Abstract
The open reading frame 8 (ORF8) protein of SARS-CoV-2 has been implicated in the onset of cytokine storms, which are responsible for the pathophysiology of COVID-19 infection. The present study investigated the potential of isolated compounds from Clerodendrum volubile leaves to stall oxidative bursts in vitro and interact with ORF8 mRNA segments of the SARS-CoV-2 whole genome using computational tools. Five compounds, namely, harpagide, 1-(3-methyl-2-butenoxy)-4-(1-propenyl)benzene, ajugoside, iridoid glycoside and erucic acid, were isolated from C. volubile leaves, and their structures were elucidated using conventional spectroscopy tools. Iridoid glycoside is being reported for the first time and is thus regarded as a new compound. The ORF8 mRNA sequences of the translation initiation sites (TIS) and translation termination sites (TTSs) encoding ORF8 amino acids were retrieved from the full genome of SARS-CoV-2. Molecular docking studies revealed strong molecular interactions of the isolated compounds with the TIS and TTS of ORF8 mRNA. Harpagide showed the strongest binding affinity for TIS, while erucic acid was the strongest for TTS. The immunomodulatory potentials of the isolated compounds were investigated on neutrophil phagocytic respiratory bursts using luminol-amplified chemiluminescence technique. The compounds significantly inhibited oxidative burst, with 1-(3-methyl-2-butenoxy)-4-(1-propenyl)benzene having the best activity. Ajugoside and erucic acid showed significant inhibitory activity on T-cell proliferation. These results indicate the potential of C. volubile compounds as immunomodulators and can be utilized to curb cytokine storms implicated in COVID-19 infection. These potentials are further corroborated by the strong interactions of the compounds with the TIS and TTS of ORF8 mRNA from the SARS-CoV-2 whole genome.
Keywords: Clerodendrum volubile, COVID-19, Cytokine storm, Iridoid glycoside, ORF8, And SARS-CoV-2
1. Introduction
Since the emergence of COVID-19 in Wuhan, China, in December 2019, there has been increased spread of the disease worldwide, resulting in a global pandemic [1]. The disease, which is caused by the novel β-coronavirus severe acute respiratory syndrome coronavirus 2 popularly regarded as SARS-CoV-2, has led to unprecedented pressure on the global health sector irrespective of a country's economic strength [2]. Over time, the virus has undergone mutations, with different variants reported in different parts of the world. These variants and mutants are Alpha (B.1.1.7) B.1.177, B.1.258; Beta (B.1.351) B.1.1.33; Gamma (P.1); Delta (B.1.617.2); Kappa (B.1.617.1) B.1.617.3; Epsilon (B.1.427/B.1.429; Zeta (P.2); Eta (B.1.525) B.1.1.20; Iota (B.1.526); and B.1.1.298 (Mink Variant). These mutations are of major health concerns, as they reportedly exacerbate the infectious rate of the virus [3]. This is depicted by the high global mortality associated with the recent waves of the disease [4,5]. Thus, there is a continuous search for novel and suitable therapies to treat and manage the disease.
SARS-CoV-2 has been reported to cause infection by triggering an excessive immune response depicted by aggravated levels of macrophage inflammatory protein 1-α, tumour necrosis factor-α (TNF-α), interleukins (IL)-2 and −7, monocyte chemoattractant protein 1, and granulocyte colony stimulating factor [[6], [7], [8]]. This process is regarded as cytokine storm and has been implicated in the pathology and severity of COVID-19 and its mortality [9,10]. Oxidative bursts in neutrophils have also been implicated in the aggravation of cytokine storm and the pathology of COVID-19 and its complication [[11], [12], [13]]. Cytokine storm activates the macrophages and neutrophils to produce respiratory bursts which generates superoxides and hydrogen peroxide, thus leading to oxidative stress [11]. Oxidative stress further aggravates the induction of cytokine storm, thus provoking a cycle of continues generation of cytokines. This has led to increasing reports on the arrest of neutrophil oxidative burst as possible therapeutic target for the treatment and management of COVID-19 [12,13].
Open reading frames (ORFs) are among the major nonstructural proteins in SARS-CoV-2, and they possess information for the synthesis and replication of genomic RNA [14]. It consists of ORF1ab, ORF3, ORF6, ORF7a, ORF8, and ORF10. The ORF8 protein has been reported for its role in glycosylation, which is important for the maturation and transportation of the SARS-CoV-2 spike (S) protein [15]. Aside from glycosylation, studies have implicated ORF8 in SAR-COV-2-mediated activation of cytokine storms in COVID-19 infection [[16], [17], [18]]. The protein is utilized by SARS-CoV-2 to elude the immune system by downregulating the surface expression of MHC-Ι [18]. This is supported by reports that ORF8 activates the IL-17 and NLRP3 signaling pathways, upregulates proinflammatory factors [16,19], and inhibits the production of IFN types I and II (IFN–I and IFN-II) [19].
Clerodendrum volubile ranks among commonly consumed vegetables for food and medicinal purposes in southern Nigeria [20,21]. In traditional medicine, the leaves are utilized in the treatment and management of arthritis, diabetes, ulcers, dropsy and rheumatism [22]. The ability of the leaves to suppress blood glucose levels and improve metabolism linked to hypoglycemia has been reported in type 2 diabetic rats [23,24]. The leaves have also been linked to improved antioxidant activities in type 2 diabetes and cancers [[23], [24], [25], [26]]. Its immunomodulatory properties have been linked to the ability of the leaves and flowers, as well as protocatechuic acid, pectolinarigenin, and harpagide 5-O-β-d-glucopyranoside (isolated from the leaves), to suppress the oxidative burst in isolated neutrophils and macrophages [6,23,27,28]. A new compound, harpagide 5-O-β-d-glucopyranoside isolated from C. volubile leaves, was proposed via in silico studies as a suitable inhibitor of initiation and termination codons of SARS-CoV-2 spike protein [6].
Targeting the ORF8 protein of SARS-CoV-2 has been suggested as a suitable therapy against COVID-19 infection [16,29,30]. The present study aimed to investigate the possible mutation trend of ORF8 in SARS-CoV-2 variants from the United Kingdom (UK), USA, India, China, Brazil and South Africa. The study also investigated the ability of compounds isolated from C. volubile leaves to interact with the initiation and termination codons of ORF8 mRNAs isolated from the whole genome of SARS-CoV-2 using computational tools. The study also investigates the ability of the isolated compounds to quench oxidative bursts in polymorpho-nuclear neutrophils (PMNs), as well as their antiproliferative activity on T-cells.
2. Materials and methods
2.1. Instrumentation
FT-IR spectra were measured on potassium bromide pellets using an FT-IR spectrophotometer. 1H and 13C NMR/DEPT (90 and 135) spectra (500 MHz and 150 MHz, respectively) and two-dimensional correlation spectroscopy (COSY), NOSEY, HSQC, and HMBC, were recorded in ppm on a Bruker AV-600 spectrometer in CD3OD. Chemical shifts were reported in δ (ppm) values as a solvent and TMS as an internal reference. The molecular weight was determined by positive mode electron ionization mass spectrometry (EI-MS) on a JEOL-MSRoute HX 110 spectrometer. The TOF-ESI-MS was recorded on a JEOL JMS-HX-110 mass spectrometer. Thin layer chromatography (TLC) was carried out on precoated silica gel 60 F254 plates (E. Merck, 0.25 mm), and spots were viewed under UV light (365 and 254 nm) and by spraying with ceric sulfate reagent.
2.2. Softwares
The computational softwares used for this work were the RNA Fold web server, RNAComposer, Maestro software, AutoDockGPU, AutoDockTools, ADMETlab platform, and MEGA X server.
2.3. Viral genome sequences and ORF8 extraction
SARS-CoV-2 ORF8 mRNA sequence was obtained from the NCBI database from the coronavirus whole genomes (ADD accession NUMBER). About 26 to 30 mRNA nucleotide sequence from both initiation and termination codons (Table 1 ) were converted into 3 D single mRNA strand and prepared for docking.
Table 1.
Sequence of initiation and termination sites of SARS-COV-2 ORF8 accessory protein mRNA.
| Sequence site | ORF8 (27894..28259) |
|---|---|
| Translation initiation site | 5′-AUGAAAUUUCUUGUUUUCUUAGGAAUC-3′ |
| Translation termination site | 5′- ACGUUCGTGUUGUUUUAGAUUUCAUCUAA-3′ |
2.4. Phylogenetic analysis and evolutionary relationships of taxa
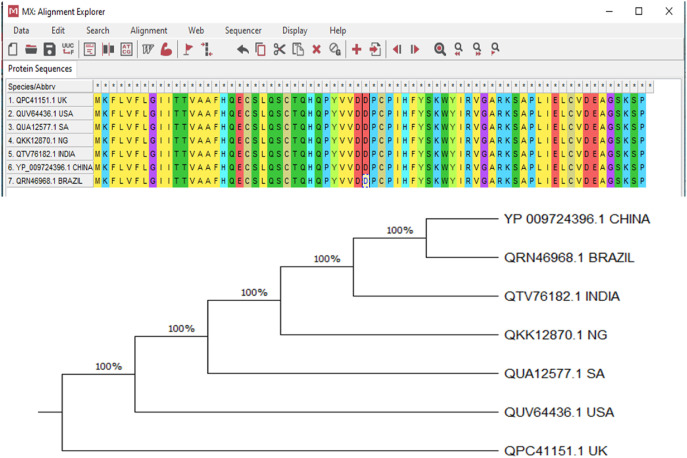
Sequences of ORF8 protein in SARS-CoV-2 variants from various countries including United Kingdom (UK), USA, India, China, Brazil and South Africa were retrieved from GenBank database. The ORF8 sequences were prepared and aligned using the CLUSTALW program [31] for the Phylogenetic analysis. Phylogenetic tree was constructed using the neighbor-joining method [32] implemented in MEGA X [33] and statistically supported by performing 1000 bootstrap replications. The evolutionary history was inferred using the UPGMA method [34]. The optimal tree is shown. (next to the branches). The evolutionary distances were computed using the Poisson correction method [35] and are in units of the number of amino acid substitutions per site. The proportion of sites where at least 1 unambiguous base is present in at least 1 sequence for each descending clade is shown next to each internal node in the tree. This analysis involved 7 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 70 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [33].
