs on the left and were determined relative to HindIII-digested lambda DNA run as a standard.
The Ura+ Met− transformants could have arisen by several mechanisms. End joining of the gapped ARS plasmid would generate unstable Ura+ Met− transformants; alternatively, gap repair events that extended to the met17-s marker (by nuclease resection or heteroduplex DNA extension) would give rise to unstable and stable Ura+ Met− transformants. In 25 of the Ura+u Met− recombinants analyzed, DNA fragments of 6.7 kb (chromosomal met17-s allele) and 7.2 kb were observed, indicative of gap repair to convert the plasmid MET17 to met17-s (Fig. 3C, 10 recombinants shown). All of the Ura+s Met− transformants analyzed had structures consistent with integration of the plasmid and duplication of the met17-s allele (Fig. 3D). Thus, all of the Ura+s transformants (21% of the total Ura+ transformants) were the result of recombinational repair to integrate the gapped plasmid. Importantly, the physical analysis demonstrated that the majority of Ura+ transformants in Rad+ cells were the result of recombinational repair rather than end joining.
In the rad51 strain, only 18% of the Ura+ transformants were Met+, compared with 49% Met+ obtained from the wild-type strain. Of these, 79% were unstable and 21% were stable and due to integration of the repaired plasmid. Physical analysis of six Ura+s Met+s transformants confirmed integration of the plasmid and the presence of MET17 heteroalleles, but, unlike for the wild-type strain, no multiple integration events were observed (data not shown). Thus, the rad51 mutation seemed not to significantly affect the ratio of crossover to noncrossover events, but the overall probability of correct repair to Met+ was greatly reduced. Physical analysis of the Ura+s Met− transformants revealed that 9 of 10 resulted from gap repair to duplicate the met17-s allele associated with integration, as observed for the wild-type strain (Fig. 3D). Again, no multiple integration events were found. The other Ura+s Met− transformant had only the chromosomal 6.7-kb fragment and most probably resulted from conversion of the ura3-1 allele (Fig. 3D, lane 5). Of 14 Ura+u Met− transformants analyzed from the rad51 strain, 11 had a 1.3-kb fragment due to nonhomologous end joining (Fig. 3C). This class was absent from Rad+ transformants. Of the 14, 2 had larger deletions and the plasmid sequences were detected only when vector sequences were used as a hybridization probe (lanes 10 and 12 and data not shown). Only 1 of the 14 Ura+u Met− transformants analyzed from the rad51 strain had a restriction pattern consistent with conversion of the plasmid MET17 to met17-s (lane 3). Thus, most of the Ura+ Met− transformants from the rad51 strain were the result of end joining rather than recombinational repair. The transformants that were generated by recombinational repair occurred by integration 60% of the time. A similar pattern was observed in the rad57 strain.
Of 34 Ura+ transformants analyzed from the rad52 strain, none were Met+. Although some Met+ transformants were generated in rad52 strains (Table 2), these were extremely rare and gap repair frequencies were determined from only one or two Met+ colonies obtained from multiple transformations. Most of the Ura+ transformants were unstable. Analysis of five unstable Ura+ Met− transformants revealed that all occurred by end joining (data not shown). The stable Ura+ transformants could have arisen by nonhomologous integration of the gapped plasmid or by conversion or reversion of the ura3 gene but were not analyzed further.
For the rad53 and rad59 strains, the percentage of Ura+ transformants that were Met+ was comparable to that in the wild type and the distribution of gap repair events associated with crossing over was also similar to that in the wild type. As predicted from the low frequency of gap repair observed with the rad51 rad59 double mutant, very few of the Ura+ transformants obtained were Met+. Six of the unstable Ura+ Met− transformants were analyzed by Southern blotting and shown to arise by end joining (data not shown). Thus, most of the Ura+ transformants obtained from the rad51 rad59 double mutant, like the rad51 and rad52 single mutants, resulted from end joining instead of homology-dependent gap repair. Although end-joining events were not expected to occur with high efficiency using the gapped substrate because the ends produced by BspEI and EcoNI are noncomplementary, these low-frequency events were recovered from recombination-deficient strains.
