Summary
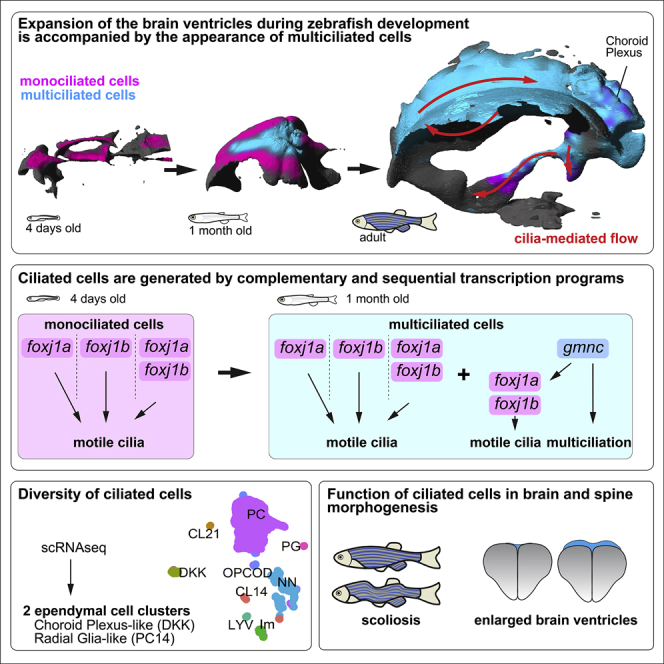
Motile cilia defects impair cerebrospinal fluid (CSF) flow and can cause brain and spine disorders. The development of ciliated cells, their impact on CSF flow, and their function in brain and axial morphogenesis are not fully understood. We have characterized motile ciliated cells within the zebrafish brain ventricles. We show that the ventricles undergo restructuring through development, involving a transition from mono- to multiciliated cells (MCCs) driven by gmnc. MCCs co-exist with monociliated cells and generate directional flow patterns. These ciliated cells have different developmental origins and are genetically heterogenous with respect to expression of the Foxj1 family of ciliary master regulators. Finally, we show that cilia loss from the tela choroida and choroid plexus or global perturbation of multiciliation does not affect overall brain or spine morphogenesis but results in enlarged ventricles. Our findings establish that motile ciliated cells are generated by complementary and sequential transcriptional programs to support ventricular development.
Keywords: cilia, multiciliated cells, foxj1, gmnc, ependymal cell, zebrafish, brain, choroid plexus, cerebrospinal fluid, scoliosis
Graphical abstract
Highlights
-
•
Glutamylated tubulin is enriched in cilia of foxj1-expressing cells in the zebrafish
-
•
Motile ciliated ependymal cells in the zebrafish forebrain are highly diverse
-
•
Gmnc drives the transition from mono- to multiciliated cells at juvenile stage
-
•
Lack of multiciliation does not impact brain and spine morphogenesis
D’Gama et al. dissect the genetic mechanisms underlying the formation of motile ciliated ependymal cell types in the zebrafish forebrain and their function in brain and spine morphogenesis. They show that an increasing diversity of ependymal lineages depends on the sequential or parallel activation of foxj1- and gmnc-dependent transcriptional programs.
Introduction
The ependyma, which is the cellular layer lining the surface of the brain ventricles and spinal canal, plays a key role in cerebrospinal fluid (CSF) dynamics (Fame and Lehtinen, 2020). In the mammalian brain, the ependyma is composed primarily of post-mitotic glia-like cells known as multiciliated ependymal cells (Del Bigio, 2010; Jiménez et al., 2014; Spassky et al., 2005; Spassky and Meunier, 2017). Ependymal cells are derived from radial glial cells and differentiate during the perinatal and postnatal periods (Spassky et al., 2005; Ortiz-Álvarez et al., 2019; Redmond et al., 2019). Like multiciliated cells (MCCs) in other tissues (Spassky and Meunier, 2017; Reiten et al., 2017), ependymal MCCs carry bundles of motile cilia on their apical surface, which beat and contribute to directional CSF flow (Worthington and Cathcart, 1963; Sawamoto et al., 2006; Faubel et al., 2016; Ringers et al., 2020). In analogy to mammals, the zebrafish ependyma is also decorated with motile ciliated cells (Ringers et al., 2020; Fame et al., 2016; Olstad et al., 2019; van Leeuwen et al., 2018; Kishimoto et al., 2011; Lindsey et al., 2012; Ogino et al., 2016; Grimes et al., 2016; Konjikusic et al., 2018). These motile ciliated cells appear as early as 28 to 32 hours post fertilization (hpf) in the embryonic brain (Fame et al., 2016; Olstad et al., 2019; van Leeuwen et al., 2018) and even earlier in the central canal of the spinal cord (Kramer-Zucker et al., 2005; Sternberg et al., 2018; Thouvenin et al., 2020). During these embryonic stages, the cells bear solitary cilia that move in a rotational manner and generate directional flow (Thouvenin et al., 2020; Olstad et al., 2019). Ependymal MCCs, analogous to mammalian ependymal cells, have only been reported in the adult zebrafish brain (Kishimoto et al., 2011; Lindsey et al., 2012; Ogino et al., 2016; Grimes et al., 2016; Konjikusic et al., 2018). Yet, it remains unclear when and where these MCCs are established and how they contribute to CSF dynamics.
Besides their role in CSF circulation, ependymal MCCs are also necessary for the adult neurogenic niche to assemble into a characteristic pinwheel-like organization (Jacquet et al., 2009; Lalioti et al., 2019; Mirzadeh et al., 2008) and for maintenance of the epithelial integrity of the ependyma (Jiménez et al., 2014; Feldner et al., 2017; Nechiporuk et al., 2007). Given this broad array of functions, abnormalities of the ependymal cells lead to a variety of neurological conditions. For instance, in mammals, ciliary defects of ependymal cells are commonly associated with ventricular defects and hydrocephalus (Brody et al., 2000; Ringers et al., 2020; Lee, 2013, Jiménez et al., 2014; Wallmeier et al., 2019; Ibañez-Tallon et al., 2004; Karimy et al., 2020). Besides this, motile cilia in the zebrafish have been shown to regulate spine morphogenesis (Zhang et al., 2018; Grimes et al., 2016) through CSF circulation and formation of a glycoprotein filament in the CSF called Reissner fiber (Cantaut-Belarif et al., 2018; Troutwine et al., 2020). To further understand how motile cilia instruct brain and spine development across species, it is now crucial to fully resolve the cellular and functional diversity of ciliated cells in the ependyma and the genetic programs driving their differentiation.
In this study, we determined the ontogeny of ependymal MCCs within the forebrain of the zebrafish. We observed that MCCs emerge at the juvenile stage, co-exist with monociliated cells, and diversify into distinct lineages through development. We dissected the transcriptional regulatory pathways directing the differentiation program of these motile ciliated cells and found that these involve the complementary and sequential activation of the master regulatory genes of motile ciliogenesis, foxj1a and foxj1b, and of multiciliation, gmnc. Finally, we characterized the role of individual motile ciliated cell lineages in the morphogenesis of the brain and body axis. We show that while critical for proper ventricular development, motile cilia regulated by foxj1b and gmnc are largely dispensable for brain and axial morphogenesis, whereas the requirement for foxj1a is more essential. Altogether, our study reveals the diversity of motile ciliated cell types within the zebrafish forebrain from a molecular, cellular, and functional standpoint and uncovers a remarkable degree of similarity as well as differences with ependymal cells of mammals.
Results
Glutamylated tubulin is a marker for cilia of foxj1-expressing cells
Previous work has suggested that glutamylated tubulin staining could be a specific marker of motile cilia in zebrafish (Pathak et al., 2007, 2011). We also observed that ciliary tubulin glutamylation was increased in a subset of cells in the nervous system, which expressed the zebrafish Foxj1 orthologs foxj1a or foxj1b (Olstad et al., 2019; Thouvenin et al., 2020). To further scrutinize the causality between foxj1 expression and axonemal tubulin glutamylation, and hence its validity as a motile cilia marker, we overexpressed Foxj1a in zebrafish embryos using a heat-shock-inducible foxj1a transgene (Yu et al., 2008; Choksi et al., 2014) and monitored the levels and distribution of glutamylated tubulin-positive cilia. We have previously shown that overexpression of Foxj1a using this strategy is sufficient to induce ectopic motile cilia in tissues that normally differentiate immotile primary cilia, such as the trunk musculature and eyes (Yu et al., 2008; Choksi et al., 2014). We observed that the short primary cilia, which normally form in these tissues, have very low to undetectable levels of ciliary glutamylated tubulin. Strikingly, staining of heat-shocked versus control animals revealed that transgenic overexpression of Foxj1a was sufficient to induce circa 1,000-fold higher glutamylated tubulin levels in cilia in the trunk and eye regions (Figures S1A, S1B, and S1F), in addition to significantly increasing ciliary length (Figures S1A, S1C, and S1G). We also observed that, in comparison to acetylated tubulin, glutamylated tubulin is not uniformly distributed across the cilium but is enriched at one end of the axoneme (Figures S1D, S1E, and S1H–S1J), juxtaposing the basal body marked by gamma-tubulin (Figure S1K) (Bosch Grau et al., 2013; Pathak et al., 2007, 2014). Taken together, these results revealed that in the zebrafish, the presence of glutamylated tubulin is a reliable marker for cilia of foxj1-expressing cells.
Motile ciliated cell abundance and onset of multiciliation correlate with expansion of brain ventricles and parenchyma during development
Having identified a reliable marker for motile cilia, we next tracked the appearance of such cilia and the transition from mono- to MCCs over the course of forebrain development. We focused on larval (4 and 14 days post fertilization [dpf]), juvenile (28 to 32 dpf), and adult stages.
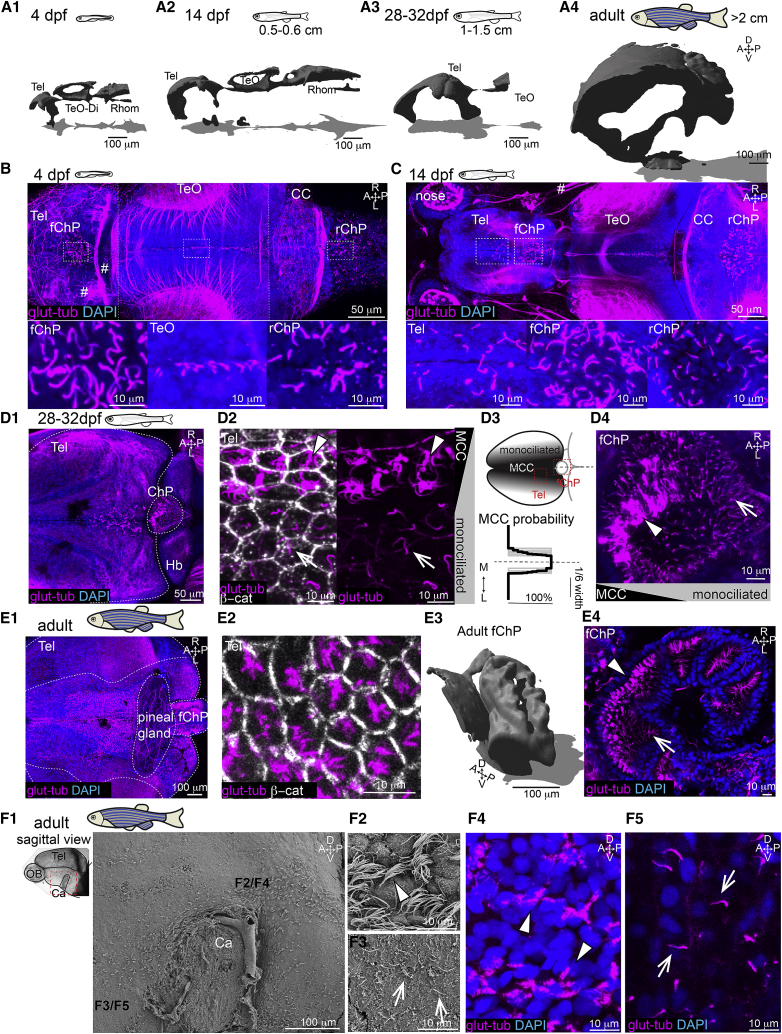
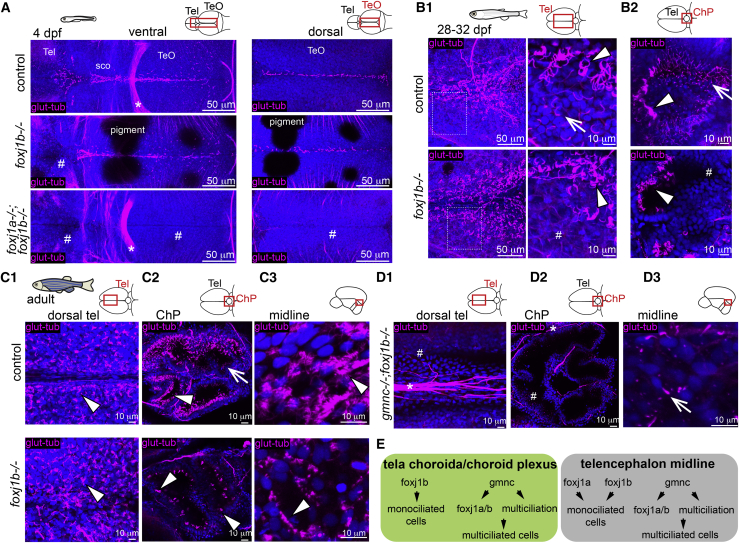
We observed that concomitant with brain volume expansion from 4 to 14 dpf (Figures 1A1 and 1A2), more cilia appeared on the dorsal telencephalon anterior to the forebrain choroid plexus (ChP) (Figures 1B and 1C). At 28 to 32 dpf, when major cognitive, learning, and social skills are acquired (Valente et al., 2012; Larsch and Baier, 2018; Dreosti et al., 2015; Hinz and de Polavieja, 2017; Jurisch-Yaksi et al., 2020; Vendrell-Llopis and Yaksi, 2015; Palumbo et al., 2020), we found that ciliated cells on the dorsal telencephalon increased not only in number but also started to harbor brushes of cilia reminiscent of MCCs (Figures 1A3, 1D1, and 1D2). At this stage, MCCs were present on the tela choroida (TC) (Folgueira et al., 2012; Nieuwenhuys, 2011; Lindsey et al., 2012), which is the epithelial layer located above the dorsal telencephalon (Figure 1D2) and in the forebrain ChP (Figure 1D4), and co-existed with monociliated cells. Interestingly, the MCCs were not randomly distributed but were enriched at the midline of the dorsal telencephalon (Figure 1D2, quantified in Figure 1D3) and anterior part of the forebrain ChP (Figure 1D4). At later developmental stages (Figures 1A4 and 1E1), MCCs covered large parts of the dorsal telencephalon yet remained enriched around the midline. Staining with a membrane marker (β-catenin) revealed that cilia do not populate the entire apical surface of these cells uniformly (Figure 1E2). Hence, they appear to be translationally polarized like mammalian ependymal MCCs (Boutin et al., 2014; Mirzadeh et al., 2010). The adult forebrain ChP, which consists of multiple connected cavities (Figure 1E3), was also composed of large numbers of MCCs, closely apposed to monociliated cells (Figure 1E4), with both cell types directing their cilia toward the lumen of the cavities.
Figure 1.
Multiciliation and ventricular/parenchymal expansion correlate during development
(A1–A4) Brain ventricles expand during development as shown upon 3D reconstruction of brain ventricles injected with Rhodamine B isothiocyanate (RITC)-dextran. (A1) 4 dpf, n = 4; (A2) 14 dpf, length: 0.5 to 0.6 cm, n = 4; (A3) 28 to 32 dpf, length: 1–1.5 cm, n = 3; and (A4) 2 to 12 months, larger than 2 cm, n = 3.
(B) At 4 dpf, single glutamylated tubulin-positive cilia are located on the forebrain choroid plexus (fChP), on the dorsal roof and ventral part of the tectal/diencephalic ventricle, and in the rhombencephalic ChP (rChP) (n = 5). Dashed lines label boundaries between confocal tiles.
(C) At 14 dpf, cilia number increases along the dorsal telencephalon, rostral to the fChP, and in the rChP. Cells are monociliated throughout the brain (n = 3).
(D1–D4) At 28 to 32 dpf, brushes of glutamylated tubulin-positive cilia appear on the dorsal telencephalon anterior to the ChP and in the ChP (n = 3; D1). (D2) The presence of monociliated (arrow) and MCCs (arrowhead) is shown upon co-staining with the membrane marker β-catenin. (D3) MCCs are located medially to monociliated cells in the tela choroida (TC; quantified in bottom panel; n = 9). (D4) The fChP comprises mono- and MCCs, arranged in an anterior-posterior manner (n = 3).