2.5. Plant material
The leaves of C. volubile were collected from local farmers at Ifon, Ondo State, Nigeria. The leaves were identified, deposited and assigned a voucher number, UBHC284, at the Herbarium, Department of Botany, University of Benin, Benin City, Nigeria. The leaves were dried under shade, blended to fine powder and stored in a zip-lock bag until further analysis.
2.6. Extraction and isolation
The powdered plant sample was extracted with methanol (MeOH) at room temperature. The extract was concentrated in vacuo using a rotary evaporator. Approximately 100 g of the concentrated extract was further dissolved in MeOH/distilled water (1:3) before liquid-liquid fractionation using chemicals of increasing polarity vis-à-vis n-hexane (Hex), n-dichloromethane (DCM), ethyl acetate (EtOAc), and butanol (BuOH). All the fractions were concentrated in vacuo and stored in glass vials at −20 °C.
The EtOAc fraction was parked into a column chromatograph loaded with silica gel for further fractionation via solvent elution. The elution process started with 100% hexane and thereafter mixtures of Hex and EtOAc (9:1) in increasing order of polarity up to 100% EtOAc. Each elute was collected in glass vials, and its purity was confirmed with thin-layer chromatography on precoated silica gel 60 F254 sheets. They were sprayed with ceric sulfate reagent, dried under hot light, and viewed under UV light (254 nm). Elutes from 100% EtOAc showed 2 overlapping spots and were further separated using reversed-phase recycling HPLC with a solvent system of EtOAc:MeOH (80:20). Harpagide was obtained after several recycling cycles and was identified by comparison of its NMR spectral data with those reported in the literature [[36], [37], [38]].
The BuOH fraction was further subjected to fractionation by VLC using DCM and DCM:EtOAc (1:1). Fraction obtained from DCM:EtOAc was loaded into a column filled with silica gel and subjected to elution starting with 100% DCM and then mixtures of DCM and MeOH (95:5) in increasing order. Each elute was collected in glass vials, and its purity was confirmed with thin-layer chromatography on precoated silica gel 60 F254 sheets. They were sprayed with ceric sulfate reagent, dried under hot light, and viewed under UV light (254 nm). Elutes obtained from DCM:MeOH (93:7) and (90:10) showed single spots each. They were subjected to NMR and mass analyses. Elutes from 90:10 were identified as erucic acid after comparison of spectral data with the literature [39]. Elutes from 93:7 were identified as 4,5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate. Elutes from DCM:MeOH (85:15) and (75:25) showed overlapping spots on TLC. They were subjected to further separation using reversed-phase recycling HPLC with a solvent system of MeOH:H2O (55:45). Ajugoside and 1-(3-methyl-2-butenoxy)-4-(1-propenyl)benzene were obtained from the respective elutes after several recycling cycles and were identified by comparison of their spectral data with those reported in the literature [[40], [41], [42], [43]].
2.7. Pharmacokinetic prediction
The pharmacokinetic properties of the phytocompounds were assessed using the ADMETlab platform (https://admet.scbdd.com/calcpre/index/) based on a previously published method [44]. This was carried out for individual physiochemical properties such as absorption, distribution, metabolism, etc. by computing the canonical SMILEs of the compounds obtained from the PubChem database into the server. The structure of 5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate was drawn using ChemDraw software (V2.42) and submitted as a sdf file. The analysis was carried out principally based on a database consisting of 288,967 compounds from peer-reviewed publications and ChEMBL, EPA and DrugBank databases. All the data were refined by the Molecular Operating Environment (MOE, version 2016) and divided into six classes (basic, A, D, M, E and T). The corresponding basic information and experimental values of these entries form the basis for prediction on a new compound established on a computational similarity check.
2.8. Drug likeness prediction
The drug likeness properties of the compounds were predicted using the same ADMETlab platform (https://admet.scbdd.com/calcpre/calc_rules/) according to a previously published method [44]. Canonical SMILEs of the compounds obtained from the PubChem database as well as the generated sdf file were computed into the server. The compounds were screened for drug-likeness properties based on numerous expert criteria considered crucial for a drug candidate. The most commonly used rules considered in this work include Lipinski's rule, Ghose's rule, Operea's rule, Veber's rule and Varma's rule.
2.9. Molecular modeling studies on TIS and TTS
Molecular modeling studies were conducted to gain deeper insights into the interacting pattern of various compounds on SARS-CoV-2 ORF8 mRNA sites. The sequences of the translation initiation sites (TISs) and translation termination sites (TTSs) were retrieved from the full genome of SARS-CoV-2 from the experiments presented in this study. Furthermore, the TIS and TTS sequences were converted into 2D configurations employing the RNA Fold web server using the default input parameters [45]. Then, the mRNA 2D structure was submitted and further converted into a single-stranded mRNA 3D structure by means of the automated web server called RNAComposer [46]. Small molecules, such as erucic acid, ajugoside, 1-(3-methyl-2-butenoxy)-4-(1-propenyl) benzene, and harpagide, were retrieved from the PubChem database [47]. The novel compound (4,5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate) was constructed using the 2D-sketcher tool in Maestro software. To perform the molecular docking studies, the recent version of docking software, i.e., AutoDockGPU [48] together with its GUI AutoDockTools (ADT) [49], was used. Earlier, both the obtained 3D mRNA structures and all the ligands were prepared and converted into the AD4-PDBQT format utilizing the scripts prepare_ligand4.py and prepare_receptor4.py, which are a part of AutoDockTools. The grid boxes of the TIS and TTS were parameterized to accommodate the whole predicted mRNA structure, as it does not contain any particular active site. For TIS, a set of grids with 66 Å × 100 Å × 48 Å along with its grid centers (13.743, −28.472, 14.243) at x, y, z coordinates with a spacing constraint of 0.669 Å. For TTS, a different set of grids with 96 Å × 50 Å × 88 Å with grid centers (−29.556, 5.025, 4.624) on their x, y, z coordinates and spacing of 0.669 Å were constructed using an AutoGrid4 element. For each ligand against both the TIS and TTS, 100 independent dockings were run. Each independent docking includes 20 million energy evaluations using the Lamarckian Genetic Algorithm Local Search (GALS) method for the conformational search. The docking poses of the ligands were established on the clusters with a root-mean-square deviation (rmsd) of 2 Å. The scoring of the binding poses was calculated based on the binding free energy (ΔGAD4).
2.10. Luminol-amplified chemiluminescence assay
Neutrophils were isolated from fresh blood samples collected from an apparently healthy volunteer after obtaining his consent as previously described [50]. The isolated neutrophils were subjected to luminol-amplified chemiluminescence assay [51]. Briefly, isolated neutrophils (1 × 106 ⁄ mL) were suspended in modified Hank's solution (MHS) and incubated with different concentrations (0.5, 5.00 and 50.00 μg/mL) of the isolated compounds for 30 min. After incubation, 1 mg/mL zymosan was added to the reaction mixture, followed by 25 μL of luminol (10−5 M). Total chemiluminescence (CL) was measured using a luminometer (Lab system Luminoskan RS, Helsinki, Finland). Control (+C) consisted of MHS containing cells only.
2.11. T-cell proliferation assay
T-cells were extracted from freshly collected blood obtained from an apparently healthy volunteer by Ficoll-Hypaque gradient centrifugation and proliferation as described previously [52]. The cells were cultured in a 96-well round bottom tissue culture plate at a concentration of 5 × 105/mL. They were then stimulated with 1.25 mg/mL phytohemagglutinin before incubating with the isolated compounds (0.5, 5.00 and 50.00 μg/mL) at 37 °C in a 5% CO2 incubator for 72 h. The reaction was pulsed with titrated thymidine 0.5 μCi/well and further incubated for 18 h. Thereafter, the cells were harvested using a cell harvester (SKATRON A.S. Flow Lab., Norway) into a glass fibre filter (Cambridge Technology USA). A liquid scintillation counter was utilized to count the cells with the aid of a liquid scintillation counter. The counts per minute (cpm) results were recorded after 120 s [27]. All studies were carried out under the ethical guidelines of the International Center for Chemical and Biological Sciences, University of Karachi, Karachi, Pakistan.
2.12. Statistical analysis
The wet experiments were repeated at least three times to address biological variability. Each treatment group consisted of three samples (n = 3). The wet experiments were repeated at least three times to address biological variability. The results are presented as the mean ± SD, following subjection to one-way analysis of variance (ANOVA). Significant differences (p < 0.05) were obtained using Tukey's HSD-multiple range post hoc test. IBM Statistical Package for the Social Sciences (SPSS) for Windows, version 23.0 (IBM Corp., Armonk, NY, USA) was used for the analyses.
3. Results and discussion
Cytokine storms have been implicated in the pathophysiology of COVID-19 infection and its complications. This is depicted by exacerbated cellular levels of proinflammatory cytokines such as TNF-α, IL-2 and IL-7 [[6], [7], [8]]. ORF8 has been reported to play a major role in SARS-CoV-2 induction of cytokine storms in COVID-19 [16,18,19]. This study investigated the ability of compounds isolated from C. volubile to suppress cytokine storms via molecular interactions with the translation initiation sites (TIS) and translation termination sites (TTS) of ORF8 mRNA from the whole genome of SARS-CoV-2 using proteomic and molecular docking tools.
