Abstract
Genetic control over a cytoskeletal network inside lipid vesicles offers a potential route to controlled shape changes and DNA segregation in synthetic cell biology. Bacterial microtubules (bMTs) are protein filaments found in bacteria of the genus Prosthecobacter. They are formed by the tubulins BtubA and BtubB, which polymerize in the presence of GTP. Here, we show that the tubulins BtubA/B can be functionally expressed from DNA templates in a reconstituted transcription-translation system, thus providing a cytosol-like environment to study their biochemical and biophysical properties. We found that bMTs spontaneously interact with lipid membranes and display treadmilling. When compartmentalized inside liposomes, de novo synthesized BtubA/B tubulins self-organize into cytoskeletal structures of different morphologies. Moreover, bMTs can exert a pushing force on the membrane and deform liposomes, a phenomenon that can be reversed by a light-activated disassembly of the filaments. Our work establishes bMTs as a new building block in synthetic biology. In the context of creating a synthetic cell, bMTs could help shape the lipid compartment, establish polarity or directional transport, and assist the division machinery.
Keywords: synthetic cell, artificial cell, cell-free gene expression, microtubule, lipid vesicle, morphogenesis
Introduction
The encapsulation of filament-forming proteins inside synthetic liposome compartments provides a route to morphogenesis that is reminiscent of shape control in living cells. Purified eukaryotic microtubules formed by the polymerization of α- and β-tubulin proteins cause liposome membranes to deform in various morphologies, including ellipsoid and lemon shapes, and two long protrusions extending from the vesicle body.1−5 Membrane shape transformation was also realized with encapsulated actin under various conditions.6−9 Prokaryotic cytoskeletal filaments made of MreB or FtsZ proteins have been reconstituted in liposomes, including when the production of the subunits was directed by a cell-free protein synthesis (CFPS).10−13 The coupling of morphogenesis with genetic information and protein expression is particularly relevant in the context of building synthetic cells,13−15 but it has not been achieved with microtubules yet.
Bacterial tubulin homologues have recently been discovered in the Prosthecobacter species.16,17 Called bacterial tubulin A and B (BtubA/B), these proteins interact to form microtubule-like structures in the presence of guanosine triphosphate (GTP).18 Bacterial microtubules (bMTs) consist of five19 or four protofilaments,20 as reported in in vivo and in vitro studies, respectively. Bacterial microtubules are thus noticeably thinner than eukaryotic microtubules, which consist of 13 protofilaments. Recent in vitro studies have shown that bMTs exhibit a dynamic instability (stochastic switching between growth and shrinkage) and a treadmilling (apparent directional movement caused by the net addition of new subunits on one end and a net removal on the other).20,21 These properties are also common to eukaryotic microtubules.
While eukaryotic microtubules are involved in essential processes, such as intracellular transport and chromosome segregation, the function of bMTs remains unclear. Prosthecobacter itself belongs to the phylum Verrucomicrobia and consists of Gram-negative bacteria, which exhibit a high degree of compartmentalization. Prosthecobacter dejongeii, for example, possess a major membrane-bounded region, containing the fibrillar nucleoid and all the ribosome-like particles as well as an intracytoplasmic membrane.22 Another distinguishing feature of Prosthecobacter is the presence of narrowed extensions of the cell wall, called prosthecae.19 Bacterial microtubules seem to be predominately located in these cell stalks, which suggests that they might be involved in their formation. It has also been proposed that BtubA/B filaments may contribute to intracellular organization.23 The only currently known protein that interacts with bMTs is BtubC (also known as bacterial kinesin light chain, Bklc), which stabilizes bMTs20 and links them to lipid membranes in vitro,23 suggesting it could play a role in the anchoring of BtubA/B filaments to membrane protrusions in vivo.
Unlike eukaryotic tubulin, BtubA/B is not dependent on protein chaperones and post-translational modifications, and it can be functionally expressed in Escherichia coli.19 Because the cytoplasm of Prosthecobacter probably resembles that of E. coli,18 CFPS platforms derived from E. coli represent a physiologically relevant environment to investigate bMTs. Not only does CFPS allow one to bypass protein purification but it also enables the continuous interrogation of the protein dynamic behavior in the course of its production.15
To better apprehend the properties of bMTs in a cytosol-like environment, while taking advantage of the versatility of cell-free assays, we reconstituted in this study BtubA/B in a CFPS system. To further mimic the membrane-rich environment in Prosthecobacter, we characterized cell-free expressed BtubA/B on supported phospholipid bilayers and within vesicle compartments. Our results demonstrate that active BtubA/B proteins can be produced from their genes in vitro. Moreover, de novo synthesized bMTs can assemble on supported membranes and inside lipid vesicles (even without BtubC), where they form cytoskeletal structures that can deform the vesicle membrane. As CFPS inside liposomes has become an attractive platform to build a synthetic cell,13,24−30 we believe that bMTs could be exploited for the spatial organization, polarization, and shape transformation of artificial cell models.
Results and Discussion
We chose the PURE system,31 a reconstituted E. coli-based translation machinery (specifically the commercially available PUREfrex2.0), as our CFPS platform. This choice was motivated by the very low levels of protease and nuclease activity, and the wide range of (membrane) proteins synthesized in an active state with this system.13,15,26−33 DNA templates for PUREfrex2.0 reactions consisted of the btubA and btubB genes from Prosthecobacter dejongeii. Both DNA constructs were sequence-optimized for (i) expression in an E. coli host by matching codon occurrence with tRNA usage, (ii) low guanine−cytosine (GC) content within the first 30 base pairs (synonymous mutations were introduced to keep the amino acid sequence unaltered), and (iii) a low propensity of intramolecular base pairing of the mRNA around (in the vicinity or involving) the start codon (Figure S1). To satisfy the latter condition, the change in Gibbs free energy (ΔG) for a few RNA molecules was calculated. Lower ΔG values represent higher melting temperatures of the RNA molecule, which is known to be potentially inhibitory of translation in the PURE system.34 Thus, we selected for each construct the DNA sequence whose corresponding mRNA has the predicted lowest melting temperature, assuming this would decrease the occurrence of inhibitory secondary structures involving the ribosome binding site and the start codon.
