Abstract
Sensing DNA damage is crucial for the maintenance of genomic integrity and cell cycle progression. The participation of chromatin in these events is becoming of increasing interest. We show that the presence of single-strand breaks and gaps, formed either directly or during DNA damage processing, can trigger the propagation of nucleosomal arrays. This nucleosome assembly pathway involves the histone chaperone chromatin assembly factor 1 (CAF-1). The largest subunit (p150) of this factor interacts directly with proliferating cell nuclear antigen (PCNA), and critical regions for this interaction on both proteins have been mapped. To isolate proteins specifically recruited during DNA repair, damaged DNA linked to magnetic beads was used. The binding of both PCNA and CAF-1 to this damaged DNA was dependent on the number of DNA lesions and required ATP. Chromatin assembly linked to the repair of single-strand breaks was disrupted by depletion of PCNA from a cell-free system. This defect was rescued by complementation with recombinant PCNA, arguing for role of PCNA in mediating chromatin assembly linked to DNA repair. We discuss the importance of the PCNA–CAF-1 interaction in the context of DNA damage processing and checkpoint control.
Sensing and signaling the presence of DNA damage to the cell cycle checkpoint machinery is crucial for the maintenance of genomic integrity and the regulation of cell cycle progression (12, 25, 61, 97). Checkpoints respond to DNA damage by halting cell cycle progression, providing time for DNA repair. This strategy avoids the replication and segregation of damaged chromosomes which could otherwise lead to genomic instability. DNA damage is caused by physical and chemical agents as well as normal cellular processes including DNA replication and oxidative stress. A variety of distinct DNA repair mechanisms involving lesion-specific DNA damage recognition proteins have been characterized in eukaryotic cells (reviewed in reference 15). The DNA damage checkpoint machinery may recognize structural perturbations in DNA and/or components of the DNA damage processing machinery during specific phases of the cell cycle. Yeast model systems have proven powerful in identifying components of mitotic DNA damage checkpoint pathways (5, 37, 43, 71, 97) which, by analogy with signal transduction pathways, consist of sensor, transducer, and effector molecules. Several checkpoint proteins have been proposed to be directly involved in DNA damage recognition based on their similarity to proteins involved in DNA metabolism, including a structural relative of a 3′-5′ exonuclease (Saccharomyces cerevisiae Rad17 [Rad17sc]) and a replication factor C (RF-C)-like protein (Rad24sc). Protein kinases such as Mec1sc and Rad53sc appear to transduce signals from DNA damage sensors to the cell cycle machinery. Significant progress has been made in delineating the protein-protein interactions and phosphorylation events occurring among some of these factors and their potential interfaces with DNA repair (96). However, the molecular nature of the links between the repair of specific DNA lesions and the DNA damage checkpoint machinery is not yet fully understood.
In addition to interconnections between DNA damage processing and the cell cycle checkpoint machinery, the way in which chromatin organization may influence both aspects is becoming of increasing interest (98). The entire genome is packaged into chromatin (90). This structure allows the compaction of DNA from the basic nucleosome unit (44) up to a higher-order organization providing a potential range of reactivity (11, 99, 100). Mutations affecting all acetylation sites in the N-terminal tail of yeast histone H4 give rise to a delay in the G2 and M phases of the cell cycle as a result of activation of the Rad9sc-dependent DNA damage checkpoint (26, 51), suggesting that DNA integrity or cell cycle progression could be monitored by a marking at the chromatin level. In addition, a mechanistic link has been observed between DNA repair and chromatin assembly. Incubation of DNA damaged by UV irradiation in repair-competent cell-free extracts revealed that de novo nucleosome assembly occurs concomitantly with nucleotide excision repair (NER) (17, 19). A general model has been proposed for NER of DNA lesions within chromatin, in which the unfolding of nucleosomal structures facilitates access of repair enzymes to DNA and is followed by a rapid refolding (reviewed in references 15, 55, and 78). The resetting of a preexisting chromatin structure during NER could relate to the mechanistic link between NER and chromatin assembly. An alternative function of de novo chromatin assembly may be to participate in the sensing of DNA damage.
The chromatin assembly pathway associated with NER is dependent on chromatin assembly factor 1 (CAF-1) (19). This three-subunit complex functions as a histone chaperone, interacting with specific forms of histone H4 and H3 (91). It is required for chromatin assembly during simian virus 40 DNA replication in vitro (35, 79, 80, 91), possibly relating to a general enrichment of this factor at replication foci in S-phase cells (39, 47, 74). Remarkably, CAF-1 can also be recruited to chromatin during the repair of UV photoproducts outside of S phase (49). This is consistent with the preservation of its capability to facilitate nucleosome formation when isolated from G1- or G2-phase nuclei (47). In addition, genetic studies of the budding yeast revealed that although none of the genes corresponding to the individual CAF-1 subunits (CAC1, CAC2, and CAC3) were essential, CAC mutants were moderately sensitive to UV irradiation and exhibited gene silencing defects (13, 14, 20, 36, 57). Together these data argue for a dual role for CAF-1 during both DNA replication and NER.
These data prompted us to explore the link(s) between CAF-1 and DNA damage processing at the biochemical level to identify partners and DNA structures critical for its recruitment. The involvement of CAF-1 in chromatin assembly during both DNA replication and NER suggested that this factor may sense, either directly or indirectly, the presence of a common nucleoprotein intermediate. This could be generated either through dual endonucleolytic cleavages in the damaged DNA strand during NER or at the 3′-hydroxyl termini of DNA replication forks. To test this hypothesis biochemically, we have created DNA substrates containing DNA damage in the form of single-strand breaks and gaps and show that they efficiently trigger the assembly of nucleosomal arrays. This nucleosome assembly pathway is dependent on CAF-1. Using the largest (p150) subunit of the CAF-1 complex as bait in a yeast two-hybrid screen, we have identified a specific interaction with PCNA. We further show that the N terminus of p150 interacts directly with specific sites on the outer front side of PCNA. To analyze the functional significance of this interaction in the context of DNA damage, we have developed an assay for factors recruited during DNA damage processing. Recruitment of PCNA and CAF-1 to damaged DNA is dependent on the number of DNA lesions and requires ATP. Furthermore, depletion of PCNA from a cell-free system disrupts chromatin assembly linked to single-strand break repair, and this defect can be rescued by complementation with recombinant PCNA. Thus PCNA, through CAF-1, can link multiple DNA repair pathways to chromatin assembly. The sliding clamp function of PCNA could account for the bidirectional propagation of nucleosomal arrays away from lesion sites during DNA repair. Our data suggest a possible mechanism for signaling the presence of DNA lesions to the DNA damage checkpoint machinery.
MATERIALS AND METHODS
Synthesis and purification of circular DNA substrates containing site-specific DNA structures.
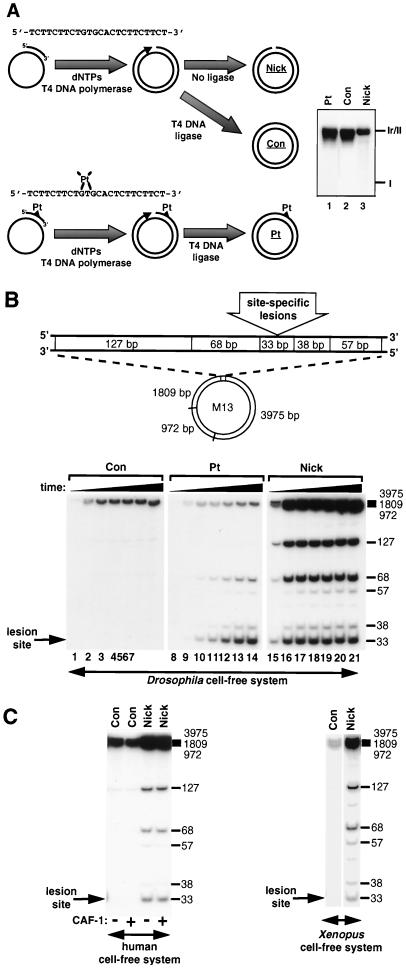
Covalently closed circular DNA molecules containing a single 1,3-intrastrand d(GpTpG)-cisplatin cross-link (Pt DNA) were prepared as described previously (56). Briefly, a 24-mer oligonucleotide containing a 1,3-intrastrand cisplatin cross-link bridging bases 10 and 12 (underlined; 5′-TCTTCTTCTGTGCACTCTTCTTCT-3′) was annealed to single-stranded M13 DNA. The 3′-OH terminus of this oligonucleotide serves as a primer for complementary-strand synthesis in the presence of T4 DNA polymerase, deoxynucleoside triphosphates (dNTPs), T4 DNA ligase, and ATP. Control DNA (Con DNA) molecules were prepared in parallel using a nonmodified oligonucleotide. Circular DNA containing a site-specific single-strand break (Nick DNA) was obtained by omitting T4 DNA ligase during the complementary-strand synthesis reaction with the nonmodified oligonucleotide. Covalently closed circular molecules and circular DNA molecules containing a single-strand break were purified by CsCl-ethidium bromide density gradient centrifugation as described previously (76).
To produce gapped circular DNA substrates, 5 μg of nick DNA was incubated with 12.5 U of T4 DNA polymerase in the absence of dNTPs for 10 min at 37°C. Gapped circular DNA substrates were purified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. Con DNA was processed in parallel as a control.