3.1. Phylogenetic and evolutionary relationships of ORF8
As shown in Fig. 1 , the ORF8 mRNAs from whole genomes of SARS-CoV-2 variants from China, Brazil, India, Nigeria, the USA, the UK and South Africa showed 100% homology and clustered with each other. This is further depicted by the alignment of their amino acid sequences. Thus, the mutation of the studied variants did not affect the ORF8 protein of the virus. There have been concerns about SARS-CoV-2 mutations, especially with their increased transmissibility and pathogenicity [53,54]. There have also been concerns about the effect of these mutations on the efficacy of vaccines and treatment for COVID-19 [55,56]. The non-effect of the viral mutations on ORF8 indicates that the efficacies of therapies targeting the protein may not be altered in the variants. Thus, these results corroborate previous reports on ORF8 as a potential therapeutic target in COVID-19 treatment [16].
Fig. 1.
Evolutionary relationship between ORF8 mRNAs of genomes of SAR-CoV-2 variants from China, Brazil, India, Nigeria, South Africa, USA and the UK.
3.2. Isolation of compounds from C. volubile leaves
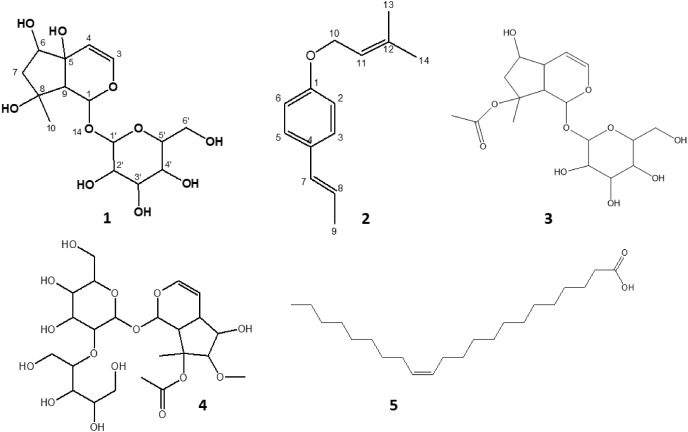
As shown in Fig. 2 , Harpagide (compound 1), 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene (compound 2), Ajugoside (compound 3), 4,5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate (compound 4), and erucic acid (compound 5) were isolated from the leaves of C. volubile.
Fig. 2.
Isolated compounds from the ethyl acetate and butanol fractions of C. volubile methanolic extract. (1) Harpagide, (2) 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene, (3) Ajugoside, (4) 4,5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate, and (5) Erucic acid.
3.2.1. Compound 1
Harpagide C15H24O10; IR (KBr); 1H NMR (500 MHz, CD3OD) δ: 6.35 (1H, d, H-3), 5.75 (1H, br, s; H-1), 4.90 (1H, d, H-4), 4.55 (1H, d, H-1ˈ), 4.02 (1H, d, H-6ˈa), 3.77–3.62 (2H, m, H-6ˈb, H-3), 3.47–3.29 (2H, m, H-4ˈ, H-5′), 3.26–3.15 (1H, dd, H-2ˈ) 2.85 (1H, br, s, H-9), 1.97 (1H, dd, H-7a), 1.85 (1H, dd, H-7b), 1.28 (3H, s, H-10); 13C NMR (150 MHz, CD3OD) δ: 94.09 (C-1), 142.00 (C-3), 104.00 (C-4), 71.94 (C-5), 78.13 (C-6), 49.42 (C-7), 93.97 (C-8), 62.76 (C-9), 23.00 (C-10), 100.03 (C-1), 76.31 (C-2ˈ) 77.47 (C-3), 72.97 (C-4ˈ), 78.16 (C-5ˈ), 64.23 (C-6ˈ). ESI-MS: m/z 387.21 corresponding to [M + Na]+ calculated for C15H24O10. HRESI-MS: m/z 387.21.
Harpagide is an iridoid glycoside that was characterized using a combination of spectral data (1H NMR, 13C NMR, FAB-BS, ESI-MS and HRESI-MS) and comparison with literature values (Ahmed et al., 2003; Venditi et al., 2017; Manguro et al., 2011). ESI-MS gave a molecular ion peak at m/z 387.21 [M + Na]+, and HRESI-MS indicated the molecular formula corresponding to C15H24O10. Compound 1 was characterized as harpagide, also known as (1S,4aS,5R,7S,7aR)-7-methyl-1-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-1,5,6,7a-tetrahydrocyclopenta[c]pyran-4a,5,7-triol, having a molecular formula of C15H24O10. Harpagide, a close relative of ajugoside, has been reportedly isolated from Stachys alopecuros of the Lamiaceae family [37,38].
3.2.2. Compound 2
1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene C14H18O; IR (KBr) V max: 3032 (Ar-CH, 2879 (C–H), 1512 (C C), 1219 (C–O), 1043 (C–O) cm−1; 1H NMR (600 MHz, CD3OD) δ: 8.17 (2H, d, H-3, H-5), 8.10 (2H, d, H-2, H-6), 5.80 (1H, m, H-11), 6.70 (1H, s, H-7), 4.59 (2H, d, H-10), 1.97 (3H, d, H-9), 1.96 (3H, s, H-13), 1.68 (3H, s, H-14); 13C NMR (150 MHz, CD3OD) δ: 153.69 (C-1), 142.00 (C-12), 141.19 (C-7), 127.0 (C-3, C-5), 104.39 (C-6), 101.27 (C-2), 62.95 (C-10), 126.00 (C-11), 22.16 (C-13), 26.52 (C-14, 21.28 (C-9), 128.0 (C-4), 124.00 (C-8). EI-MS: m/z 202 [M]+ calculated for CHO; FAB-MS: m/z 201 [M − H]+ calculated for C12H15O2.
The structure of compound 2 was established using a combination of spectral data and comparison with the literature [42,43]. The IR had no O–H stretching, confirming the absence of any hydroxyl group. Aromatic C–H stretching was observed at 3032 cm−1, while aliphatic C–H stretching and C C vibrations were observed at 2879 and 1512 cm−1, respectively. EI-MS indicated the molecular ion peak at m/z 202 and a prominent peak at m/z 135 corresponding to C9H10O following the cleavage of an ether, methylbut-2-ene (-C5H10). DEPT-135 also shows only one methylene at δ 62.95 and three methyl groups. Compound 3 was identified as 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene with the molecular formula C12H16O2. 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene was detected in Ocotea bofo Kunth (Lauraceae) Calyces (Guerrini et al., 2006) and Illicium verum extract [43,57,58].
3.2.3. Compound 3
Ajugoside (C17H26O10), an iridoid glycoside, was isolated for the second time from C. volubile leaves, and the 1H NMR and m/z data have already been published [41].
3.2.4. Compound 4
4,5-dihydroxy-6-(hydroxymethyl)-5-methoxy-3-(1,3,4,5-tetrahydroxypentan-2-yloxy)tetrahydro-2H-pyran-2-yloxy)-5-hydroxy-7-methyl-1,4a,5,6,7,7a-hexahydrocyclopenta[c]pyran-7-yl acetate (C23H38O15), 1H NMR (CD3OD, 500 MHz): Aglycone δH 6.40 (d, 1H), 4.63 (t, 1H), 2.20 (q, 1H), 3.25 (q, 1H), 1.97 (s, 1H), 1.53 (s, 3H), 1.90 (s, 3H), 2.80 (t, 1H), 4.80 (d, 1H). Sugar moiety 4.70 (d, 1H), 3.58 (d, 1H), 3.32 (tt, 2H), 3.27 (s, 3H), 3.40–3.45 (m, 2H), 3.72 (q, 1H), 3.51–3.58 (m, 6H), 1.95–2.00 (m, 6OH), 3.2 (q, 1H), 3.61 (t, 1H). 13C NMR (CD3OD, 150 MHz): δC 173.2 (CO), 142.0 (1), 101.3 (1), 100.0 (1), 94.1 (1), 88.0 (1), 83.5 (1), 78.6 (1), 78.16 (1), 78.0 (1), 77.1 (1), 74.8 (1), 73.8 (1), 71.9 (1), 71.7 (1), 64.7 (1), 64.4 (1), 62.9 (1), 47.5 (1), 28.44 (1), 22.17 (1). EI-MS (+) calculated for aglycone C12H18O6 [M − 2H]+ m/z 256.3.
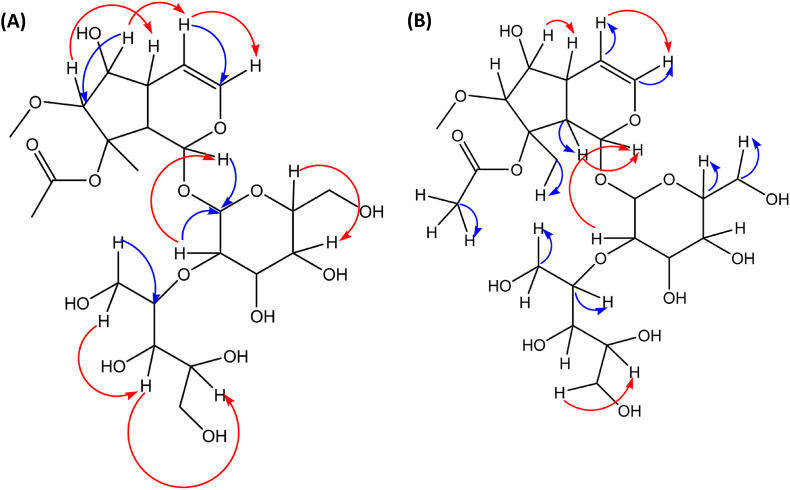
The proposed structure of the compound was established using various NMR techniques to be iridoid glycoside. The distortionless enhancement by polarization transfer (DEPT) 90 and 135 experiments confirms the presence of fifteen methine protons, three methylene protons and three methyl protons. Two quaternary carbons were also observed. Broad band decoupled carbon-13 NMR showed the presence of twenty-three carbons, including the carbonyl of the acetate. The disappearance of the signals at 173 and 88 ppm in the DEPT spectrum confirms the presence of quaternary carbons. The data suggests an iridoid. The aglycone moiety of the compound had a NMR signal similar to that of ajugoside. Major differences were obtained in the sugar moiety as well as the methoxy of the aglycone. Following the details of the data presented, the compound was assigned an iridoid glycoside, as shown in Fig. 2. The atom connectivities are established by two-dimensional NMR spectrometries, which include double quantum filtered proton-proton correlation spectroscopy (DQF 1H–1H COSY), nuclear Overhauser effect spectroscopy (NOESY), heteronuclear single quantum coherence (HSQC) and long-range heteronuclear multiple bond correlation (HMBC), as indicated in Fig. 3 A and B.