(E1–E4) In the adult brain, MCCs are enriched in the medial part of the TC above the telencephalon (E1) and in the ChP (E4). (E2) Cilia do not cover the entire apical surface of MCCs as shown upon co-staining with β-catenin (n = 4). (E3) Adult fChP consists of multiple interconnected cavities, as shown upon 3D reconstruction of ventricles injected with RITC-dextran (n = 4), and contains mono- and MCCs (E4) (n = 3).
(F1–F5) Mono- and MCCs are present on the adult telencephalic/diencephalic midline in the region surrounding the anterior commissure (Ca) highlighted in red. Scanning electron microscopy (n = 3) and immunostaining with glutamylated tubulin (n = 3) show the presence of MCCs (F2 and F4) and monociliated cells (F3 and F5). Location of (F2)–(F5) is indicated in (F1).
A, anterior; P, posterior; D, dorsal; V, ventral; M, medial; L, lateral; Tel, telencephalon; TeO, optic tectum; Rhomb, rhombencephalon; CC, cerebellum; MCCs, multiciliated cells. Arrowheads show MCCs, and arrows show monociliated cells.
See also Figure S1.
Next, using scanning electron microscopy and glutamylated tubulin staining, we observed that ciliated cells were also present along the ventral part of the telencephalon on the midline surrounding the anterior commissure (Figures 1F1 and 1F2). Ciliated cells carried either a ciliary brush (Figure 1F4) or a single cilium (Figure 1F5).
Altogether, our observations show that as the brain parenchyma and ventricular cavities expanded in size, the number of motile ciliated cells increased and the multiciliation program was initiated.
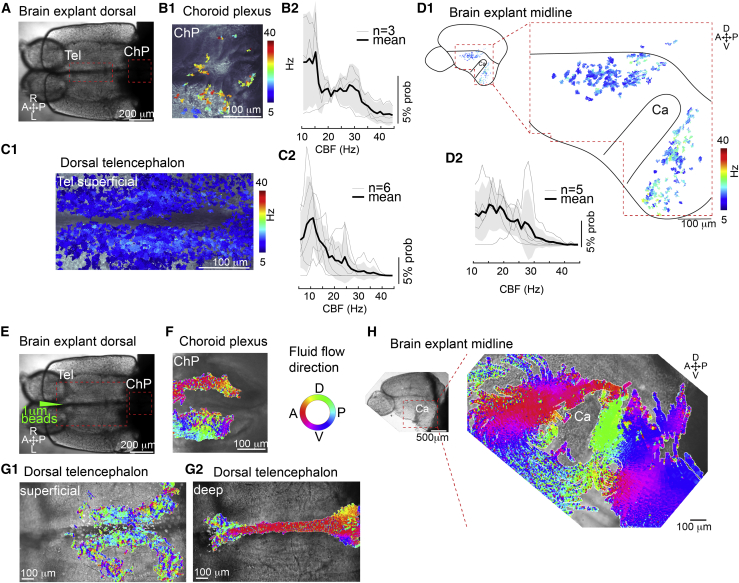
Cilia on cells of the telencephalon and ChP are motile and generate directional CSF flow
To confirm that glutamylated tubulin-positive cilia are motile, we performed video microscopy on adult brain explants using transmission microscopy (Figure 2A), followed by a Fourier-based analysis (Reiten et al., 2017; Olstad et al., 2019). We observed beating cilia in the ChP (Figure 2B1), TC (Figure 2C1), and ventral telencephalon (Figure 2D1) at the locations where glutamylated tubulin-positive cilia were present. Although beat frequencies largely varied (Figures 2B2, 2C2, and 2D2), ciliary beating was sufficient to elicit directional fluid flow revealed by particle tracking of injected fluorescent beads (Figures 2E–2H; Videos S1, S2, and S3). In particular, we observed a clear directionality in the dorsal telencephalic ventricle, with flow from anterior to posterior in the superficial part (Figure 2G1; Video S1) and posterior to anterior in the deeper region (Figure 2G2; Video S2). In the ventral telencephalon, we also observed that flow is directionally organized, going rostrally anterior to the commissure and caudally posterior to the commissure (Figure 2H). Thus, cilia identified using glutamylated tubulin staining are motile and generate directional fluid flow.
Figure 2.
Cilia in the telencephalon, TC, and ChP are motile and generate CSF flow
(A) Dorsal view of an adult brain explant.
(B–D) Cilia in the forebrain ChP (B), TC (C), and telencephalic midline (D) are motile as shown by analysis of high-speed video microscopy using a pixel-based Fourier analysis.
(B1–D1) Map of ciliary beating with ciliary beat frequency (CBF) color-coded in the ChP (B1), TC (C1), and hemisphere explant anterior and posterior to the Ca (D1).
(B2–D2) Probability histogram showing the frequency of the pixel-based analysis and the average ± SEM for ChP (B2), TC (C2), and telencephalic hemisphere (D2). n = number of brain explants.
(E) Dorsal view of an adult brain explant injected with 1 μm fluorescent beads.
(F–H) Directional fluid flow in the ChP (F; n = 4), dorsal telencephalon (G; n = 5), and telencephalic midline (H; n = 5) is color-coded.
Tel, telencephalon; A, anterior; P, posterior; D, dorsal; V, ventral; R, right; L, left.
Fluid flow measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Fluid flow is measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Fluid flow is measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Motile ciliated cells express Foxj1 orthologs foxj1a and foxj1b differentially until the initiation of the multiciliation program by gmnc
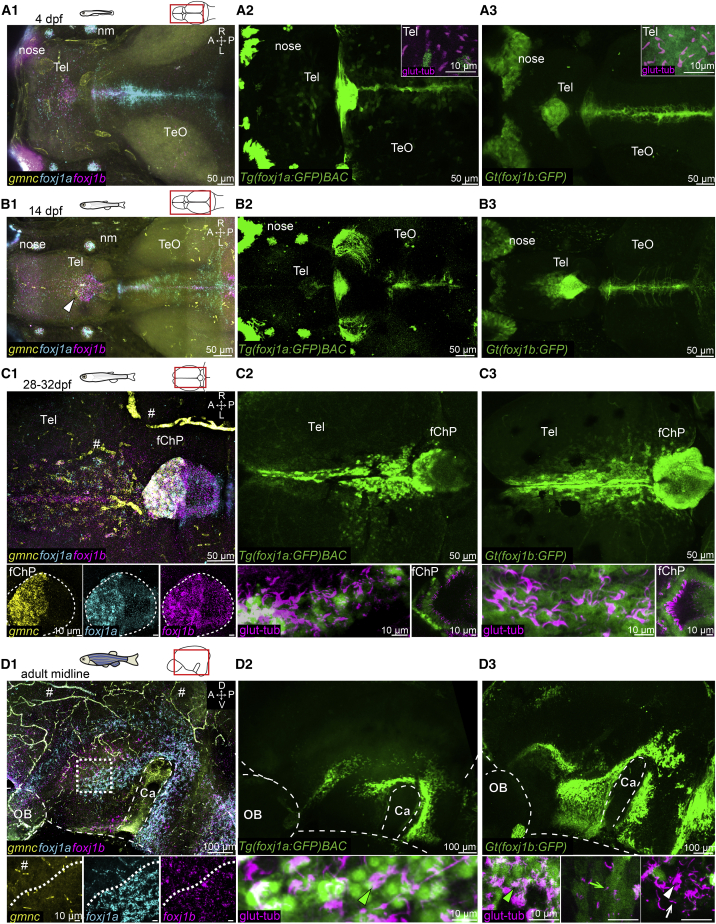
To elucidate the genetic identity of the motile ciliated cells, we analyzed the expression of the master regulator of motile ciliogenesis foxj1 (Yu et al., 2008; Stubbs et al., 2008) and of multiciliation gemc1/gmnc (Arbi et al., 2016; Kyrousi et al., 2015; Terré et al., 2016; Zhou et al., 2015; Lewis and Stracker, 2020). We performed multiplexed hybridization chain reaction (HCR) for the two zebrafish foxj1 genes foxj1a and foxj1b and gmnc simultaneously. We also characterized the expression pattern of foxj1a, using a bacterial artificial chromosome (BAC)-based reporter transgenic strain Tg(foxj1a:gfp)BAC vcc41, and foxj1b, using the gene trap line Gt(foxj1b:GFP)tsu10Gt, which contains GFP inserted within the first intron of foxj1b (Olstad et al., 2019; Tian et al., 2009).
Using these tools, we detected a differential expression of foxj1a and foxj1b in the developing brain, confirming our previous observations with the larval brain (Olstad et al., 2019). In particular, we identified a more prominent expression of foxj1b than foxj1a in the dorsal telencephalon and ChP from 4 dpf (Figures 3A1–3A3) to 14 dpf (Figures 3B1–3B3) when only monociliated cells were present. In agreement with the lack of multiciliation until 3 to 4 weeks, we did not observe gmnc expression at 4 dpf (Figure 3A1) and only few gmnc puncta at 14 dpf (Figure 3B1, arrowhead). In contrast, we identified a major increase of gmnc signal at 1 month in locations with MCCs, such as the medio-posterior TC and anterior forebrain ChP (Figure 3C1). In line with increased gmnc expression, we observed elevated foxj1a levels (Figures 3C1 and 3C2), suggesting that foxj1 levels and activity of the multiciliation program might be correlated.
Figure 3.
Ciliated cells in the TC, telencephalon, and ChP express foxj1a, foxj1b, and gmnc to different extents
(A–D) Expression of foxj1a, foxj1b, and gmnc in the brain using multiplex HCR (A1–D1) and two transgenic lines, Tg(foxj1a:gfp)BACvcc41 (A2–D2) and Gt(foxj1b:GFP)tsu10Gt (A3–D3).
(A1–A3) At 4 dpf, foxj1b is expressed in the dorsal telencephalon, nose, neuromasts, and midbrain; foxj1a is expressed in the nose, neuromasts, and midbrain; and no gmnc HCR signal is detected in the brain. Cells with a solitary glutamylated tubulin-labeled cilium (magenta in A2 and A3 insets) on the dorsal telencephalon express primarily foxj1b.
(B1–B3) At 14 dpf, there is an expansion of foxj1b-expressing cells anterior to the ChP (B1 and B3). foxj1a expression remains low in the dorsal telencephalon and ChP (B1 and B2). Few gmnc puncta are present in the anterior part of the ChP (arrowhead in B1).
(C1–C3) At 1 month, gmnc and foxj1a are highly expressed in the anterior ChP and dorso-medial TC (C1 and C2). (C1 and C3) foxj1b is expressed in both the anterior and posterior ChP. (C2 and C3) foxj1a:GFP and foxj1b:GFP cells bear glutamylated tubulin-positive cilia.
(D1–D3) On the midline of the adult telencephalon/diencephalon, foxj1a and foxj1b are mainly expressed in the ventral part of the brain, surrounding the Ca. (D1) foxj1a- and foxj1b-expressing domains are not fully overlapping. The dotted line in inset indicates a sharp boundary for foxj1a expression but not for foxj1b. There is very low gmnc expression. (D2 and D3) Immunostaining with glutamylated tubulin shows that foxj1a:GFP- and foxj1b:GFP- expressing cells harbor multiple cilia (arrowhead) or a solitary cilium (arrow). (D3) Ciliated cells with multiple cilia (white arrowhead) or a single cilium (white arrow) are foxj1b:GFP-negative.
Number of datasets: A1 = 4, B1 = 4, C1 = 4, D1 = 3, A2 = 4, B2 = 3, C3 = 4, D3 = 4, A3 = 4, B3 = 3, C3 = 4, and D3 = 3.
A, anterior; P, posterior; D, dorsal; V, ventral; M, medial; L, lateral; Tel, telencephalon; TeO, optic tectum; OB, olfactory bulb; nm, neuromast. Note that there is unspecific HCR signal associated with blood cells and vasculature (indicated by the hashtag symbol).
See also Figure S2.
To further assess the interdependency between multiciliation and foxj1 levels, we turned to the Tg(foxj1a:gfp)BAC vcc41 and Gt(foxj1b:GFP)tsu10Gt transgenic lines. We observed that MCCs, bearing brushes of glutamylated tubulin-positive cilia, expressed foxj1a:GFP and foxj1b:GFP more intensely than monociliated cells (Figures 3C2 and 3C3; Figures S2A1–S2A3). This was particularly evident in the 1-month ChP, where only the anterior and multiciliated portion of the ChP expressed foxj1a (Figures 3C1 and 3C2). foxj1a expression remained sparse in the adult ChP (Figures S2A and S2B). Taken together, our results suggest a sequential and complementary expression of the two Foxj1 orthologs in the TC and ChP, with foxj1b expressed from 4 dpf onward and foxj1a induced when ciliated cells begin multiciliation.
To ascertain the location of ciliated cells in relation to CSF, we injected a fluorescent dye within the ventricles of brain explants from Gt(foxj1b:GFP)tsu10Gt transgenic animals. Using this approach, we confirmed that ciliated cells are located in the TC and not in the brain parenchyma. We also observed that foxj1b:GFP-expressing cells in the ChP surrounded the CSF-filled cavity (Figures S2D and S2E).
To discern whether ciliated cells within the telencephalic midline expressed foxj1a and foxj1b similar to the TC and ChP, we performed HCR on the midline of the adult telencephalic parenchyma and scrutinized the expression patterns of Tg(foxj1a:gfp)BAC vcc41 and Gt(Foxj1b:GFP)tsu10Gt transgenic animals. We observed that foxj1a- and foxj1b-expressing cells were located mostly on the ventral part of the telencephalon and in the preoptic nucleus of the diencephalon (Figure 3D1), in the region where we observed ciliated cells by scanning electron microscopy (Figure 1F1). While foxj1a and foxj1b HCR patterns were mainly overlapping at the surface of the parenchyma, we also observed cells expressing only foxj1b (Figure 3D1). Upon staining with glutamylated tubulin antibody, we identified multiciliated foxj1a:GFP- and foxj1b:GFP-positive cells (Figures 3D2 and 3D3). We also observed that some foxj1b:GFP-positive cells were non-ciliated or monociliated (Figure 3D3) and that a proportion of mono- and MCCs were not foxj1b:GFP-positive (Figure 3D3), suggesting that these cells might only express foxj1a.
In sum, our results show that motile ciliated cells express foxj1a, foxj1b, and gmnc differentially depending on the development stage and their location within the telencephalon, thereby implying that ependymal cells have diverse identities.
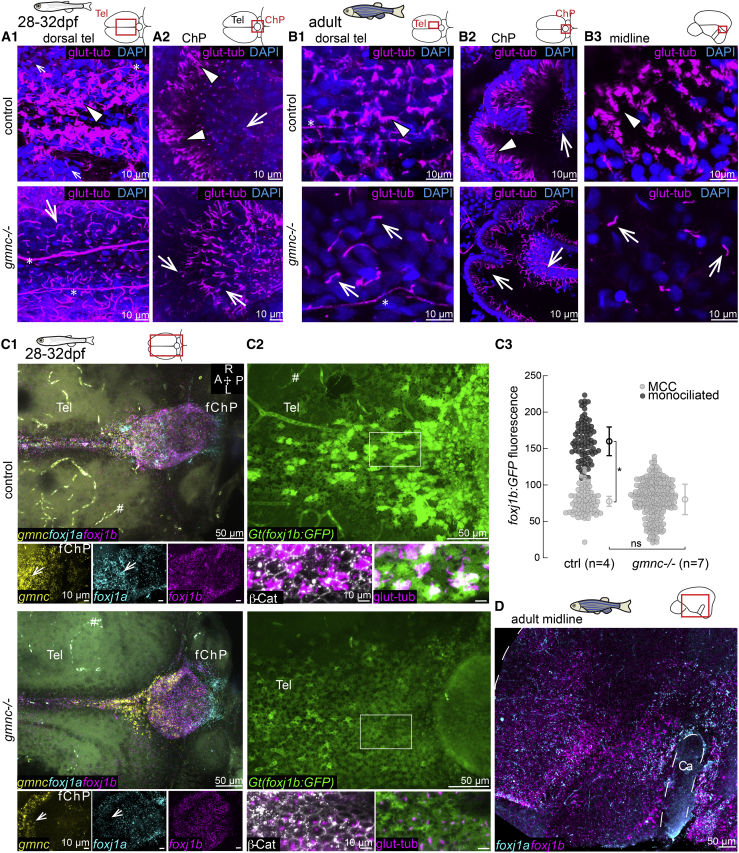
Multiciliation is driven by a genetic program involving gmnc
We next sought to identify the importance of gmnc for the transition from mono- to multiciliation. We analyzed gmnc mutants (Zhou et al., 2015) at 1 month (Figure 4A), when multiciliation had occurred, as well as in adult stages (Figure 4B). We observed that ciliated cells in all parts of the telencephalon and ChP of gmnc mutants did not harbor ciliary brushes but instead single cilia. Similar results were obtained by scanning electron microscopy analysis of the rhombencephalic ventricle (Figure S3). Since ciliated cells in gmnc mutants retained a glutamylated tubulin-positive solitary cilium, we examined whether they maintained the expression of foxj1. We imaged the brain of 1-month-old gmnc mutants stained for HCR as well as gmnc mutants carrying the Gt(foxj1b:GFP)tsu10Gt transgene. We observed that foxj1a was reduced in gmnc mutants, in particular, in the anterior ChP at 1 month (Figure 4C1). In contrast, foxj1b remained expressed in the ciliated cells despite the absence of gmnc activity (Figures 4C1 and 4C2) albeit at reduced and more homogeneous levels as compared to controls (Figures 4C2 and 4C3). In the telencephalic/diencephalic midline, expression of both foxj1a and foxj1b was maintained (Figure 4D). These findings establish that while gmnc induces multiciliation in motile ciliated cells of the telencephalon and ChP and increases expression levels of foxj1, it is not required for motile ciliogenesis, per se.