To prepare randomly nicked circular DNA substrates, pUC19 DNA (10 μg) was incubated in a 650-μl reaction mixture containing 10 mM HEPES-KOH (pH 7.6), 50 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, and 6.5 U of DNase I (Boehringer Mannheim) at 25°C for 1 min. pUC19 was also incubated in the absence of DNase I as a control. Reactions were terminated by adding EDTA to a final concentration of 100 mM before purification of DNA by ethanol precipitation.
Analysis of repair and chromatin assembly in vitro.
Analysis of DNA repair synthesis and chromatin assembly in the Drosophila cell-free system was performed as described previously (17). Briefly, circular DNA substrates containing site-specific DNA lesions were incubated with Drosophila preblastoderm embryo extracts under reaction conditions which suppress the assembly of regularly spaced nucleosomes onto nondamaged DNA (17, 54). For analysis of DNA repair synthesis and chromatin assembly in a human cell-free system, we used a cytosolic extract derived from HeLa cells grown on plates (49). This cytosolic extract contains only trace amounts of the p60 and p150 subunits of CAF-1 and can be complemented by the addition of purified recombinant CAF-1 complex (91) to promote chromatin assembly linked to DNA repair (49). The location and extent of DNA synthesis observed during repair of each site-specific lesion were assessed by digestion of the DNA flanking the lesion site into small restriction fragments. The accumulation of extensively supercoiled DNA molecules (form I) is proportional to the number of nucleosomes formed on a circular DNA molecule and thus represents a simple assay for nucleosome assembly (23). Quantification of supercoiling on the total DNA population was performed with a PhosphorImager (Molecular Dynamics) in conjunction with Imagequant and Microsoft Excel software. The relative efficiency of extensive supercoiling was calculated as the percentage of highly supercoiled (form I) topoisomers compared to the entire population of topoisomers (total). The identity of distinct topological forms of DNA (I, Ir, II, III, and gapped) was confirmed by agarose gel electrophoresis in the presence of either chloroquine or ethidium bromide. Micrococcal nuclease (MNase) digestion analysis was used to assess the formation of regular nucleosomal arrays. The size of MNase digestion products was determined by comparison with a 123-bp ladder (GIBCO-BRL).
Purified proteins.
CAF-1 complex purified from baculovirus-infected insect cells (91) was a generous gift from A. Verreault (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). Recombinant PCNA and mutants were constructed and purified as described elsewhere (32). The correct folding and trimerization of all mutants was confirmed by native gel electrophoresis. This is particularly important for the LAPK251 mutant, which has a tendency to aggregate upon storage at high concentration (46).
GST pull-down assays and depletion of PCNA.
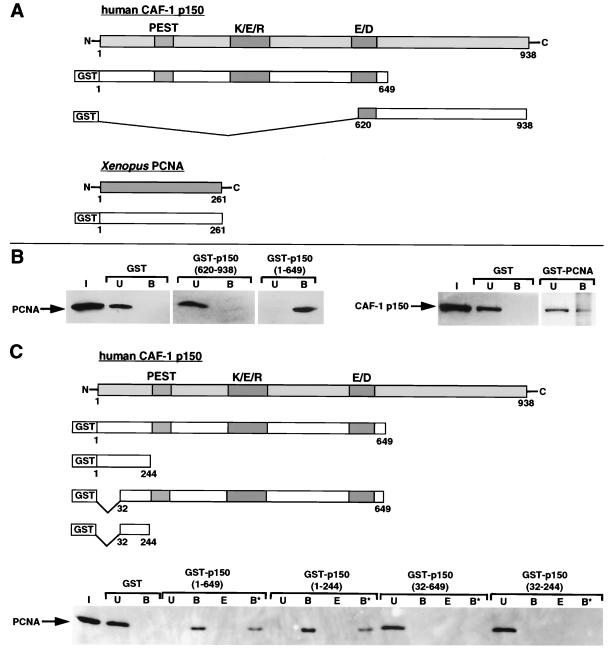
Glutathione S-transferase (GST) fusion proteins were constructed as shown in Fig. 5A and C. Constructs in which GST was fused to portions of human CAF-1 p150 (containing amino acids 1 to 649, 1 to 244, 32 to 244, 32 to 649, and 620 to 938) were derived from plasmid pPK8 (35). The various GST-p150 constructs, GST-Xenopus PCNA (isolated in the two-hybrid screen), and GST were produced as soluble proteins in Escherichia coli BL21 (DE3)pLysS cells (based essentially on the method described in reference 81). Approximately 10 μg of GST fusion proteins bound to 100 μl of glutathione-Sepharose 4B resin (Pharmacia-Amersham Biotech) was used in each GST pull-down experiment. HeLa nuclear extract (30 μl), recombinant wild-type human PCNA protein (500 ng), or recombinant mutant human PCNA proteins (LAPK251, SHV43, QLGI125, and VDK188; 500 ng of each) were mixed with the GST fusion proteins bound to the resin in a final volume of 100 μl with binding buffer (20 mM HEPES-KOH [pH 7.4], 150 mM KCl, 1 mM EDTA, 0.025% NP-40, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 10 μg each of leupeptin and pepstatin per ml). Total proteins bound to the GST chimeras were resuspended in 1× Laemmli buffer. Proteins still bound after extraction with an equal volume of binding buffer containing 600 mM KCl were resuspended in 1× Laemmli buffer.
FIG. 5.
The amino terminus of CAF-1 p150 interacts directly with PCNA. (A) Scheme of GST fusion proteins. (B) Pull-down assay from HeLa nuclear extract using GST-p150(620-938), GST-p150(1-649), and GST-PCNA (Xenopus open reading frame). GST alone was used as a control. Input (I), unbound (U), and bound (B) fractions of proteins were revealed by Western blotting with monoclonal antibodies against PCNA and the p150 subunit of CAF-1. Twice the amount of input fraction was loaded compared to the bound and unbound fractions. (C) Pull-down of recombinant human PCNA by GST-p150(1-649), GST-p150(1-244), GST-p150(32-649), and GST-p150(32-244). GST alone was used as a control. PCNA present in the input (I), unbound (U), bound (B), 0.6 M KCl elution (E), and 0.6 M KCl-resistant (B*) fractions was revealed by Western blotting with a specific monoclonal antibody (PC10). Equal amounts of each fraction were loaded.
For the depletion of PCNA from the Drosophila cell-free system, approximately 15 μg of GST-p150(1-244) or GST bound to 150 μl of glutathione-Sepharose 4B resin was incubated with 300 μl of undiluted Drosophila preblastoderm embryo extract for 1.5 h at 4°C. The unbound supernatant was used directly for reactions without prior freezing. The efficient depletion of endogenous PCNA from the unbound supernatant was confirmed by Western blotting (data not shown).
Assay for protein factors recruited to DNA containing single-strand breaks or UV photoproducts.
Linearized pUC19 biotinylated on one end was coupled to Dynabeads M-280 (Dynal SA) essentially as described previously (72). For the induction of single-strand breaks, aliquots of bead-linked DNA were resuspended in 650-μl reaction mixtures containing 10 mM HEPES-KOH (pH 7.6), 50 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, and 6.5 U of DNase I (Boehringer Mannheim) and incubated at 25°C for 2, 4, and 6 min. One aliquot was incubated in the absence of DNase I as a control. Reactions were terminated by adding EDTA to a final concentration of 100 mM and then washed and stored in 2 M NaCl–1 mM EDTA–10 mM Tris (pH 7.5) at 4°C. For the induction of UV photoproducts, bead-linked DNA (10 μg) was resuspended in buffer A (40 mM HEPES [pH 7.8], 40 mM KCl, 0.05% NP-40) in petri dishes cooled on ice and irradiated with 5 J/cm2 using a germicidal UV-C (254 nm) lamp essentially as described elsewhere (18). Bead-linked DNA was incubated in the absence of UV-C light as a control. For incubation in a human cell-free system derived from HeLa cells (49), 600 ng of each bead-linked DNA substrate was washed once in buffer A containing 1 mg of bovine serum albumin per ml and once in buffer A alone. Bead-linked DNA was then resuspended in 50-μl reaction mixtures containing 200 μg of proteins from the cytosolic extract, 20 μg of proteins from the nuclear extract, 5 mM MgCl2, 40 mM HEPES-KOH (pH 7.8), 0.5 mM dithiothreitol, 4 mM ATP, 20 μM each dGTP, dATP, and dTTP, 8 μM dCTP, 40 mM phosphocreatine, and 4 μg of creatine phosphokinase (type 1; Sigma). Reaction mixtures were incubated at 37°C on a rotating wheel (Dynal SA) to keep bead-linked DNA in suspension. In experiments where ATPγS was used in place of ATP, we omitted dNTPs, phosphocreatine, and creatine phosphokinase from the reaction mixture. Reactions were terminated by concentration of bead-linked DNA on a magnet and four successive washes with buffer A. Bound proteins were eluted by boiling for 5 min in 1× Laemmli buffer.
Western blot analysis of proteins.
Protein analysis by sodium dodecyl sulfate-polyacrylamide gels and Western blotting was as described by Martini et al. (49). Monoclonal antibody 48, against the largest subunit of human CAF-1, p150 (79), was used at a dilution of 1:2,000. Rabbit polyclonal antibody 1, directed against the p60 subunit of CAF-1 (47), was used at a dilution of 1:400 to reveal both phosphorylated and nonphosphorylated forms. A monoclonal antibody against PCNA (PC10; Dako) was used at a dilution of 1:100. A monoclonal antibody against the largest subunit (p140) of human RF-C, a gift from B. Stillman's laboratory (Cold Spring Harbor Laboratory) was used at a dilution of 1:300. The primary antibodies were detected using horseradish peroxidase (Jackson ImmunoResearch Laboratories)- or alkaline phosphatase (Promega)-conjugated secondary antibodies in conjunction with a Supersignal substrate detection kit (Pierce) or a 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium-driven color reaction (Promega), respectively.