Proteins BtubA/B (Figure 1A) were first separately expressed from linear DNA templates in PUREfrex2.0, supplemented with FluoroTect GreenLys reagent to fluorescently label the gene products through cotranslational incorporation of lysine residues conjugated to a fluorophore (Figure 1B). An analysis of samples by denaturing polyacrylamide gel electrophoresis (PAGE) confirmed the in vitro production of each Btub protein (Figure 1C, Figure S2). The concentration of expressed BtubA/B was estimated by using a standard curve with purified proteins, where values of ∼13 μM (BtubA) and ∼17 μM (BtubB) were obtained (Figure S3). A subsequent coexpression from an equimolar amount of the btubA and btubB genes yielded ∼3 μM of BtubA and ∼5 μM of BtubB (Figure S3). It is unclear why the total concentration of synthesized proteins is lower in coexpression assays compared to separate expression of single genes.
Figure 1.
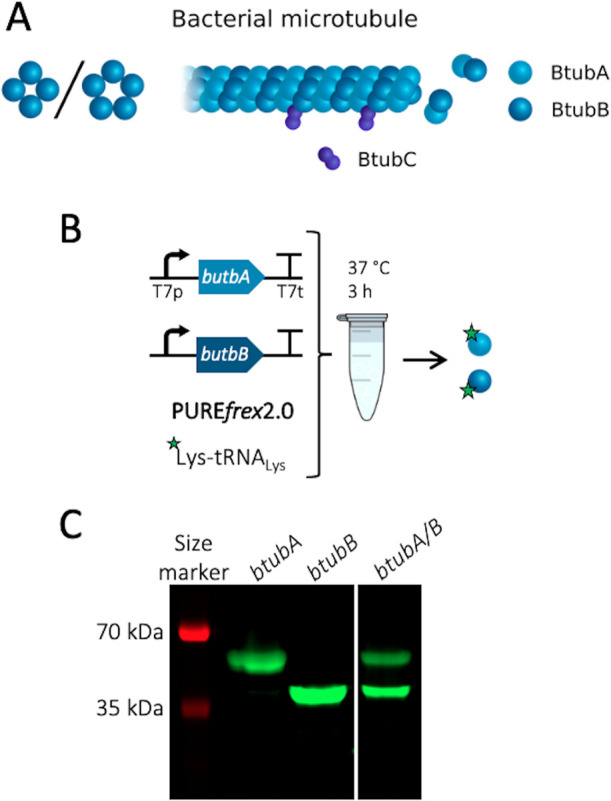
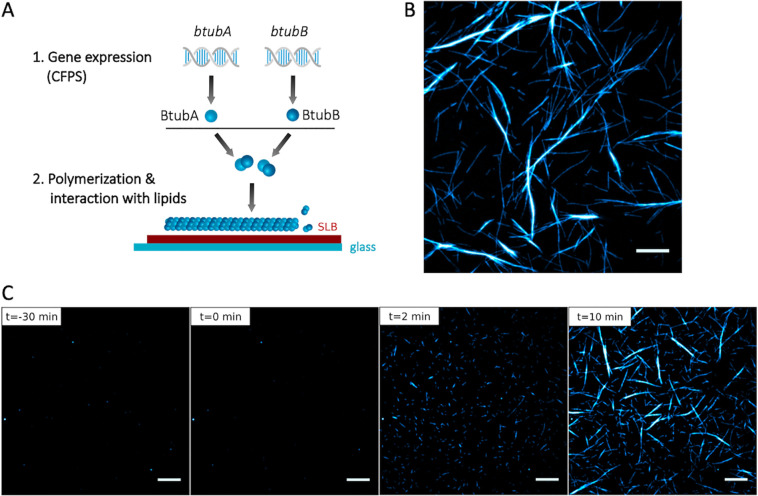
Cell-free expression of BtubA/B. (A) Schematic of a bacterial microtubule consisting of the BtubA/B subunits arranged in four to five protofilaments. The protein BtubC binds to the outside of the filament, predominantly interacting with BtubB. (B) Schematic of CFPS reaction. PUREfrex2.0 was supplemented with the GreenLys reagent to fluorescently label the synthesized proteins through boron dipyrromethene (BODIPY)-conjugated lysine residues. (C) SDS-PAGE analysis of the gene expression products. The gene names of the expressed DNA templates are indicated on the lanes. The uncropped gel is shown in Figure S2.
Next, we examined the activity of purified and cell-free synthesized bMTs on supported lipid bilayers (SLBs). The membrane composition in SLB assays consisted of ∼50 mol % of phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) lipids, which are also found in the membrane of Prosthecobacter.35 Purified BtubA/B proteins recombinantly expressed in E. coli cells were investigated in a PUREfrex2.0 background to closely emulate the molecular and ionic complexity of the bacterial cytosol. Bacterial microtubules were successfully assembled on top of an SLB from 2.5 μM purified BtubA/B doped with Atto488-labeled subunits for visualization. The bMTs localized exclusively on the SLB and not on the bare glass areas (Figure 2A). Filaments of BtubA/B appeared to stably interact with the membrane without the need for anchoring proteins such as BtubC. The protein BtubC synthesized in PUREfrex2.0 was able to bind to lipid membranes and to promote the recruitment of BtubA/B (Figure S4 and Supporting Information Note 1). Moreover, bMTs formed bundles of multiple protofilaments over time (Figure 2A and Movie 1), suggesting lateral interactions, as recently supported by cryo-electron microscopy data.20 The critical BtubA/B concentration for the assembly of bMTs was reported to be 2.5–5 μM at the potassium concentration present in the PURE system.6 Here, bMTs were observed on SLBs already at concentrations of ∼1 μM of purified tubulin in PUREfrex2.0 (Figure S5). This result suggests that, in the arguably more physiological conditions used here (in particular, the higher molecular crowding), the critical concentration for BtubA/B polymerization is lower. Similarly, the use of the PURE system as a reconstitution medium shifted the activity range of prokaryotic filaments in SLB assays.13 Differences due to the types of activity assays (high-speed pelleting of BtubA/B and filaments grown from seeds in ref (21)) may also explain this change.
