Abstract
The deletion of the long arm of chromosome 20 is a characteristic cytogenetic marker of myeloid disorders. Rarely, it is also found in lymphoproliferative diseases, including multiple myeloma (MM). The role of 20q- in MM is not fully understood. In the cases of MM which co-exist with primary or therapy-related dysplasia, this anomaly is mostly linked to the occurrence of myeloid neoplasms. On the other hand 20q- is found as an isolated anomaly in cases with MM that have no dysplastic features or is not accompanied with other hematological diseases which suggests that the 20q deletion is also important for the development of MM. This report describes an isolated 20q- anomaly in a case of a light chain myeloma co-existing with myelodysplastic syndrome (MDS). Fluorescent in situ hybridization (FISH) experiments have demonstrated the presence in the patient's bone marrow of a basic clone (stemline) with deletion of the PTPRT gene (located at 20q13.11) and two sidelines: one with deletion of the PTPRT and MAPRE1 genes (located at 20q11.12) found in the mature granulocytes and one with deletion of PTPRT and duplication of MAPRE1 found in the myeloma cells. These data have indicated that 20q- has appeared in the multipotent precursor cells and affects both myeloid and lymphoid lineage by two different molecular mechanisms - one possibly related to the pathogenesis of the MDS and another to the pathogenesis of the MM.
Keywords: 20q deletion, Multiple myeloma, Myelodysplastic syndrome, PTPRT, MAPRE1
1. Introduction
The deletion of the long arm of chromosome 20 is accepted as myeloid cytogenetic marker, because it is a common finding in myelodysplastic syndrome (MDS), acute myeloid leukemia (AML) and Philadelphia negative chronic myeloproliferative neoplasms [1]. Rarely, 20q- is also observed in B-lymphoproliferative diseases, including multiple myeloma (MM) [2]. The role of 20q- in MM especially as a sole anomaly in the karyotype is not fully understood. Part of MM cases with isolated 20q- were associated with de novo or therapy-related (t) -MDS / t-AML, indicating that the anomaly is related to the occurrence of myeloid disorders. The latter is supported by the fact that 20q- in patients with coexisting MM and MDS is frequentely present in the myeloid line as a sole anomaly and is not detected in the plasma cells [3, 4]. On the other hand, isolated 20q- was found in patients with MM who were not associated with myeloid neoplasia or dysplastic changes in the hematopoietic tissue, which suggested that 20q - may also be important for the appearance of MM [5, 6]. In this report is present coexistence of a MM and MDS in a case with isolated 20q- anomaly and evidence that the 20q deletion is possibly linked to the pathogenesis of both diseases by two different molecular mechanisms.
2. Case report
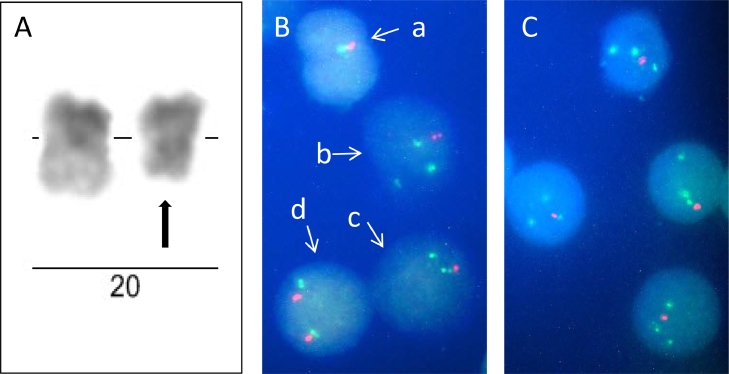
The patient, a 69-year-old male, was admitted to the clinic in January 2019 with evidence of moderate macrocytic and hyperchromatic anemia, neutropenia with two years duration and complaints of episodic bone pain and tingling of the fingers. His-hematological data were as follows: hemoglobin levels (HBG) 101 g/l, hematocrit (HCT) 28.5%, red blood cell count (RBC) 2.79 × 1012/l, white blood cell count (WBC) 4 x 109/l (neutrophil count 1.61 × 109/l), platelet count (PLT) 220 × 109/l, mean corpuscular volume (MCV) 101 fl and mean