RESULTS
Repair of site-specific single-strand DNA breaks.
Nick DNA molecules were constructed (Fig. 1A) by adapting a method previously used to obtain the Pt substrate (56). A nonmodified Con substrate was obtained in parallel. Omitting T4 DNA ligase from the complementary-strand synthesis reaction results in the formation circular DNA molecules containing a single-strand break. This reaction is favored by the fact that T4 DNA polymerase does not exhibit significant strand displacement or exonuclease activities in the presence of high concentrations of dNTPs. Restriction fragment end-labeling analysis of the damaged DNA strand in purified nick DNA revealed the presence of a site-specific single-strand break in the majority of DNA molecules with no missing bases (data not shown). The migration of the three DNA substrates on an agarose gel illustrates their different topologies. The Pt and Con DNA substrates consist of a distribution of topoisomers concentrated around the most relaxed molecules (Fig. 1A, lanes 1 and 2) as expected for closed circular molecules obtained after ligation. In contrast, a single band was observed for the nick DNA (Fig. 1A, lane 3).
FIG. 1.
Extensive DNA synthesis associated with repair of a site-specific single-strand DNA break. (A) Synthetic 24-mer oligonucleotides (nonmodified or containing a 1,3-intrastrand cisplatin-cross-link) were annealed to single-stranded M13 DNA and elongated by T4 DNA polymerase in the presence of dNTPs. Con and Pt DNA substrates were covalently closed with T4 DNA ligase. The omission of T4 DNA ligase resulted in the formation of circular DNA molecules containing a site-specific single-strand DNA break (Nick). The synthesis of Pt and Con DNA substrates in vitro results in a range of topoisomers distributed around the most relaxed molecules (lanes 1 and 2). This was confirmed by agarose gel electrophoresis in the presence of either chloroquine or ethidium bromide (data not shown). In contrast, nick DNA consists of form II and runs as a single band (lane 3). (B) Con, Pt, and nick DNA substrates were incubated in a Drosophila cell-free system, and site-specific DNA synthesis was localized by restriction enzyme digestion of each DNA substrate. A schematic representation of the restriction fragments flanking the lesion site is shown at the top, and the sizes (in base pairs) of labeled restriction fragments are indicated alongside each gel. Incubation times were 1 min (lanes 1, 8, and 15), 5 min (lanes 2, 9, and 16), 15 min (lanes 3, 10, and 17), 45 min (lanes 4, 11, and 18), 90 min (lanes 5, 12, and 19), 180 min (lanes 6, 13, and 20), and 360 min (lanes 7, 14, and 21). (C) Con and nick DNA substrates were incubated for 6 h with either cytosolic extract derived from human cells (with [+] or without [−] recombinant CAF-1 complex) or Xenopus egg extract at 37 or 23°C, respectively.
To analyze how these DNA substrates were processed in a Drosophila cell-free system, we determined the location and extent of DNA synthesis during repair of each site-specific lesion. After incubation in the extract in the presence of radiolabeled dNTP precursors, DNA flanking the lesion site was digested into small restriction fragments (Fig. 1B). Repair of the nick led to DNA synthesis in the 33-, 68-, and 127-bp restriction fragments (lanes 15 to 21). This indicates that little 5′-3′ nick translation is initiated at the single-strand break and implies that 3′-5′ degradation of the damaged DNA strand from the site of the single-strand break precedes DNA resynthesis. Some background DNA synthesis was visible in the high-molecular-weight restriction fragments of Con DNA, but no signal was detectable in the five smallest restriction fragments (Fig. 1B, lanes 1 to 7), arguing for the specificity of the signal for single nick molecules. No further increase in DNA repair synthesis was observed after 15 min (Fig. 1B, compare lanes 17 to 21). At this time point, >95% of single-strand breaks had been rejoined (data not shown). These data demonstrate that single-strand breaks are efficiently processed through a defined mechanism which involves extensive DNA synthesis. Remarkably, this defined processing of single-strand breaks was also observed in human and Xenopus cell-free systems (Fig. 1C). This mechanism is distinct from NER of a 1,3-intrastrand cisplatin cross-link (Fig. 1B, lanes 10 to 14) which resulted in a short (∼30-nucleotide patch of DNA synthesis spanning the 33- and 68-bp restriction fragments, consistent with previous reports (17, 19, 56). Thus, we have defined a conserved mechanism for the processing and repair of single-strand breaks in three different cell-free systems.
Single-strand DNA breaks trigger efficient nucleosome assembly in the absence of extensive DNA synthesis.
To examine whether nucleosome formation occurred during the defined DNA damage processing reactions described above, we further used the Drosophila cell-free system under conditions which suppress chromatin assembly on nonrepaired DNA molecules (17, 54). Since the accumulation of supercoiled DNA molecules is proportional to the number of nucleosomes formed on circular DNA molecules (23), nucleosome formation was initially followed by a standard supercoiling assay. Repair of nick DNA in the Drosophila cell-free system was accompanied by the preferential accumulation of supercoiled DNA molecules relative to undamaged Con DNA (Fig. 2A). This is consistent with the suppression of chromatin assembly on nonrepaired DNA molecules under our experimental conditions. Transient gapped circular intermediates detected at early reaction times (Fig. 2A, lane 11, labelled DNA) most likely represent intermediates generated by 3′-5′ degradation of the damaged DNA strand away from the site of a single-strand break prior to the DNA synthesis (Fig. 1B). Comparison of the relative efficiency of supercoiling associated with the repair of the different site-specific DNA lesions (assigned as the percentage of highly supercoiled molecules [I] compared to the entire population [total]) revealed that single-strand breaks were reproducibly twofold more efficient in triggering chromatin assembly than 1,3-intrastrand cisplatin cross-links (Fig. 2B, compare lanes 1 and 3, total DNA, and graph). The low levels of supercoiling on control DNA (Fig. 2B, lane 2, total DNA) presumably reflect nucleosome assembly initiated by single-strand breaks arising during the preparation of the Con DNA substrate or during incubation in the cell-free system. Importantly, the nucleosome assembly machinery seems to exhibit a specificity for single-strand breaks arising during DNA damage processing since the transient nick formed during the rapid relaxation of supercoiled undamaged control DNA by topoisomerase I activities in the Drosophila cell-free system does not trigger chromatin assembly (17).
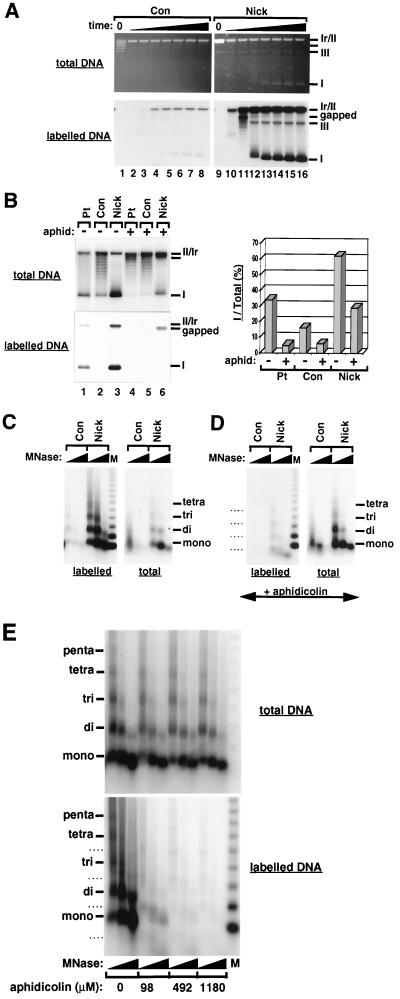
FIG. 2.
Single-strand breaks trigger chromatin assembly in the absence of extensive DNA synthesis. (A) Supercoiling analysis of chromatin assembly on DNA substrates containing site-specific single-strand breaks after incubation in a Drosophila cell-free system. Incubation times were 1 min (lanes 2 and 10), 5 min (lanes 3 and 11), 15 min (lanes 4 and 12), 45 min (lanes 5 and 13), 90 min (lanes 6 and 14), 180 min (lanes 7 and 15), and 360 min (lanes 8 and 16). Lanes 1 and 9 contain untreated Con and nick DNA, respectively. The migration of relaxed/nicked circular DNA (Ir/II), linear DNA (III), supercoiled DNA (I), and labeled gapped circular molecules is indicated. (B) Supercoiling analysis of chromatin assembly using DNA substrates containing site-specific DNA lesions after incubation in a Drosophila cell-free system for 6 h in the presence or absence of aphidicolin (590 μM). The graph shows the quantification of the number of extensively supercoiled topoisomers (I) relative to the total topoisomer population as a percentage. The migration of relaxed/nicked circular DNA (Ir/II), linear DNA (III), supercoiled DNA (I), and labeled gapped circular molecules is indicated. (C) MNase digestion analysis of nucleosomal arrays formed on nick DNA after 6 h incubation in a Drosophila cell-free system. Two digestions with increasing amounts of MNase are shown for each reaction. The regularly spaced DNA bands corresponding to mono-, di-, tri-, and tetranucleosomal DNA are indicated. A 123-bp ladder was used as a molecular weight marker (M). (D) Effect of aphidicolin (590 μM) on nucleosomal arrays formed using nick DNA in a 6-h reaction with the Drosophila cell-free system (MNase digestion analysis was performed under conditions identical to those used for panel C). (E) Formation of nucleosomal arrays using nick DNA in a 6-h reaction with the Drosophila cell-free system in the presence of increasing amounts of aphidicolin. The distinct pattern of MNase digestion products seen in the presence of aphidicolin on labeled Nick DNA molecules in panels D and E is indicated by dashed lines. This pattern is thought to result from the assembly of nucleosomal arrays adjacent to gaps in the damaged DNA strand.