Fig. 3.
(A) Significant HMBC (blue lines) and NOESY (red lines); and (B) Significant HSQC (blue lines) and 1H–1HCOSY (red lines) of compound 4.
3.2.5. Compound 5
Erucic acid (13-Docosenoic acid; C22H42O2), IR (KBr): 3500 (O–H), 2075 (C O), 1636 (aliphatic C C) cm−1.1H NMR (CD3OD, 500 MHz): δ 9.93 (OH, s), δ 5.38 (1H, m), δ 5.37 (1H, m), 2.18 (2H, tt), δ 1.67–1.28 (m, 34H, CH2), 0.89 (t, 3H, CH3). 13C NMR (CD3OD, 150 MHz): δ 190.1 (CO), 130.8 (2), 33.6 (1), 30.8–30.4 (12), 28.1–22.5 (5), 14.4 (1). LRMS (ESI) calcd for C22H42O2 [M]+ m/z 338.0 found m/z 338. HRMS (ESI) for C22H42O2 found m/z 338.3472.
The FT-IR spectrum of erucic acid revealed peaks at 3500, 2075, and 1636 cm−1, which correspond to hydroxyl, carbonyl and olefinic C C stretching vibrations. The 1H and 13C NMR spectral data were also in agreement with the proposed structures. The 1H NMR spectrum of erucic acid indicated the presence of a carboxylic acid functional group at 9.93 ppm (OH, s), with one double bond at 5.38 ppm (1H, m). Other proton signals were seen at 2.18 ppm (2H, tt) and 1.67–1.28 ppm overlap (m, 34H, CH2). The terminal methyl signal was observed upfield 0.89 ppm. The H-decoupled 13C NMR spectrum of the compound revealed the presence of twenty-two carbons. The carbonyl carbon resonated at 190.1 ppm, while the alkene carbons resonated at 130.8 ppm. Other carbons appeared at 33.6 (1), 30.8–30.4 (12), 28.1–22.5 (5), and 14.4 (1). The ESI mass spectrum of erucic acid showed a molecular ion [M]+ at m/z 338, in agreement with the molecular formula C22H42O2 and literature data [39].
3.3. Pharmacokinetic prediction
As shown in Table 2 , the compounds exhibited moderate solubility based on the distribution coefficient and poor bilayer permeability as well as aqueous solubility based on the partition coefficient. The compounds indicated the potential to be absorbed by human epithelial colorectal adenocarcinoma cells, with the exception of compound 2. Compounds 2–4 showed an ability to cross human intestinal cells, which may be attributed to the attached sugars and –OH groups [59,60]. Plasma protein binding prediction suggested that the compounds to be transported in the bloodstream were not bound to plasma proteins, and all, with the exception of compound 3, could cross the blood-brain barrier (BBB). The ability of these compounds to cross the BBB makes them good candidates for the treatment of neurodegenerative diseases [61,62]. The metabolism of the compounds in consideration of Cytochrome 1A2 and 3A4 illustrates that the compounds do not interact with these enzymes except for compound 5, which is predicted to inhibit cyt1A2. This suggests that the compounds may not cause a drug-drug interaction when administered with drugs metabolized by these enzymes [63]. However, compound 2 was predicted to be a substrate for cyt3A4, thus insinuating a drug-drug interaction with drugs metabolized by the enzyme. The predicted excretion properties of the compounds revealed a very low clearance rate and half-life time. The compounds were also predicted to be nonhERG blockers, nonmutagenic and hepatotoxic (except for compounds 1 and 3). The predicted hepatoxicity may require caution in the consumption of compounds 2, 4 and 5.
Table 2.
Predicted Pharmacokinetic profile of the selected compounds.
| Category | Property (unit) | Predicted Result |
Inference/Reference Range | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| Basic physicochemical property | LogP (partition coefficient) (log mol/L) | −3.464 | 4.065 | −2.008 | −3.951 | 7.669 | Optimal: 0< LogP <3 LogP <0: poor lipid bilayer permeability. LogP >3: poor aqueous solubility. |
| LogD7.4 (Distribution coefficient D) (log mol/L) |
1.243 |
1.833 |
1.069 |
1.364 |
1.767 |
<1: High Solubility; 1 to 3: Moderate Solubility; ≥3: Low Solubility. |
|
| Absorption | Papp (Caco-2 permeability) (cm/s) | −6.455 | −4.272 | −6.22 | −6.935 | −4.87 | Optimal: higher than −5.15 or −4.70 |
| HIA (Human Intestinal Absorption) (%) |
0.181 |
0.78 |
0.183 |
0.115 |
0.794 |
>0.5: HIA positive <0.5: HIA negative |
|
| Distribution | PPB (Plasma protein binding) (%) | 20.96 | 86.9 | 24.3 | 21.9 | 84.12 | 90%: Significant with drugs that are highly protein-bound and have a low therapeutic index. |
| BBB (Blood brain barrier) (%) |
0.635 |
0.938 |
0.453 |
0.058 |
0.91 |
≥0.1: BBB positive. <0.1: BBB negative. |
|
| Metabolism | CYP1A2-Inhibitor | 0.008 | 0.475 | 0.009 | 0.019 | 0.968 | >0.5: An inhibitor <0.5: Non-inhibitor |
| CYP1A2-Substrate | 0.198 | 0.476 | 0.226 | 0.22 | 0.118 | >0.5: Substrate <0.5: Non-substrate |
|
| CYP3A4-Inhibitor | 0.038 | 0.017 | 0.015 | 0.131 | 0.006 | >0.5: An inhibitor <0.5: Non-inhibitor |
|
| CYP3A4-Substrate |
0.348 |
0.584 |
0.35 |
0.346 |
0.03 |
>0.5: Substrate <0.5: Non-substrate |
|
| Excretion | Clearance (mL/min/kg) | 1.288 | 1.938 | 1.133 | 0.71 | 1.13 | Range: >15 high; 5< Cl < 15: moderate; <5: low. |
| T1/2 (Half life) (H) |
0.864 |
1.587 |
0.723 |
1.098 |
1.736 |
Range: >8H: high; 3h < Cl < 8H: moderate; <3H: low. |
|
| Toxicity | hERG (hERG blockers) | 0.458 | 0.447 | 0.311 | 0.474 | 0.437 | >0.5: A Blocker <0.5: Non-blocker |
| H-HT (Human Hepatotoxicity) | 0.618 | 0.43 | 0.722 | 0.626 | 0.038 | >0.5: HHT positive <0.5: HHT negative |
|
| AMES (Ames mutagenicity) | 0.418 | 0.176 | 0.49 | 0.422 | 0.016 | >0.5: Positive <0.5: Negative |
|
1 = Harpagide, ECA = Erucic Acid, AJD = Ajugoside, IGD = Iridoid Glycoside and MBB = 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene.
3.4. Drug likeness prediction
The benchmarks outlined by Lipinski, Ghose, Oprea, Vaber and Verma were used to predict the druggable character of the compounds. The compounds showed robust compliance with all the standards except compounds 1 (33%) and 3 (33.9%) for Veber's rule, as well as compounds 2 (0%) and 5 (33%) for Oprea's rule (Table 3 ). In drug and bioactive compounds structures, presence of hydrogen bond acceptors (0-10) and donors (0–5) help determine water solubility, membrane transport, distribution, and drug-receptor interactions [44,64]. In addition to those parameters, mass and size of a compound also affects its systemic absorption and distribution [65]. Polar surface area allows prediction of transport properties of a drug and a value of <125 indicate an efficient bioavailability of a compound [44]. Number of aromatic ring in a compound also play a central role in drug distribution and receptor interaction as it affect solubility [66]. Drug-receptor interactions is heavily determined by different conformational stability of a compound which in turn also depend on the number of rotatable bonds present. Therefore, compound with these visible physicochemical properties could potentially be explored during drug selection, design, and testing and at in vitro/in vivo experiments, and clinical trials.
Table 3.