Figure 4.
gmnc is required for multiciliation in ependymal cells
(A and B) Absence of MCCs in 1-month-old (A1 and A2; n = 3 ctrl; 3 mutants) and adult brains of gmnc mutants (B1–B3; n = 3 ctrl; 3 mutants). Arrowheads show MCC in controls. All ciliated cells in gmnc mutant are monociliated (arrow). (A1) 1-month dorsal telencephalon and (A2) ChP. (B1) Adult dorsal telencephalon. (B2) Anterior portion of the adult ChP. (B3) Adult midline above the Ca.
(C1–C3) gmnc enhances the expression of foxj1a and foxj1b in the TC and ChP. (C1) HCR revealed a gmnc-dependent foxj1a expression, particularly in the anterior ChP (arrow in insets). n = 4 controls; 6 mutants. The hashtag symbol shows nonspecific signal in blood vessels. (C2) foxj1b:GFP remained expressed in gmnc mutant with reduced and more homogeneous levels (n = 4 controls; 7 mutants). β-catenin and glutamylated tubulin staining was used for quantification of GFP levels in C3. (C3) Monociliated cells express less foxj1b:GFP than MCCs and monociliated cells in gmnc mutant express similar foxj1b:GFP levels as control monociliated cells. All data points are plotted; mean and SD of n fish; ∗p < 0.05 calculated on the average fluorescence per fish using rank sum test.
(D) HCR shows that foxj1a and foxj1b remained expressed in the telencephalic midline of gmnc mutant (n = 3).
Asterisk indicates axons. A, anterior; P, posterior; R, right; L, left; Tel, telencephalon.
See also Figure S3.
foxj1b induces monociliation in the medial TC and ChP and plays overlapping roles with foxj1a in MCC formation
Since foxj1b is expressed in a large number of ciliated cells in the telencephalon and ChP from early development, we sought to identify the function of foxj1b in these cells. For this, we generated a mutant allele, foxj1bsq5719, lacking most of the coding sequence (Figure S3A). Homozygous mutants are viable and do not exhibit obvious morphological abnormalities other than otolith defects in the embryonic inner ear, consistent with our earlier work using morpholinos and the hypomorphic foxj1b allele produced by the transgenic insertion Gt(foxj1b:GFP)tsu10Gt (Yu et al., 2011). We then investigated the presence of ciliated cells in the brain of these mutants from larval to adult stages. At 4 dpf, we observed that foxj1b mutants lacked cilia in the dorsal telencephalon but not in cells surrounding the optic tectum (Figure 5A). To identify whether these remaining ciliated cells were dependent on the expression of the paralog foxj1a, we generated foxj1a;foxj1b double mutant larvae. We observed a complete loss of cilia from the brains of double foxj1a;foxj1b mutant larvae (Figure 5A, bottom panel), confirming that formation of glutamylated tubulin-positive cilia is instructed by different combinations of foxj1 genes.
Figure 5.
foxj1a and foxj1b play diverse roles in the formation of motile ciliated cells
(A) Lack of cilia in the dorsal telencephalon in foxj1b mutant (middle; n = 9) and throughout the larval ventricular system in double foxj1a/b mutant (n = 6) at 4 dpf. Maximum projection at two different depths is shown.
(B1 and B2) MCCs (arrowhead) are not affected in the foxj1b mutant on the 1-month dorsal telencephalon (B1; n = 6) and ChP (B2; n = 6). Cilia remain absent in the monociliated cells (arrow) located laterally to the MCCs as shown in the magnified inset.
(C1–C3) MCCs are not affected in the adult medial TC (C1; n = 3), ChP (C2; n = 4), and midline of the telencephalon/diencephalon (C3; n = 3). (C2) Monociliated cells are particularly affected in the ChP.
(D1–D3) Loss of cilia in the medial TC (D1) and ChP (D2) of adult gmnc;foxj1b double mutants (n = 4). (D3) Single cilia remained in the telencephalic midline of double mutants (arrow) (n = 4).
(E) Diverse genetic programs instruct the formation of ciliated cells in the TC/ChP and in the telencephalon midline.
TeO, optic tectum; sco, sub-commissural organ. Arrows indicate monociliated cells, arrowheads indicate MCCs, and asterisks indicate axon tracts. Cilia loss is indicated by hashtag symbols.
See also Figures S4 and S5.
We next analyzed the brains of 2-week-old foxj1b mutants (Figure S4B) prior to multiciliation and observed that cilia in the dorsal telencephalon were lacking, as seen in 4-dpf larvae. Surprisingly, at 1 month of age, foxj1b mutants harbored similar numbers of MCCs as control animals both in the TC and ChP (Figures 5B1 and 5B2). At this stage, we observed that monociliated cells adjacent to the MCC in the medial TC (Figure 5B1) and in the posterior part of the forebrain ChP remained affected (Figure 5B2, indicated by hash symbol). Further analyses of the adult brain by confocal (Figures 5C1–5C3) and scanning electron microscopy (Figure S4B) analyses revealed that MCCs were not particularly disturbed in foxj1b mutants. However, cilia in monociliated cells, in particular those in the ChP, were largely absent even at adult stages (Figure 5C2). Altogether, these results indicate that foxj1b is mainly required for ciliogenesis of monociliated cells in the TC and ChP but is not essential for MCC differentiation.
Since foxj1b mutation leads to ciliary defects in mono- but not MCCs, we hypothesized that foxj1a expression induced upon multiciliation is sufficient for MCC formation. We first confirmed that neither foxj1a nor gmnc levels were affected in the foxj1b mutants using HCR (Figure S4D). Next, to test the impact of foxj1a in MCC ciliation, we generated foxj1a+/−;foxj1b−/− mutants. In contrast to foxj1a−/−, which are embryonically lethal due to severe body curvature (Olstad et al., 2019, Zhang et al., 2018), we were able to identify foxj1a+/−;foxj1b−/− viable adults. In the brains of these mutants, we observed reduced numbers of cilia in the ChP compared to wild-type control or foxj1b−/− (Figure S5A). In the TC, we also observed reduced numbers of MCCs but with a variable degree of penetrance (Figure S5B). However, large numbers of MCCs remained in the telencephalic/diencephalic midline (Figure S5C). These results suggest that foxj1a and foxj1b play important and overlapping roles for MCC ciliation in the telencephalon and that one copy of the foxj1a gene is sufficient for MCC differentiation in the TC and midline.
To test whether foxj1a activity induced by gmnc is sufficient for MCC differentiation, we generated gmnc;foxj1b double mutants. In these animals, which are viable, we observed dramatic cilia loss from the medial TC and ChP (Figures 5D1 and 5D2). By contrast, solitary cilia persisted in the telencephalic midline (Figure 5D3), most likely associated with the cells that exclusively express foxj1a, which remains expressed in the midline in gmnc mutant (Figure 4D). In sum, these results highlight that the diversity of motile ciliated cells within the zebrafish brain ventricles expands during development and depends on the sequential or parallel activation of foxj1- and gmnc-dependent transcriptional programs (Figure 5E).
foxj1a and foxj1b are expressed in two ependymal cell lineages and in different subsets of neuronal progenitors
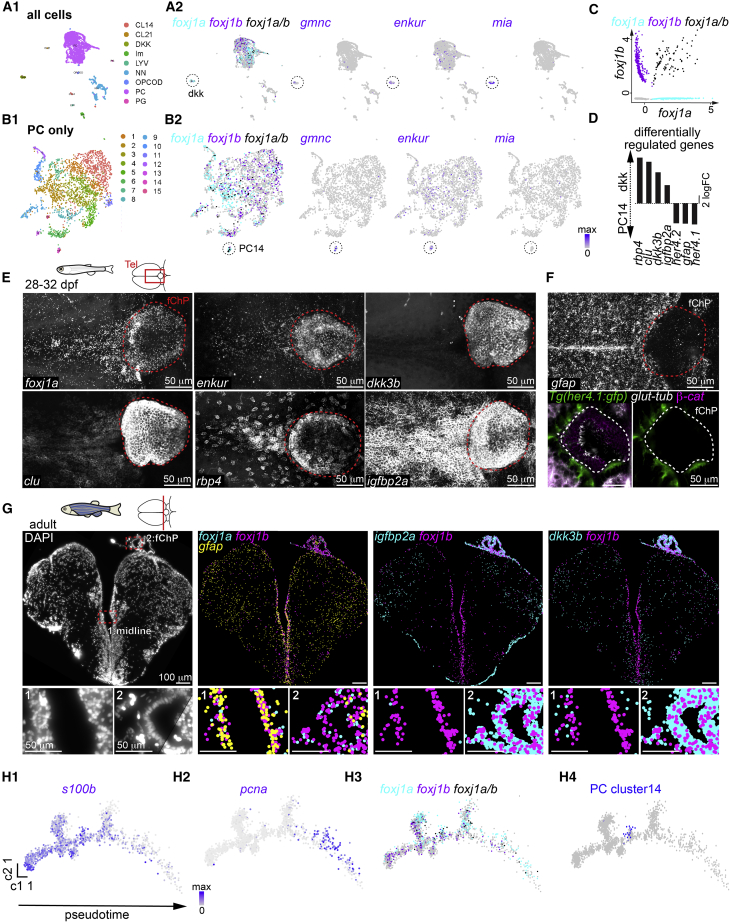
To further unravel the genetic diversity of ependymal lineages, we performed single-cell transcriptomic analysis of an adult telencephalon using a protocol described previously (Cosacak et al., 2019). We labeled cells located in close proximity to the ventricle by injecting a fluorescent cell tracer in 1-year-old animals (Kizil and Brand, 2011). We then fluorescence-activated cell sorted (FACS) telencephalic cells, performed single-cell sequencing (Figure S6A), and recovered 3,158 single cells belonging to various cell types including neurons, immune cells, oligodendrocytes, and progenitor-like cells (PCs) (Figures 6A1, 6B1, S6B, and S6C).
Figure 6.
Diversity of motile ciliated cells in the forebrain
(A and B) Single-cell RNA sequencing analysis of all cells (A1) or all progenitor-like cells (PCs; B1) revealed the presence of two potential ependymal clusters, dkk (A2) and PC14 (B2), based on the expression of foxj1a/b (foxj1a-only cells in cyan, foxj1b-only cells in magenta, and foxj1a/b-co-expressing cells in black) gmnc, enkur, and mia.
(C) Scatterplot representing the expression levels of foxj1a and foxj1b in all cells.
(D) Selection of differentially regulated genes between the dkk and PC14 ependymal clusters.
(E) The dkk cluster markers dkk3b (n = 5), clu (n = 13), rbp4 (n = 4), and igfbp2a (n = 8) are expressed in the 1-month-old TC and ChP, similar to foxj1a and enkur, as shown by HCR.
(F) The PC14 markers gfap (n = 4) and her4 (n = 3) are excluded from the ChP at 1 month. (Top) HCR for gfap shows expression in radial glia and absence of signal in ChP. (Bottom) Immunostaining of Tg(her4:gfp) with membrane marker β-catenin and cilia marker glutamylated tubulin revealed the absence of her4:gfp signal in ciliated cells of the ChP.
(G) Molecular cartography on an adult telencephalic cryosection (DAPI; left) showed that foxj1a and foxj1b colocalized with gfap in the midline (inset 1) and not in the ChP (inset 2) and that igfbp2a and dkk3b are enriched in the ChP (inset 2) as compared to the midline (inset 1). n = 7 sections from one brain.
(H1–H4) Pseudotime analysis of the PCs plotted using Monocle algorithm showed a progression from quiescent (s100b; H1) to a proliferative stage (pcna; H2). (H3) foxj1b is expressed more in quiescent progenitors, while foxj1a is expressed more in proliferative progenitors. (H4) PC14 ependymal cells (indicated in blue) branch out from quiescent progenitors.
To identify potential multiciliated ependymal clusters, we searched for cells expressing foxj1a, foxj1b, gmnc, and previously reported ependymal markers from mice (Shah et al., 2018) and zebrafish (Cosacak et al., 2019), including enkur (Sigg et al., 2017) and mia (Figures 6A2 and 6B2). Based on these criteria, we identified two potential clusters of ependymal-like cells: the PC cluster 14 (PC14) and the dkk cluster (Figures 6A and 6B). Interestingly, one of these ependymal clusters, the PC14, but not the dkk cluster, is highly similar to PCs. We also identified that only the dkk cluster, and not the PC14, expressed the ependymal cell marker mia (Figures 6A2 and 6B2). Strikingly, we observed that foxj1a and foxj1b expression was not limited to these two ependymal cell clusters, but the genes were broadly expressed in PC (Figures 6A2 and 6B2). Most cells expressed either foxj1a or foxj1b, with approximately 15% expressing both foxj1a and foxj1b (Figure 6C). gmnc was also expressed in both ependymal-like clusters (Figures 6A2 and 6B2). Since gmnc is expressed transiently when the cells initiate multiciliation (Terré et al., 2016; Arbi et al., 2016; Zhou et al., 2015), our results suggest that these two populations of ependymal cells are most likely generated independently and do not represent cells at different developmental stages. Moreover, we observed that all gmnc-expressing cells were either foxj1a-and/or foxj1b-positive (Figure S6D), confirming previous observations that gmnc induces foxj1 expression (Zhou et al., 2015; Stubbs et al., 2012; Chong et al., 2018; Terré et al., 2016; Arbi et al., 2016; Kyrousi et al., 2015). Next, we parsed the differentially regulated genes between these two ependymal clusters and identified a total of 257 genes (Table S1). Among these, radial glia/astroglia markers (her4.1, her4.2, and gfap [Diaz Verdugo et al., 2019]) were enriched in the PC14, while other genes, including those encoding secretory proteins present in CSF (rbp4 [Chang et al., 2016]), markers of ChP (igfbp2a [Khan, 2019] and clu [Jiao et al., 2011]), or mammalian ependymal cells (mia [Shah et al., 2018]), were enriched in the dkk ependymal cluster (Figure 6D and S6E). To further validate whether motile ciliated cells of the dkk ependymal cluster correspond to the ChP, we analyzed the expression pattern of 6 of these genes. We identified that many dkk cluster markers (clu, rbp4, igfbp2a, and dkk3b) were expressed in the TC and the ChP of a 1-month-old juvenile (Figure 6E) and that these cells were devoid of the PC14 markers gfap and her4.1 (Figure 6F). Next, we used Molecular Cartography, a highly multiplexed method from Resolve Biosciences, to spatially detect and quantify mRNAs at single-cell resolution using combinatorial single-molecule fluorescent in situ hybridization on adult telencephalic cryosections. Using this approach, we identified that foxj1-expressing ependymal cells in the midline were enriched in a PC14 marker (gfap) and devoid of dkk markers and cells in the ChP were enriched in dkk markers (dkk3b and igfbp2a; Figure 6G). Altogether, our data suggest that the dkk cluster most probably corresponds to ependymal cells of the ChP and TC, while PC14 corresponds to ependymal cells of the telencephalic midline.
We then sought to unravel the identity of “non-ependymal” foxj1-expressing PCs. We plotted all PCs on a cell-trajectory using Monocle (Figures 6H1–6H4) (Qiu et al., 2017). Using this approach, we were able to monitor the progression of PCs from a quiescent (shown by expression of, for example, s100b; Figure 6H1) to a more proliferative neuroblast state (shown by expression of, for example, pcna; Figure 6H2). We observed that foxj1b is expressed preferentially in quiescent progenitors, while foxj1a is expressed at higher levels in proliferative neuroblasts (Figure 6H3). This analysis also revealed that the PC14 branches out from radial glia-like quiescent progenitors (Figure 6H4), in agreement with similar deductions in the mouse brain (Spassky et al., 2005; Ortiz-Álvarez et al., 2019; Redmond et al., 2019). Altogether, our single-cell RNA sequencing data have delineated that there are at least two different ependymal cell clusters with different origins and that foxj1a and foxj1b are expressed not only in ependymal cells but also in neuronal progenitors.