We next assessed whether extensive DNA synthesis was important for triggering nucleosome assembly during single-strand break processing. In the presence of aphidicolin, most DNA synthesis (>85%) associated with the processing of single-strand breaks was inhibited (Fig. 2B, compare lanes 3 and 6, labelled DNA). Under these conditions, the repair of single-strand breaks, occurring either by direct ligation or via a very limited DNA synthesis reaction, gave rise to extensive supercoiling on a significant number of molecules (29% of total [Fig. 2B, lane 6, total DNA and graph]). However, gapped molecules generated in these reactions, if assembled into chromatin, would not be revealed in a supercoiling assay. We thus performed a partial MNase digestion under similar conditions. These data confirmed that the supercoiling observed when our nick DNA was used as input in the reaction (Fig. 2A and B) was due to the deposition of regularly spaced nucleosomes (Fig. 2C, total DNA). Most importantly, the formation of regular nucleosomal arrays in the presence of aphidicolin was not significantly affected (compare Fig. 2C and D, total DNA) even at very high aphidicolin concentrations (Fig. 2E), except for a distinctly shifted MNase digestion pattern on labeled DNA (Fig. 2D and E, labelled DNA). This shift is likely to result from the assembly of nucleosomal arrays adjacent to gaps in the damaged DNA strand (data not shown). Thus, our data support the hypothesis that even incomplete repair events including gapped DNA intermediates can trigger nucleosome assembly. These observations are consistent with our findings in assays using NER substrates in which chromatin assembly could occur without completion of DNA repair synthesis during NER (17).
We conclude that (i) a unique single-strand DNA break in a circular molecule can directly trigger efficient nucleosome assembly and (ii) extensive DNA synthesis is dispensable for this event to occur.
Gapped DNA triggers efficient nucleosome assembly.
To directly test whether single-strand gaps are able to trigger nucleosome assembly, we constructed gapped circular DNA substrates by treatment of nick DNA with T4 DNA polymerase in the absence of dNTPs as depicted in Fig. 3. This resulted in a heterogeneous population of gaps 5′ to the original single-strand break. We then tested whether these substrates could trigger nucleosome assembly in the Drosophila cell-free system. Repair of gapped DNA molecules was accompanied by the formation of regular nucleosomal arrays as detected by MNase digestion (Fig. 3, − aphidicolin). In the presence of aphidicolin, these gaps were not repaired (data not shown). However, the MNase assay established that regularly spaced nucleosomal arrays could be formed adjacent to these gaps (Fig. 3, + aphidicolin). Thus, both single-strand breaks and gaps are able to trigger nucleosome assembly in the absence of extensive DNA synthesis and ligation. Although extensive DNA synthesis and ligation are dispensable for the initiation of chromatin assembly, our data do not exclude that CAF-1 (and additional components of the nucleosome assembly machinery) may be recruited during the later stages of DNA damage processing, as shown during postreplicative CAF-1-dependent nucleosome assembly (34) via the PCNA marking of newly replicated DNA (74). We conclude that 3′-hydroxyl and/or 5′-phosphoryl termini within duplex DNA, either directly or in conjunction with protein factors, are the critical intermediates responsible for triggering nucleosome assembly during DNA damage processing.
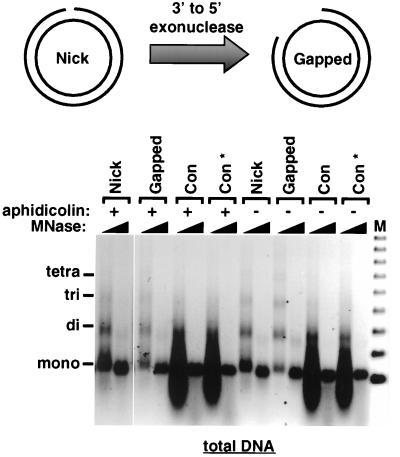
FIG. 3.
Single-strand gaps trigger chromatin assembly in the absence of extensive DNA synthesis or ligation. Gapped circular DNA substrates were prepared from nick DNA by exploiting the 3′-to-5′ exonuclease activity of T4 DNA polymerase in the absence of dNTPs. Con DNA was treated in parallel as a control (designated Con*). Gapped circular, nick, and Con DNA substrates were incubated in the Drosophila cell-free system for 6 h in the presence or absence of aphidicolin and then subjected to MNase digestion analysis. Two different MNase digests are shown for each reaction. The oligonucleosomal DNA bands corresponding to mono-, di-, tri-, and tetranucleosomal DNA are indicated. A 123-bp ladder was used as a molecular weight marker (M).
CAF-1 promotes nucleosome assembly during the processing of single-strand breaks.
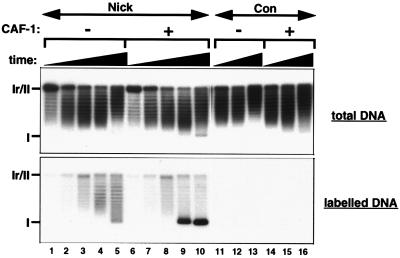
We then tested whether the histone chaperone CAF-1 participates in nucleosome assembly during single-strand break repair. Cytosolic extract derived from HeLa cells, containing only trace amounts of CAF-1, which can be complemented by the addition of purified recombinant CAF-1 complex was used as previously described (49). Control DNA was poorly assembled into chromatin either in the presence or absence of CAF-1 (Fig. 4, lanes 11 to 16). Repaired single-nick DNA was only partially supercoiled in the absence of CAF-1 (Fig. 4, lanes 1 to 5). This limited chromatin assembly is likely to be due to trace amounts of endogenous CAF-1 in the extract. The addition of CAF-1 complex to the cytosolic extract favored the preferential supercoiling of repaired nick DNA (Fig. 4, compare labeled form I DNA in lanes 5 and 10). We note that this labeled fraction represented only 4% of the total topoisomers (Fig. 4, lane 10, total DNA) due to the low efficiency of single-strand break processing in the human cell-free system. The extent of supercoiling detected revealed that CAF-1 can promote extensive nucleosome assembly triggered by a unique single-strand break. This is in agreement with the data obtained for the Drosophila cell-free system (see reference 17 and above). We could thus demonstrate for the first time a role for CAF-1 in promoting nucleosome loading events from a single nick in the human in vitro system.
FIG. 4.
CAF-1 stimulates nucleosome assembly during single-strand break repair. For kinetic analysis of supercoiling during single-strand break repair in a human cell-free system, nick and Con DNA substrates were incubated with HeLa cytosolic extract which contains only trace amounts of endogenous CAF-1 either in the absence or in the presence of exogenous purified CAF-1 complex (1 ng/μl). Incubation times were 1 min (lanes 1 and 6), 5 min (lanes 2 and 7), 15 min (lanes 3, 8, 11, and 14), 45 min (lanes 4, 9, 12, and 15), and 180 min (lanes 5, 10, 13, and 16). The migration of relaxed/nicked circular DNA (Ir/II) and supercoiled DNA (I) molecules is indicated.
The first 31 amino acids of CAF-1 p150 are necessary for strong binding to PCNA.
We searched for CAF-1-interacting proteins which might mediate the recruitment of CAF-1 to single-strand breaks. To facilitate the detection of transient and low-affinity CAF-1 interactions in vivo, we used a yeast two-hybrid screen with human CAF-1 p150 as bait and a Xenopus oocyte (stage I to VI) cDNA library as prey. Among the 44 positive clones obtained (P. Grandi and G. Almouzni, unpublished results), 4 contained the complete open reading frame of Xenopus PCNA (GenBank accession no. P18248). These data are consistent with the observation that CAF-1 and PCNA can be coimmunoprecipitated in vitro (74). Other interactors for PCNA include a variety of factors involved in replication, repair, recombination, cell cycle regulation, and methylation (9, 33, 93). Full-length Xenopus PCNA and two portions of human CAF-1 p150 including the N- and C-terminal regions were individually fused to GST (Fig. 5A). The fusion of full-length Xenopus PCNA to GST was able to bind to human CAF-1 p150 in the crude cell extract (Fig. 5B), consistent with the interaction observed in our two-hybrid screen. GST-p150(1-649) was able to bind to, and quantitatively deplete, human PCNA from the crude nuclear extract (Fig. 5B, compare input and bound fractions). In contrast, no interaction was detected between GST-p150(620-938) and human PCNA in the crude nuclear extract (Fig. 5B). To show that the interaction between CAF-1 p150 (amino acids 1 to 649) and PCNA was direct, we then performed a GST pull-down assay using purified recombinant human PCNA. GST fusions with defined regions within amino acids 1 to 649 of CAF-1 p150 were tested to delineate the domain important for PCNA binding (schematic in Fig. 5C). Our data showed that both GST-p150(1-649) and GST-p150(1-244) could interact directly with human PCNA (Fig. 5C, compare lanes I [input] and B [bound]). Furthermore, this interaction was stable at 0.6 M KCl (Fig. 5C, compare lanes B, E, and B*). In contrast, neither GST-p150(32-244) nor GST-p150(32-649) was able to interact with human PCNA (Fig. 5C). These data revealed that a short amino acid sequence at the N terminus of CAF-1 p150 was responsible for strong binding to PCNA. Thus, CAF-1 interacts directly with PCNA, even between different species, and the first 31 amino acids at the N terminus of CAF-1 p150 are necessary for strong binding to PCNA.