Figure 2.
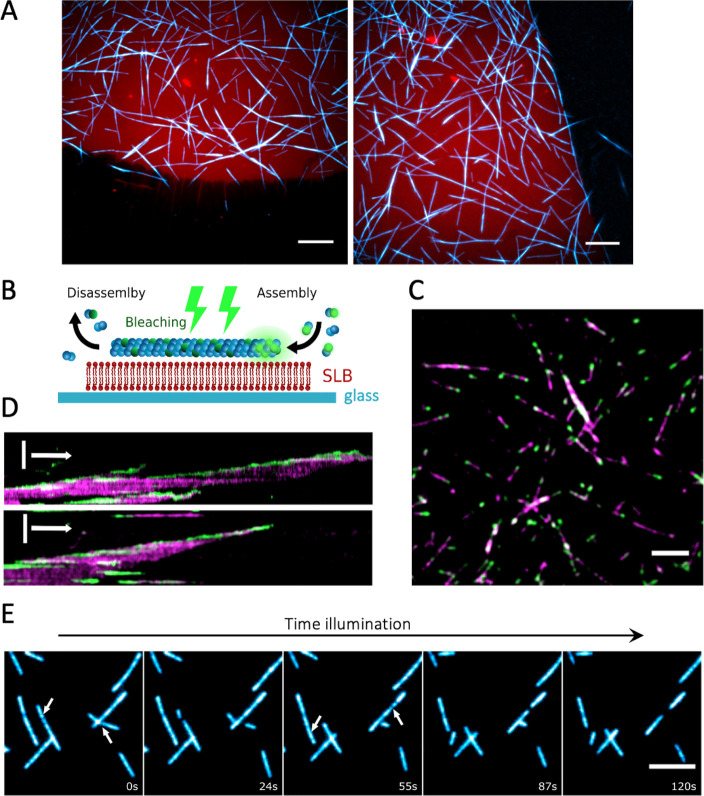
Dynamics of bMTs formed by purified proteins on SLBs. (A) Fluorescence microscopy images showing the selective binding of bMTs (cyan, Atto488-BtubA/B) onto an SLB (red, DHPE-TexasRed). Concentration of bacterial tubulins was 2.5 μM. Scale bars: 10 μm. (B) Schematic of the dual color labeling assay to study bMT dynamics on an SLB. Purified BtubA/B proteins labeled with either Atto488 or Atto565 were used. Atto488-labeled bacterial tubulins were bleached continuously during imaging. The only active Atto488 fluorophores are located at the plus end of the filament, where new subunits incorporate. (C) Bleached filaments display fluorescent ends that originate from the continuous addition of fresh, unbleached Atto488-BtubA/B (green). Atto565-BtubA/B is colored in magenta. Scale bar: 5 μm. (D) Kymographs show bMT dynamics during a photobleaching. Data from two different bMTs are shown. Color coding is the same as in (C). Addition of subunits at the growing end can be seen as the continuous upward drift of the green signal over time. Vertical scale bar: 5 μm. Horizontal arrow: 1 min. (E) Time series images show bMT disassembly events during a high-intensity illumination. Arrows indicate the locations at which the filaments break apart. Illumination time points are appended on each image. Scale bar: 5 μm.
Single filaments appear to undergo a directional movement along their longitudinal axis (Movie 1). This behavior is presumably caused by a simultaneous growth and shrinkage on the two opposite ends of the filament, a phenomenon known as treadmilling, as was previously also observed for purified BtubA/B filaments in simple buffers.20,21 To validate this hypothesis, we used a dual color labeling of tubulin with the Atto488 or Atto565 dye and bleached one of the fluorophores (Atto488) during imaging (Figure 2B). Continuous illumination yielded filaments displaying extensions with fluorescent extremities that originate from the addition of tubulin subunits to the growing ends (Figure 2C,D, Movie 2). Further, single fluorescent spots on the bleached filaments remained immobile. These observations demonstrate that bMTs undergo a treadmilling behavior on SLBs in a solution compatible with CFPS.
When the bMTs on the SLB were imaged at a high illumination intensity, the filaments could break apart, either completely disassembling or with generated fragments detaching from the membrane (Figure 2E). This effect was dependent on the presence of the fluorophore used for labeling (Atto488 or Atto565), suggesting a dye-specific photochemical reaction. If the excitation was performed using a laser with a wavelength outside the excitation range of the fluorophore, a prolonged illumination was not accompanied by filament breaking. Dual labeling experiments confirmed that the physical integrity of bMTs was altered, as opposed to photobleaching effects (Movie 3).
BtubA and BtubB proteins were separately expressed with PUREfrex2.0, mixed with a trace amount of labeled purified bacterial tubulin (100 nM), and the solution was added to an SLB (Figure 3A,B). The bMT assembly and recruitment to the SLB were immediately observed (Figure 3B). To rule out the possibility that filament assembly might be caused by the low amount of labeled purified tubulin, we also imaged the SLB with expressed BtubA and 100 nM of labeled tubulin for 30 min, which should give ample time for bMTs to assemble. No bMTs were observed in the absence of expressed BtubB, but they readily appeared within a few seconds upon the addition of expressed BtubB (Figure 3C, Movie 4). As with the purified proteins, dynamic instability and treadmilling on the membrane as well as a bundling of the microtubules were observed (Movie 4).
Figure 3.