Drug likeness Properties of the Selected Compounds.
| S/N | Name of Rule | Property | Rules | Predicted Result and Percentages Matches for Erucic Acid | Predicted Result and Percentages Matches for Ajugoside | Predicted Result and Percentages Matches for Harpagide | Predicted Result and Percentages Matches for Iridroid Glycoside | Predicted Result and Percentages Matches for 1-(3-Methyl-2-butenoxy)-4-(1-propenyl)benzene | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lipinki's rule | Molecular weight | ≤500 | 338.6 | 75% | 390.4 | 100% | 364.4 | 75% | 510.5 | 25% | 202.3 | 100% |
| Lipophilicity (logP) | ≤5 | 7.7 | −2.01 | −3.5 | −3.95 | 4.1 | |||||||
| Hydrogen bond acceptor | ≤10 | 1 | 10 | 10 | 14 | 1 | |||||||
| Hydrogen bond donors | ≤5 | 1 | 5 | 7 | 8 | 0 | |||||||
| 2 | Ghose's rule | Lipophilicity (logP) | −5.6<logP < −0.4 | 7.7 | 75% | −2.01 | 75.0% | −3.5 | 75% | −3.95 | 50% | 4.1 | 100% |
| Molecular weight | 160<MW < 480 | 338.6 | 390.4 | 364.4 | 510.5 | 202.3 | |||||||
| Molar refractivity | 40<MR < 130 | 105.6 | 86.64 | 78.6 | 110.6 | 66.1 | |||||||
| Total number of atoms | 20<atoms<70 | 66 | 53 | 49 | 69 | 33 | |||||||
| 3 | Oprea's rule | Number of rings | ≥3 | 0 | 33% | 3 | 66.7% | 3 | 66.7% | 3 | 100% | 1 | 0% |
| Number of rigid bonds | ≥18 | 4 | 24 | 24 | 27 | 11 | |||||||
| No. of rotatable bonds | ≥6 | 19 | 5 | 3 | 10 | 4 | |||||||
| 4 | Veber's rule | No. of rotatable bonds | ≥10 | 19 | 66.7% | 5 | 33.9% | 3 | 33% | 10 | 33% | 5 | 100% |
| TPSA | ≤140 | 37.3 | 155.1 | 169.3 | 225.1 | 9.2 | |||||||
| Hydrogen bond donor | ≤12 | 1 | 5 | 7 | 8 | 0 | |||||||
| Hydrogen bond acceptor | ≤12 | 1 | 10 | 10 | 14 | 1 | |||||||
| 5 | Varma's rule | Molecular weight | ≤500 | 338.6 | 80% | 390.4 | 100% | 364.4 | 60% | 510.5 | 40% | 202.3 | 100% |
| TPSA | ≤125 | 37.3 | 155.1 | 169.3 | 225.1 | 9.2 | |||||||
| Hydrogen bond donor | ≤9 | 1 | 5 | 7 | 8 | 0 | |||||||
| Hydrogen bond acceptor | ≤9 | 1 | 10 | 10 | 14 | 3 | |||||||
| No. of rotatable bonds | ≥6 | 19 | 5 | 3 | 10 | 4 | |||||||
3.5. Molecular docking studies
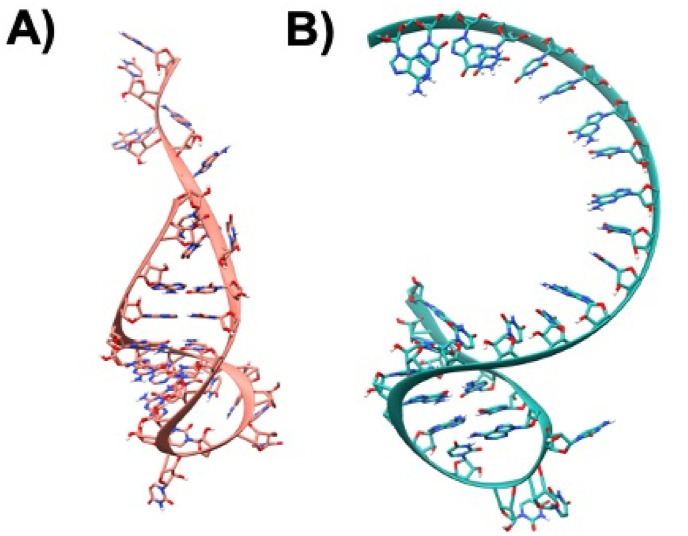
To study the binding modes and interaction patterns of the various small molecules against translation initiation sites (TISs) and translation termination sites (TTS) of ORF8 mRNA from the whole genome of SARS-CoV-2 at the molecular level, molecular modeling studies were performed. In this attempt, the 3D structures of TIS and TTS were modeled using RNA Fold and RNA Composer, as shown in Fig. 4 A and B. The structures were visualized using molecular visualization software - UCSF Chimera [67].
Fig. 4.
A) Modeled single-stranded mRNA 3D-structure of TIS, the atoms are shown in Ribbons and Sticks and the structure is colored in Salmon B) Modeled single-stranded mRNA 3D-structures of TTS atoms are depicted in Ribbons and Sticks and the structure is colored in sea-green. The images were rendered using UCSF Chimera software.
3.5.1. Identification of binding sites in TIS-SARS-CoV-2
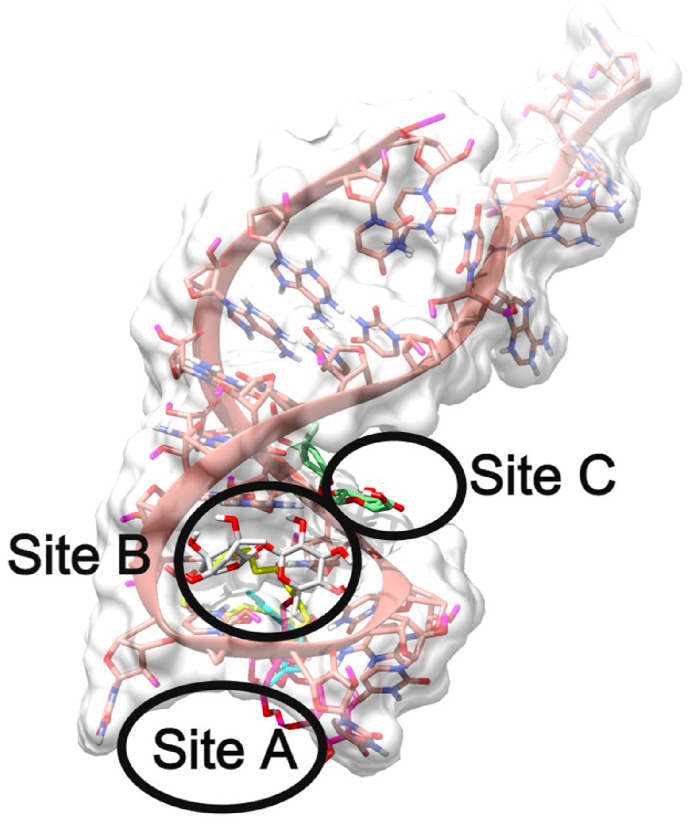
In the docking analysis of various compounds against the TIS mRNA structure, it is interesting to note that all the compounds were docked at three different binding sites. The novel binding sites identified in this docking study are named Site A, Site B, and Site C, as shown in Fig. 5 .
Fig. 5.
Representation of global binding sites of various ligands bound at the various sites of TIS mRNA 3D-structure colored in Salmon. The surface of the TIS is shaded in white and the various binding sites are labeled as Site A, Site B, Site C.
3.5.2. Binding site A
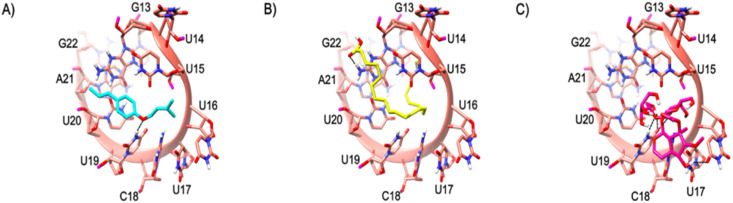
The Site A of TIS is lined by various nucleotides such as G13, U14, U15, U16, U17, C18, U19, U20, A21 and G22. The major part of the binding site is by the nucleotide ‘U’. This is the most dominating residue in this site and out of 5-compounds 3 of them are bound at this site. Compounds 2, 4 and 5 were bound in Site A. The enoxy part of Compound 2 formed a H-bond by accepting from the side chain NH of the U19 nucleotide as shown in Fig. 6 A. The acidic scaffold present at the end of the Compound 5 also formed a H bond by accepting the H from the side chain NH of the A21 as shown in Fig. 6B. The novel compound (compound 4) was tightly bound to this site making various H-bond interactions with various nucleotides such as exchanging of H-atom from the same nucleotide U19 as its side chain both accept and donate H-bonds with the ligand. Another H-bond interaction could be seen with the U17 side chain O atom accepting an H from the OH of compound 4 as shown in Fig. 6C. Although the binding energy of the novel compound (Compound 4) is comparatively less when compared to the rest of the ligands, the interaction pattern is tighter. The binding energies of the compounds are presented in Table 4 .
Fig. 6.
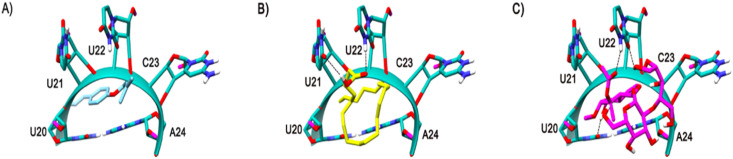
The mRNA structure of TIS is colored in Salmon. The binding site nucleotides of TIS are labeled. Compounds 2, 5, and 4 are shaded in cyan, yellow, and magenta respectively. The H-bonds are shown in black dashed lines.
Table 4.
Table depicting the binding energies of various compounds docked against TIS mRNA of SARS-CoV-2.
| Compound | Binding Energy (kcal/mol) | Preferred Binding Site |
|---|---|---|
| Compound 1 | −4.69 | Binding Site B |
| Compound 2 | −4.37 | Binding Site A |
| Compound 3 | −4.79 | Binding Site C |
| Compound 4 | −1.69 | Binding Site A |
| Compound 5 | −2.81 | Binding Site A |
3.5.3. Binding site B
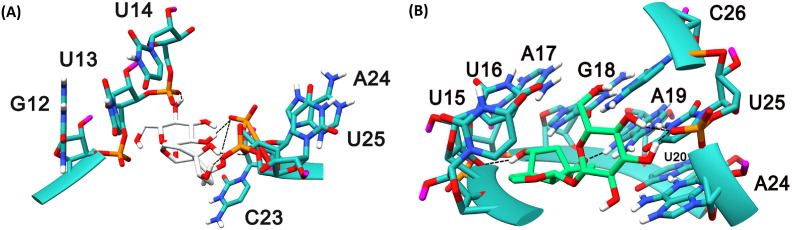
The binding site B is lined by the nucleotides such as U12, U13, U14, U15, U16, U17, C18, and U19 . This site is heavily dominated by U. Except one nucleotide, the others are Uracil. Compound 1 bounded here and was tightly bound to the binding site B by various interactions with the TIS such as the backbone O atom of U12 accepting multiple H-bonds from the multiple OH atoms of the 5,7-triol, whereas similar pattern of H-bond formation with 3,4-triol could be seen with the backbone O atom of C18. The sidechain of U19 accepts a H-bond from the OH of 2-Hydroxymethyl, while the U13 NH side chain donates a H-bond to the O atom of oxane in Compound 1 as shown in Fig. 7 (A) .