CAF-1 p150 interacts with the outer front side of PCNA.
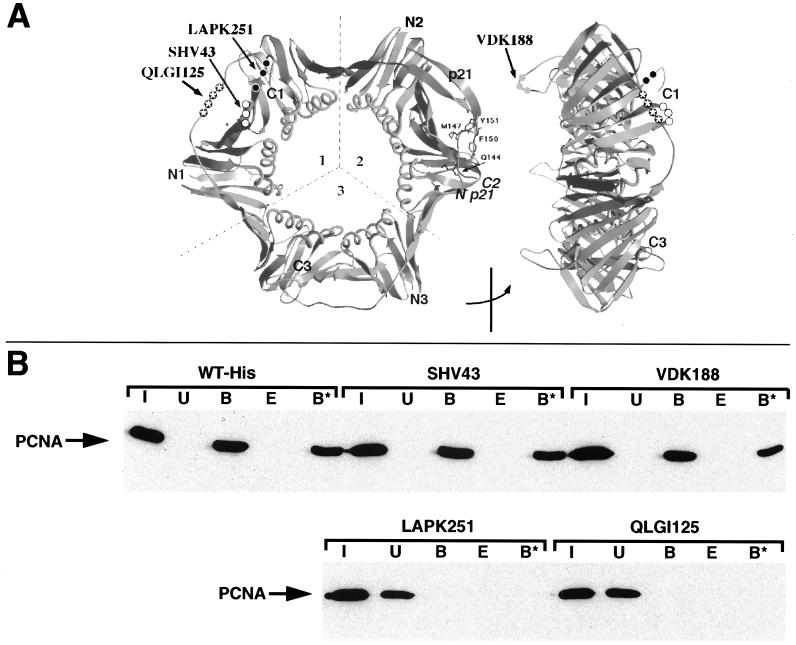
Specific amino acid residues of PCNA are involved in interactions with a variety of proteins including DNA polymerases δ and ɛ, RF-C, FEN-1, and p21 (9, 33, 93). The effect of mutations in human PCNA on the interaction with CAF-1 p150 (amino acids 1 to 649) was tested in the GST pull-down assay using four recently designed histidine-tagged PCNA mutants (created via alanine substitutions), each of which is still able to form a trimeric toroidal ring structure (Fig. 6A) (32, 46). We first verified that GST-p150(1-649) interacted equally well with histidine-tagged and untagged wild-type PCNA (compare Fig. 6B and 5C). The interaction observed between GST-p150(1-649) and the PCNA mutants SHV43 and VDK188 (Fig. 6B) was comparably resistant to 0.6 M KCl extraction. In contrast, two PCNA mutations, QLGI125 and LAPK251, completely abolished interaction of PCNA with CAF-1 p150 (Fig. 6B). Three of the PCNA mutants (SHV43, QLGI125, and LAPK251) were made in loops which form a hydrophobic pocket on the outer front side of PCNA. Only two of these (QLGI125 and LAPK251) were critical for the interaction with CAF-1 p150. The neutral effect of SHV43 on the interaction of CAF-1 p150 with PCNA leaves open the possibility for this site to be occupied by RF-C. The QLGI125 mutation has been shown to be part of the binding site for DNA polymerase δ, p21, and FEN-1 (10, 24, 32, 103). Importantly, another critical CAF-1 p150 binding site (LAPK251) on the surface of PCNA lies within the most highly conserved region in PCNA (62). This mutation renders PCNA defective for the stimulation of DNA polymerase ɛ (46). Together these data demonstrate that CAF-1 p150 directly interacts with specific sites on the PCNA domain connecting loop, on the side which faces forward in relation to DNA synthesis (32).
FIG. 6.
CAF-1 interacts with specific sites on the outer front side of PCNA. (A) Front and side views of the homotrimeric PCNA toroid showing positions of the mutations used in the GST pull-down assay. (B) GST pull-down of wild-type and mutant recombinant human PCNAs by GST-p150(1-649). Equivalent amounts of C-terminally histidine-tagged wild-type (WT-His) and mutant (LAPK251, QLGI125, SHV43, and VDK188; created via alanine substitutions) PCNA proteins were used in each experiment. Input (I), unbound (U), bound (B), 0.6 M KCl elution (E), and 0.6 M KCl-resistant (B*) fractions of PCNA proteins were revealed by Western blotting with a monoclonal antibody against PCNA.
Single-strand DNA breaks lead to the recruitment of PCNA and CAF-1.
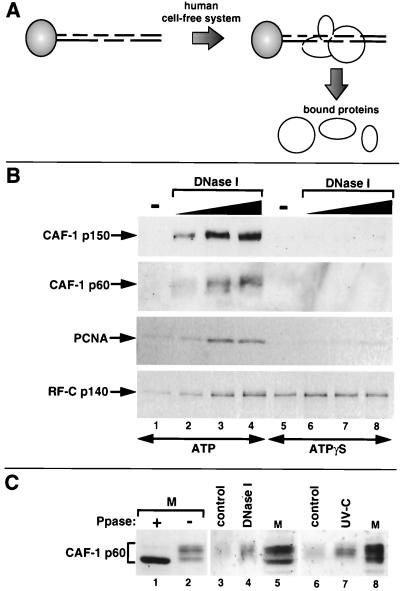
To test the functional significance of the interaction between PCNA and CAF-1 p150 in the context of damaged DNA, we developed a novel assay for factors which are recruited during single-strand break repair (Fig. 7A). Linearized plasmid DNA was linked to paramagnetic beads, and single-strand breaks were introduced using DNase I. Control experiments confirmed that single-strand breaks induced by DNase I in plasmid DNA efficiently triggered chromatin assembly (data not shown; see also Fig. 8A). Bead-linked DNA substrates containing either no damage or different amounts of single-strand breaks were incubated in a human cell-free system, and bound proteins were eluted and analyzed by Western blotting. In the absence of single-strand breaks, the binding of CAF-1 p150, CAF-1 p60, and PCNA was barely detectable after 5 min of incubation (Fig. 7B, lane 1). In contrast, the binding of all three factors was stimulated by the presence of DNase I-induced single-strand breaks and was proportional to the number of DNA lesions induced (Fig. 7B, lanes 2 to 4). Furthermore, this binding was significantly reduced by the substitution of ATP with ATPγS (Fig. 7B, compare lanes 6 to 8 with lanes 2 to 4). This was further confirmed in an experiment in which the ATP was completely removed by dialysis of the extract (data not shown). These observations suggest that hydrolysis of the γ-phosphate bond of ATP may contribute to the stable loading of PCNA onto DNA during single-strand break processing. Neither the omission of exogenous dNTPs not the addition of aphidicolin affected the amount of PCNA and CAF-1 binding (data not shown), consistent with single-strand breaks rather than extensive DNA synthesis triggering chromatin assembly. Purified CAF-1 and PCNA by themselves or in combination displayed a very low affinity for both undamaged and damaged linear DNA under these reaction conditions (data not shown). This implies that additional factors, possibly including an ATP-utilizing activity, are necessary for the recruitment of these two proteins to nicked DNA. The RF-C complex can load PCNA onto DNA in an ATP-dependent manner (7, 86). The binding of the largest subunit of the RF-C complex (p140) was observed on bead-linked linear DNA (Fig. 7B, lane 1) and was stimulated by the presence of single-strand breaks (Fig. 7B, lanes 2 to 4). This is consistent with the specific affinity of this RF-C subunit for both 3′-hydroxyl and 5′-phosphoryl termini within duplex DNA (2, 88). We note that the substitution of ATP by ATPγS increased the amount of RF-C p140 bound to DNA (compare lanes 1 and 5). This is in agreement with previous observations of the stabilization of RF-C on linear DNA molecules by ATPγS (66, 92). Based on these properties, the RF-C complex is a good candidate for PCNA loading in these experiments although we cannot formally exclude that other factors may be involved. Interestingly, we noted that the p60 subunit of CAF-1 bound to DNase I-treated bead-linked DNA consisted largely of a single population. Since CAF-1 p60 is known to exist in various phosphorylated forms (47, 49), we analyzed the migration of CAF-1 p60 bound to either DNase I-treated or UV-C-irradiated bead-linked DNA in comparison to HeLa cell extract which had been treated with λ phosphatase (Fig. 7C). These data demonstrate that the slower-migrating forms of CAF-1 p60 bound to damaged DNA consist mainly of phosphorylated forms. This is reminiscent of the preferential recruitment of phosphorylated forms of CAF-1 p60 which was previously observed during repair of UV photoproducts in vivo (49), thus further validating our in vitro assay. Our present data using a novel assay for the specific recruitment of proteins to defined DNA lesions provide a molecular basis for the recruitment of CAF-1 through an interaction with PCNA during DNA repair.
FIG. 7.