Loss of foxj1b and gmnc does not impact body axis and brain morphogenesis but does affect the size of brain ventricles
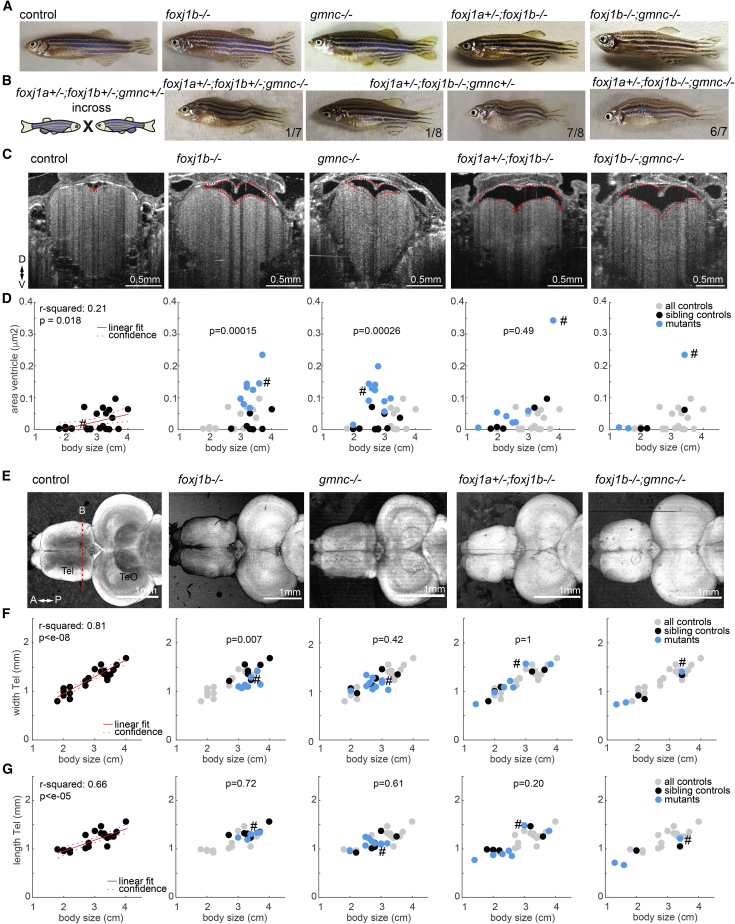
Previous work using zebrafish revealed that motile cilia defects on brain MCCs induce severe scoliosis of the spine (Grimes et al., 2016). Subsequent analysis attributed this deformity to aberrations in Reissner fiber formation, impaired CSF flow, catecholamine transport, and urotensin II-related peptide gene expression in spinal CSF-contacting neurons (Troutwine et al., 2020; Vesque et al., 2019; Rose et al., 2020; Zhang et al., 2018; Lu et al., 2020). Surprisingly, we did not observe axial malformations among foxj1b, gmnc, foxj1a+/−;foxj1b−/−, or foxj1b;gmnc double mutant adults (Figure 7A; embryonic lethality of foxj1a−/− animals prevented us from analyzing the homozygous mutants as adults). Thus, our findings suggest that global loss of multiciliation is not the etiological basis of scoliosis nor is the lack of cilia from foxj1b-expressing cells. To further understand the contribution of motile cilia in scoliosis, we generated progeny from intercrosses of triple foxj1a;foxj1b;gmnc heterozygous mutants. We now observed scoliosis in a significant proportion of these animals (approximately 20%). Upon genotypic evaluation, we found a variable penetrance of the scoliotic phenotype. In general, we identified a higher prevalence of scoliosis among foxj1a+/−;foxj1b−/− animals with one or both mutant alleles of gmnc, but not all fishes of these genotypes showed axial deformations (Table S2; Figure 7B). Taken together, our data revealed that ciliary defects caused by the combined loss of foxj1a, foxj1b, and gmnc is necessary but not sufficient to induce axial deformation, underscoring a critical requirement of foxj1a in this process.
Figure 7.
gmnc−/−, foxj1b−/−, foxj1a+/−;foxj1b−/−, and gmnc−/−;foxj1b−/− mutants do not display major body and brain malformations but have enlarged telencephalic ventricles
(A) Pictures of adult zebrafish showing absence of scoliosis. foxj1anw3 allele was analyzed.
(B) Occurrence of scoliosis in some of the progeny of triple foxj1a; foxj1b; gmnc heterozygous incross. Note that the phenotype is not fully penetrant. The numbers indicate the ratio of animals showing the phenotype among all genotyped individuals. foxj1asq5717 allele was analyzed.
(C) OCT images of anaesthetized adult zebrafish revealed enlarged telencephalic ventricles (highlighted with dashed red line) in animals with cilia defects. Transverse section taken as indicated by red dashed line in (D).
(D) Quantification of ventricular size in all animals as a function of body length.
(E) SD projection of OCT images of adult brain explants showing absence of overt brain malformations in cilia mutants.
(F and G) Quantification of the telencephalic width (F) and length (G) as a function of body length.
(D, F, and G) R-squared indicate the linear relationship between body length and the measurements as shown in red. Left: all controls pooled. Others: all controls in gray, sibling controls in black, and mutants in blue. p value based on rank sum between sibling controls indicated in black and blue. Examples shown are highlighted in the scatterplot with a hashtag symbol.
D, dorsal; V, ventral; A, anterior; P, posterior; Tel, telencephalon; TeO, optic tectum.
Impaired motile cilia are also associated with hydrocephalus both in mice and human ciliopathy patients (Brody et al., 2000; Ringers et al., 2020; Lee, 2013; Jiménez et al., 2014; Wallmeier et al., 2019; Ibañez-Tallon et al., 2004; Karimy et al., 2020; Boon et al., 2014; Wallmeier et al., 2014). To test for such a hydrocephalus phenotype, we used optical coherent tomography (OCT) (Date et al., 2019; Dur et al., 2020) and visualized the ventricular size in anesthetized adult zebrafish in vivo, since dissected brain explants often caused the ventricular system to collapse (Figure S6A). In line with data from mammalian studies, we observed that foxj1b and gmnc mutants have significantly enlarged ventricles. The trend was similar for foxj1a+/−;foxj1b−/− mutants and foxj1b;gmnc double mutants, although the results for these genotypes were not statistically significant due to limitations in numbers of animals and age distribution (Figures 7C and 7D). Finally, to assess the impact of impaired motile cilia on overall zebrafish brain development, we imaged adult zebrafish brain explants by OCT. Our morphometric analysis showed no overt brain malformations between various motile cilia mutants and control animals (Figures 7E–7G and S7E).
Thus, the lack of multiciliation or loss of cilia in specific subpopulations of ependymal cells results in ventricular defects but not in overt brain malformation, as commonly observed in hydrocephalic mice with cilia defects.
Discussion
We have characterized the processes regulating the formation of motile ciliated cells in the ependyma of the zebrafish brain and described their role in CSF flow and ventricular and axial development. We showed that as the brain and its ventricles expand, MCCs appear progressively on the TC, ChP, and parenchymal surface in the midline. In addition, our work has revealed extensive heterogeneity in the genetics, development, anatomy, and functional properties of the motile ciliated cells. Finally, we showed that this heterogenous population of ciliated cells collectively generate a stereotypical pattern of CSF flow, contributing to proper development of the brain ventricles.
We relied on various methods to detect the presence of motile cilia, primarily based on the expression of foxj1 and gmnc genes, immunostaining with cilia markers, and video microscopy. Importantly, our work has established the causality between foxj1 activity and glutamylation of axonemal tubulin, detectable using the GT335 anti-glutamylated tubulin antibody. This discovery has important implications, as it allows for the detection of potential motile cilia based primarily on an antibody staining, at least in the zebrafish. However, it is to be noted that not all foxj1-expressing cells have an actively beating cilium. For example, hair cells of the inner ear and lateral line sense organs have non-motile cilia that are also marked by glutamylated tubulin (Figure S2F). Thus, glutamylated tubulin positivity cannot be used solely as a marker of cilia motility. High levels of ciliary glutamylated tubulin most likely arises from enzymes glutamylating tubulin, such as ttll6, being targets of Foxj1 (Choksi et al., 2014). Ttll6 was previously shown to be expressed in mouse ependymal cells (Bosch Grau et al., 2013; Pathak et al., 2011) and to regulate ciliary beating in zebrafish and mice (Bosch Grau et al., 2013; Pathak et al., 2011). Together, these data suggest that glutamylation of ependymal cilia is conserved across species and is important for ciliary motility.
In the zebrafish, two paralogous Foxj1 genes exist: foxj1a and foxj1b (Yu et al., 2008; Aamar and Dawid, 2008). Using a combination of mutants and transgenic lines described here and in our previous study (Olstad et al., 2019), we showed that in the larval brain, there are three cell populations that express different combinations of these genes. This diversity further expands as the animals develop. For instance, in the TC, the two Foxj1 orthologs are expressed at different development stages, with foxj1b from early development and foxj1a later in MCCs. This raises the question as to why foxj1a is needed later in development in this population of cells. It is possible that foxj1a is expressed because Gmnc eventually induces the expression of both Foxj1 orthologs (Zhou et al., 2015; Stubbs et al., 2012; Chong et al., 2018; Terré et al., 2016; Arbi et al., 2016; Kyrousi et al., 2015; Li et al., 2020) without discriminating between them. Alternatively, MCCs could require both foxj1 genes to transcribe enough motile cilia-specific gene transcripts for multiciliation. Since foxj1b mutants do not show discernible MCC defects in the TC or the midline telencephalon, it is possible that foxj1a plays a more important role in these cells. It is presently unclear why the brain requires such a diversity of ciliated cells and whether the two Foxj1 orthologs are contributing to such diversity. Our previous work revealed that both foxj1 genes have the same efficacy at generating ectopic motile cilia-like cilia when overexpressed (Yu et al., 2008). Likewise, we showed here that both genes are equally important for ciliogenesis in monociliated cells where only one paralog is expressed. It will be interesting to identify whether foxj1a and foxj1b have specific transcriptional programs, in addition to a common one, and if their unique targets could contribute to ciliary diversity.
Our single-cell transcriptomics analysis revealed that, beyond MCCs, neuronal progenitors also express foxj1. We found that foxj1b is expressed more in quiescent stem cells, foxj1a more in proliferative neuroblasts, and a small percentage of cells express both genes. This observation correlates with the description that radial glial cells harbor cilia with motile cilia-like features in the adult zebrafish telencephalon (Kishimoto et al., 2011). Foxj1 was also shown to be expressed in neuronal progenitors in the embryonic and adult neurogenic niche of the mouse (Jacquet et al., 2009, 2011). Our single-cell data have also revealed that some ependymal MCCs are, in many aspects, similar to PCs and probably derived from quiescent PCs. This relates well to data obtained from the mouse, where ependymal cells and quiescent neural stem cells share a large number of markers (Shah et al., 2018) and radial glia act as progenitors of the ependymal cells during embryonic development (Redmond et al., 2019; Ortiz-Álvarez et al., 2019; Spassky et al., 2005).
We also showed that motile ciliated cells on the dorsal telencephalon are not located in the brain parenchyma but instead on the TC, which is the epithelial layer located above the telencephalon (Folgueira et al., 2012; Lindsey et al., 2012), and in the ChP. Based on our observations, ciliated cells in the TC and ChP rely on similar transcriptional program, suggesting that these two structures may have more features in common. In fact, the TC is a fold of the pia mater that gives rise to the ChP prior to the formation of the plexus with the blood vasculature (García-Lecea et al., 2008; Bill and Korzh, 2014). Therefore, it is not surprising that these two structures could rely on the same ciliogenic transcriptional program. While at the larval stage the TC/ChP comprise a flat monolayer, it eventually folds into a structure composed of several cavities as development progresses, with cells pointing their cilia toward the cavities. Interestingly, there remains a spatial organization of mono- versus MCCs in the adult ChP. The inverted configuration of the zebrafish forebrain ChP is different from that of mammals, where ependymal cells point outward in the ventricles and bathe in CSF (Ghersi-Egea et al., 2018). These different configurations may result from the eversion of the neural tube in the zebrafish, in contrast to evagination of the mammalian brain (Jurisch-Yaksi et al., 2020), and the necessity to increase the surface area of the ependymal layer with limited space. Similar to the zebrafish, cells of the mammalian ChP are multiciliated (Nonami et al., 2013; Banizs et al., 2005; Li et al., 2020) and express Foxj1 (Lim et al., 1997; Li et al., 2020). Cilia on the mouse ChP have also been shown to be motile, particularly at the perinatal period (Narita et al., 2010; Nonami et al., 2013; Banizs et al., 2005). Yet, it is currently thought that these cilia do not contribute significantly to CSF flow (Nonami et al., 2013; Narita et al., 2012) but are rather involved in other cellular processes (Narita et al., 2010; Banizs et al., 2005).
Upon loss of the central regulator of multiciliation gmnc, we found that zebrafish ependymal MCCs differentiate a single cilium, implying that Gmnc activity does not impact motile ciliogenesis, per se, but only the process of multiciliation. This contrasts with mouse ependymal cells, where loss of Gmnc/Gemc1 completely abolishes ciliogenesis and prevents the maturation of ependymal cells (Kyrousi et al., 2015; Terré et al., 2016) and MCCs of the ChP (Li et al., 2020). Thus, ciliary defects in the gmnc mutant zebrafish brain resembles more closely the phenotype of Mcidas knockout mice, where multiciliated precursor cells are still specified but generate a single motile-like cilium (Lu et al., 2019; Li et al., 2020). These results are also similar to our earlier report of gmnc loss of function in the zebrafish embryonic kidneys where MCC precursors assemble a single motile cilium (Zhou et al., 2015). We argue that these contrasting effects are due to the way the ciliogenic transcription factors are deployed during ciliated cell development in the different organisms. In zebrafish embryonic kidneys and brain, MCC fate is instituted by gmnc acting on cells that also express foxj1a or foxj1b, respectively, and have already differentiated a single motile cilium. Gmnc modifies this monociliated program to drive multiciliogenesis, in part by upregulating the expression of the foxj1 genes and, on the other hand, by activating genes for multiple basal body production. Consequently, on loss of gmnc, these cells by default remain monociliated. By contrast, during MCC formation in the mouse, Gmnc/Gemc1 functions at the top of the hierarchy of ciliary transcription factors, activating both the expression of Foxj1 and other regulatory genes involved in ciliation and multiple basal body production (Kyrousi et al., 2015; Terré et al., 2016; Lewis and Stracker, 2020). Consequently, in this context, loss of Gmnc/Gemc1 completely impairs motile ciliogenesis. All of these data illustrate how combining several motile cilia-related transcription factors, and their differential deployment in one or more cascades, can generate a high diversity of ciliated cell types.
Defects in ependymal cells can have dramatic consequences on brain development. In mice, ependymal dysfunction ultimately results in hydrocephalus with enlarged ventricles and thinning of the brain parenchyma (Lee, 2013; Jiménez et al., 2014; Ringers et al., 2020; Ibañez-Tallon et al., 2004), typically leading to the premature death. By contrast, in humans, the prevalence of hydrocephalus upon ciliary dysfunction is rather low (Wallmeier et al., 2019; Lee, 2013; Ringers et al., 2020). Our foxj1b and gmnc mutant zebrafish show enlarged ventricles but no severe brain malformation. It remains to be understood whether this difference in hydrocephalus prevalence among species relates to differences in susceptibility to aqueduct stenosis, the degree of cilia loss in the different conditions, overall CSF dynamics, and/or in additional genetic predisposition. Additionally, defects in motile cilia have been associated with scoliotic malformations of the spine in zebrafish at larval, juvenile, and adult stages (Grimes et al., 2016; Ringers and Jurisch-Yaksi, 2020; Zhang et al., 2018; Cantaut-Belarif et al., 2018; Vesque et al., 2019; Bearce and Grimes, 2021). We did not observe scoliosis in foxj1b and gmnc, as well as foxj1a+/−;foxj1b−/− and foxj1b;gmnc, mutant animals, suggesting that spine morphogenesis may rely primarily on foxj1a. In support of this view, we report a higher prevalence of scoliosis in adult foxja+/−;foxj1b−/− animals with at least one mutant copy of gmnc.