Fig. 7.
The mRNA structure of TIS is colored in Salmon. The binding site nucleotides of TIS are labeled. Compounds 1 (A) and 3 (B) are shaded in white and green colors, respectuvely. The H-bonds are shown in black dashed lines.
3.6. Binding site C
The binding site C is lined by the nucleotides continued by Binding site B, such as U20, A21, G22, and G23. This site was not typically dominated by any particular nucleotide. Compound 3 was tightly bound to the binding site C by having various interactions with TIS such as the backbone O atom of U20 donating a H-bond, a similar pattern of multiple acceptance of H-bond formation was seen with the backbone O atom of A21 from the 3,4,5-triols present in 2-(hydroxymethyl) oxane-3,4,5-triol scaffold. The Oxygen atom present in the acetate scaffold of the ligand also compound accepts an H-bond from the side chain NH of G22 as shown in Fig. 7 (B).
3.6.1. Identification of binding sites in TTS-SARS-CoV-2
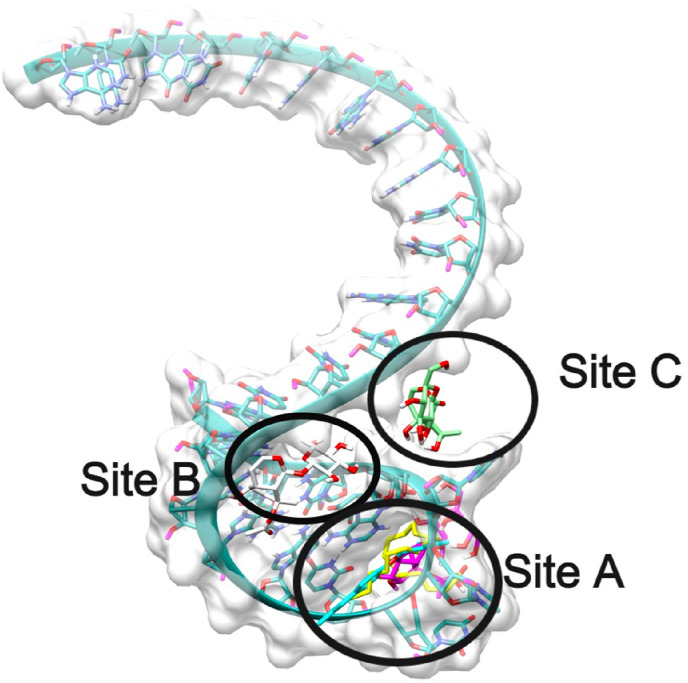
Similar to TIS identification of binding sites, blind docking analysis on TTS revealed 3-binding sites, all compounds docked were bound at only these 3-sites. The unique binding sites identified in this docking study are named as Site A, Site B, Site C as shown in Fig. 8 .
Fig. 8.
Representation of global binding sites of various ligands bound at the various sites of TTS mRNA 3D-structure colored in sea-green. The surface of the TTS is shaded in white and the various binding sites are labeled as Site A, Site B, Site C.
3.6.2. Binding site A
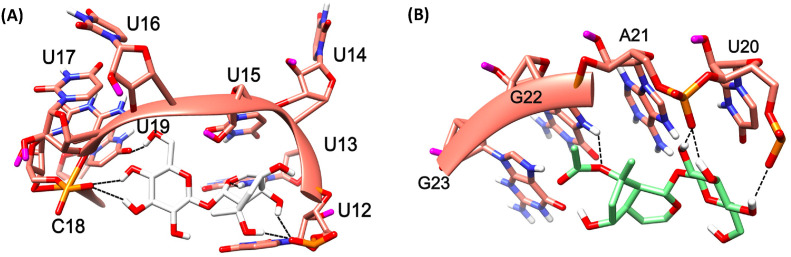
The Site A of TTS is lined by various nucleotides such as U20, U21, U22, C23, and A24. Unlike TIS, there is no specific nucleotide in abundance regarding this binding site. Indeed, similar to TIS, out of the 5 compounds 3 of them (compounds 2, 4 and 5) bounded at this site, even the compounds are the same that are bound to this TIS Site A. Compound 2 binding pose is shown in Fig. 9 A. There was no specific H-bond interaction seen with this compound. In Compound 5, the acidic scaffold at the end also formed a H bond by accepting the H atom from the side chain NH of the U22 and donated a H-bond to the side chain O atom of U21, as shown in Fig. 9B. The novel compound, (compound 4) bounded tightly to this site making various H-bond interactions with various nucleotides via exchange of H-atom from the same nucleotide U22 as its side chain O atom accepts and side chain NH donate H-bonds with the ligand. Another H-bond interaction is seen with the U20 side chain O atom accepting an H from the OH of compound 4 as shown in Fig. 9C. Although the binding energy of the compound 4 is comparatively less when compared to the rest of the ligands, the interaction pattern is tighter which is also very similar in case of these compounds docked against TIS. The binding energies of the compounds are shown in Table 5 .
Fig. 9.
The mRNA structure of TTS is colored in Sea-green. The binding site nucleotides of TTS are labeled. Compound 2, 5, and 4 are shaded in cyan, yellow, and magenta colors respectively. The H-bonds are shown in black dashed lines.
Table 5.
Table depicting the binding energies of various compounds docked against TTS mRNA of SARS-CoV-2.
| Compound | Binding Energy (kcal/mol) | Preferred Binding Site |
|---|---|---|
| Compound 1 | −4.42 | Binding Site B |
| Compound 2 | −4.66 | Binding Site A |
| Compound 3 | −5.45 | Binding Site C |
| Compound 4 | −1.58 | Binding Site A |
| Compound 5 | −2.14 | Binding Site A |
3.6.3. Binding site B
The binding site B is lined by the nucleotides such as G12, U13, U14, C23, A24, and U25. It is noticeable that there is a presence of U in abundance at this site. Compound 1 bounded tightly to the binding site B by having various interactions with the TTS such as the 2-(hydroxymethyl) oxane-3,4,5-triol scaffold is involved in an H-bond interaction with the U14 backbone O atom by donating a H-atom to it. The backbone O atom of A24 accepting multiple H-bonds from the OH atoms involving 1,4a,5,7-tetrol scaffolds of the ligand, whereas similar pattern of H-bond formation is seen with the backbone O atom of U25. The backbone O atom of U25 is accepts a H-bond from the OH atom of compound 1 is shown in Fig. 10 (A) .
Fig. 10.
The mRNA structure of TTS is colored in Sea-green. The binding site nucleotides of TTS are labeled. Compounds 1 (A) and 3 (B) are shaded in white and green colors, respectively. The H-bonds are shown in black dashed lines.
3.6.4. Binding site C
The binding site C is lined by the nucleotides continued by Binding site B, such as U15, U16, A17, G18, A19, U20, A24, U25, and C26. There were a couple of nucleotides that are present in both binding sites B and C such as A24 and U25, and participates in interactions with both compounds 1 and 3. Compound 3 bounded tightly to the binding site C by having various interactions with TIS such as the backbone O atom of U20 donating a H-bond, the similar pattern of multiple acceptance of H-bond formation is seen with the backbone O atom of U25 4,5 OH atoms of the 2-(hydroxymethyl) oxane-3,4,5-triol scaffold. The backbone O atom of U16 also seem to be accepting an H-bond from the 3-hydroxy OH atom and Compound 3 accepts an H-bond from the side chain NH of A19 as shown in Fig. 10 (B).
These molecular interactions of the compounds with the binding sites of the TIS and TTS of ORF8 mRNA indicate potentials of the compounds to impair and/or prevent the recruitment of initiating and terminating factors around mRNA sequence. Thereby arresting the translation of ORF8 protein mRNA and ultimately suppressing the initiation of cytokine storm which has been implicated in the pathophysiology of COVID-19 infection.