PCNA and CAF-1 are corecruited to DNA containing single-strand breaks. (A) Schematic of an assay for factors which are recruited to magnetic bead-linked DNA containing single-strand breaks induced by DNase I. (B) Bead-linked DNA substrates containing different amounts of DNase I-induced single-strand breaks were incubated in a human cell-free system in the presence of either ATP (4 mM) or ATPγS (4 mM). Specific proteins bound to DNA were detected by Western blotting with antibodies against CAF-1 p150, CAF-1 p60, PCNA, and RF-C p140. (C) Preferential binding of slower-migrating forms of CAF-1 p60 to bead-linked DNA containing either single-strand breaks (lane 4) or UV photoproducts (lane 7). HeLa nuclear extract (lanes M) was loaded alongside the proteins bound to bead-linked DNA to reveal the multiple phosphorylated forms of CAF-1 p60 (indicated by a bracket) present in the cell extract (47, 49). The HeLa nuclear extract shown in lane 1 was preincubated with λ phosphatase (Ppase) to reveal the migration of dephosphorylated CAF-1 p60.
FIG. 8.
PCNA mediates chromatin assembly linked to single-strand break repair. (A) Analysis of supercoiling during single-strand break repair in the Drosophila cell-free system. Supercoiled circular plasmid DNA (lane 1) was treated with DNase I to induce single-strand breaks (lane 2). These DNA substrates were incubated for 3 h at 23°C with either nondepleted extract (lanes 3 and 4), extract depleted with GST alone (lanes 5 and 6), or extract depleted with GST-p150(1-244) (lanes 7 to 10). Recombinant human PCNA was added to the reactions in lanes 9 and 10 to a final concentration of 8 ng/μl. The migration of relaxed/nicked circular DNA (Ir/II) and supercoiled DNA (I) molecules is indicated. (B) Bead-linked DNA substrates containing DNase I-induced single-strand breaks or undamaged control molecules were incubated in the Drosophila cell-free system as described for panel A for 1 min at 23°C. Bead-linked DNA substrates were equilibrated and washed in a slightly higher ionic strength buffer (40 mM HEPES-KOH [pH 7.8], 100 mM KCl, 0.05% NP-40) for reactions with the Drosophila cell-free system. PCNA binding to the bead-linked DNA substrates was detected by Western blotting. The recombinant human PCNA containing a C-terminal histidine tag migrates more slowly than endogenous Drosophila PCNA.
PCNA mediates chromatin assembly linked to single-strand break repair.
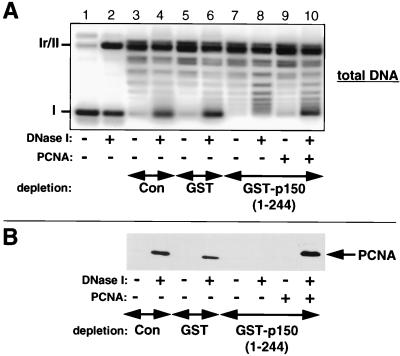
To directly test the importance of the PCNA interaction with CAF-1 for chromatin assembly linked to single-strand break repair, we exploited the strong and specific interaction between the N-terminal part of CAF-1 p150(1-244) and PCNA as a means to deplete PCNA from the Drosophila cell-free system. The depleted extract was then used in a supercoiling assay linked to single-strand break repair (Fig. 8A). Preferential supercoiling was observed when circular DNA substrates containing DNase I-induced single-strand breaks were incubated with nondepleted (lane 4) or mock-depleted (lane 6) extract. In contrast, the depletion of PCNA with GST-p150(1-244) led to a loss of extensive supercoiling in this assay (lane 8). The addition of recombinant human PCNA rescued this defect in supercoiling (lane 10). The supercoiling defect associated with PCNA depletion is paralleled by the loss of PCNA loading onto bead-linked DNA containing DNase I-induced single-strand breaks (Fig. 8B). Similar results were obtained by depleting PCNA from the human cell-free system (data not shown). These data argue for a role of PCNA in mediating chromatin assembly linked to single-strand break repair.
DISCUSSION
Single-strand breaks and gaps in the genome are generated following exposure to DNA-damaging agents and during the replication and recombination of DNA, particularly if dNTP pools are perturbed. These structures are thought to be critical in signaling to DNA damage checkpoint pathways (30, 45, 59). The detection and processing of DNA lesions, together with the checkpoint signaling elicited, operate within the nucleus in a chromatin environment. It is thus critical to perceive how chromatin dynamics may influence these events.
In this report we show that the processing of single-strand breaks and gaps can trigger chromatin assembly. The reaction is stimulated by a chromatin assembly factor, CAF-1. The largest subunit (p150) of this histone chaperone can interact directly with PCNA, and critical regions for this interaction on both proteins have been defined. We discuss how the recruitment of an assembly factor (CAF-1) to nicks through an interaction with PCNA (or PCNA-like molecules) may be important in sensing or signaling DNA structural perturbations in the genome.
Molecular interactions involved in triggering chromatin assembly during DNA damage processing.
Our observation of a high efficiency of nucleosome assembly triggered by single-strand breaks or gaps even in the absence of extensive DNA synthesis or ligation demonstrates that 3′-hydroxyl and/or 5′-phosphoryl termini within duplex DNA are critical DNA structural intermediates responsible for triggering nucleosome assembly. We further show that the sensing of single-strand breaks during DNA repair by the chromatin assembly machinery is mediated by PCNA. The yeast two-hybrid system allowed us to demonstrate an interaction between PCNA and the largest subunit (p150) of CAF-1. Our biochemical observation of a direct interaction between specific sites on the outer front side of PCNA and the N terminus of CAF-1 p150 is consistent with their functioning in a common complex. Interestingly, the N-terminal part of CAF-1 p150 interacts with the same region of the PCNA surface as a number of other proteins, including DNA polymerase δ, FEN-1 nuclease, cyclin-dependent kinase inhibitor p21, and DNA ligase I (9, 33, 93), raising the possibility of competition between the binding of these different factors. Since the first 296 amino acids of CAF-1 p150 are not required for nucleosome assembly coupled to simian virus 40 DNA replication in vitro (35), a reaction which is mediated by the interaction of CAF-1 with PCNA (74), our data raise an intriguing paradox. One possible explanation could be that CAF-1 p150 contains multiple PCNA binding sites which may be important for chromatin assembly coupled to distinct DNA transactions. The functional significance of the N-terminal PCNA binding site in CAF-1 p150 during DNA repair is currently under investigation. Several PCNA binding proteins have been reported to possess a conserved PCNA binding motif (32, 93, 94, 102). We have identified a sequence (QARLPF) which closely resembles this motif within the first 31 amino acids of the human CAF-1 p150 N terminus. This motif is conserved in the N termini of mouse CAF-1 p150 (GenBank accession no. CAB55497) and Xenopus CAF-1 p150 (Grandi and Almouzni, unpublished results), suggesting that the interaction of PCNA with the N terminus of CAF-1 p150 may be conserved among higher eukaryotes. Although the homologues of the largest subunit of CAF-1 in Schizosaccharomyces pombe and S. cerevisiae do not contain the conserved QARLPF motif, they both possess amino acid sequences which conform to the reported PCNA binding consensus sequence (32, 93, 94, 102). Analysis of mutations affecting the PCNA binding capacity of CAF-1 p150 in these different organisms should help to elucidate the physiological roles of CAF-1.
We observed that the recruitment onto DNA containing single-strand breaks of both PCNA and CAF-1 was dependent on the presence of ATP, possibly implying additional factors in the loading process. Among the known PCNA-interacting proteins, RF-C is able to load PCNA onto DNA containing single-strand breaks (66), and the largest RF-C subunit (p140) possesses specific DNA binding domains for both 3′-hydroxyl and 5′-phosphoryl termini within duplex DNA (2, 88). The RF-C complex is thought to facilitate the opening of the PCNA ring structure in the presence of ATP, while closure of the PCNA ring structure around a DNA molecule and dissociation of RF-C is stimulated by ATP hydrolysis. Alternative mechanisms for the recruitment of PCNA could involve the direct interaction of PCNA with structure-specific endonucleases involved in excision repair, such as FEN-1 (8, 41) and XPG (21). Both of them interact directly with the outer front side of PCNA. PCNA staining in repair-defective fibroblasts provides evidence that a single NER incision (3′) is sufficient for PCNA binding in vivo (1, 53), and thus PCNA may associate with the XPG endonuclease prior to loading onto the primer terminus for PCNA-dependent DNA repair synthesis. Analysis of the recruitment of these factors during the processing of various DNA lesions in assays similar to those presented in this paper should be useful to sort out these issues.
Since PCNA is involved in a wide range of DNA transactions including replication (4, 6, 7, 40, 67–69, 82, 86, 95), excision repair (16, 31, 50, 60, 75, 89), recombinational repair of double-strand breaks (28, 29), and sister chromatid cohesion (77), our observations raise the issue of how CAF-1 may participate in these various processes. PCNA is stably associated with newly replicated DNA in vitro and is required for CAF-1-dependent chromatin assembly consistent with the colocalization of PCNA and CAF-1 at replication foci in vivo (74). An important function for CAF-1-mediated chromatin assembly pathway during DNA damage processing in vivo is supported by our observations that both the p150 and p60 subunits of CAF-1 are recruited to chromatin after UV irradiation of human cells (49). This recruitment, in parallel with PCNA, occurred outside of S phase, thus distinguishing it from the role of CAF-1 during DNA replication. These in vivo data can be explained in light of the recruitment of both CAF-1 and PCNA to DNA containing either single-strand breaks or UV photoproducts (this study). The main question then is whether the dual role of CAF-1 during DNA replication and repair serves a similar purpose. The viability of CAC deletion mutants in S. cerevisiae together with their UV-sensitive phenotype (13, 14, 20, 36, 57) raises important questions concerning the function of CAF-1. Although redundant chromatin assembly pathways might exist, CAF-1 may have a more specialized role during replication and repair perhaps as part of DNA damage surveillance/checkpoint mechanism. In light of the recent observations that DNA damage in the form of double-strand breaks in S. cerevisiae can lead to the redistribution of Sir (silent information regulator) and Ku proteins (48, 52), it will be of interest to examine whether CAF-1 can be recruited to sites of double-strand breaks.