Gap repair of the CEN/ARS and nonreplicative plasmids.
To further determine whether the RAD51 pathway is involved in only a particular noncrossover pathway, CEN ARS and nonreplicating plasmids were used as substrates for gap repair. These plasmids are constrained to remain episomal (CEN ARS) or to integrate (nonreplicating plasmid) during gap repair. If the hypothesis that the RAD51 pathway is required only for noncrossover events is correct, we would expect to obtain no Met+ transformants in the rad51 and rad57 strains with the CEN ARS plasmid and wild-type levels of Met+ transformants with the nonreplicating plasmid. The gap repair frequency from the chromosomal donor with the CEN ARS plasmid was lower than observed with the ARS plasmid (17 × 10−2) and was reduced further when the nonreplicative plasmids were used in wild-type strains (8.2 × 10−2) (Table 4). In contrast to our expectations, there was a greater reduction in gap repair when the nonreplicative plasmid rather than the CEN/ARS plasmid was used in all of the rad mutants tested. Thus, crossing over is dependent on RAD51, RAD57, and RAD59. As found for the ARS plasmid, gap repair of the CEN/ARS and nonreplicative plasmids was defective in these mutants when using either chromosomal or plasmid templates (Table 4).
TABLE 4.
Gap repair efficienciesa with the CEN-ARS and integrating (NON) gapped plasmids
| Relevant genotype | Gap repair frequency (10−2) for CEN-ARS x chromosome | Fold decreaseb | Gap repair frequency (10−2) for CEN-ARS x plasmid | Fold decrease | Gap repair frequency (10−2) for NON x chromosome | Fold decrease | Gap repair frequency (10−2) for NON x plasmid | Fold decrease |
|---|---|---|---|---|---|---|---|---|
| RAD | 17 | 1 | 37 | 1 | 8.1 | 1 | 7.2 | 1 |
| rad51 | 0.66 | 26 | 0.32 | 120 | 0.05 | 150 | 0.04 | 180 |
| rad57 | 0.97 | 18 | 1.2 | 30 | 0.08 | 99 | 0.14 | 51 |
| rad59 | 3.8 | 4 | 2.9 | 12 | 0.39 | 21 | 0.32 | 22 |
Gap repair frequencies were determined from the number of Ura+ Met+ transformants obtained from the gapped plasmid divided by the number of Ura+ Met+ transformants from the uncut plasmid (see Materials and Methods).
Decrease relative to RAD.
As expected, most of the Ura+ transformants obtained from the gapped CEN/ARS plasmid showed mitotic instability of the plasmid markers (Table 5). In all of the strains, approximately equal numbers of Met+ and Met− transformants were recovered. The rare stable Ura+ Met− transformants were probably due to conversion of the chromosomal ura3-1 allele by the plasmid URA3 marker. More than 99% of the Ura+ transformants obtained from the gapped nonreplicative plasmid showed mitotic stability indicative of integration of the plasmid (Table 5). Again, approximately equal numbers of Met+ and Met− transformants were recovered from the wild-type and rad59 strains, but there was a significant decrease in the number of Met+ transformants recovered from the rad51 and rad57 mutants. Analysis of repair events between the gapped ARS substrate and plasmid donor indicated even fewer crossover events than were observed from the chromosomal donor (data not shown), but these could be the result of secondary recombination events. For this reason, the products of recombination between plasmid substrates and plasmid donors were not analyzed in detail because the backbones of the vectors are homologous and could give aberrant results due to secondary recombination events between direct repeats generated by integration.
TABLE 5.