In conclusion, our comprehensive analysis has identified the major motile ciliated ependymal cell types of the zebrafish brain longitudinally through the process of development and has dissected the genetic mechanisms underlying their differentiation. We believe that these data have laid the foundation for more detailed investigations aimed at unravelling how these different ciliated lineages regulate specific aspects of brain development and physiology as well as their contribution to proper morphogenesis of the body axis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal glutamylated tubulin (GT335) | Adipogen | Cat# AG-20B-0020-C100; RRID: AB_2490210 |
| GFP Polyclonal Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat # A-21311, RRID:AB_221477 |
| Alexa fluor plus Goat anti mouse 488 | Thermo Fisher Scientific | Cat # A32723; RRID:AB_2633275 |
| Alexa fluor plus Goat anti rabbit 555 | Thermo Fisher Scientific | Cat# A32732; RRID:AB_2633281 |
| Beta catenin | cell signaling technologies | Cat# 9562; RRID:AB_331149 |
| Alexa fluor plus Goat anti mouse 555 | Thermo Fisher Scientific | Cat# A32727, RRID:AB_2633276 |
| Chemicals, peptides, and recombinant proteins | ||
| Dapi | Thermo Fisher Scientific | Cat# D1306; RRID:AB_2629482 |
| x2 power up SYBR master mix | Thermo Fisher Scientific | Cat# A25742 |
| Microamp optical 96 well reaction plate (Applied biosystems) | Thermo Fisher Scientific | Cat# N8010560 |
| 70 kDa rhodamine B isothiocyanate-dextran | Sigma-Aldrich | Cat# R9379 |
| SPHERO Fluorescent Yellow Particles 1% w/v, F = 1 μm | Spherotech | Cat# FP-1552-2 |
| Phosphate buffered saline | Thermofisher | Cat# BR0014G |
| Triton X-100 | Merck | Cat# 1086031000 |
| Bovine Serum Albumin (BSA) | PanReac AppliedChem | Cat# A1391 |
| Dimethyl sulfoxide (DMSO) | Sigma | Cat# D8418 |
| Glycerol | VWR | Cat# 24387.292 |
| Acetone | VWR | Cat# 20066.296 |
| Formaldehyde solution (PFA) | Sigma | Cat# F8775-25ml |
| MS-222 | Sigma | Cat# E10621-60G |
| Proteinase K from tritirachium album | Sigma | Cat# P2308-25MG |
| Tris | Sigma | Cat# 252859-100G |
| CellTracker Red CMTPX Dye | Thermofisher | Cat# C34552 |
| Neg-50 Frozen Section Medium | VWR | Cat# 6502 |
| Critical commercial assays | ||
| multiplex HCRv3 reagents | Molecular Instruments | https://www.molecularinstruments.com/ |
| Deposited data | ||
| Single cell RNA data | This paper | GEO: GSE161834 |
| Raw and analyzed data | This paper | Mendeley Data: https://doi.org/10.17632/ygy3pnbnxn.1 |
| Experimental models: Organisms/strains | ||
| T2BGSZ10 Gt(Foxj1b:GFP) | Meng lab, Tsinghua University (Tian et al., 2009) | ZFIN: ZDB-ALT-110301-1 |
| Gmnc mutant (gmncsq34) | Roy lab, A∗STAR (Zhou et al., 2015) | ZFIN: ZDB-ALT-160901-7 |
| Foxj1b mutant (foxj1bsq5719) | Roy lab, A∗STAR | This study |
| Foxj1a mutant (foxj1anw3) | Yaksi lab, NTNU (Olstad et al., 2019) | ZFIN: ZDB-ALT-190620-14 |
| Foxj1a mutant (foxj1asq5717) | Roy lab, A∗STAR (Zhang et al., 2018) | ZFIN: ZDB-ALT-190529-1 |
| Tg(foxj1a:gfp)BACvcc41 | Kikuchi lab, NCVC | This study |
| Tg(Hs:foxj1a)sq5713Tg | Roy lab, A∗STAR (Yu et al., 2008) | ZFIN: ZDB-ALT-141111-1 |
| Tg(her4.1:GFP) | Yeo et al., 2007 | ZFIN: ZDB-ALT-070612-3 |
| Nacre mitfab692 | Lister et al., 1999 | ZFIN:ZDB-ALT-010919-2 |
| Oligonucleotides | ||
| Gmnc genotyping crispr deletion Forward TTGTGATTGTCTCATGCTGTTG | IDT | N/A |
| Gmnc genotyping crispr deletion Reverse AAAAATTCCAGTTTGTCAAGGC | IDT | N/A |
| Foxj1b genotyping crispr deletion Forward TCTTCAGACCAGCAAAGACAGT | IDT | N/A |
| Foxj1b genotyping crispr WT allele Reverse CTCCATCCTCAGTGCCAACC | IDT | N/A |
| Foxj1b genotyping crispr deletion Mutant allele Reverse CGGC TCTGCGTATCTGTAGT |
IDT | N/A |
| foxj1anw3 genotyping Forward GCTGGTCAGGCTGTCGTCTAAA | IDT | N/A |
| foxj1anw3 genotyping Reverse CGCTATCGAGGAAGGACAGGATTT | IDT | N/A |
| foxj1bsq5719 genotyping Forward GAGCTAACCAAATCA ATCGGGG | IDT | N/A |
| foxj1bsq5719 genotyping WT allele Reverse CGTAAGTTCGCGCAGGATTG | IDT | N/A |
| foxj1bsq5719 genotyping Mutant allele Reverse ATCCAGTAGAGCGTCCCA GT | IDT | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | Schindelin et al., 2012 | |
| CiliaQ | Wachten lab, University of Bonn | Hansen et al., 2020 |
| Other | ||
| Pressure injector | Eppendorf | Femtojet 4i |
| Confocal microscope | Zeiss | Examiner Z1; Olympus Fluoview |
| Optical Coherence Tomography | Thorlabs | Telesto 1300nm SD-OCT and LK-4 objective |
| Step One Real Time PCR system | Thermofisher | Cat# 4376357 |
| Cryostat | Thermo Scientific | CryoStar NX70 |
| Molecular Cartography | Resolve Biosciences | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nathalie Jurisch-Yaksi (nathalie.jurisch-yaksi@ntnu.no).
Materials availability
Transgenic and mutant fish lines generated in this paper will be shared by their creators (S.R: foxj1b mutant, S.P.H, K.K: Tg(foxj1a:GFP)BAC) upon request and completion of a material transfer agreement.
Experimental model and subject details
Zebrafish maintenance and strains
The animal facilities and maintenance of the zebrafish, Danio rerio, were approved by the NFSA (Norwegian Food Safety Authority) and the Singapore National Advisory Committee on Laboratory Animal Research. All the procedures were performed on zebrafish larvae of different developmental stages post fertilization in accordance with the European Communities Council Directive, the Norwegian Food Safety Authorities and the Singapore National Advisory Committee on Laboratory Animal Research. Embryonic, larval and adult zebrafish were reared according to standard procedures of husbandry at 28.5°C, unless mentioned otherwise. For our experiments, the following fish lines were used: T2BGSZ10 Gt(foxj1b:GFP) (Tian et al., 2009), gmnc mutant (Zhou et al., 2015), foxj1a mutants (two different alleles generated in two different laboratories were used; Olstad et al., 2019 and Zhang et al., 2018), Tg(hs:foxj1a) (Yu et al., 2008), foxj1b mutant and Tg(foxj1a:gfp)BAC (see below).
Animals were analyzed irrespective of their gender. Note that for zebrafish younger than 2-3 months, gender is not yet apparent. All animals from 1 month of age were selected according to their body size, which is reported in the manuscript, to ensure reproducibility of the results. Mutants were obtained either from heterozygous incross, heterozygous crossed with homozygous, or homozygous incross. We did not observe an impact of the parents’ genotype on the phenotype of the progeny. Controls were siblings with a control genotype (heterozygous or wild-type). For homozygous incross, controls were the progeny of a cross of wild-type siblings of the homozygous parents. Animals were genotyped prior to the experiments and their genotype was re-confirmed following the experiments. Animals were either in the AB or pigmentless mitfa−/− background.
CRISPR/Cas9 mediated mutation of foxj1b
For generating a mutation in the foxj1b gene, we used two gRNAs targeting regions within exon 2 and exon 3. The gRNA sequences are as follows: Exon 2 5′-AAGCCGGACACTACATAACG-3′ and exon 3 5′-GGTCCACCCGCAATATTACG-3′. This resulted in a large deletion of 804 nucleotides, as well as addition of 4 nucleotides at the 5′ gRNA target site in exon 2. As a consequence, the mutant Foxj1b protein is expected to be highly truncated and consist of only 140 amino acids (bereft of the forkhead as well as the transactivation domains), of which 120 retains identity with wild-type Foxj1b, and is likely to be completely non-functional.
Generation of Tg (foxj1a:gfp)BACvcc41 transgenic strain
The TgBAC(foxj1a:EGFP)vcc41 construct was generated by inserting the EGFP expression cassette into the CH211-151D17, after the foxj1a translational start codon, using Red/ET recombineering (GeneBridges, Heidelberg, Germany). The final construct was purified using the BACMAX DNA Purification kit (Epicenter, Madison, WI, USA), linearized with SfiI, and injected into the single cell-stage zebrafish eggs to establish stable transgenics.
Method details
Genotyping
For genotyping, the samples were subjected to gDNA isolation using 100 μl PCR lysis buffer (containing 10 mM tris pH7.5, 50 mM EDTA, 0.2% Triton X-100 and 0.1mg/ml Proteinase K) overnight at 50°C. To stop the reaction the samples were heated to 95°C for 10 minutes. The samples were then centrifuged at 13000 rpm for 2 minutes. The supernatant containing gDNA was used for qpcr. For performing qpcr, 5 μl SYBR green pcr master mix (Thermofisher) was mixed with 0.5 μl each of forward and reverse primer and 4 μl water to make a 10 μl reaction mixture. This reaction was added to a 96well qpcr plate (Thermofisher) and 2 μl of extracted gDNA were mixed with this reaction. The samples were then analyzed based on their melting curves as wt, het or homozygous or by gel electrophoresis.
Ectopic expression of foxj1a by heat shock
Tg(Hs:foxj1a) embryos at ∼18 hpf were heat-shocked at 37°C in water bath for one hour. The heat-shocked embryos were further incubated at 28°C until 24 hpf, before proceeded for fixation and antibody staining. Controls included both non-heat shocked transgenic as well as heat shocked non-transgenic wild-type.
Antibody staining and confocal imaging
Immunostaining of the brain
Larvae or juvenile fishes were first euthanized and then fixed in a solution containing 4% paraformaldehyde solution (PFA), 1% DMSO and 0.3% Triton X-100 in PBS (0.3% PBSTx) for at least 2 h at room temperature or 4°C overnight. Stainings were performed on cut heads to improve the penetration of the antibodies. Dissected adult brains were fixed in 4%PFA in PBS. For 1-month old juveniles, the skin above the brain was dissected out for better penetration of the antibodies. The samples were washed with 0.3% PBSTx after fixing to remove any traces of the fixing solution. For permeabilization, samples were incubated for 10 min (larvae) or 1h (juvenile/adult) at −20°C (larvae) with acetone. Subsequently, samples were washed with 0.3% PBSTx (3x10 min) and blocked in 0.1% BSA/0.3% PBSTx for 2 h (larvae) or 4 h (adult) at room temperature. Samples were incubated with glutamylated tubulin (GT335, 1:400, Adipogen) for staining cilia and beta-catenin antibody (1:400, 9562 Cell Signaling antibodies) overnight at 4°C. On the second day, samples were washed (0.3% PBSTx, 3x1 h) and subsequently incubated with the secondary antibody (Alexa-labeled GAM488 plus, and GAM555 plus Thermo Scientific, 1:1,000) and 0.1% DAPI overnight at 4°C. Anti-gfp tagged polyclonal antibody coupled with Alexa fluor 488 (Thermo Scientific) was used to enhance the GFP signal if needed. On the third day after incubation with the secondary antibody the larvae were washed (0.3% PBSTx, 3x1 h) and transferred to a series of increasing glycerol concentrations (25%, 50% and 75% made in PBS). After staining the larvae were stored in 75% glycerol at 4°C and imaged using a Zeiss Examiner Z1 confocal microscope with a 20x plan NA 0.8 objective.
Immunostaining on foxj1a overexpressed embryos
The following primary antibodies were used: rabbit anti-acetylated tubulin (1:500, Cell Signaling), mouse anti-glutamylated tubulin (1:500, Adipogen). Primary antibodies were diluted in PBDT (1% (w/v) BSA, 1% DMSO, 0.5% Triton X-100, PBS base), and incubated with the embryos at 4°C for overnight. After extensive washes with PBDT, the embryos were then incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen, 1:500) at 4°C for overnight. DAPI staining were subsequently performed at room temperature for 30 min, and the stained embryos were thoroughly washed with PBDT, before mounted in 70% glycerol. Confocal imaging was performed with Olympus Fluoview Upright Confocal Microscope, with a 100x plan NA 1.45 objective.
Hybridization chain reaction (HCR)
Samples were fixed in 2 ml of 4% PFA for 24 h at 4 ◦C. The next day embryos/larvae were washed 3 × 5 min with 1 ml of 1 × Dulbeccos phosphate-buffered saline (dPBS) to stop the fixation. For 4, 14 dpf and dissected adult brains, the samples were directly transferred to chilled methanol for overnight incubation at −20°C. For 1-month old juveniles, the skin above the brain was dissected out for better penetration of HCR probes. Samples were then transferred to methanol at −20°C. The next day, the samples were rehydrated 5 mins each with a series of methanol washes (75%, 50% and 25% methanol made in dPBS), and then washed for 5 minutes with PBST (0.1% tween-20 made in dPBS). The samples were then treated with 1 ml of proteinase K (30 μg/ml) for 45 min at room temperature. After this treatment, the samples were quickly washed with two 1ml washes of PBST without incubation. After this wash, the samples were fixed with 4% PFA for 20 minutes at RT. The samples were then washed 5 × 5 min with 1 ml of PBST.
Around 10-15 embryos/larvae, 3-4 one-month juvenile, or single adult brain were transferred to a 1.5ml tube respectively. The animals were then pre-hybridized with 500 μl of probe hybridization buffer for 30 min at 37 ◦C. The probes were prepared by adding 2 pmol of each probe set to 500 μl of probe hybridization buffer at 37 ◦C. The pre-hybridization solution was discarded, and the probe solution was added to the tubes and incubated overnight (12–16 h) at 37 ◦C. The next day the embryos/larvae were washed 4 × 15 min with 500 μl of probe wash buffer at 37 ◦C. The embryos/larvae were then washed 2 × 5 min with 5 × SSCT at room temperature.
The samples were then treated with 500 μl of amplification buffer for 30 min at room temperature. The amplifiers were prepared separately by using 30 pmol of hairpin h1 and 30 pmol of hairpin h2 by snap cooling 10 μl of 3 μM stock (heat at 95 ◦C for 90 s and cool to room temperature in a dark drawer for 30 min). The HCR hairpin solution was prepared by adding snap-cooled h1 hairpins and snap-cooled h2 hairpins to 500 μl of amplification buffer at room temperature. The pre-amplification solution was discarded and the samples incubated with hairpin solution for 12–16 h in the dark at room temperature. The next day the samples were washed with 500 μl of 5 × SSCT at room temperature for 2 × 5 min, 2 × 30 mins and 1 × 5 mins respectively. The samples were then washed 3 × 5 min with dPBS, after which they were treated with a series of increasing concentrations of glycerol (25, 50 and 75%) glycerol made in dPBS. After staining the larvae were stored in 75% glycerol at 4°C and imaged using a Zeiss Examiner Z1 confocal microscope with a 20x plan NA 0.8 objective.
Molecular cartography
Sample fixation
∼6 months old zebrafish were euthanized in cold water chilled on ice prior to dissecting the whole brain in aCSF. Brains were fixed and stabilized according to Resolve guidelines. Briefly, tissue specimens were incubated at room temperature in PAXgene® fixation solution for 4 hours. Next, PAXgene® fixation solution was replaced with PAXgene® tissue stabilizer at room temperature for approximately 48 hours. Tissue was then transferred into a suitable vial filled with 30% Sucrose (w/v in 1X PBS) solution. For cryoprotection the tissue specimens were incubated 24 hr at 4°C until the samples sink to the bottom of the vial.
Cryo-embedding and cryo-sectioning
A small amount of optimal cutting temperature (OCT) mounting medium (Neg-50 Frozen Section Medium, VWR) was poured into a Peel-A-Way mold to form a uniform layer across the entire bottom of the mold. Tissue specimens were removed from the sucrose solution and dabbed it on an absorbent sheet to remove excess solution. Tissue specimen was placed into the middle of the cryo-embedding medium. Using forceps, the cryomold was placed on the top of the liquid nitrogen in the gas phase until the medium was completely white. The cryo-embedded block was then kept in dry ice until sectioning. The cryo-embedded block was removed from the Peel-A-Way mold and cut to a small cube and placed on a room temperature specimen disk using a small amount of OCT mounting medium. Together, the block and disk were placed inside the cooled cryostat (CryoStar NX70, Thermo Scientific) at −20°C so that the OCT could freeze, adhering the embedded brain to the specimen disk. Coronal sections were collected by positioning the embedded brain anterior side up. Initially, 40 μm sections were taken to trim the block until the region of interest. Once the region of interest was reached, 10 μm serial sections were collected by mounting onto a precooled coverslip. After all sections were collected, the samples were sent to Resolve GmbH, Germany for their analysis.