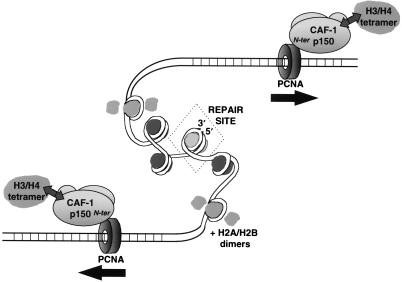
Importantly, PCNA is a homotrimer and forms a toroidal ring structure in solution (38, 73) which is able to slide along the DNA helix (65, 66, 84). These properties might account for the bidirectional propagation of nucleosomal arrays over relatively large distances away from various repair sites including lesions subject to NER (17) and single-strand breaks (Fig. 2 and 4 and data not shown). A simple scheme in Fig. 9 integrates our findings for the CAF-1–PCNA interaction in the context of a link between chromatin assembly and DNA damage. A possible implication of this process would be the establishment of a repressive chromatin structure. This may relate to the chromatin-mediated repression of basal transcription in vivo (3). In support of this hypothesis, a PCNA mutation in Drosophila, mus209, results in the suppression of position-effect variegation (i.e., heterochromatin-mediated transcriptional silencing) (27, 101), and genetic studies of cac mutants in S. cerevisiae have revealed defects in the silencing of telomeric and mating-type loci (13, 14, 36, 57). The repression of undesirable DNA transactions may allow time for the DNA damage checkpoint machinery to elicit an appropriate DNA damage response. Since we found that DNA repair intermediates prior to the completion of repair are able to trigger chromatin assembly, it is tempting to speculate that the potentially large scale changes in chromatin structure associated with the propagation of nucleosomal arrays might even be an integral part of the DNA damage sensing and signaling process (e.g., by transmitting and perhaps amplifying the presence of DNA damage to larger or distinct domains of the genome).
FIG. 9.
Model for bidirectional propagation of nucleosomal arrays facilitated by the interaction of CAF-1 with PCNA sliding clamps during DNA damage processing. The initiation of this nucleosome assembly process is dependent on the presence of single-strand breaks and gaps produced either by direct DNA damage or during excision repair. The recruitment to repair sites of PCNA and the histone chaperone CAF-1 (together with H3/H4) requires ATP. The amino terminus of CAF-1 p150 is engaged in a stable interaction with specific sites on the outer front side of PCNA. The ability of the PCNA toroidal ring to slide along the DNA helix provides a possible mechanism for nucleosomal arrays to propagate bidirectionally from sites of repair (indicated by the arrows). The hatched nucleosome represents an undefined structure at the repair site.
PCNA sliding clamps: communicators between DNA damage processing and CAF-1-dependent chromatin assembly.
The direct interaction between PCNA and CAF-1 p150 may provide a means to coordinate a variety of signals produced during the DNA damage response through chromatin changes. Links between PCNA and the products of several checkpoint genes including p21 have been identified (85). The observation that CAF-1 p150 interacts with the outer front side of PCNA, a site known to interact with several other proteins, raises the possibility that CAF-1 might compete for access to this site during the DNA damage response. However, this issue is complicated by the availability of three equivalent binding sites per PCNA homotrimer.
Our observation that PCNA links distinct DNA damage processing reactions to a CAF-1-dependent chromatin assembly pathway suggests that the initial sensing of single-strand breaks and gapped intermediates may be performed by the clamp loader protein RF-C. This protein, along with a number of clamp loader-like proteins, has been implicated in several cell cycle checkpoint pathways (12, 58, 70, 87). Remarkably, two distinct S. pombe DNA damage checkpoint proteins, Rad1sp (83) and Rad9sp (22), as well as their homologues in higher eukaryotes contain PCNA-like motifs. Comparative modeling of Rad1sp and PCNA homologues revealed that the Rad1 family of cell cycle checkpoint proteins could form a sliding clamp structure similar to PCNA (83). Rad1 of humans exhibits 3′-5′ exonuclease activity (63) and interacts with the clamp loader-like factor Rad17 (64). Genetic studies in both budding and fission yeast have led to the proposal that the Rad1sp/Rad17sc 3′-5′ exonuclease may play a central role in sensing DNA damage and generating a cell cycle-inhibitory signal (37, 45). We are currently examining whether the 3′-5′ processing of single-strand breaks that we observed in this study reflects an analogous reaction in higher eukaryotes. Based on our observation of a specific interaction between the N terminus of CAF-1 and the outer front side of PCNA, we speculate that the p150 subunit of CAF-1 might be recruited by sliding clamp-like checkpoint proteins via interaction with a region resembling the outer front side of PCNA. Sequence alignment of the PCNA protein with the human homologue of Rad9sp (42) revealed that a critical region for the interaction with CAF-1 p150, i.e., the LAPK251 region of PCNA (Fig. 6A), is highly conserved (data not shown). This would provide a connection between distinct DNA damage checkpoint proteins and a common CAF-1-dependent chromatin assembly. Furthermore, it would support our prediction that the propagation of chromatin structures during DNA damage processing (Fig. 9) is highly conserved and important for signaling to the cell cycle machinery. In summary, the functional consequences of linking a common chromatin assembly pathway to distinct DNA damage processing events may include: (i) formation of a structure which is sensed by the cell cycle checkpoint machinery; (ii) repression of, or interference with, DNA transactions; and (iii) resetting of a preexisting chromatin structure. Consequently, the sliding clamp structure of PCNA and PCNA-like molecules appears to have been selected during evolution as a versatile communicator molecule in a variety of DNA damage processing reactions providing links between the DNA repair, chromatin assembly, and cell cycle checkpoint machinery.
ACKNOWLEDGMENTS
We are most grateful to B. Stillman (Cold Spring Harbor Laboratory) and his laboratory for their generous collaboration in providing antibodies as well as recombinant human CAF-1. We gratefully acknowledge F. Bunz (Cold Spring Harbor Laboratory) for making and characterizing the RF-C p140 monoclonal antibody and J. Moreau (Institut Jacques Monod) for providing the Xenopus laevis cDNA library used in the two-hybrid screen. We thank D. M. J. Roche for sharing her expertise and unpublished data on the analysis of chromatin assembly in the human cell-free system. We thank all members of our laboratory for help and advice and E. Moustacchi and E. Bailly for critical reading of the manuscript.
J.G.M. was supported first by an EMBO long-term fellowship and then by a European Union Training, Mobility and Research (TMR) fellowship. P.G. was supported by an EMBO long-term fellowship. Z.O.J. and U.H. were supported by the Swiss National Science Foundation (grant 31-43138.35/2) and by the Canton of Zürich. This work was supported by the Association pour la Recherche sur le Cancer, La Ligue Nationale contre le Cancer, Fondation de la Recherche Medicale, and a TMR Network grant from the European Union (G.A.).