Phenotypes of Ura+ transformants derived from recombination between the gapped CEN ARS plasmid or the gapped integrating plasmid and chromosomal met17-s donor
| Relevant genotype | No. of Ura+ transformantsa | % of Ura+ transformantsa that were:
|
|||||
|---|---|---|---|---|---|---|---|
| Ura+u Met+u | Ura+u Met− | Ura+s Met+s | Ura+s Met− | Ura+s Met+u | Met+b | ||
| Gapped CEN ARS plasmid | |||||||
| RAD | 1,020 | 45 | 55 | 0 | 0.4 | 0 | 45 |
| rad51 | 132 | 42 | 52 | 0 | 5 | 1 | 43 |
| rad57 | 288 | 55 | 41 | 0 | 4 | 0 | 55 |
| rad59 | 288 | 60 | 40 | 0 | 0.4 | 0 | 60 |
| Gapped integrating plasmid | |||||||
| RAD | 1,153 | 0 | 0 | 51 | 49 | 0 | 51 |
| rad51c | 45 | 0 | 0 | 16 | 84 | 0 | 16 |
| rad57c | 173 | 0 | 0 | 31 | 69 | 0 | 31 |
| rad59 | 288 | 0 | 0 | 50 | 50 | 0 | 50 |
Number of Ura+ transformants analyzed, the other numbers are given as percentages of the total Ura+.
Percentage of Ura+ transformants that were Met+.
Significantly different from RAD as determined by chi-square analysis.
DISCUSSION
An assay for plasmid gap repair during yeast transformation was developed to distinguish between homologous recombination events resulting in crossover (integration) or noncrossover products by genetic methods. The assay is based on the repair of a gap within a plasmid-borne MET17 gene by a chromosomal or plasmid template containing a nonsense mutation near the 3′ end of the MET17 ORF. A series of plasmids was constructed that contain the selectable/counterselectable marker URA3 and the colony color marker MET17 but differ in their ability to replicate in yeast. These features can be used to distinguish between crossover and noncrossover modes of gap repair by growth on various media (Fig. 2). The goals in the establishment of this assay were to assess the relationship between gene conversion and crossing over and to determine the basis for RAD51-independent mitotic recombination events. Several hypotheses have been put forward to explain RAD51-independent events. First, Sugawara et al. (60) suggested that RAD51, RAD54, and RAD57 are not directly involved in recombination but instead are required to facilitate access to chromatin templates. Second, Rattray and Symington (45) proposed that RAD51, RAD54, RAD55, and RAD57 are required for gene conversion but play a less important role in events that can be resolved as crossovers. As described in detail below, our results are inconsistent with either hypothesis. Instead, our results support the idea that strand invasion or strand annealing between repeated sequences can occur in the absence of RAD51.
RAD51 is required for recombination from chromosomal or plasmid donors.
Using the ARS plasmid substrate, the gap repair frequency was reduced 98-fold with a chromosomal template and 110-fold with a plasmid template in rad51 strains (Table 2). Similar reductions were observed in rad57 mutants, consistent with other studies indicating that these genes function in the same pathway. This pattern was also evident with the CEN/ARS and integrating vectors (Table 4). Therefore, in this DSB repair system, RAD51 is required for repair even when the donor locus is expressed and on a plasmid. This contrasts with the results of Sugawara et al. (60), who found efficient repair of DSBs from expressed plasmid donor templates in rad51 mutants but a requirement for RAD51 when the donor was chromosomal or transcriptionally silent and on a plasmid. Although in the plasmid gap repair system described here the recipient sequences are in the form of naked transformed DNA, the donors are in the same configuration as those used in the study by Sugawara et al. (60). Thus, donor accessibility is expected to be similar in the two systems. We cannot exclude the possibility that RAD51 plays an additional role in protection of the broken ends and that this role is more important when the recipient DNA is naked instead of in chromatin.