3.7. Oxidative burst and T-cell proliferation
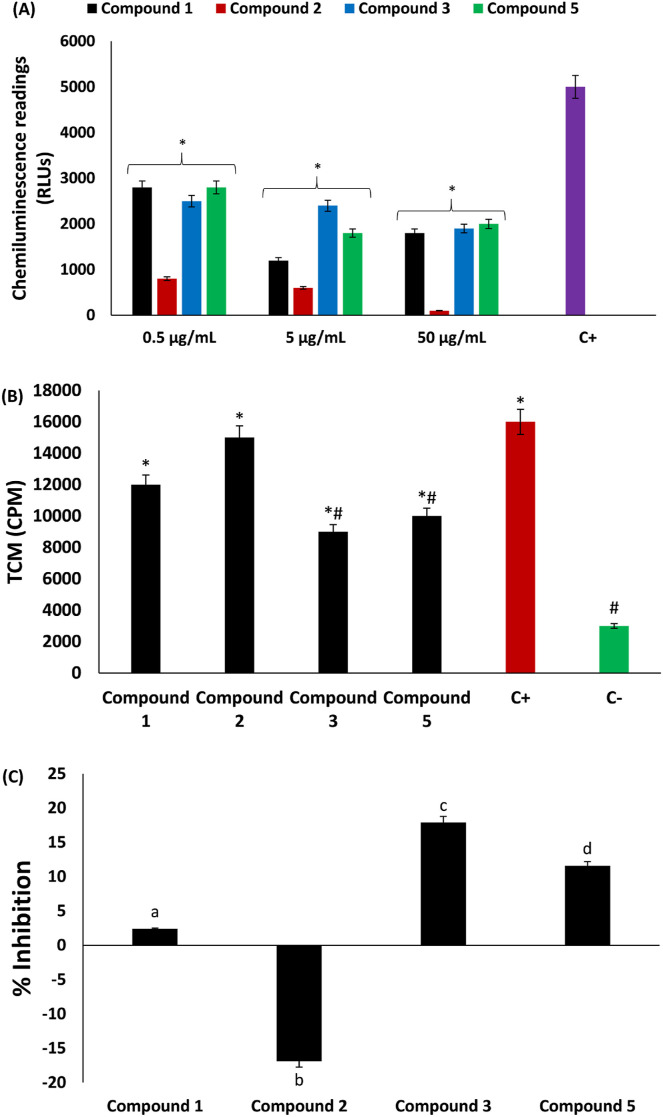
The role of the phagocytic respiratory oxidative burst in innate immunity and the generation of inflammatory cytokines have been reported [6,68]. The phagocytic respiratory oxidative burst is characterized by exacerbated ROS production arising from amplified nonmitochondrial oxygen consumption from glucose dehydrogenation through the hexose monophosphate shunt [23,69]. This is depicted in the present study by the increased production of ROS in PMNs following stimulation with zymosan, as shown in Fig. 11 A. ROS production was significantly (p < 0.05) inhibited dose-dependently in neutrophils incubated with the isolated compounds, with compound 2 having the best activity. Inhibition of oxidative phagocytic respiratory burst can contribute to suppressed production of inflammatory cytokines and therefore arrest cytokine storms. Thus, the isolated compounds have the potential to suppress cytokine storms and hence may be beneficial in the treatment and management of COVID-19 infections.
Fig. 11.
Effect of compounds isolated from C. volubile leaves on (A) oxidative burst of neutrophils; (B) and (C) T-cell proliferation. Values = mean ± SD; n = 3. +C = positive control; –C = negative control; RLUs = relative light units; TCP = T-cell proliferation; CPM = counts per minute. *Statistically significant (p < 0.05) compared to positive control; #Statistically significant (p < 0.05) compared to negative control. abcValues with different letter above the bars are significantly (p < 0.05) different from each other.
The proliferation of T cells has also been linked to the activation of cytokine storms in COVID-19 infections [[70], [71], [72]]. Studies have therefore suggested that targeting T-cell proliferation may be a therapeutic mechanism against COVID-19. In the present study, however, only compounds 3 and 5 significantly (p < 0.05) inhibited T-cell proliferation, as shown in Fig. 11B and C. Thus, these compounds have the potential to suppress cytokine production.
4. Conclusion
Taken together, these results indicate the potential of compounds isolated from C. volubile as immunomodulatory agents which can be utilized to curb cytokine storms implicated in COVID-19 infection. These potentials are further corroborated by the strong interactions of the isolated compounds with the translation initiation and translation termination sites of ORF8 mRNA from the whole genome of SARS-CoV-2 at the molecular level. Further studies are recommended on the immunomodulatory properties of the new compound iridoid glycoside and its potential in drug development for the treatment and management of COVID-19 infection.
Declaration of competing interest
None Declared.
Acknowledgement
OE acknowledges The World Academy of Science (TWAS) for 2012 ICCBS-TWAS fellowship at the H.E.J. Research Institute of Chemistry, ICCBS, University of Karachi, Karachi, Pakistan.
References
- 1.Parvez M.K., Jagirdar R.M., Purty R.S., Venkata S.K., Agrawal V., Kumar J., Tiwari N. COVID-19 pandemic: understanding the emergence, pathogenesis and containment. World Acad. Sci. J. 2020;2(5) 1-1. [Google Scholar]
- 2.Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. Jama. 2020;323(8):709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 3.Chen J., Wang R., Wang M., Wei G.-W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James N., Menzies M., Radchenko P. COVID-19 second wave mortality in Europe and the United States. Chaos. 2021;31(3) doi: 10.1063/5.0041569. [DOI] [PubMed] [Google Scholar]
- 5.Fisayo T., Tsukagoshi S. Three waves of the COVID-19 pandemic. Postgrad. Med. 2021;97(1147) doi: 10.1136/postgradmedj-2020-138564. 332-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erukainure O.L., Atolani O., Muhammad A., Katsayal S.B., Ebhuoma O.O., Ibeji C.U., Mesaik M.A. Targeting the initiation and termination codons of SARS-CoV-2 spike protein as possible therapy against COVID-19: the role of novel harpagide 5-O-β-D-glucopyranoside from Clerodendrum volubile P Beauv.(Labiatae) J. Biomol. Struct. Dyn. 2020;3:1–14. doi: 10.1080/07391102.2020.1840439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B., Huang S., Yin L. The cytokine storm and COVID‐19. J. Med. Virol. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Collaboration H.A.S. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cecchini R., Cecchini A.L. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020;143:110102. doi: 10.1016/j.mehy.2020.110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang C.-C., Korinek M., Cheng W.-J., Hwang T.-L. Targeting neutrophils to treat acute respiratory distress syndrome in coronavirus disease. Front. Pharmacol. 2020;11:572009. doi: 10.3389/fphar.2020.572009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masso-Silva J.A., Moshensky A., Lam M.T., Odish M., Patel A., Xu L., et al. Increased peripheral blood neutrophil activation phenotypes and NETosis in critically ill COVID-19 patients: a case series and review of the literature. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai P.-H., Wang M.-L., Yang D.-M., Liang K.-H., Chou S.-J., Chiou S.-H., Lin T.-H., Wang C.-T., Chang T.-J. Genomic variance of Open Reading Frames (ORFs) and Spike protein in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Chin. Med. Assoc. 2020;83(8):725. doi: 10.1097/JCMA.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sicari D., Chatziioannou A., Koutsandreas T., Sitia R., Chevet E. Role of the early secretory pathway in SARS-CoV-2 infection. J. Cell Biol. 2020;219(9) doi: 10.1083/jcb.202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., Xing N., Wang Y., Xue W., Xiong Y. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. Iscience. 2021;24(4):102293. doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Q., Wang B., Mao J. The pathogenesis and treatment of theCytokine Storm'in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., Xia B., Ma X., Yang T., Yu F. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118(23) doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramasamy S., Subbian S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin. Microbiol. Rev. 2021;34(3) doi: 10.1128/CMR.00299-20. e00299-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erukainure O.L., Sanni O., Islam M.S. In: Polyphenols: Mechanisms of Action in Hum. Health Dis. Watson R., Preedy V., Zibadi S., editors. Elsevier; 2018. Clerodendrum volubile: phenolics and applications to health. [Google Scholar]
- 21.Erukainure O., Oke O., Ajiboye A., Okafor O. Nutritional qualities and phytochemical constituents of Clerodendrum volubile, a tropical non-conventional vegetable. Int. Food Res. J. 2011;18(4):1393–1399. [Google Scholar]
- 22.Burkill H.M., Kew . The Useful Plants of West Tropical Africa. second ed. Royal Botanic Gardens; 1985. [Google Scholar]
- 23.Erukainure O.L., Hafizur R.M., Choudhary M.I., Adhikari A., Mesaik A.M., Atolani O., Banerjee P., Preissner R., Muhammad A., Islam M.S. Anti-diabetic effect of the ethyl acetate fraction of Clerodendrum volubile: protocatechuic acid suppresses phagocytic oxidative burst and modulates inflammatory cytokines. Biomed. Pharmacother. 2017;86:307–315. doi: 10.1016/j.biopha.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Erukainure O.L., Hafizur R., Kabir N., Choudhary I., Atolani O., Banerjee P., Preissner R., Chukwuma C.I., Muhammad A., Amonsou E. Suppressive effects of clerodendrum volubile P beauv.[labiatae] methanolic extract and its fractions on type 2 diabetes and its complications. Front. Pharmacol. 2018;9:8. doi: 10.3389/fphar.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erukainure O.L., Zaruwa M.Z., Choudhary M.I., Naqvi S.A., Ashraf N., Hafizur R.M., Muhammad A., Ebuehi O.A., Elemo G.N. Dietary fatty acids from leaves of clerodendrum volubile induce cell cycle arrest, downregulate matrix metalloproteinase-9 expression, and modulate redox status in human breast cancer. Nutr. Cancer. 2016;68(4):634–645. doi: 10.1080/01635581.2016.1156714. [DOI] [PubMed] [Google Scholar]
- 26.Erukainure O.L., Ashraf N., Naqvi A.S., Zaruwa M.Z., Muhammad A., Odusote A.D., Elemo G.N. Fatty acids rich extract from clerodendrum volubile suppresses cell migration; abates oxidative stress; and regulates cell cycle progression in glioblastoma multiforme (U87 MG) cells. Front. Pharmacol. 2018;9:251. doi: 10.3389/fphar.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erukainure O.L., Mesaik M.A., Atolani O., Muhammad A., Chukwuma C.I., Islam M.S. Pectolinarigenin from the leaves of Clerodendrum volubile shows potent immunomodulatory activity by inhibiting T− cell proliferation and modulating respiratory oxidative burst in phagocytes. Biomed. Pharmacother. 2017;93:529–535. doi: 10.1016/j.biopha.2017.06.060. [DOI] [PubMed] [Google Scholar]