REFERENCES
- 1.Aboussekhra A, Wood R D. Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp Cell Res. 1995;221:326–332. doi: 10.1006/excr.1995.1382. [DOI] [PubMed] [Google Scholar]
- 2.Allen B L, Uhlmann F, Gaur L K, Mulder B A, Posey K L, Jones L B, Hardin S H. DNA recognition properties of the N-terminal DNA binding domain within the large subunit of replication factor C. Nucleic Acids Res. 1998;26:3877–3882. doi: 10.1093/nar/26.17.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almouzni G, Wolffe A P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 4.Bauer G A, Burgers P M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley N J, Carr A M. DNA structure-dependent checkpoints in model systems. Biol Chem. 1997;378:1267–1274. [PubMed] [Google Scholar]
- 6.Bravo R, Frank R, Blundell P A, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-δ. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 7.Burgers P M J. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ɛ. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 8.Chen J, Chen S, Saha P, Dutta A. p21cip1/Waf1 disrupts the recruitment of human FEN-1 by proliferating cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci USA. 1996;93:11597–11602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox L S. Who binds wins: competition for PCNA rings out cell-cycle changes. Trends Cell Biol. 1997;7:493–498. doi: 10.1016/S0962-8924(97)01170-7. [DOI] [PubMed] [Google Scholar]
- 10.Eissenberg J C, Ayyagari R, Gomes X V, Burgers P M. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol. 1997;17:6367–6368. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elgin S C R. Chromatin structure and gene expression. Oxford, England: IRL Press; 1995. [Google Scholar]
- 12.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enomoto S, McCune-Zierath P, Geraminejad M, Sanders M, Berman J. Rlf2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–363. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. pp. 291–294. [Google Scholar]
- 16.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox L S, Lane D P, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 17.Gaillard P-H, Moggs J G, Roche D M J, Quivy J-P, Becker P B, Wood R D, Almouzni G. Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J. 1997;16:6281–6289. doi: 10.1093/emboj/16.20.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard P-H, Roche D R, Almouzni G. Nucleotide excision repair coupled to chromatin assembly. Methods Mol Biol. 1998;119:231–243. doi: 10.1385/1-59259-681-9:231. [DOI] [PubMed] [Google Scholar]
- 19.Gaillard P H, Martini E M, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 20.Game J C, Kaufman P D. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gary R, Ludwig D L, Cornelius H L, MacInnes M A, Park M S. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 22.Gaspari T, Carr A M. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- 23.Germond J E, Rouvière-Yaniv J, Yaniv M, Brutlag D. Nicking-closing enzyme assembles nucleosome-like structures in vitro. Proc Natl Acad Sci USA. 1979;76:3779–3783. doi: 10.1073/pnas.76.8.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulbis J M, Kelman Z, Hurwitz J, Odonnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 25.Hartwell L, Kastan M. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 26.Hartzog G A, Winston F. Nucleosomes and transcription: recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- 27.Henderson D S, Banga S S, Grigliatti T A, Boyd J B. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 1994;13:1450–9. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson H S, Glover D M. Chromosome fragmentation resulting from an inability to repair transposase-induced DNA double-strand breaks in PCNA mutants of Drosophila. Mutagenesis. 1998;13:57–60. doi: 10.1093/mutage/13.1.57. [DOI] [PubMed] [Google Scholar]
- 29.Holmes A M, Haber J E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 30.Huang L-C, Clarkin K C, Wahl G M. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson R E, Kovvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 32.Jónsson Z O, Hindges R, Hübscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jónsson Z O, Hübscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 34.Kamakaka R T, Bulger M, Kaufman P D, Stillman B, Kadonaga J T. Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor I. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I—a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 37.Kitazono A, Matsumoto T. “Isogaba Maware”: quality control of genome DNA by checkpoints. Bioessays. 1998;20:391–399. doi: 10.1002/(SICI)1521-1878(199805)20:5<391::AID-BIES6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 38.Krishna T S R, Kong X-P, Gary S, Burgers P M, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 39.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 40.Lee S-H, Kwong A D, Pan Z-Q, Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase δ. J Biol Chem. 1991;266:594–602. [PubMed] [Google Scholar]
- 41.Li X, Li J, Harrington J, Lieber M R, Burgers P M J. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 42.Lieberman H B, Hopkins K M, Nass M, Demetrick D, Davey S. A human homologue of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci USA. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 45.Lydall D, Weinert T. Yeast checkpoint genes in DNA-damage processing—implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 46.Maga G, Jónsson Z O, Stucki M, Spadari S, Hübscher U. Dual mode of interaction of DNA polymerase ɛ with proliferating cell nuclear antigen in primer binding and DNA synthesis. J Mol Biol. 1999;285:259–267. doi: 10.1006/jmbi.1998.2314. [DOI] [PubMed] [Google Scholar]
- 47.Marheineke K, Krude T. Nucleosome assembly and intracellular localisation of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 48.Martin S G, Laroche T, Suka N, Grunstein M, Gasser S M. Relocalisation of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 49.Martini E, Roche D M J, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin following UV irradiation of human cells. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumoto Y, Kim K, Bogenhagen D F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol Cell Biol. 1994;14:6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Megee P C, Morgan B A, Smith M M. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 52.Mills K D, Sinclair D A, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 53.Miura M, Nakamura S, Sasaki T, Takasaki Y, Shiomi T, Yamaizumi M. Roles of XPG and XPF/ERCC1 endonucleases in UV-induced immunostaining of PCNA in fibroblasts. Exp Cell Res. 1996;226:126–132. doi: 10.1006/excr.1996.0210. [DOI] [PubMed] [Google Scholar]
- 54.Moggs J G, Almouzni G. Assays for chromatin remodelling during DNA repair. Methods Enzymol. 1999;304:333–351. doi: 10.1016/s0076-6879(99)04020-3. [DOI] [PubMed] [Google Scholar]
- 55.Moggs J G, Almouzni G. Chromatin rearrangements during nucleotide excision repair. Biochimie. 1999;81:45–52. doi: 10.1016/s0300-9084(99)80037-6. [DOI] [PubMed] [Google Scholar]
- 56.Moggs J G, Yarema K J, Essigmann J M, Wood R D. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 57.Monson E K, de Bruin D, Zakian V A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mossi R, Hübscher U. Clamping down on clamps and clamp loaders, the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- 59.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nichols A F, Sancar A. Purification of PCNA as a nucleotide excision repair protein. Nucleic Acids Res. 1992;20:3559–3564. doi: 10.1093/nar/20.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurse P. Checkpoint pathways come of age. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 62.O'Donnell M, Onrust R, Dean F B, Chen M, Hurwitz J. Homology in accessory proteins of replicative polymerases—E. coli to humans. Nucleic Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker A E, Van de Weyer I, Laus M C, Oostveen I, Yon J, Verhasselt P, Luyten W H M L. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 64.Parker A E, Van de Weyer I, Laus M C, Verhasselt P, Luyten W H M L. Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J Biol Chem. 1998;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- 65.Podust L M, Podust V N, Floth C, Hubscher U. Assembly of DNA polymerase delta and epsilon holoenzymes depends on the geometry of the DNA template. Nucleic Acids Res. 1994;22:2970–2975. doi: 10.1093/nar/22.15.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Podust L M, Podust V N, Sogo J M, Hubscher U. Mammalian DNA polymerase auxiliary proteins: analysis of replication factor C-catalyzed proliferating cell nuclear antigen loading onto circular double-stranded DNA. Mol Cell Biol. 1995;15:3072–3081. doi: 10.1128/mcb.15.6.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Podust V N, Hübscher U. Lagging strand DNA synthesis by calf thymus DNA polymerases α, β, δ and ɛ in the presence of auxiliary proteins. Nucleic Acids Res. 1993;21:841–846. doi: 10.1093/nar/21.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prelich G, Kostura M, Marshak D R, Mathews M B, Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature. 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 69.Prelich G, Tan C-K, Kostura M, Mathews M B, So A G, Downey K M, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-δ auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 70.Reynolds N, Fantes P A, MacNeill S A. A key role for replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 1999;27:462–469. doi: 10.1093/nar/27.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sandaltzopoulos R, Blank T, Becker P B. Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm Drosophila chromatin. EMBO J. 1994;13:373–379. doi: 10.1002/j.1460-2075.1994.tb06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schurtenberger P, Egelhaaf S U, Hindges R, Maga G, Jónsson Z O, May R P, Glatter O, Hübscher U. The solution structure of functionally active human proliferating cell nuclear antigen determined by small-angle neutron scattering. J Mol Biol. 1998;275:123–132. doi: 10.1006/jmbi.1997.1435. [DOI] [PubMed] [Google Scholar]
- 74.Shibahara K-I, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 75.Shivji M K K, Kenny M K, Wood R D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 76.Shivji M K K, Moggs J G, Kuraoka I, Wood R D. Dual incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol Biol. 1998;113:313–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- 77.Skibbens R V, Corson L B, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smerdon M J, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 79.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 80.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 81.Studier F W, Rosemberg A H, Dunn J J, Dubendorff J. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 82.Tan C-K, Castillo C, So A G, Downey K M. An auxiliary protein for DNA polymerase-δ from fetal calf thymus. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 83.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 84.Tinker R L, Kassavetis G A, Geiduschek E P. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J. 1994;13:5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tournier S, Leroy D, Goubin F, Ducommun B, Hyams J S. Heterologous expression of the human cyclin-dependent kinase inhibitor p21Cip1 in the fission yeast, Schizosaccharomyces pombe reveals a role for PCNA in the chk1+ cell cycle checkpoint pathway. Mol Biol Cell. 1996;7:651–662. doi: 10.1091/mbc.7.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 87.Turner J, Hingorani M M, Kelman Z, O'Donnell M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 89.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 90.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 91.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 92.Waga S, Stillman B. Cyclin-dependent kinase inhibitor p21 modulates the DNA primer-template recognition complex. Mol Cell Biol. 1998;18:4177–4187. doi: 10.1128/mcb.18.7.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 94.Warbrick E, Lane D P, Glover D M, Cox L S. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: potential mechanism to coordinate DNA replication and DNA repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 95.Waseem N H, Labib K, Nurse P, Lane D P. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992;11:5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 97.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Gen Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 98.Weinert T, Lundblad V. Forever hopeful relations: chromatin, telomeres and checkpoints. Nat Genet. 1999;21:151–152. doi: 10.1038/5930. [DOI] [PubMed] [Google Scholar]
- 99.Wolffe A. Chromatin: structure and function. 2nd ed. London, England: Academic Press; 1995. [Google Scholar]
- 100.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 101.Yamamoto Y, Girard F, Bello B, Affolter M, Gehring W J. The cramped gene of Drosophila is a member of the Polycomb-group, and interacts with mus209, the gene encoding proliferating cell nuclear antigen. Development. 1997;124:3385–3394. doi: 10.1242/dev.124.17.3385. [DOI] [PubMed] [Google Scholar]
- 102.Zhang P, Mo J-Y, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee M Y W T. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J Biol Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 103.Zhang P, Sun Y, Hsu H, Zhang L, Zhang Y, Lee M Y W T. The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. J Biol Chem. 1998;273:713–719. doi: 10.1074/jbc.273.2.713. [DOI] [PubMed] [Google Scholar]