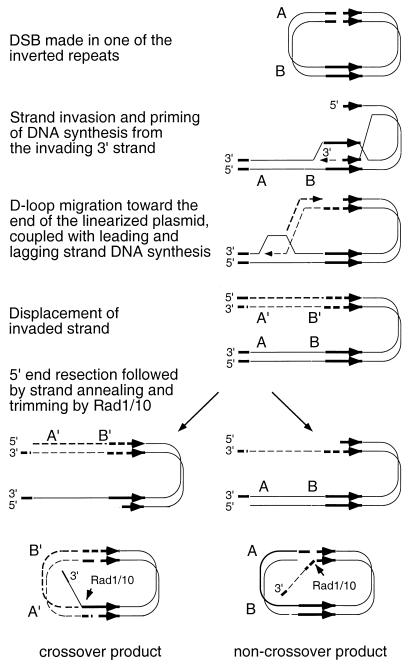
Most of the RAD51-independent recombination events detected in previous studies occurred between plasmid-borne inverted repeats (4, 19, 60). Thus, an alternative explanation to donor accessibility for RAD51-independent recombination is that this pathway operates efficiently between closely spaced inverted repeats. Repair of the HO-induced DSB in an inverted-repeat plasmid could occur by one-ended strand invasion to prime DNA synthesis followed by strand annealing (Fig. 4). The two main features of this model, strain invasion to prime DNA synthesis to the end of a DNA duplex and strand annealing, are both known to occur in the absence of RAD51 (19, 27). The prediction from this model is that RAD51-independent recombination of inverted repeats should be reduced in rad1 or rad10 strains because the Rad1/10 endonuclease is required to trim intermediates formed during SSA (18). The rate of spontaneous recombination of a chromosomal inverted repeat was reduced synergistically in a rad1 rad51 double mutant, consistent with this prediction (45). Repair of the HO-induced DSB in one of the plasmid inverted-repeat substrates described by Sugawara et al. (60) was RAD51 dependent. In that case, the donor DNA was the HMR locus, which is assembled in heterochromatin. We would argue that RAD51-independent strand invasion, DNA synthesis, or strand annealing is prevented by heterochromatin. Although repair of a chromosomal DSB is generally RAD51 dependent, Sugawara et al. (60) reported RAD51-independent repair when the donor was expressed and on a plasmid. Since the donor plasmid in that particular experiment contained two MAT alleles, only one of which was MATα-inc (refractory to HO cleavage), repair of the chromosomal break could have occurred by single-strand annealing from the linearized donor plasmid. When a donor plasmid with only the MATα-inc allele was used, repair of the chromosomal DSB was RAD51 dependent (N. Sugawara and J. Haber, personal communication). Although this supports the idea that intermolecular DSBR events require RAD51, an alternative explanation is that RAD51 is not required if the donor sequence is linearized (N. Sugawara and J. Haber, personal communication).
FIG. 4.
Model for RAD51-independent recombination of inverted repeats. Following introduction of a DSB in one of the repeats, one end is resected to produce a 3′ single-stranded tail that invades the other repeat, possibly through the action of Rad52. DNA synthesis is primed from the invading strand and proceeds to the end of the DNA molecule (the other side of the break). The linear molecule formed contains short sequences corresponding to the 5′ end of the inverted repeat at its ends. If the linear intermediate is degraded by a 5′-3′ exonuclease so that complementary single-stranded regions are revealed, then strand annealing can occur to form two types of products. One has the same structure as a reciprocal exchange, and the other has the same structure as the parental plasmid. The inverted repeats are shown by thick arrows, DNA synthesized during repair is shown by dashed lines, and sequences between the inverted repeats are designated A and B.
The RAD51-dependent pathway is biased toward noncrossover products.