Cell-free expressed BtubA/B assemble into dynamic bMTs on SLBs. (A) Schematic of CFPS and BtubA/B polymerization on supported lipid membranes. (B) Fluorescence microscopy image of bMTs reconstituted from separately expressed BtubA and BtubB. Before the addition to an SLB the mixed solution was supplemented with a small fraction of purified Atto488-BtubA/B for imaging. Scale bar: 10 μm. (C) Cell-free synthesized BtubA was mixed with 100 nM Atto488-BtubA/B and incubated on an SLB for 30 min. No filament was observed. At time t = 0, separately expressed BtubB was added on top of the SLB, triggering the immediate formation of short filaments that developed into longer and thicker bundles. Scale bars: 10 μm.
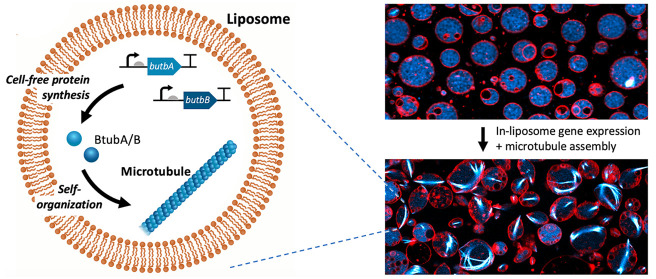
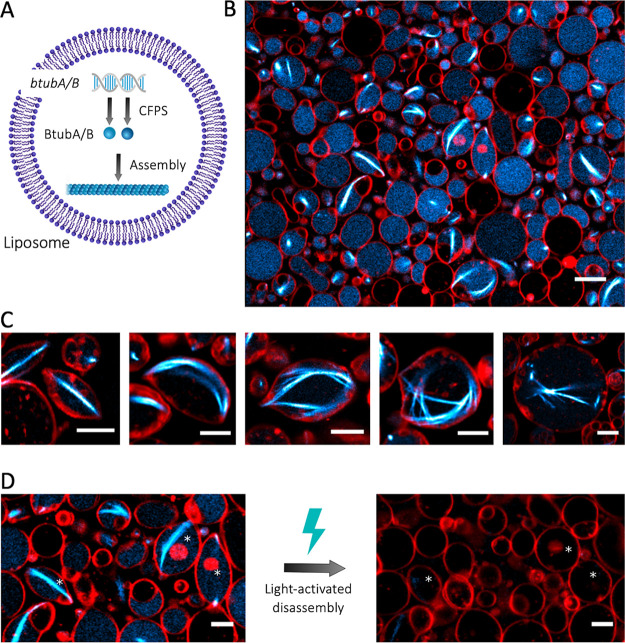
The compartmentalization of bMTs in cell-sized lipid vesicles provides a unique platform to study their self-organization in a closed volume as well as their ability to exert pushing forces and deform the membrane. Large and giant liposomes were produced by glass bead-assisted lipid film swelling.36,37 PUREfrex2.0, the btubA/B DNA constructs, a mix of DnaK chaperone to promote protein folding,15 and 100 nM of Atto488-labeled purified BtubA/B were coencapsulated (Figure 4A). A slightly higher concentration of the btubA gene compared to the btubB gene (3.75 vs 2.5 nM) was used to compensate for the lower amount of expressed BtubA versus BtubB when an equimolar concentration of the two templates is used (Figure 1C). Liposomes that sedimented in a glass chamber were incubated at 37 °C to trigger gene expression and imaged at various time points by fluorescence confocal microscopy. Initially, liposomes that had encapsulated the labeled tubulin displayed an even fluorescence in their lumen (Figure S6). During the course of the gene expression, filament structures appeared, with the length and bundling propensity increasing over time (Figure 4B, Figure S6). This internal cytoskeleton could clearly be attributed to an in situ synthesis of BtubA/B, as the small fraction of purified labeled tubulin did not yield filaments until an accumulation of sufficient expression products (Figure S6) nor in control samples where the btubA/B genes were omitted (Figure S7). The number of liposomes exhibiting cytoskeletal structures, the time of incubation until the first filaments appear (typically 1 h), and the number of filaments per liposome differed from one experiment to the other, in particular, when using different DNA batches. Yet, the emergence of bMT networks was a robust observation. In the absence of the DnaK mix, the synthesized BtubA/B could not develop into cytoskeletal structures, suggesting that chaperones are needed to prevent synthesized proteins from aggregating or misfolding in liposomes.38 A similar observation was made when active Min proteins were expressed in PUREfrex2.0.15 A variety of BtubA/B filament and liposome morphologies was seen: straight or curved bundles or meshes, located across the lumen or near the membrane (Figure 4C). Most liposomes containing bMTs exhibited morphological changes. Vesicles that were originally spherical showed local protrusions or a global elongation (Figure 4B,C). Such deformation events were likely the result of the pushing force exerted by bMTs that grew on the liposome membrane, a phenomenon that is well-documented for eukaryotic microtubules and actin filaments.1,39 Similar results were obtained upon a direct encapsulation of purified BtubA/B (6.6 μM final concentration in PUREfrex2.0), with the only noticeable difference that the average occurrence of filaments was higher with purified proteins, and a few cross-shaped liposomes were observed (Figure 5). This result suggests that the concentration of expressed active BtubA/B is lower than ∼7 μM but higher than 1 μM in bMT-containing vesicles (Supporting Information Note 2). Liposome-to-liposome heterogeneity regarding the internal concentration of labeled tubulin, expression efficiency, and cytoskeleton features were likely the consequence of a varying encapsulation efficiency and stochastic effects, as previously reported for other reconstituted biological systems.13,15,26,30,36,37
Figure 4.
CFPS and the assembly of bMTs inside liposomes. (A) Schematic of liposome-compartmentalized gene expression and synthesis of BtubA/B that self-organize into bMTs. (B) Fluorescence confocal microscopy images of liposomes (red, DHPE-TexasRed) with encapsulated Atto488-BtubA/B (100 nM, cyan) after 4 h of CFPS reaction. In situ synthesized bMTs (cyan) are visible in several liposomes. Scale bar: 10 μm. (C) Examples of different bMT cytoskeletal structures showing clear membrane deformation. Samples were observed after 5 h of incubation at 37 °C. Scale bars: 5 μm. (D) Breaking of bMTs and relaxation of liposomes into a spherical shape was triggered by the exposure of samples to a high laser intensity. Asterisks indicate liposomes whose shape was modified by the light-activated disassembly of bMTs. Scale bars: 5 μm.