Molecular cartography
Paxgene Fixed samples were used for Molecular Cartography according to the manufacturer’s instructions (protocol 3.0; available for download from Resolve’s website to registered users), starting with the aspiration of ethanol and the addition of buffer BST1 (step 6 and 7 of the tissue priming protocol). Briefly, tissues were primed followed by overnight hybridization of all probes specific for the target genes (see below for probe design details and target list). Samples were washed the next day to remove excess probes and fluorescently tagged in a two-step color development process. Regions of interest were imaged as described below and fluorescent signals removed during decolorization. Color development, imaging and decolorization were repeated for multiple cycles to build a unique combinatorial code for every target gene. The experimental process through which the combinatorial code is decoded from the raw images is described below.
Probe design
The probes were designed using Resolve’s proprietary design algorithm. The probe-design was performed at the gene-level. For every targeted gene all full-length protein-coding transcript sequences from the ENSEMBL database were used as design targets if the isoform had the GENCODE annotation tag ‘basic’ (Frankish et al., 2019; Yates et al., 2020). To speed up the process, the calculation of computationally expensive parts - especially the off-target searches, the selection of probe sequences - was not performed randomly but limited to sequences with high success rates. To filter highly repetitive regions, the abundance of k-mers was obtained from the background transcriptome using Jellyfish (Marçais and Kingsford, 2011). Every target sequence was scanned once for all k-mers, and those regions with rare k-mers were preferred as seeds for full probe design. A probe candidate was generated by extending a seed sequence until a certain target stability was reached. A set of simple rules was applied to discard sequences that were found experimentally to cause problems. After these fast screens, every kept probe candidate was mapped to the background transcriptome using ThermonucleotideBLAST (Gans and Wolinsky, 2008) and probes with stable off-target hits were discarded. Specific probes were then scored based on the number of on-target matches (isoforms), which were weighted by their associated APPRIS level (Rodriguez et al., 2018), favoring principal isoforms over others. A bonus was added if the binding-site was inside the protein-coding region. From the pool of accepted probes, the final set was composed by greedily picking the highest scoring probes.
Imaging
Samples were imaged on a Zeiss Celldiscoverer 7, using the 50x Plan Apochromat water immersion objective with an NA of 1.2 and the 0.5x magnification changer, resulting in a 25x final magnification. Standard CD7 LED excitation light source, filters, and dichroic mirrors were used together with customized emission filters optimized for detecting specific signals. Excitation time per image was 1000 ms for each channel. For DAPI, we used excitation time of 20 ms per image. A z stack was taken at each region with a distance per z-slice according to the Nyquist-Shannon sampling theorem. The custom CD7 CMOS camera (Zeiss Axiocam Mono 712, 3.45 μm pixel size) was used.
For each region, a z stack per fluorescent color (two colors) was imaged per imaging round. A total of eight imaging rounds were done for each position, resulting in 16 z stacks per region. The completely automated imaging process per round (including water immersion generation and precise relocation of regions to image in all three dimensions) was achieved through a custom python script using the scripting API of the Zeiss ZEN software (Open application development).
Spot segmentation
The algorithms for spot segmentation were written in Java and are based on the ImageJ library functionalities. Only the iterative closest point algorithm is written in C++ based on the libpointmatcher library (https://github.com/ethz-asl/libpointmatcher).
Preprocessing
First, all images were corrected for background fluorescence. A target value for the allowed number of maxima was determined based upon the area of the slice in μm2 multiplied by the factor 0.5. This factor was empirically optimized. The brightest maxima per plane were determined, based upon an empirically optimized threshold. The number and location of the respective maxima was stored. This procedure was done for every image slice independently. Maxima that did not have a neighboring maximum in an adjacent slice (called z-group) were excluded. The resulting maxima list was further filtered in an iterative loop by adjusting the allowed thresholds for (Babs-Bback) and (Bperi-Bback) to reach a feature target value (Babs: absolute brightness, Bback: local background, Bperi: background of periphery within 1 pixel). This feature target values were based upon the volume of the 3D-image. Only maxima still in a z-group of at least 2 after filtering were passing the filter step. Each z-group was counted as one hit. The members of the z-groups with the highest absolute brightness were used as features and written to a file. They resemble a 3D-point cloud.
Final signal segmentation and decoding
To align the raw data images from different imaging rounds, images had to be corrected. To do so the extracted feature point clouds were used to find the transformation matrices. For this purpose, an iterative closest point cloud algorithm was used to minimize the error between two point-clouds. The point clouds of each round were aligned to the point cloud of round one (reference point cloud). The corresponding point clouds were stored for downstream processes. Based upon the transformation matrices the corresponding images were processed by a rigid transformation using trilinear interpolation.
The aligned images were used to create a profile for each pixel consisting of 16 values (16 images from two color channels in 8 imaging rounds). The pixel profiles were filtered for variance from zero normalized by total brightness of all pixels in the profile. Matched pixel profiles with the highest score were assigned as an ID to the pixel.
Pixels with neighbors having the same ID were grouped. The pixel groups were filtered by group size, number of direct adjacent pixels in group, number of dimensions with size of two pixels. The local 3D-maxima of the groups were determined as potential final transcript locations. Maxima were filtered by number of maxima in the raw data images where a maximum was expected. Remaining maxima were further evaluated by the fit to the corresponding code. The remaining maxima were written to the results file and considered to resemble transcripts of the corresponding gene. The ratio of signals matching to codes used in the experiment and signals matching to codes not used in the experiment were used as estimation for specificity (false positives).
Data analysis
The location of single spots were uploaded in ImageJ using the Polylux plugin (provided by Resolve) and merged to the DAPI channel.
Brain ventricle injections and imaging
Prior to injections, all larvae/juveniles were anaesthetized in 0.01% MS-222 in AFW. Injections were done on larvae/juvenile embedded in 2% low-melting point agarose in AFW and 0.01% MS-222 in AFW. For animals older than 2 months, animals were first euthanized by hypothermia. The brain was then dissected in artificial cerebrospinal fluid (aCSF) and mounted on sylgard using a metal pin (Kermen et al., 2020a, 2020b). The aCSF was composed of the following chemicals diluted in reverse osmosis–purified water: 131 mM NaCl, 2 mM KCl, 1.25 mM KH2PO4, 2 mM MgSO47H2O, 10 mM glucose, 2.5 mM CaCl2, and 20 mM NaHCO3. The injection mixture contained 70 kDa rhodamine B isothiocyanate-dextran (RITC-dextran; Sigma-Aldrich, R9379) dissolved in aCSF at a final concentration of 10 mg/ml. The needles used for the injections were pulled with a Sutter Instrument Co. Model P-2000, from thin-walled glass capillaries (1.00 mm; VWR®), using the following settings: heat = 785, filament = 4, velocity = 40, delay = 220, pull = 70. The needle tip was cut open with forceps. A pressure injector (Eppendorf Femtojet 4i) was used to inject between 1 (larvae) −15 (adult) nl of solution (depending on the developmental stage) in the telencephalic ventricle. The pressure and time used for the injection were calibrated for each needle using a 0.01 mm calibration slide for microscopy. After injection, the larvae, juvenile or brain explant were immediately transferred to the confocal microscope (Zeiss Examiner Z1), and imaged with a 20x water-immersion objective (Zeiss, NA 1.0, Plan-Apochromat) at room temperature. In order to avoid bodily movement during the recordings, larvae/juvenile were maintained in AFW containing 0.01% MS-222. The laser power was corrected according to the depth of imaging. Multiple images were acquired per sample and stitch using the 3D stitching plugin in Fiji/ImageJ. Prior to 3D reconstruction, the 3D stacks were Gaussian blurred and reduced in size. 3D reconstructions were done using the 3D viewer plugin in Fiji/ImageJ (threshold levels were adjusted for each sample), exported as stl file, and projected in 2D in Photoshop.
Image processing
To generate whole brain images shown in Figure 1 and 3, confocal stacks were stitched in Fiji/ImageJ using the deprecated 3D stitching plugin (Preibisch et al., 2009). For Figure 1A2, adjacent maximum projected stacks were aligned manually in Photoshop. To avoid a pixelated appearance of zoom-in inset, the pixel resolution of the inset was increased in Fiji/ImageJ by doubling the pixel number. The colors for Figures S2D and S2E were modified in Photoshop. For all panels, maximum projections are shown unless described differently.
Scanning electron microscopy
For gmnc and foxj1b mutants, 6 sets of WT and mutant zebrafish brain samples (dissected midline along the sagittal plane) were fixed by immersion in 4% Formaldehyde and 2% Glutaraldehyde in 0.1M cacodylate buffer, post-fixed with 1% osmium tetroxide in distilled water and dehydrated in ethanol series up to 100% Ethanol. Dehydrated samples were dried using critical point drying (Leica EM CPD030), mounted onto aluminum stubs with double sided carbon tape and sputter coated with 4 nm layer of platinum (Leica EM SCD050). Scanning electron microscopy analysis was performed using JSM 6701F SEM (JEOL) operating at 5kV.
Recording of ciliary beating
To measure ciliary beating in adult brain, we performed light transmission microscopy on a brain explant perfused with aCSF bubbled with carbogen (95% O2/5% CO2). Recordings were obtained using a Manta camera and a custom designed software as previously described (Reiten et al., 2017). Recordings were obtained on an upright Olympus microscope with a 40x water immersion objective at circa 100Hz frame rate for 30 s. The resolution with the 40x objective was 1pixel = 0.314 μm. Multiple recordings were performed per brain explant along the dorsal telencephalon or along the telencephalic midline.
Recording of fluid flow
To measure the directionality of fluid flow, we injected 1 μm beads in the telencephalic ventricle of a brain explant as described above. The injection mixture contained fluorescent beads (SPHERO Fluorescent Yellow Particles 1% w/v, F = 1 mm) diluted in 1/6 in RITC-dextran containing aCSF. Fluid flow was measured using an epifluorescence microscope, a 10x water immersion objective and an ocular camera using an acquisition speed of 24-50 frames per second for at least 1 minute. The resolution with the 10x objective was 1pixel = 1.13 μm.
Single-cell sequencing, reads alignment and analyses
2 female and 1 male 12 months old fish were injected with cell tracking dye (CMPTx, Cat# C34552; Invitrogen) (Bhattarai et al., 2016). 5-6 min post-injection, fish were euthanized and telencephalon dissected followed by dissociation and Fluorescent Associated Cell sorted (Cosacak et al., 2019, 2020).
To prepare the cells for droplet-based sequencing, around 15000 cells were flow-sorted into a Bovine serum albumin (BSA) coated tubes containing 2 μl saline solution with 0.04% BSA. Subsequently, the single cell suspension was carefully mixed with reverse transcription mix before loading the cells on the 10 X Genomics Chromium systems. During the encapsulation, the released polyadenylated RNA bound to the barcoded bead, which was encapsulated with the cell. Following the guidelines of the 10x Genomics user manual, the droplets were directly subjected to reverse transcription, the emulsion was broken and cDNA was purified using Dyna beads MyOne Silane beads. After the amplification of cDNA with 10 cycles, purification and quantification was performed. Half of the material of the cDNA was fragmented for five minutes and dA-tailed, followed by adaptor ligation step and an indexing PCR of 10 cycles in order to generate libraries. After quantification, the libraries were sequenced on an Illumina NovaSeq machine using an S1 flowcell in PE mode (R1: 29 cycles; I1: 8 cycles; R2: 93 cycles).
The raw sequencing data was then processed with the ‘count’ command of the Cell Ranger software (v3.1.0) provided by 10X Genomics. The option of ‘–expect-cells’ was set to 5,500 (all other options were used as per default). To build the reference for Cell Ranger, zebrafish genome (GRCz11) as well as gene annotation (Ensembl 95) were downloaded from Ensembl and the annotation was filtered with the ‘mkgtf’ command of Cell Ranger (options: ‘–attribute = gene_biotype:protein_coding–attribute = gene_biotype:lincRNA –attribute = gene_biotype:antisense’). Genome sequence and filtered annotation were then used as input to the ‘mkref’ command of Cell Ranger to build the appropriate CellRanger Reference.
In order to analyze the single-cell data, in total 3808 cells recovered after filtering by cell ranger. Then, outlier cells with 5-fold difference between total number of reads (nCount_RNA) and total number of features (nFeature_RNA) were removed from analysis. We further removed cells; with less than 500 genes, with more than 6% mitochondrial RNA genes. The remaining 3158 cells were used for downstream analysis with Seurat (3.2.0). We performed 3 iterative clustering; (i) clustering of all cells to identify the main cell types as done previously (Cosacak et al., 2019), (ii) the clustering of Progenitor Cells (PC) to identify subtype of PC cells and (iii) the clustering of Foxj1 positive (foxj1a and/or foxj1b) cells.
All cells were clustered by scaling to 10e4 and using top 2000 variable genes. nCount_RNA and nFeature_RNA were regressed out and dims = 50 and resolution = 1 were used to identify clusters. The following markers were used to identify main cell clusters; Progenitor Cells (PC) (fabp7a), Neurons (NN) (sv2a), Immune Cells (Im) (lcp1, wasb, pfn1), Oligodendrocytes (OPC) (olig1/2, aplnra/b), Pineal Gland (PG) (exorh), DKK (dkk3b), LYV (lyve1) and unassigned clusters (14 and 21; named as CL14 and CL21, respectively).
The PC cells were further clustered as above parameters; except with dims = 30. Foxj1 cells were similarly clustered, except dims = 30 and resolution = 0.6. Additionally, PC cells and DKK cells were clustered together, as DKK cells contains foxj1a/b and gmnc genes.
We use monocle2 (Qiu et al., 2017) to perform cell trajectory for all clustering of PC and foxj1 cells as described previously (Cosacak et al., 2019). In brief, we use the scaled data and variable genes from Seurat and set SizeFactor 1.0 to prevent any further normalization.
The raw data for single-cell sequencing has been deposited on Gene Expression Omnibus under the following accession number: GSE161834
OCT imaging
Adult zebrafish of various genotype were anaesthetized with 0.01% MS222 in bubbled and pH adjusted (using bicarbonate buffer) artificial fish water. Following loss of balance, anaesthetized animals were placed in a plastic container and immobilized using wet kim wipes. Animals were imaged using a Telesto OCT (Thorlabs, laser 1300nm) with the LK04 objective (resolution of 20μm). Following image acquisition, animals were euthanized and the brain explanted prior to be re-imaged in the OCT.
Brains of 3 additional foxj1b;gmnc double mutants were dissected and inspected using a stereomicroscope. None of these animals showed major brain malformations or size differences. These quantifications were not included in Figure 7 because photos were made in fixed samples using a stereomicroscope.
Quantification and statistical analysis
CiliaQ analysis
Confocal images were analyzed using the CiliaQ workflow (Hansen et al., 2020; Hansen et al., 2021). First, images were pre-processed and segmented with CiliaQ Preparator (Settings: Gaussian Blur with sigma 1.0, Hysteresis threshold with low threshold delivered by algorithm Huang and high threshold delivered by algorithm Triangle, Threshold determined in a maximum-intensity-projection). Second, images were edited with CiliaQ Editor by a trained observer: incompletely segmented cilia were connected manually, multiple adjacent cilia connected to one object were separated, long motile cilia at the pronephros were removed from the analysis. Third, images were analyzed with CiliaQ to quantify the cilia in the images (settings: minimum cilium size 25 voxel, cilia touching X Y or Z borders excluded, Gauss XY sigma 2.0 voxel, Gauss Z sigma 0.0 voxel). Because pixel size differed between images (either 0.124 μm or 0.079 μm, stack interval in all images: 0.4 μm), profiles from images with higher pixel size were scaled to the lower pixel size by linear interpolation (using the R function approxfun()). Next, Ciliary glutamylated-Tubulin intensity profiles were aligned with a custom-written algorithm in R. The integrated intensity of the first third and the last third of the CiliaQ-derived intensity profiles was compared. If the integrated intensity of the last third was higher than the integrated intensity of the first third, the profile was reversed. Graphical representations and statistical analysis were done using GraphPad. The type of statistical test performed depended on the results from Normality and Lognormality tests (Shapiro-Wilk, Kolmogorov-Smirnov) were passed is indicated in the respective figure legend
Quantification of ciliary beating
We aligned all light-transmission brain ventricle recordings using a custom-written alignment algorithm (Reiten et al., 2017), adapted from Bergen et al. (1992), which corrects for occasional x-y drifts. We assumed that beating cilia are manifested in the acquired recordings as periodic changes in pixel intensity. To extract those periodic changes, we performed the MATLAB fast Fourier Transform algorithm for every pixel in the recording. From the resulting power spectra, we determined the primary frequency as the frequency of the highest peak between a lower frequency cut off (see table) and half the frequency of acquisition. Those primary frequencies were then rendered in the original pixel locations to form a frequency map. Beating cilia do not cover the entire recording and should span a minimum number of pixels. To segment signal and noise regions, we used a custom standard deviation (SD) thresholding-algorithm. In brief, we moved a 3x3 kernel across the entire frequency map and any pixel belonging to a 3x3 kernel whose SD was below 2 (see table) was considered as signal, whereas pixels belonging only to kernels with an SD above 2 were considered as noise. To remove small signal islands, we listed all connected pixels in the frequency map using the bwconncomp MATLAB function, and removed those islands that were smaller than 500 pixels (see table). Altogether, with SD-thresholding algorithm and by removing small signal islands, we were able to restrict the analysis only to those regions with beating cilia.