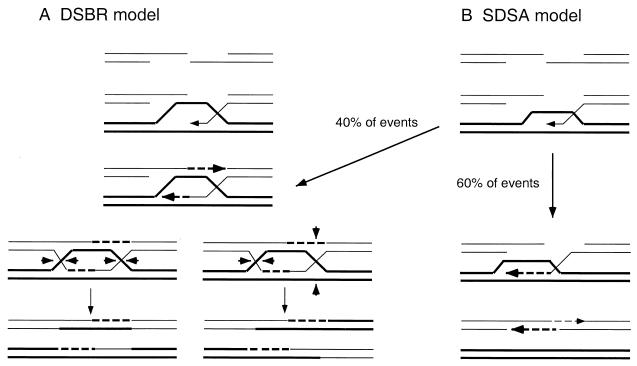
Previous studies of spontaneous mitotic recombination with an ade2 inverted repeat recombination substrate led to the suggestion that the RAD51 pathway (including RAD54, RAD55, and RAD57) is primarily required for gene conversion. In this study, DSBR events associated or unassociated with crossing over were differentiated in two ways. First, repair of the ARS plasmid substrate results in integration (crossover) or the plasmid remains episomal (noncrossover). These two classes can be distinguished by genetic and physical tests. Second, repair of the nonreplicative plasmid can occur only by integration, and repair of the CEN ARS plasmid to yield stable transformants can occur only by a noncrossover mechanism. In wild-type strains, repair of the ARS plasmid yielded 76% episomal and 21% integration events (Table 3). While this is different from the 50% integration reported by Orr-Weaver and Szostak (37), it is consistent with recent studies of gap repair in S. cerevisiae and in other organisms (12, 35, 39, 44). Studies in Drosophila and U. maydis have led to the proposal of the synthesis-dependent strand annealing (35) and migrating D-loop (12) models to explain the low incidence of crossing over associated with DSBR. In these models, strand invasion and DNA synthesis primed from the invading 3′ end occur as described for the DSBR model, but instead of forming a Holliday junction intermediate, the invading strand is displaced and can then pair with the single-stranded tail on the other side of the break (Fig. 5). Crossover events can occur if the displacement loop pairs with the other side of the break, or by appropriate strand cleavages such that a Holliday junction is formed. To account for the level of crossing over observed in this study, we suggest that the strand invasion intermediate is converted to a double Holliday structure 40% of the time. The differences in the level of associated crossing over found in different studies could reflect locus-specific effects on the ratio of these intermediates.
FIG. 5.
Models for the recombinational repair of DSBs. (A) The DSBR model. After formation of a DSB, the 5′ ends are resected to form 3′ single-stranded tails. One of the 3′ single-stranded tails invades a homologous duplex and primes DNA synthesis. The displaced strand from the donor duplex pairs with single-stranded DNA at the other side of the break and is the template for DNA synthesis. After ligation, a double Holliday structure is formed and can be resolved to yield noncrossover or crossover products. (B) The synthesis-dependent strand-annealing (SDSA)/migrating D-loop model. The first two steps are the same as those in the DSBR model, but most of the time a double Holliday junction intermediate is not formed. Instead, the invading strand that has been extended by DNA synthesis is displaced from the donor duplex and can anneal to the single-stranded tail on the other side of the break. The resulting gaps are filled by DNA synthesis and ligation to yield a noncrossover product. To account for the crossover products recovered from plasmid gap repair, we propose that 40% of the events form a Holliday junction intermediate and, of these, 50% resolve to generate crossover products. Dashed lines represent newly synthesized DNA.
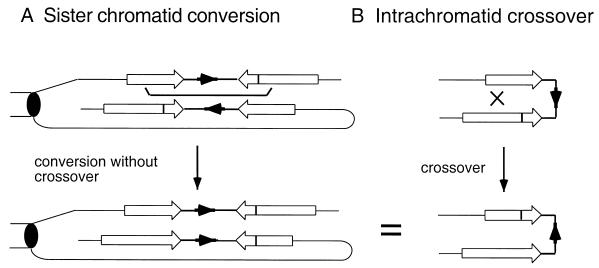
Inversion events observed with the ade2-inverted repeat can be explained by a G2 conversion model for recombination (Fig. 6) (9). If sister chromatids pair such that one reporter is in the opposite orientation to the other, recombination by gene conversion would generate viable products but a single crossover would create inviable dicentric and acentric products. A sister chromatid gene conversion event initiated within one pair of repeats that terminated within the second pair would create an inversion of the intervening DNA even though no true crossover (by Holliday junction resolution) had occurred. It is possible that most of the inversion events observed in Rad+ strains with the ade2 system occur by this mechanism of long conversion tracts, and this class of events may predominate in rad51 mutants, giving the observed bias in the products recovered (46). Alternatively, inversions might occur by RAD51-independent strand annealing of partially replicated sister chromatids during S phase.