Figure 5.
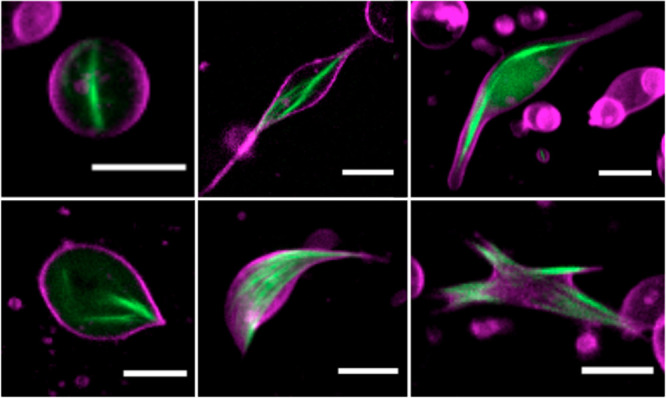
Bacterial microtubules (green, 100 nM Atto488-BtubA/B) formed by 6.6 μM of purified tubulins inside liposomes (magenta). Scale bars: 5 μm.
Finally, we sought to demonstrate the reversible nature of liposome deformations by the light-activated bMT disassembly mechanism that we observed on SLBs (Figure 2E). The exposure of filament-containing liposomes to an intense illumination triggered the bMT collapse and subsequent vesicle relaxation to a spherical shape (Figure 4D). Similar observations have been reported with actin filaments7−9 and with eukaryotic microtubules.5 In some cases, sphericality was not fully restored, which might be caused by the high packing of liposomes or by surface tethering over an extended area, both effects altering the mechanical properties of the vesicles. Overall, these results demonstrate that liposome-confined, de novo synthesized bMTs can self-organize into cytoskeletal structures that can deform the liposome through forces that push on the membrane. Moreover, light-activated bMT disassembly can be exploited for the reversible shape control of single targeted liposomes within a population.
Currently, quantitative insights about the physical parameters describing bMT mechanical properties are missing. The bending rigidity of bMTs is probably lower than that of eukaryotic microtubules due to their smaller diameter.19,20 The BtubA/B filament and bundle morphologies share similarities with those reported with encapsulated actin filaments bundled by linker proteins.6,8 Therefore, we assume that the rigidity of bMTs lies in the range between eukaryotic microtubules and actin filaments. Other relevant parameters that remain to be determined include the critical force for buckling, the pushing force of a growing bMT, and the number of filaments per bundle. This information will help in the design of liposome experiments, where the membrane tension and BtubA/B expression levels may be adjusted to modulate the vesicle shape and organization of cytoskeletal structures.
This work expands the scope of bMT applications in synthetic biology. Bacterial microtubules have structural and biochemical properties that give them decisive advantages for bioengineering and for an implementation in artificial cells compared to eukaryotic microtubules and actin: a lower threshold of monomer concentration for polymerization (∼1 vs ∼25 μM for eukaryotic microtubules2), an ability to form bundles without additional cofactors, and suitability for cell-free gene expression (Supporting Information Figure 8 and Note 3). Other prokaryotic protein filaments have already been reconstituted in liposomes. MreB expressed in liposomes assembles into cytoskeletal networks without inducing a membrane deformation,10 unless molecular crowding at the vesicle membrane is applied.11 Rings of FtsZ-FtsA can form budding necks and constrict liposomes.13 Here, we show that bMTs expand the repertoire of protein filaments and the range of associated functions. The endowment of liposomes with a DNA-programmed mechanism for elongation and membrane remodelling, as realized here with bMTs, might be instrumental to support processes such as polarization and compartment division. For instance, a bMT-mediated elongation of vesicles may promote pole-to-pole oscillations of the reconstituted Min system15 or the positioning of the Z-ring.13 The prospect for discovering new bMT-interacting partners (natural or engineered), especially end-tracking proteins, could further extend their role in intravesicular transport, segregation of synthetic chromosomes, or polarization.
Methods
Preparation of DNA Constructs
To ensure a high expression level, we adjusted the DNA sequence of the three btub genes of Prosthecobacter dejongeii (btubA, btubB, btubC) with respect to their GC content and intramolecular base pairing for the first 30 bp after the start codon. For each gene, we listed all possible DNA sequences of the first 30 bp encoding the same amino acid sequence, selected the sequences with the lowest GC content, and calculated the predicted melting temperature of their corresponding RNA sequence. We computed the conformations of the RNA sequences with the highest value for change in free Gibbs energy (ΔG) regarding the intramolecular base pairing of the 30 bp before and 30 bp after the start codon (60 bp total) using mfold.40 If sequences had similar ΔG values, the sequence with the least number of base pairs at the ribosome binding site (RBS) and start codon was chosen. An example of the calculated RNA structures is shown in Figure S1. The sequence-optimized DNA fragments, including a T7 promotor, RBS, and T7 terminator, were synthesized and cloned in the pUC57 plasmid (GenScript). Linear templates were generated by a polymerase chain reaction (PCR) using forward and reverse primers 5′-CAGTCACGACGTTGTAAAACGAC-3′ and 5′-CACACAGGAAACAGCTATGAC-3′, respectively. The PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega), following the manufacturer’s protocol. The concentration and purity of the DNA constructs were determined by spectrophotometry using the NanoDrop 2000 (Thermo Scientific). Samples were also analyzed by electrophoresis on 1% agarose gels.
Purification and Labeling of BtubA/B
BtubA/B proteins were copurified and labeled as described in ref (21), except that C41(DE3) cells were used for the expression instead of BL21(DE3). The concentration of purified bacterial tubulin was determined from an absorbance at 280 nm (extinction coefficient 103 754.2 M–1 cm–1).