Since we recorded from opaque explanted tissue, we encountered several additional sources of noise that would hinder frequency detection: not all movement could be resolved by alignment; wandering tissue particles occasionally interfered with the ciliary signal; and the opaque tissue limits the detection of a ciliary signal such that that its peak frequency is overshadowed by a power spectral low frequency shoulder, reminiscent of pink noise (Ward and Greenwood, 2007).
To ensure the origin of a signal, we interpreted the raw recordings. All ciliary signals should be constituted by visible ciliary beating. When a signal did not meet this requirement, we attempted to remove it by either pre-processing the recording with a high-pass filter – designed using designfilt and applied with filtfilt in MATLAB (see table) - or by manually masking a spatially separated noisy signal using a custom-written script in MATLAB. Occasionally, we detected signals from two distinct brain regions in one recording. Here, we segmented the recording as well. Recordings without a ciliary signal or with remaining noise were excluded from the analysis.
For a given recording, we collected the primary frequencies in a histogram and normalized its count with the number of signal pixels. We first averaged all histograms for a given brain region and fish, and then across fish to obtain one histogram per brain region: tela choroida, choroid plexus, and midline.
Settings
| Tela Choroida | Midline | Choroid Plexus | |
|---|---|---|---|
| Lower frequency cut off | 5.5 [Hz] | 10 [Hz] | 10 [Hz] |
| Standard deviation | 2 | 2 | 2 |
| Spatial resolution | 0.31445 [μm / pixel] | 0.31445 [μm / pixel] | 0.31445 [μm / pixel] |
| Minimum number of pixels | 500 | 500 | 500 |
Filter specifications
| Dura | Midline | Choroid Plexus | |
|---|---|---|---|
| Type | highpassiir | highpassiir | highpassiir |
| StopbandFrequency | 2 | 4 | 4 |
| PassbandFrequency | 5 | 10 | 10 |
| StopbandAttenuation | 2 | 2 | 2 |
| PassbandRipple | 1 | 1 | 1 |
| Sample Rate∗ | ∼100Hz | ∼100Hz | ∼100Hz |
| DesignMethod | butter | butter | butter |
| MatchExactly | passband | passband | passband |
∗ varies slightly per recording
To produce the Figures 2B1, 2C1, and 2D1, transmission microscopy and ciliary beating heatmaps images were generated by MATLAB in png format from individual recordings. The transmission microscopy images were first aligned manually in Photoshop using overlapping information from neighboring images. Then the ciliary beating frequency map were aligned to the transmission microscopy images and their background was removed using blending options in Photoshop.
Quantification of fluid flow direction
Injected fluorescent particles were tracked using the trackmate plugin in Fiji/ImageJ (Tinevez et al., 2017) using the LoG detector for 8-10 pixel blobs and a threshold of 10-50 depending on the recording. Other parameters were initial search radius: 15,0, max search radius: 15,0, max frame gap: 2. The tracks were then imported in MATLAB. Only tracks containing more than 20 data points and covering a distance longer than 10 pixels were included in the further analysis. The direction between two consecutive data point was then plotted using the surf function in MATLAB.
Quantification of fluorescence intensity on confocal images
To identify the GFP levels in gmnc;foxj1b:GFP brains, stained samples were imaged using the same settings on the confocal microscopy. No correction of laser power was applied. Individual cells were manually segmented using the membrane marker beta-catenin signal on Fiji/ImageJ on individual optical sections. Cells were categorized as mono- or multiciliated cells based on the glutamylated tubulin immunostaining. A total of 40 cells were analyzed per animal. The average intensity in each region of interest was recovered and imported in MATLAB. All datapoints were plotted. Statistical analyses were done on the average value per sample using MATLAB.
Quantification of brain size measured by OCT
Brain size and ventricle area were measured using the polygonal selection tool and the straight-line selection tool on resliced images in Fiji. Linear regressions were calculated in MATLAB using fitlm function.
Statistical analyses and figure assembly
The statistical tests used and description of the numbers of observations are indicated in each figure legend. Figures were assembled using Illustrator.
Acknowledgments
We thank M. Wulliman and A.S. Mathuru for constructive suggestions and critical reading of the manuscript, the Research Support Centre A∗STAR Microscopy Platform for scanning electron microscopy analysis, CMCB Flow Cytometry core for flow cytometry, DRESDEN-concept Genome Center in TU Dresden for single-cell sequencing, and Thorlabs for loaning us an OCT device (Thorlabs 1300 nm SD-OCT system TELESTO). We thank V. Nguyen, A. Nygaard, and the Trondheim fish facility team for their technical support. This work was supported by an NTNU strategy grant (N.J.Y.); RCN FRIPRO grants 314189 (N.J.Y.) and 239973 (E.Y.); ERC starting grant 335561 (E.Y.); Helse Midt-Norge Samarbeidsorganet grant (E.Y. and N.J.Y.); a Boehringer Ingelheim Fonds fellowship (C.R. and J.N.H.); a postdoctoral fellowship from the National Research Foundation (NRF) of Singapore (Q.T.); a Singapore International Graduate Award (SINGA) (D.R.); funds from the Agency for Science, Technology and Research (A∗STAR) Science Award (C.K.A.L.); research funds from the A∗STAR of Singapore (S.R.); and German Center for Neurodegenerative Diseases (DZNE) within Helmholtz Association (C.K.). Work in the Yaksi laboratory is supported by NTNU and Kavli Foundation.
Author contributions
Conceptualization, supervision, project administration, funding acquisition, visualization, manuscript writing, N.J.-Y. and S.R.; supervision, funding acquisition, manuscript writing, E.Y.; methodology, investigation, formal analysis, visualization, P.P.D., T.Q., A.K., D.R, C.R., D.W., E.W.O., Y.L.C., C.K.A.L., and D.R.; CiliaQ analysis, J.N.H. and D.W.; single-cell sequencing, M.I.C. and C.K.; scanning electron microscopy, C.P.N. and D.L.; Molecular Cartography, F.A.H., N.K., A.G., and B.S.N.; generation of the Tg(foxj1a:GFP)BAC line, S.P.H. and K.K.; editing of the manuscript, all authors.
Declaration of interests
N.K., A.G., and B.S.N. are full-time employees of Resolve Biosciences GmbH.
Published: October 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109775.
Contributor Information
Sudipto Roy, Email: sudipto@imcb.a-star.edu.sg.
Nathalie Jurisch-Yaksi, Email: nathalie.jurisch-yaksi@ntnu.no.
Supplemental information
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is GSE161834. Microscopy data reported in this paper are deposited in Mendeley Data https://doi.org/10.17632/ygy3pnbnxn.1. Other microscopy data will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information and code required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aamar E., Dawid I.B. Isolation and expression analysis of foxj1 and foxj1.2 in zebrafish embryos. Int. J. Dev. Biol. 2008;52:985–991. doi: 10.1387/ijdb.072477ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbi M., Pefani D.E., Kyrousi C., Lalioti M.E., Kalogeropoulou A., Papanastasiou A.D., Taraviras S., Lygerou Z. GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep. 2016;17:400–413. doi: 10.15252/embr.201540882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banizs B., Pike M.M., Millican C.L., Ferguson W.B., Komlosi P., Sheetz J., Bell P.D., Schwiebert E.M., Yoder B.K. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 2005;132:5329–5339. doi: 10.1242/dev.02153. [DOI] [PubMed] [Google Scholar]
- Bearce E.A., Grimes D.T. On being the right shape: Roles for motile cilia and cerebrospinal fluid flow in body and spine morphology. Semin. Cell Dev. Biol. 2021;110:104–112. doi: 10.1016/j.semcdb.2020.07.005. [DOI] [PubMed] [Google Scholar]
- Bergen J.R., Anandan P., Hanna K.J., Hingorani R. In: European Conference on Computer Vision. Sandini G., editor. ECCV’92. 1992. Hierarchical model-based motion estimation; pp. 237–252. [Google Scholar]
- Bhattarai P., Thomas A.K., Cosacak M.I., Papadimitriou C., Mashkaryan V., Froc C., Reinhardt S., Kurth T., Dahl A., Zhang Y., Kizil C. IL4/STAT6 Signaling Activates Neural Stem Cell Proliferation and Neurogenesis upon Amyloid-β42 Aggregation in Adult Zebrafish Brain. Cell Rep. 2016;17:941–948. doi: 10.1016/j.celrep.2016.09.075. [DOI] [PubMed] [Google Scholar]
- Bill B.R., Korzh V. Choroid plexus in developmental and evolutionary perspective. Front. Neurosci. 2014;8:363. doi: 10.3389/fnins.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon M., Wallmeier J., Ma L., Loges N.T., Jaspers M., Olbrich H., Dougherty G.W., Raidt J., Werner C., Amirav I. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Commun. 2014;5:4418. doi: 10.1038/ncomms5418. [DOI] [PubMed] [Google Scholar]
- Bosch Grau M., Gonzalez Curto G., Rocha C., Magiera M.M., Marques Sousa P., Giordano T., Spassky N., Janke C. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 2013;202:441–451. doi: 10.1083/jcb.201305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C., Labedan P., Dimidschstein J., Richard F., Cremer H., André P., Yang Y., Montcouquiol M., Goffinet A.M., Tissir F. A dual role for planar cell polarity genes in ciliated cells. Proc. Natl. Acad. Sci. USA. 2014;111:E3129–E3138. doi: 10.1073/pnas.1404988111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody S.L., Yan X.H., Wuerffel M.K., Song S.K., Shapiro S.D. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- Cantaut-Belarif Y., Sternberg J.R., Thouvenin O., Wyart C., Bardet P.L. The Reissner Fiber in the Cerebrospinal Fluid Controls Morphogenesis of the Body Axis. Curr. Biol. 2018;28:2479–2486.e4. doi: 10.1016/j.cub.2018.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.T., Lehtinen M.K., Sive H. Zebrafish cerebrospinal fluid mediates cell survival through a retinoid signaling pathway. Dev. Neurobiol. 2016;76:75–92. doi: 10.1002/dneu.22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S.P., Babu D., Lau D., Yu X., Roy S. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development. 2014;141:3410–3419. doi: 10.1242/dev.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y.L., Zhang Y., Zhou F., Roy S. Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo. Dev. Biol. 2018;443:165–172. doi: 10.1016/j.ydbio.2018.09.013. [DOI] [PubMed] [Google Scholar]
- Cosacak M.I., Bhattarai P., Reinhardt S., Petzold A., Dahl A., Zhang Y., Kizil C. Single-Cell Transcriptomics Analyses of Neural Stem Cell Heterogeneity and Contextual Plasticity in a Zebrafish Brain Model of Amyloid Toxicity. Cell Rep. 2019;27:1307–1318.e3. doi: 10.1016/j.celrep.2019.03.090. [DOI] [PubMed] [Google Scholar]
- Cosacak M.I., Bhattarai P., Kizil C. Alzheimer’s disease, neural stem cells and neurogenesis: cellular phase at single-cell level. Neural Regen. Res. 2020;15:824–827. doi: 10.4103/1673-5374.268896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date P., Ackermann P., Furey C., Fink I.B., Jonas S., Khokha M.K., Kahle K.T., Deniz E. Visualizing flow in an intact CSF network using optical coherence tomography: implications for human congenital hydrocephalus. Sci. Rep. 2019;9:6196. doi: 10.1038/s41598-019-42549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bigio M.R. Ependymal cells: biology and pathology. Acta Neuropathol. 2010;119:55–73. doi: 10.1007/s00401-009-0624-y. [DOI] [PubMed] [Google Scholar]
- Diaz Verdugo C., Myren-Svelstad S., Aydin E., Van Hoeymissen E., Deneubourg C., Vanderhaeghe S., Vancraeynest J., Pelgrims R., Cosacak M.I., Muto A. Glia-neuron interactions underlie state transitions to generalized seizures. Nat. Commun. 2019;10:3830. doi: 10.1038/s41467-019-11739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreosti E., Lopes G., Kampff A.R., Wilson S.W. Development of social behavior in young zebrafish. Front. Neural Circuits. 2015;9:39. doi: 10.3389/fncir.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dur A.H., Tang T., Viviano S., Sekuri A., Willsey H.R., Tagare H.D., Kahle K.T., Deniz E. In Xenopus ependymal cilia drive embryonic CSF circulation and brain development independently of cardiac pulsatile forces. Fluids Barriers CNS. 2020;17:72. doi: 10.1186/s12987-020-00234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame R.M., Lehtinen M.K. Emergence and Developmental Roles of the Cerebrospinal Fluid System. Dev. Cell. 2020;52:261–275. doi: 10.1016/j.devcel.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame R.M., Chang J.T., Hong A., Aponte-Santiago N.A., Sive H. Directional cerebrospinal fluid movement between brain ventricles in larval zebrafish. Fluids Barriers CNS. 2016;13:11. doi: 10.1186/s12987-016-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubel R., Westendorf C., Bodenschatz E., Eichele G. Cilia-based flow network in the brain ventricles. Science. 2016;353:176–178. doi: 10.1126/science.aae0450. [DOI] [PubMed] [Google Scholar]
- Feldner A., Adam M.G., Tetzlaff F., Moll I., Komljenovic D., Sahm F., Bäuerle T., Ishikawa H., Schroten H., Korff T. Loss of Mpdz impairs ependymal cell integrity leading to perinatal-onset hydrocephalus in mice. EMBO Mol. Med. 2017;9:890–905. doi: 10.15252/emmm.201606430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueira M., Bayley P., Navratilova P., Becker T.S., Wilson S.W., Clarke J.D. Morphogenesis underlying the development of the everted teleost telencephalon. Neural Dev. 2012;7:32. doi: 10.1186/1749-8104-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans J.D., Wolinsky M. Improved assay-dependent searching of nucleic acid sequence databases. Nucleic Acids Res. 2008;36:e74. doi: 10.1093/nar/gkn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Lecea M., Kondrychyn I., Fong S.H., Ye Z.R., Korzh V. In vivo analysis of choroid plexus morphogenesis in zebrafish. PLoS ONE. 2008;3:e3090. doi: 10.1371/journal.pone.0003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi-Egea J.F., Strazielle N., Catala M., Silva-Vargas V., Doetsch F., Engelhardt B. Molecular anatomy and functions of the choroidal blood-cerebrospinal fluid barrier in health and disease. Acta Neuropathol. 2018;135:337–361. doi: 10.1007/s00401-018-1807-1. [DOI] [PubMed] [Google Scholar]
- Grimes D.T., Boswell C.W., Morante N.F., Henkelman R.M., Burdine R.D., Ciruna B. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. 2016;352:1341–1344. doi: 10.1126/science.aaf6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.N., Kaiser F., Klausen C., Stüven B., Chong R., Bönigk W., Mick D.U., Möglich A., Jurisch-Yaksi N., Schmidt F.I., Wachten D. Nanobody-directed targeting of optogenetic tools to study signaling in the primary cilium. eLife. 2020;9:e57907. doi: 10.7554/eLife.57907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.N., Rassmann S., Stüven B., Jurisch-YaksiI N., Wachten D. CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E. Soft Matter. 2021;44:18. doi: 10.1140/epje/s10189-021-00031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz R.C., de Polavieja G.G. Ontogeny of collective behavior reveals a simple attraction rule. Proc. Natl. Acad. Sci. USA. 2017;114:2295–2300. doi: 10.1073/pnas.1616926114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez-Tallon I., Pagenstecher A., Fliegauf M., Olbrich H., Kispert A., Ketelsen U.P., North A., Heintz N., Omran H. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 2004;13:2133–2141. doi: 10.1093/hmg/ddh219. [DOI] [PubMed] [Google Scholar]
- Jacquet B.V., Salinas-Mondragon R., Liang H., Therit B., Buie J.D., Dykstra M., Campbell K., Ostrowski L.E., Brody S.L., Ghashghaei H.T. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet B.V., Muthusamy N., Sommerville L.J., Xiao G., Liang H., Zhang Y., Holtzman M.J., Ghashghaei H.T. Specification of a Foxj1-dependent lineage in the forebrain is required for embryonic-to-postnatal transition of neurogenesis in the olfactory bulb. J. Neurosci. 2011;31:9368–9382. doi: 10.1523/JNEUROSCI.0171-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S., Dai W., Lu L., Liu Y., Zhou J., Li Y., Korzh V., Duan C. The conserved clusterin gene is expressed in the developing choroid plexus under the regulation of notch but not IGF signaling in zebrafish. Endocrinology. 2011;152:1860–1871. doi: 10.1210/en.2010-1183. [DOI] [PubMed] [Google Scholar]