FIG. 6.
Models for inversions between ade2 inverted repeats (9, 46). (A) In G2 cells, the inverted repeats on different chromatids (open arrows) can pair such that the intervening DNA sequences (shown as solid arrows) are in an antiparallel configuration. A gene conversion event initiated within one repeat that extends to the other repeat could result in the inversion of the intervening DNA. (B) A reciprocal crossover between inverted repeats located on the same chromatid inverts the intervening DNA sequences, but the product is indistinguishable from a G2 conversion event. The inverted repeats are shown by open boxes with arrowheads, the short repeat corresponds to a truncation of the 5′ end, and the long repeat contains a point mutation shown by the vertical line.
Aberrant events in rad51 mutants.
The proportion of repair events associated with crossing over was increased in rad51 and rad57 mutants compared to the wild-type strain. Although this difference was not significant for the Met+ events, the Met− events showed a significant bias toward integration of the plasmid. Eighty percent of the Ura+ Met− transformants that occurred by recombinational repair of the ARS plasmid in the rad51 strain were the result of integration of the plasmid and duplication of the met17-s allele. A similar alteration in the ratio of crossover to noncrossover recombinants was observed during plasmid gap repair in rad51 mutants of U. maydis (13). Although crossover events were more frequently recovered when the ARS plasmid was used, integration of the nonreplicative plasmid was reduced more than 100-fold in the rad51 mutant (Table 4), indicating an important role for RAD51 in reciprocal exchange. The rare crossover events that occur in rad51 mutants could potentially occur by two sequential break-induced replication events primed from the plasmid ends to duplicate the met17-s locus and both chromosome arms (27, 33).
Integration of the gapped ARS and nonreplicative plasmids yielded Met+ and Met− products. Analysis of the stable Ura+ Met− transformants derived from the gapped ARS plasmid revealed that most had arisen by duplication of the met17-s allele (Fig. 3). These events could reflect exonuclease digestion from the EcoNI site beyond the SnaBI site, heteroduplex extension of a Holliday junction past the SnaBI site, or extensive DNA synthesis primed from the invading 3′ end at the distal side of the gap past the chromosomal met17-s mutation. Heteroduplex DNA resulting from any of these events could be repaired to duplicate the met17-s allele. In rad51 and rad57 strains, there was a significant increase in the number of stable Ura+ Met− transformants derived from the ARS and integrating gapped plasmids. This is consistent with more extensive degradation of the 5′ end of the DSB site in rad51 and rad57 mutants (60). Alternatively, it could reflect a difference in the strand invasion and replication step by the RAD51-independent pathway. This bias toward Met− products was not observed in the rad mutants when the CEN/ARS plasmid was used. Repair of the gapped CEN/ARS plasmid is constrained to occur by a noncrossover mechanism, and this might be mechanistically different from events that result in integration. Conversion to the Holliday junction intermediate (Fig. 5) may involve different processing steps, giving rise to the altered distribution of Met+ and Met− products recovered from the CEN/ARS and nonreplicative plasmids.
Gap repair is partially dependent on RAD53.
RAD53 is classified as a member of the RAD52 epistasis group but is thought to affect recombination indirectly by transducing signals induced by DNA damage and stalled replication complexes to downstream targets (2, 61, 68). RAD53 encodes an essential, cell cycle-regulated protein kinase. rad53-21 is a conditional mutation that confers defects in the replication checkpoint and damage-induced phosphorylation of the Dun1 protein kinase (2). Consistent with a previous study (14), we found a small (two to fourfold) reduction in the efficiency of gap repair in the rad53-21 strain. However, recombination occurred at much higher frequencies than were observed for the other rad mutants, suggesting that phosphorylation of Rad proteins by Rad53/Spk1 is unlikely to play a significant role in recombinational repair.
Plasmid gap repair is dependent on RAD52.