Bulk CFPS
Bulk CFPS reactions were performed with PUREfrex2.0 (GeneFrontier Corporation) and 5 nM of linear DNA template according to the supplier’s protocol. When btubA and btubB genes were coexpressed, 2.5 nM of each DNA construct was used. For a visualization of gene products by PAGE, 1 μL of GreenLys solution (FluoroTect, Promega) was supplemented to a PUREfrex2.0 mix (total volume 10 μL), and the reactions were performed in PCR tubes at 37 °C for 3 h. Samples with added sodium dodecyl sulfate (SDS, 20% w/v final) were incubated at 90 °C for 10 min and loaded on a 12% SDS-PAGE gel that was run first for 20 min at 100 V and then for 40 min at 160 V. Fluorescently labeled proteins were visualized on a fluorescence gel imager (Typhoon, Amersham Biosciences) using a 488 nm laser and a 515–535 nm band-pass emission filter. Subsequently, the gel was stained with InstantBlue (expedeon) overnight and imaged with a ChemiDocTM imaging system (Bio-Rad).
Estimation of the Concentration of Synthesized BtubA/B
Preran PUREfrex2.0 samples that contained expressed bacterial tubulin were loaded onto a 10% stain-free gel, either undiluted or fivefold diluted. Purified bacterial tubulin was loaded in concentrations ranging from 0.125 to 8 μM, and a calibration curve was generated from these samples using Fiji41 to quantify band intensities.
Imaging Chambers
Experiments involving SLB or liposome imaging were performed in self-made glass chambers. Three glass slides of 1 mm thickness were glued together with NOA 61 UV glue (Norland Products). Several holes of 3 mm diameter were created with a diamond drill, and a 150 μm thick coverslip (Menzel-Gläser) was glued to the bottom with NOA 61. Before use, the chambers were cleaned by sequential washing steps that included 10 min of sonication each in chloroform/methanol (1:1 volume), 2% Hellmanex III (Hellma), and 1 M KOH, ethanol, and Milli-Q water. In the case of SLB experiments, the glass chambers were additionally treated every second experiment with an acid piranha solution. For some liposome experiments, aluminum chambers were used, which were fabricated in the same manner as described for the glass chambers and were cleaned following the same protocol, except that the KOH and piranha treatments were omitted.
Preparation of Lipid-Coated Beads
All stock lipids in chloroform were supplied by Avanti Polar Lipids, except for Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (DHPE-TexasRed), which was from Invitrogen. A lipid mixture consisting of ∼50 mol % 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 36 mol % 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE), 12 mol % 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG), 2 mol % 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (18:1 cardiolipin), 0.2 mol % DHPE-TexasRed, and 1 mass% 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(poly(ethylene glycol))-2000] (DSPE-PEG-biotin), was prepared in a 10 mL round-bottom glass flask. A solution of 100 mM rhamnose in methanol was added (2.5:1 chloroform-to-methanol volume ratio), followed by 0.6 g of 212–300 μm glass beads (acid washed, Sigma-Aldrich). The solvent was removed by rotary evaporation at 200 mbar for 2 h at room temperature. The lipid-coated beads were aliquoted, desiccated overnight, and stored under argon at −20 °C.
Preparation of SUVs
Small unilamellar vesicles (SUVs) had the same lipid composition as described for the preparation of lipid-coated beads, except that DSPE-PEG-biotin was omitted. A lipid film of 500 μg was formed at the bottom of a glass vial by a gentle evaporation of chloroform. Lipids were resuspended with 400 μL of Milli-Q water (1.25 mg mL–1 final concentration), and the solution was vortexed for 2 min. The sample was extruded using a mini extruder (Avanti Polar Lipids) equipped with 250 μL Hamilton syringes, two filters (drain disc 10 mm diameter, Whatman), and a polycarbonate membrane with a pore size of 0.2 μm (first extrusion) and 0.03 μm (second extrusion). The SUV stock solution was stored at −20 °C until use.
SLB Experiments
Imaging glass chambers were treated with oxygen plasma (Harrick Plasma) for 15 min to activate the surface. Six microliters of SUV solution was added to the chamber and supplemented with 12 μL of 6 mM CaCl2. The chamber was covered with a coverslip, placed on a 0.5 mm thick adhesive silicone sheet (Life Technologies), and incubated for 30 min at 37 °C. The formed SLB was washed four times with MRB80 buffer (80 mM K-Pipes, 4 mM MgCl2, 1 mM EGTA, pH 6.8) and incubated for 10 min with 0.5 mg mL–1 k-Casein in MRB80 buffer. For experiments with purified BtubA/B, 20 μL of a solution containing PUREfrex2.0, 0.05% (w/v) methylcellulose, 4 μL of MRB80, 3 μL of Milli-Q water, and purified unlabeled and Atto488/Atto561-labeled BtubA/B were added to the glass chamber. The imaging was performed at either 25 or 30 °C. For experiments with cell-free synthesized BtubA/B, 8.5 μL of a preran PUREfrex2.0 sample that contained expressed BtubA was mixed with 1.5 μL of 1 μM labeled BtubA/B-Atto488; the sample was added to the SLB and imaged for 30 min at 30 °C. Then, 5 μL of a preran PUREfrex2.0 sample that contained expressed BtubB was added during a total internal reflection fluorescence (TIRF) imaging. The setup consisted of an Ilas2 system (Roper Scientific) on a Nikon Ti-E inverted microscope with a Nikon CFI Plan Apochromat 100 × NA1.45 TIRF oil objective and two Evolve 512 EMCCD cameras (Photometrics) for a simultaneous dual-acquisition. The system was operated with MetaMorph 7.8.8.0 (Molecular Device).