- Jiménez A.J., Domínguez-Pinos M.D., Guerra M.M., Fernández-Llebrez P., Pérez-Fígares J.M. Structure and function of the ependymal barrier and diseases associated with ependyma disruption. Tissue Barriers. 2014;2:e28426. doi: 10.4161/tisb.28426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurisch-Yaksi N., Yaksi E., Kizil C. Radial glia in the zebrafish brain: Functional, structural, and physiological comparison with the mammalian glia. Glia. 2020;68:2451–2470. doi: 10.1002/glia.23849. [DOI] [PubMed] [Google Scholar]
- Karimy J.K., Reeves B.C., Damisah E., Duy P.Q., Antwi P., David W., Wang K., Schiff S.J., Limbrick D.D., Jr., Alper S.L. Inflammation in acquired hydrocephalus: pathogenic mechanisms and therapeutic targets. Nat. Rev. Neurol. 2020;16:285–296. doi: 10.1038/s41582-020-0321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F., Darnet L., Wiest C., Palumbo F., Bechert J., Uslu O., Yaksi E. Stimulus-specific behavioral responses of zebrafish to a large range of odors exhibit individual variability. BMC Biol. 2020;18:66. doi: 10.1186/s12915-020-00801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F., Lal P., Faturos N.G., Yaksi E. Interhemispheric connections between olfactory bulbs improve odor detection. PLoS Biol. 2020;18:e3000701. doi: 10.1371/journal.pbio.3000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. IGFBP-2 Signaling in the Brain: From Brain Development to Higher Order Brain Functions. Front. Endocrinol. (Lausanne) 2019;10:822. doi: 10.3389/fendo.2019.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto N., Alfaro-Cervello C., Shimizu K., Asakawa K., Urasaki A., Nonaka S., Kawakami K., Garcia-Verdugo J.M., Sawamoto K. Migration of neuronal precursors from the telencephalic ventricular zone into the olfactory bulb in adult zebrafish. J. Comp. Neurol. 2011;519:3549–3565. doi: 10.1002/cne.22722. [DOI] [PubMed] [Google Scholar]
- Kizil C., Brand M. Cerebroventricular microinjection (CVMI) into adult zebrafish brain is an efficient misexpression method for forebrain ventricular cells. PLoS ONE. 2011;6:e27395. doi: 10.1371/journal.pone.0027395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konjikusic M.J., Yeetong P., Boswell C.W., Lee C., Roberson E.C., Ittiwut R., Suphapeetiporn K., Ciruna B., Gurnett C.A., Wallingford J.B. Mutations in Kinesin family member 6 reveal specific role in ependymal cell ciliogenesis and human neurological development. PLoS Genet. 2018;14:e1007817. doi: 10.1371/journal.pgen.1007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker A.G., Olale F., Haycraft C.J., Yoder B.K., Schier A.F., Drummond I.A. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Kyrousi C., Arbi M., Pilz G.A., Pefani D.E., Lalioti M.E., Ninkovic J., Götz M., Lygerou Z., Taraviras S. Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development. 2015;142:3661–3674. doi: 10.1242/dev.126342. [DOI] [PubMed] [Google Scholar]
- Lalioti M.E., Kaplani K., Lokka G., Georgomanolis T., Kyrousi C., Dong W., Dunbar A., Parlapani E., Damianidou E., Spassky N. GemC1 is a critical switch for neural stem cell generation in the postnatal brain. Glia. 2019;67:2360–2373. doi: 10.1002/glia.23690. [DOI] [PubMed] [Google Scholar]
- Larsch J., Baier H. Biological Motion as an Innate Perceptual Mechanism Driving Social Affiliation. Curr. Biol. 2018;28:3523–3532.e4. doi: 10.1016/j.cub.2018.09.014. [DOI] [PubMed] [Google Scholar]
- Lee L. Riding the wave of ependymal cilia: genetic susceptibility to hydrocephalus in primary ciliary dyskinesia. J. Neurosci. Res. 2013;91:1117–1132. doi: 10.1002/jnr.23238. [DOI] [PubMed] [Google Scholar]
- Lewis M., Stracker T.H. Transcriptional regulation of multiciliated cell differentiation. Semin. Cell Dev. Biol. 2020;110:51–60. doi: 10.1016/j.semcdb.2020.04.007. [DOI] [PubMed] [Google Scholar]
- Li Q., Han Z., Singh N., Terré B., Fame R.M., Arif U., Page T.D., Zahran T., Abdeltawab A., Huang Y. GEMC1-MCIDAS transcriptional program regulates multiciliogenesis in the choroid plexus and acts as a barrier to tumorigenesis. bioRxiv. 2020 2020.11.22.393298. [Google Scholar]
- Lim L., Zhou H., Costa R.H. The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc. Natl. Acad. Sci. USA. 1997;94:3094–3099. doi: 10.1073/pnas.94.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey B.W., Darabie A., Tropepe V. The cellular composition of neurogenic periventricular zones in the adult zebrafish forebrain. J. Comp. Neurol. 2012;520:2275–2316. doi: 10.1002/cne.23065. [DOI] [PubMed] [Google Scholar]
- Lister J.A., Robertson C.P., Lepage T., Johnson S.L., Raible D.W. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Lu H., Anujan P., Zhou F., Zhang Y., Chong Y.L., Bingle C.D., Roy S. Mcidas mutant mice reveal a two-step process for the specification and differentiation of multiciliated cells in mammals. Development. 2019;146:dev172643. doi: 10.1242/dev.172643. [DOI] [PubMed] [Google Scholar]
- Lu H., Shagirova A., Goggi J.L., Yeo H.L., Roy S. Reissner fibre-induced urotensin signalling from cerebrospinal fluid-contacting neurons prevents scoliosis of the vertebrate spine. Biol. Open. 2020;9:bio052027. doi: 10.1242/bio.052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Merkle F.T., Soriano-Navarro M., Garcia-Verdugo J.M., Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Han Y.G., Soriano-Navarro M., García-Verdugo J.M., Alvarez-Buylla A. Cilia organize ependymal planar polarity. J. Neurosci. 2010;30:2600–2610. doi: 10.1523/JNEUROSCI.3744-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita K., Kawate T., Kakinuma N., Takeda S. Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic. 2010;11:287–301. doi: 10.1111/j.1600-0854.2009.01016.x. [DOI] [PubMed] [Google Scholar]
- Narita K., Kozuka-Hata H., Nonami Y., Ao-Kondo H., Suzuki T., Nakamura H., Yamakawa K., Oyama M., Inoue T., Takeda S. Proteomic analysis of multiple primary cilia reveals a novel mode of ciliary development in mammals. Biol. Open. 2012;1:815–825. doi: 10.1242/bio.20121081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk T., Fernandez T.E., Vasioukhin V. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5-/- mice. Dev. Cell. 2007;13:338–350. doi: 10.1016/j.devcel.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuys R. The development and general morphology of the telencephalon of actinopterygian fishes: synopsis, documentation and commentary. Brain Struct. Funct. 2011;215:141–157. doi: 10.1007/s00429-010-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami Y., Narita K., Nakamura H., Inoue T., Takeda S. Developmental changes in ciliary motility on choroid plexus epithelial cells during the perinatal period. Cytoskeleton (Hoboken) 2013;70:797–803. doi: 10.1002/cm.21132. [DOI] [PubMed] [Google Scholar]
- Ogino T., Sawada M., Takase H., Nakai C., Herranz-Pérez V., Cebrián-Silla A., Kaneko N., García-Verdugo J.M., Sawamoto K. Characterization of multiciliated ependymal cells that emerge in the neurogenic niche of the aged zebrafish brain. J. Comp. Neurol. 2016;524:2982–2992. doi: 10.1002/cne.24001. [DOI] [PubMed] [Google Scholar]
- Olstad E.W., Ringers C., Hansen J.N., Wens A., Brandt C., Wachten D., Yaksi E., Jurisch-Yaksi N. Ciliary Beating Compartmentalizes Cerebrospinal Fluid Flow in the Brain and Regulates Ventricular Development. Curr. Biol. 2019;29:229–241.e6. doi: 10.1016/j.cub.2018.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Álvarez G., Daclin M., Shihavuddin A., Lansade P., Fortoul A., Faucourt M., Clavreul S., Lalioti M.E., Taraviras S., Hippenmeyer S. Adult Neural Stem Cells and Multiciliated Ependymal Cells Share a Common Lineage Regulated by the Geminin Family Members. Neuron. 2019;102:159–172.e7. doi: 10.1016/j.neuron.2019.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo F., Serneels B., Pelgrims R., Yaksi E. The Zebrafish Dorsolateral Habenula Is Required for Updating Learned Behaviors. Cell Rep. 2020;32:108054. doi: 10.1016/j.celrep.2020.108054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Obara T., Mangos S., Liu Y., Drummond I.A. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Austin C.A., Drummond I.A. Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J. Biol. Chem. 2011;286:11685–11695. doi: 10.1074/jbc.M110.209817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Austin-Tse C.A., Liu Y., Vasilyev A., Drummond I.A. Cytoplasmic carboxypeptidase 5 regulates tubulin glutamylation and zebrafish cilia formation and function. Mol. Biol. Cell. 2014;25:1836–1844. doi: 10.1091/mbc.E13-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.-A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond S.A., Figueres-Oñate M., Obernier K., Nascimento M.A., Parraguez J.I., López-Mascaraque L., Fuentealba L.C., Alvarez-Buylla A. Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell Rep. 2019;27:429–441.e3. doi: 10.1016/j.celrep.2019.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiten I., Uslu F.E., Fore S., Pelgrims R., Ringers C., Diaz Verdugo C., Hoffman M., Lal P., Kawakami K., Pekkan K. Motile-Cilia-Mediated Flow Improves Sensitivity and Temporal Resolution of Olfactory Computations. Curr. Biol. 2017;27:166–174. doi: 10.1016/j.cub.2016.11.036. [DOI] [PubMed] [Google Scholar]
- Ringers C., Jurisch-Yaksi N. Development: How the Reissner Fiber Keeps Our Back Straight. Curr. Biol. 2020;30:R705–R708. doi: 10.1016/j.cub.2020.04.073. [DOI] [PubMed] [Google Scholar]
- Ringers C., Olstad E.W., Jurisch-Yaksi N. The role of motile cilia in the development and physiology of the nervous system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190156. doi: 10.1098/rstb.2019.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.M., Rodriguez-Rivas J., Di Domenico T., Vázquez J., Valencia A., Tress M.L. APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res. 2018;46(D1):D213–D217. doi: 10.1093/nar/gkx997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C.D., Pompili D., Henke K., Van Gennip J.L.M., Meyer-Miner A., Rana R., Gobron S., Harris M.P., Nitz M., Ciruna B. SCO-Spondin Defects and Neuroinflammation Are Conserved Mechanisms Driving Spinal Deformity across Genetic Models of Idiopathic Scoliosis. Curr. Biol. 2020;30:2363–2373.e6. doi: 10.1016/j.cub.2020.04.020. [DOI] [PubMed] [Google Scholar]
- Sawamoto K., Wichterle H., Gonzalez-Perez O., Cholfin J.A., Yamada M., Spassky N., Murcia N.S., Garcia-Verdugo J.M., Marin O., Rubenstein J.L. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.T., Stratton J.A., Stykel M.G., Abbasi S., Sharma S., Mayr K.A., Koblinger K., Whelan P.J., Biernaskie J. Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell. 2018;173:1045–1057.e9. doi: 10.1016/j.cell.2018.03.063. [DOI] [PubMed] [Google Scholar]
- Sigg M.A., Menchen T., Lee C., Johnson J., Jungnickel M.K., Choksi S.P., Garcia G., 3rd, Busengdal H., Dougherty G.W., Pennekamp P. Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Dev. Cell. 2017;43:744–762.e11. doi: 10.1016/j.devcel.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N., Meunier A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- Spassky N., Merkle F.T., Flames N., Tramontin A.D., García-Verdugo J.M., Alvarez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg J.R., Prendergast A.E., Brosse L., Cantaut-Belarif Y., Thouvenin O., Orts-Del’Immagine A., Castillo L., Djenoune L., Kurisu S., McDearmid J.R. Pkd2l1 is required for mechanoception in cerebrospinal fluid-contacting neurons and maintenance of spine curvature. Nat. Commun. 2018;9:3804. doi: 10.1038/s41467-018-06225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J.L., Oishi I., Izpisúa Belmonte J.C., Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J.L., Vladar E.K., Axelrod J.D., Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terré B., Piergiovanni G., Segura-Bayona S., Gil-Gómez G., Youssef S.A., Attolini C.S., Wilsch-Bräuninger M., Jung C., Rojas A.M., Marjanović M. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016;35:942–960. doi: 10.15252/embj.201592821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenin O., Keiser L., Cantaut-Belarif Y., Carbo-Tano M., Verweij F., Jurisch-Yaksi N., Bardet P.L., van Niel G., Gallaire F., Wyart C. Origin and role of the cerebrospinal fluid bidirectional flow in the central canal. eLife. 2020;9:e47699. doi: 10.7554/eLife.47699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Zhao L., Zhao X., Zhang M., Meng A. A zebrafish gene trap line expresses GFP recapturing expression pattern of foxj1b. J. Genet. Genomics. 2009;36:581–589. doi: 10.1016/S1673-8527(08)60150-2. [DOI] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Troutwine B.R., Gontarz P., Konjikusic M.J., Minowa R., Monstad-Rios A., Sepich D.S., Kwon R.Y., Solnica-Krezel L., Gray R.S. The Reissner Fiber Is Highly Dynamic In Vivo and Controls Morphogenesis of the Spine. Curr. Biol. 2020;30:2353–2362.e3. doi: 10.1016/j.cub.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente A., Huang K.H., Portugues R., Engert F. Ontogeny of classical and operant learning behaviors in zebrafish. Learn. Mem. 2012;19:170–177. doi: 10.1101/lm.025668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen L.M., Evans R.J., Jim K.K., Verboom T., Fang X., Bojarczuk A., Malicki J., Johnston S.A., van der Sar A.M. A transgenic zebrafish model for the in vivo study of the blood and choroid plexus brain barriers using claudin 5. Biol. Open. 2018;7:bio030494. doi: 10.1242/bio.030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell-Llopis N., Yaksi E. Evolutionary conserved brainstem circuits encode category, concentration and mixtures of taste. Sci. Rep. 2015;5:17825. doi: 10.1038/srep17825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesque C., Anselme I., Pezeron G., Cantaut-Belarif Y., Eschstruth A., Djebar M., Santos D.L., Ribeuz H.L., Jenett A., Khoury H. Loss of the Reissner Fiber and increased URP neuropeptide signaling underlie scoliosis in a zebrafish ciliopathy mutant. bioRxiv. 2019 2019.12.19.882258. [Google Scholar]
- Wallmeier J., Al-Mutairi D.A., Chen C.T., Loges N.T., Pennekamp P., Menchen T., Ma L., Shamseldin H.E., Olbrich H., Dougherty G.W. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Genet. 2014;46:646–651. doi: 10.1038/ng.2961. [DOI] [PubMed] [Google Scholar]
- Wallmeier J., Frank D., Shoemark A., Nöthe-Menchen T., Cindric S., Olbrich H., Loges N.T., Aprea I., Dougherty G.W., Pennekamp P. De Novo Mutations in FOXJ1 Result in a Motile Ciliopathy with Hydrocephalus and Randomization of Left/Right Body Asymmetry. Am. J. Hum. Genet. 2019;105:1030–1039. doi: 10.1016/j.ajhg.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L.M., Greenwood P.E. 1/f noise. Scholarpedia. 2007;2:1537. [Google Scholar]
- Worthington W.C., Jr., Cathcart R.S., 3rd Ependymal cilia: distribution and activity in the adult human brain. Science. 1963;139:221–222. doi: 10.1126/science.139.3551.221. [DOI] [PubMed] [Google Scholar]
- Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S.Y., Kim M., Kim H.S., Huh T.L., Chitnis A.B. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Yu X., Ng C.P., Habacher H., Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- Yu X., Lau D., Ng C.P., Roy S. Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development. 2011;138:487–494. doi: 10.1242/dev.057752. [DOI] [PubMed] [Google Scholar]
- Zhang X., Jia S., Chen Z., Chong Y.L., Xie H., Feng D., Wu X., Song D.Z., Roy S., Zhao C. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat. Genet. 2018;50:1666–1673. doi: 10.1038/s41588-018-0260-3. [DOI] [PubMed] [Google Scholar]
- Zhou F., Narasimhan V., Shboul M., Chong Y.L., Reversade B., Roy S. Gmnc Is a Master Regulator of the Multiciliated Cell Differentiation Program. Curr. Biol. 2015;25:3267–3273. doi: 10.1016/j.cub.2015.10.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluid flow measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Fluid flow is measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Fluid flow is measured upon injection of 1 μm diameter particles in a brain explant using an epifluorescence microscope. Anterior to the left, posterior to the right.
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is GSE161834. Microscopy data reported in this paper are deposited in Mendeley Data https://doi.org/10.17632/ygy3pnbnxn.1. Other microscopy data will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information and code required to reanalyze the data reported in this paper is available from the lead contact upon request.