Repair of the gapped ARS plasmid occurred at very low frequency in the rad52 strain, and no products were obtained that resulted from recombinational repair. This is consistent with other assays for mitotic recombination, which have shown a strong dependence on RAD52 function, and with our suggestion that RAD52 is an essential component of RAD51-dependent and RAD51-independent recombination pathways (3, 4).
RAD59 is required for RAD51-dependent and RAD51-independent recombination.
RAD59 encodes a Rad52 homologue and was identified by its requirement for RAD51-independent recombination using an ade2 inverted-repeat substrate (4). A synergistic decrease in spontaneous recombination of chromosomal inverted repeats was observed in rad51 rad59 double mutants. In this study, we show a defect in repair of the gapped ARS plasmid in rad59 mutants and a synergistic decrease in rad51 rad59 double mutants. The observation that 95% of repair events require RAD59 suggests that Rad59 plays an important role in the RAD51 pathway, possibly as an accessory factor for Rad52. The synergistic decrease in gap repair efficiency observed in the rad51 rad59 double mutant is consistent with observations obtained with the ade2 inverted repeat and indicates an important role for RAD59 in both spontaneous and DSB-induced recombination. Ninety-nine percent of the repair events involving the gapped ARS plasmid require RAD51, indicating that most repair events are mediated by the RAD51 pathway. However, some repair can occur in the absence of RAD51, and these residual events require RAD52 and RAD59. This observation supports the hypothesis that Rad52 or the Rad52-Rad59 complex promotes strand invasion in vivo, albeit inefficiently. This type of recombinational repair might be initiated by annealing of transiently single-stranded regions of the substrates. Such events are likely to be favored during replication or by transcriptional activation of sequences in close proximity, such as repeats. The idea that proteins without RecA homology can catalyze strand invasion is not without precedent. The bacteriophage lambda β protein is important for lambda recombination and catalyzes strand annealing and strand exchange in vitro (22, 24). Recent studies have shown that β protein and RecT, a homologue of β protein, can promote recA-independent gene replacement in E. coli, suggesting a role in strand invasion in vivo (71, 72). β protein forms ring structures on DNA similar to those formed by the yeast and human Rad52 proteins (41, 56, 67). The observation that Rad52 catalyzes single-strand annealing in vivo and in vitro is consistent with the structural analogy to lambda β protein (34, 59). Thus it seems reasonable to propose that Rad52, or the Rad52/Rad59 heterodimer catalyze an alternate pathway for strand invasion in vivo. However, this appears to be inefficient under most circumstances.
In summary, homology-dependent repair of plasmid DSBs from either chromosomal or plasmid donor sequences occurs predominantly by the RAD51/RAD52 pathway. The low levels of repair that occur in the absence of RAD51 are mechanistically different from repair events generated by the RAD51 pathway and require RAD52 and RAD59.
ACKNOWLEDGMENTS
We thank members of the Symington laboratory, W. K. Holloman, and C. S. H. Young for helpful discussions through the course of this work, and we thank H. Klein and R. Rothstein for yeast strains.
This work was supported by Public Health Service grant NIH GM54099 from the National Institutes of Health.