CFPS in Liposomes
Twenty micrograms of lipid-coated beads was added to 20 μL of a swelling solution that consisted of PUREfrex2.0, 1 μL of DnaK mix (GeneFrontier Corporation), 100 nM Atto488-BtubA/B, and 3.75 nM of btubA and 2.5 nM of btubB DNA constructs. The sample was incubated on ice for 2 h, during which the tube was gently manually rotated a few times. Four freeze–thaw cycles were applied by dipping the tube into liquid nitrogen, followed by thawing at room temperature. An imaging glass chamber was incubated for 5 min with a mix of bovine serum albumin (BSA) and BSA-biotin (1:1 molar ratio, 1 mg mL–1, Thermo Fisher Scientific), followed by an incubation with Neutravidin (1 mg mL–1, Sigma-Aldrich). Next, 4 μL of liposome sample along with 12 μL of dilution buffer (PUREfrex2.0 Solution I and Milli-Q water (7:4 volume ratio) supplemented with 83 mg L–1 proteinase K) were added to the imaging chamber. The fluorescence imaging was performed with a confocal microscope (A1+ from Nikon, 100× oil immersion objective) by using the 488- and 561 nm laser lines for an excitation of Atto488-BtubA/B and DHPE-TexasRed, respectively. Samples were incubated at 37 °C during the image acquisition.
Acknowledgments
The authors thank V. Volkov for assistance with the establishing of bacterial microtubules in the lab and for useful discussions. Microscopy measurements were performed at the Kavli Nanolab Imaging Center Delft. This work was financially supported by The Netherlands Organization for Scientific Research (NWO/OCW) through the “NanoFront–Frontiers of Nanoscience” Gravitation grant and the “BaSyC–Building a Synthetic Cell” Gravitation grant (024.003.019).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.1c00278.
Gene sequences, vesicle floatation assay, discussion of the interaction of BtubA/B/C with the vesicle membrane, discussion of the relationship between the concentration of expressed BtubA/B and the liposome morphology, discussion of the comparison between eukaryotic and bacterial MTs, mfold analysis of RNA structure around the start codon, gel electrophoresis analysis of cell-free protein expression, quantitation of BtubA/B expression levels, control experiments with bMTs on SLB and in liposomes, fluorescence images of eukaryotic MTs inside liposomes (PDF)
TIRF imaging of purified bMTs doped with Atto488-labelled BtubA/B on a planar lipid bilayer. Experimental conditions are as described in Figure 2A (AVI)
TIRF imaging of purified bMTs doped with Atto488- and Atto565-labelled BtubA/B on a planar lipid bilayer. Experimental conditions are as described in Figure 2B–D (AVI)
TIRF imaging of purified bMTs doped with Atto488- and Atto565-labelled BtubA/B on a planar lipid bilayer exposed to intense illumination. Experimental conditions are as described in Figure 2E (AVI)
TIRF imaging of cell-free expressed bMTs doped with Atto488-labelled BtubA/B on a planar lipid bilayer. Experimental conditions are as described in Figure 3 (AVI)
Author Contributions
All authors designed the experiments. J.K. and A.D. performed the experiments. J.K. and C.D. wrote the manuscript with inputs from all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Hotani H.; Miyamoto H. Dynamic features of microtubules as visualized by dark-field microscopy. Adv. Biophys. 1990, 26, 135–156. 10.1016/0065-227X(90)90010-Q. [DOI] [PubMed] [Google Scholar]
- Kuchnir Fygenson D.; Elbaum M.; Shraiman B.; Libchaber A. Microtubules and vesicles under controlled tension. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 1997, 55, 850–859. 10.1103/PhysRevE.55.850. [DOI] [Google Scholar]
- Fygenson D. K.; Marko J. F.; Libchaber A. Mechanics of microtubule-based membrane extension. Phys. Rev. Lett. 1997, 79, 4497–4500. 10.1103/PhysRevLett.79.4497. [DOI] [Google Scholar]
- Emsellem V.; Cardoso O.; Tabeling P. Vesicle deformation by microtubules: A phase diagram. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 1998, 58, 4807–4810. 10.1103/PhysRevE.58.4807. [DOI] [Google Scholar]
- Hayashi M.; Nishiyama M.; Kazayama Y.; Toyota T.; Harada Y.; Takiguchi K. Reversible morphological control of tubulin-encapsulating giant liposomes by hydrostatic pressure. Langmuir 2016, 32, 3794–3802. 10.1021/acs.langmuir.6b00799. [DOI] [PubMed] [Google Scholar]
- Tsai F.-C.; Koenderink G. H. Shape control of lipid bilayer membranes by confined actin bundles. Soft Matter 2015, 11, 8834–8847. 10.1039/C5SM01583A. [DOI] [PubMed] [Google Scholar]
- Tanaka S.; Takiguchi K.; Hayashi M. Repetitive stretching of giant liposomes utilizing the nematic alignment of confined actin. Commun. Phys. 2018, 1, 18. 10.1038/s42005-018-0019-2. [DOI] [Google Scholar]
- Bashirzadeh Y.; Wubshet N. H.; Liu A. P. Confinement geometry tunes fascin-actin bundle structures and consequently the shape of a lipid bilayer vesicle. Front Mol. Biosci. 2020, 7, 610277. 10.3389/fmolb.2020.610277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschel T.; Kelley C. F.; Holz D.; Adeli Koudehi M.; Vogel S. K.; Burbaum L.; Mizuno N.; Vavylonis D.; Schwille P. Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 2021, 12, 2254. 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y. T.; Nakadai T.; Shin J.; Uryu K.; Noireaux V.; Libchaber A. Assembly of MreB filaments on liposome membranes: a synthetic biology approach. ACS Synth. Biol. 2012, 1, 53–59. 10.1021/sb200003v. [DOI] [PubMed] [Google Scholar]
- Garenne D.; Libchaber A.; Noireaux V. Membrane molecular crowding enhances MreB polymerization to shape synthetic cells from spheres to rods. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 1902–1909. 10.1073/pnas.1914656117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusato T.; Horie F.; Matsubayashi H. T.; Amikura K.; Kuruma Y.; Ueda T. De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth. Biol. 2018, 7, 953–961. 10.1021/acssynbio.7b00350. [DOI] [PubMed] [Google Scholar]