- 28.Erukainure O.L., Mesaik A.M., Muhammad A., Chukwuma C.I., Manhas N., Singh P., Aremu O.S., Islam M.S. Flowers of Clerodendrum volubile exacerbate immunomodulation by suppressing phagocytic oxidative burst and modulation of COX-2 activity. Biomed. Pharmacother. 2016;83:1478–1484. doi: 10.1016/j.biopha.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han N., Hwang W., Tzelepis K., Schmerer P., Yankova E., MacMahon M., et al. Identification of SARS-CoV-2–induced pathways reveals drug repurposing strategies. Sci. Advances. 2021;7 doi: 10.1126/sciadv.abh3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Prot. Bioinform. 2003 doi: 10.1002/0471250953.bi0203s00. 2.3. 1-32.3. 22. [DOI] [PubMed] [Google Scholar]
- 32.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sneath P.H., Sokal R.R. 1973. Numerical Taxonomy. The Principles and Practice of Numerical Classification. [Google Scholar]
- 35.Zuckerkandl E., Pauling L. Evolving Genes and Proteins. Elsevier; 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
- 36.Ahmed B., Al-Rehaily A.J., Al-Howiriny T.A., El-Sayed K.A., Ahmad M.S. Scropolioside-D2 and harpagoside-B: two new iridoid glycosides from Scrophularia deserti and their antidiabetic and antiinflammatory activity. Biol. Pharm. Bull. 2003;26(4):462–467. doi: 10.1248/bpb.26.462. [DOI] [PubMed] [Google Scholar]
- 37.Venditti A., Frezza C., Lorenzetti L.M., Maggi F., Serafini M., Bianco A. Reassessment of the polar fraction of Stachys alopecuros (L.) benth. subsp. divulsa (ten.) grande (Lamiaceae) from the monti sibillini national park: a potential source of bioactive compounds. J. Intercult. Ethnopharmacol. 2017;6(2):144. doi: 10.5455/jice.20170327073801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manguro L.O.A., Lemmen P., Hao P. Iridoid glycosides from underground parts of Ajuga remota. Record Nat. Prod. 2011;5:147–157. [Google Scholar]
- 39.Vargas‐Lopez J., Wiesenborn D., Tostenson K., Cihacek L. Processing of crambe for oil and isolation of erucic acid. J. Am. Oil Chem. Soc. 1999;76:801–809. [Google Scholar]
- 40.Charami M.T., Lazari D., Karioti A., Skaltsa H., Hadjipavlou‐Litina D., Souleles C. Antioxidant and antiinflammatory activities of Sideritis perfoliata subsp. perfoliata (Lamiaceae) Phytother Res. 2008;22(4):450–454. doi: 10.1002/ptr.2333. [DOI] [PubMed] [Google Scholar]
- 41.Erukainure O.L., Ebuehi O.A., Choudhary I.M., Adhikari A., Hafizur R.M., Perveen S., Muhammad A., Elemo G.N. Iridoid glycoside from the leaves of Clerodendrum volubile beauv. shows potent antioxidant activity against oxidative stress in rat brain and hepatic tissues. J. Diet. Suppl. 2014;11(1):19–29. doi: 10.3109/19390211.2013.859213. [DOI] [PubMed] [Google Scholar]
- 42.Guerrini A., Sacchetti G., Muzzoli M., Moreno Rueda G., Medici A., Besco E., Bruni R. Composition of the volatile fraction of Ocotea bofo Kunth (Lauraceae) calyces by GC-MS and NMR fingerprinting and its antimicrobial and antioxidant activity. J. Agric. Food Chem. 2006;54(20):7778–7788. doi: 10.1021/jf0605493. [DOI] [PubMed] [Google Scholar]
- 43.Ladha D., Shah N., Ghelichkhah Z., Obot I., Khorrami Dehkharghani F., Yao J.Z., Macdonald D. Experimental and computational evaluation of illicium verum as a novel eco‐friendly corrosion inhibitor for aluminium. Mater. Corros. 2018;69(1):125–139. [Google Scholar]
- 44.Dong J., Wang N.-N., Yao Z.-J., Zhang L., Cheng Y., Ouyang D., Lu A.-P., Cao D.-S. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminf. 2018;10:1–11. doi: 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattei E., Pietrosanto M., Ferrè F., Helmer-Citterich M. Web-Beagle: a web server for the alignment of RNA secondary structures. Nucleic Acids Res. 2015;43:W493–W497. doi: 10.1093/nar/gkv489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biesiada M., Purzycka K.J., Szachniuk M., Blazewicz J., Adamiak R.W. RNA Structure Determination. Springer; 2016. Automated RNA 3D structure prediction with RNAComposer; pp. 199–215. [DOI] [PubMed] [Google Scholar]
- 47.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos-Martins D., Solis-Vasquez L., Tillack A.F., Sanner M.F., Koch A., Forli S. Accelerating AutoDock4 with GPUs and gradient-based local search. J. Chem. Theor. Comput. 2021;17(2):1060–1073. doi: 10.1021/acs.jctc.0c01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aliyu M., Odunola O.A., Farooq A.D., Mesaik A.M., Choudhary M.I., Azhar M., Asif M.M., Erukainure O.L. Antioxidant, mitogenic and immunomodulatory potentials of acacia honey, Nutr. Ther Metab. 2014;32(2):68–78. [Google Scholar]
- 51.Helfand S.L., Werkmeister J., Roder J.C. Chemiluminescence response of human natural killer cells. I. The relationship between target cell binding, chemiluminescence, and cytolysis. J. Exp. Med. 1982;156(2):492–505. doi: 10.1084/jem.156.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T‐cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noh J.Y., Jeong H.W., Shin E.-C. SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern. Signal Transduct Target Ther. 2021;6(1):203. doi: 10.1038/s41392-021-00623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou W., Wang W. Fast-spreading SARS-CoV-2 variants: challenges to and new design strategies of COVID-19 vaccines. Signal Transduct Target Ther. 2021;6(1):226. doi: 10.1038/s41392-021-00644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S.-G., Zhou B.-G., Li M.-Y., Liu S., Hua R.-M., Lin H.-F. Chemical composition of Illicium verum fruit extract and its bioactivity against the peach–potato aphid, Myzus persicae (Sulzer) Arthropod Plant Interact. 2017;11:203–212. [Google Scholar]
- 58.Wang Z., Wang L., Li T., Zhou X., Ding L., Yu Y., Yu A., Zhang H. Rapid analysis of the essential oils from dried Illicium verum Hook. f. and Zingiber officinale Rosc. by improved solvent-free microwave extraction with three types of microwave-absorption medium. Anal. Bioanal. Chem. 2006;386(6):1863–1868. doi: 10.1007/s00216-006-0778-6. [DOI] [PubMed] [Google Scholar]
- 59.Németh K., Plumb G.W., Berrin J.-G., Juge N., Jacob R., Naim H.Y., Williamson G., Swallow D.M., Kroon P.A. Deglycosylation by small intestinal epithelial cell β-glucosidases is a critical step in the absorption and metabolism of dietary flavonoid glycosides in humans. Eur. J. Nutr. 2003;42(1):29–42. doi: 10.1007/s00394-003-0397-3. [DOI] [PubMed] [Google Scholar]
- 60.Hollman P.C. Absorption, bioavailability, and metabolism of flavonoids. Pharm. Biol. 2004;42(1):74–83. [Google Scholar]
- 61.Pardridge W.M. The blood-brain barrier and neurotherapeutics. NeuroRx. 2005;2(1):1. doi: 10.1602/neurorx.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pardridge W.M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raish M., Ahmad A., Ansari M.A., Alkharfy K.M., Ahad A., Al-Jenoobi F.I., Al-Mohizea A.M., Khan A., Ali N. Effects of sinapic acid on hepatic cytochrome P450 3A2, 2C11, and intestinal P-glycoprotein on the pharmacokinetics of oral carbamazepine in rats: potential food/herb-drug interaction. Epilepsy Res. 2019;153:14–18. doi: 10.1016/j.eplepsyres.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 64.Kumar B.P., Soni M., Bhikhalal U.B., Kakkot I.R., Jagadeesh M., Bommu P., Nanjan M. Analysis of physicochemical properties for drugs from nature. Med. Chem. Res. 2010;19:984–992. [Google Scholar]
- 65.Doak B.C., Over B., Giordanetto F., Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol. 2014;21(9):1115–1142. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 66.Cheng A.C., Coleman R.G., Smyth K.T., Cao Q., Soulard P., Caffrey D.R., Salzberg A.C., Huang E.S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007;25(1):71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 67.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(12):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Cirulli A., Loria M.P., Dambra P., Di Serio F., Ventura M.T., Amati L., Jirillo E., Sabba C. Patients with hereditary hemorrhagic telangectasia (HHT) exhibit a deficit of polymorphonuclear cell and monocyte oxidative burst and phagocytosis: a possible correlation with altered adaptive immune responsiveness in HHT. Curr. Pharmaceut. Des. 2006;12(10):1209–1215. doi: 10.2174/138161206776361336. [DOI] [PubMed] [Google Scholar]
- 69.Allen R.C. Role of oxygen in phagocyte microbicidal action. Environ. Health Perspect. 1994;102(10):201–208. doi: 10.1289/ehp.94102s10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo X.H., Zhu Y., Mao J., Du R.C. T cell immunobiology and cytokine storm of COVID‐19. Scand. J. Immunol. 2021;93(3) doi: 10.1111/sji.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Omarjee L., Janin A., Perrot F., Laviolle B., Meilhac O., Mahe G. Targeting T-cell senescence and cytokine storm with rapamycin to prevent severe progression in COVID-19. Clin. Immunol. 2020;216:108464. doi: 10.1016/j.clim.2020.108464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fara A., Mitrev Z., Rosalia R.A., Assas B.M. Cytokine storm and COVID-19: a chronicle of pro-inflammatory cytokines. Open Biol. 2020;10(10):200160. doi: 10.1098/rsob.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]