REFERENCES
- 1.Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr Genet. 1995;27:298–305. doi: 10.1007/BF00352096. [DOI] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Davis A P, Symington L S. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai Y, Symington L S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson F E, Baumann P, West S C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 7.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 8.Bouton A H, Smith M M. Fine-structure analysis of the DNA sequence requirements for autonomous replication of Saccharomyces cerevisiae plasmids. Mol Cell Biol. 1986;6:2354–2363. doi: 10.1128/mcb.6.7.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–6537. doi: 10.1128/mcb.18.11.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cost G J, Boeke J D. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast. 1996;12:939–941. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C939::AID-YEA988%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Fan H Y, Cheng K K, Klein H L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson D O, Rice M C, Rendi M H, Kotani H, Kmiec E B, Holloman W K. Interaction between Ustilago maydis REC2 and RAD51 genes in DNA repair and mitotic recombination. Genetics. 1997;145:243–251. doi: 10.1093/genetics/145.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser V M, Glasunov A V, Tevzadze G G, Perera J R, Shestakov S V. Genetic control of plasmid DNA double-strand gap repair in yeast, Saccharomyces cerevisiae. Curr Genet. 1990;18:1–5. doi: 10.1007/BF00321107. [DOI] [PubMed] [Google Scholar]
- 15.Glasunov A V, Glaser V M. The influence of mutation rad57-1 on the fidelity of DNA double-strand gap repair in Saccharomyces cerevisiae. Curr Genet. 1999;34:430–437. doi: 10.1007/s002940050417. [DOI] [PubMed] [Google Scholar]
- 16.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinnen A, Hicks J B, Fink G R. Transformation of yeast. Proc Natl Acad Sci USA. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov E L, Haber J E. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R, Symington L S. Functional differences and interactions among the yeast RecA homologues Rad51, Rad55 and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerjan P, Cherest H, Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986;14:7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kmiec E, Holloman W K. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 23.Lewis L K, Kirchner J M, Resnick M A. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Karakousis G, Chiu S K, Reddy G, Radding C M. The beta protein of phage lambda promotes strand exchange. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 25.Liefshitz B, Parket A, Maya R, Kupiec M. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovett S T, Mortimer R K. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone R E, Esposito R E. The RAD52 gene is required for homothallic interconversion of mating type and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA. 1980;77:503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald J P, Levine A S, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald J P, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milne G T, Jin S, Shannon K, Weaver D T. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrow D M, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 37.Orr-Weaver T L, Szostak J W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orr-Weaver T L, Szostak J W, Rothstein R J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paques F, Leung W Y, Haber J E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park M S, Ludwig D L, Stigger E, Lee S H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 41.Passy S I, Yu X, Li Z, Radding C M, Egelman E H. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perera J R, Glasunov A V, Glaser V M, Boreiko A V. Repair of double-strand breaks in plasmid DNA in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1988;213:421–424. doi: 10.1007/BF00339611. [DOI] [PubMed] [Google Scholar]
- 43.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 44.Plessis A, Dujon B. Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene. 1993;134:41–50. doi: 10.1016/0378-1119(93)90172-y. [DOI] [PubMed] [Google Scholar]
- 45.Rattray A J, Symington L S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rattray A J, Symington L S. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reaume S E, Tatum E L. Spontaneous and nitrogen mustard-induced nutritional deficiencies in Saccharomyces cerevisiae. Arch Biochem. 1949;22:331–338. [PubMed] [Google Scholar]
- 48.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roman H. Studies of gene mutation in Saccharomyces. Cold Spring Harbor Symp Quant Biol. 1956;21:175–185. doi: 10.1101/sqb.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Roth D B, Porter T N, Wilson J H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiestl R H, Zhu J, Petes T D. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schild D, Calderon I L, Contopoulou C R, Mortimer R K. Cloning of yeast recombination repair genes and evidence that several are nonessential genes. In: Friedberg E C, Bridges B A, editors. Cellular responses to DNA damage. New York, N.Y: Alan R. Liss, Inc.; 1983. pp. 417–427. [Google Scholar]
- 53.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 54.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 55.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 56.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 57.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 59.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: Homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 61.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 62.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 63.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 64.Sung P, Robberson D L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–462. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 65.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 66.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Dyck E, Hajibagheri N M, Stasiak A, West S C. Visualisation of human rad52 protein and its complexes with hRad51 and DNA. J Mol Biol. 1998;284:1027–1038. doi: 10.1006/jmbi.1998.2203. [DOI] [PubMed] [Google Scholar]
- 68.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 69.Wilson T E, Grawunder U, Lieber M R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 70.Winston F, Chumley F, Fink G R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- 71.Yokochi T, Kusano K, Kobayashi I. Evidence for conservative (two-progeny) DNA double-strand break repair. Genetics. 1995;139:5–17. doi: 10.1093/genetics/139.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Buchholz F, Muyrers J P P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 73.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]