- Godino E.; Lopez J. N.; Zarguit I.; Doerr A.; Jimenez M.; Rivas G.; Danelon C. Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. Commun. Biol. 2020, 3, 539. 10.1038/s42003-020-01258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourian Z.; Scott A.; Danelon C. Toward the assembly of a minimal divisome. Systems and Synthetic Biology 2014, 8, 237–247. 10.1007/s11693-014-9150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godino E.; Lopez J. N.; Foschepoth D.; Cleij C.; Doerr A.; Castella C. F.; Danelon C. De novo synthesized Min proteins drive oscillatory liposome deformation and regulate FtsA-FtsZ cytoskeletal patterns. Nat. Commun. 2019, 10, 4969. 10.1038/s41467-019-12932-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati G.; Lenzi P.; Franco V. ‘Epixenosomes’: Peculiar epibionts of the protozoon ciliate Euplotidium itoi: Do their cytoplasmic tubules consist of tubulin?. Micron 1993, 24, 465–471. 10.1016/0968-4328(93)90025-V. [DOI] [Google Scholar]
- Petroni G.; Spring S.; Schleifer K.-Z.; Verni F.; Rosati G. Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 1813–1817. 10.1073/pnas.030438197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag C. A.; Staley J. T.; Erickson H. P. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J. Cell Biol. 2005, 169, 233–238. 10.1083/jcb.200410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M.; Ladinsky M. S.; McDowall A. W.; Petroni G.; Jensen G. J. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011, 9, e1001213 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.; et al. Four-stranded mini microtubules formed by prosthecobacter BtubAB show dynamic instability. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E5950–58. 10.1073/pnas.1705062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Celis C.; et al. Bacterial tubulins A and B exhibit polarized growth, mixed-polarity bundling, and destabilization by GTP hydrolysis. J. Bacteriol. 2017, 199, e00211 10.1128/JB.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-C.; Webb R. I.; Janssen P. H.; Sangwan P.; Romeo T.; Staley J. T.; Fuerst J. A. Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 2009, 9, 5. 10.1186/1471-2180-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akendengue L.; Trepout S.; Grana M.; Voegele A.; Janke C.; Raynal B.; Chenal A.; Marco S.; Wehenkel A. M. Bacterial kinesin light chain (Bklc) links the Btub cytoskeleton to membranes. Sci. Rep. 2017, 7, 45668. 10.1038/srep45668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V.; Libchaber A. A vesicle bioreactor as a step towards an artificial cell assembly. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 17669–17674. 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi P. L.; Ferri F.; Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften 2006, 93, 1–13. 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- van Nies P.; Westerlaken I.; Blanken D.; Salas M.; Mencía M.; Danelon C. Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat. Commun. 2018, 9, 1583. 10.1038/s41467-018-03926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhanu S.; Ueda T.; Kuruma Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 2019, 10, 1325. 10.1038/s41467-019-09147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S.; Matsuura T.; Sunami T.; Kazuta Y.; Yomo T. In vitro evolution of α-hemolysin using a liposome display. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 16796–1680. 10.1073/pnas.1314585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A.; Noga M. J.; de Graaf P.; Westerlaken I.; Yildirim E.; Danelon C. Cell-free phospholipid biosynthesis by gene-encoded enzymes reconstituted in liposomes. PLoS One 2016, 11, e0163058 10.1371/journal.pone.0163058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanken D.; Foschepoth D.; Serrao A. C.; Danelon C. Genetically controlled membrane synthesis in liposomes. Nat. Commun. 2020, 11, 4317. 10.1038/s41467-020-17863-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y.; Inoue A.; Tomari Y.; Suzuki T.; Yokogawa T.; Nishikawa K.; Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Doerr A.; Foschepoth D.; Forster A. C.; Danelon C. In vitro synthesis of 32 translation-factor proteins from a single template reveals impaired ribosomal processivity. Sci. Rep. 2021, 11, 1898. 10.1038/s41598-020-80827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J.; Hansen G. P.; Venancio-Marques A.; Baigl D. Cell-free preparation of functional and triggerable giant proteoliposomes. ChemBioChem 2013, 14, 2243–2247. 10.1002/cbic.201300501. [DOI] [PubMed] [Google Scholar]
- Sadler F. W.; Dodevski I.; Sarkar C. A. RNA thermometers for the PURExpress system. ACS Synth. Biol. 2018, 7, 292–296. 10.1021/acssynbio.7b00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Park B.; Woo S.-G.; Lee J.; Park J. Prosthecobacter algae sp. nov., isolated from activated sludge using algal metabolites. Int. J. Syst. Evol. Microbiol. 2014, 64, 663–667. 10.1099/ijs.0.052787-0. [DOI] [PubMed] [Google Scholar]
- Nourian Z.; Roelofsen W.; Danelon C. Triggered gene expression in fed-vesicle microreactors with a multifunctional membrane. Angew. Chem., Int. Ed. 2012, 51, 3114–3118. 10.1002/anie.201107123. [DOI] [PubMed] [Google Scholar]
- Blanken D.; van Nies P.; Danelon C. Quantitative imaging of gene-expressing liposomes reveals rare favorable phenotypes. Phys. Biol. 2019, 16, 45002. 10.1088/1478-3975/ab0c62. [DOI] [PubMed] [Google Scholar]
- Fernandez C.; Giraldo R. Modulation of the aggregation of the prion-like protein RepA-WH1 by chaperones in a cell-free expression system and in cytomimetic lipid vesicles. ACS Synth. Biol. 2018, 7, 2087–2093. 10.1021/acssynbio.8b00283. [DOI] [PubMed] [Google Scholar]
- Cortese J. D.; Schwab B. 3rd; Frieden C.; Elson E. L. Actin polymerization induces a shape change in actin-containing vesicles. Proc. Natl. Acad. Sci. U. S. A. 1989, 86, 5773–5777. 10.1073/pnas.86.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.