corpuscular hemoglobin 36.2 pg. Biochemical examinations were normal – total protein 62.70 g/l, albumin 38 g/l, creatinine 83 µmol/l, urea 5.2 mmol/l, beta2 microglobulin 2.03 mg/ml, iron 16.7 µmol/l, calcium 2.43 mmol/l and phosphate 1.04 mmol/l. C-reactive protein was slightly elevated - 5.3 mg/L (reference range 0–5.0 mg/L). Serum electrophoresis and immunofixation electrophoresis (IFE) of serum using anti G, A, M, kappa and lambda chain antisera did not reveal M-protein. IFE of urine (24 h) was positive for free kappa light chain (2.5 g/l) and serum light chain assay showed increased kappa free chain 4292 mg/l (normal range 3.3–19.4 mg/L), normal lambda free chain 6.27 mg/l and abnormal kappa:lambda (κ:λ) ratio 684.53 (normal range 0.26–1.65). Pathological findings in the patient's bone X-ray pictures were not identified. Bone marrow aspiration biopsy revealed hypercellularity with 66% myeloma plasma cells, 1% blasts and trilineage dysplasia. More than 10% dysplasia was determined in each of the three lineages: 62% in erythroblasts (megaloblastoid erythroblasts with nuclear budding, nuclear cytoplasmic asynchrony caused by impaired haemoglobinization and binucleated cells), 27% in granulocytes (hypogranulation or agranularity, single cell with a pseudo-Pelger-Huet anomaly and ring form of the nucleus) and 16% in megakaryocytes (multinucleated megakaryocytes with separated nuclei and hypo- or non-lobated micromegakaryocytes). The test of ring sideroblasts was negative. Conventional banding analysis of bone marrow cells showed karyotype of 46,XY,del(20)(q11.2q13.1)[5] / 46,XY[25] (Fig. 1A). Metaphase FISH with specific DNA probes for the genes MAPRE1 and PTPRT located at 20q11.21 and 20q12 respectively confirmed the deletion of 20q - 46,XY.ish del(20)(q11.2q13.1) (MAPRE1-,PTPRT-). Interphase FISH with the same probes demonstrated that three cytogenetic clones exist in the bone marrow with different 20q anomalies – one with deletion only of the PTPRT gene (clone №1): nuc ish(MAPRE1 × 2,PTPRTx1), one with deletion of both genes (clone №2): nuc ish(MAPRE1 × 1,PTPRTx1) and one with duplication of the MAPRE1 gene and deletion of the PTPRT gene (clone №3): nuc ish(MAPRE1 × 3,PTPRTx1). The 20q- anomaly from the first and third clone was observed only in the mononuclear cells, while the anomalies of the second clone in the mononuclear cells as well as in granulocytes (metamyelocytes, bands and segments) (Fig. 1B). Interphase FISH examination on purified CD138+ cells revealed that the myeloma cells carried the deletion of the PTPRT gene and the duplication of the MAPRE1 gene (Fig. 1C). Additionally the patient's bone marrow was tested with the MAFB/IGH t(14;20) fusion probe. No MAFB/IGH fusion was seen, but in all of the three clones deletion of MAFB was found - nuc ish 14q32(IGHx2),20q12(MAFBx1). Diagnosis of light chain myeloma associated with refractory cytopenia with multilineage dysplasia was made and chemotherapy with CVelDex regimen – bortezomib 1.3 mg/m2 (i.v.) (D1, D4, D8, D11); Endoxan – 300 mg/m2 (i.v.) (D1, D4); Dexamethasone 40 mg (D1–D4, D8–D11); Zolendronic acid was carried out. After six cycles of chemotherapy, partial response to the selected therapy was achieved - the plasmocytic infiltration of the bone marrow decreased to 11%, the free kappa light chain in urine (24 h) to 0.01 g, the serum free kappa light chain to 562 mg/L and the kappa:lambda (κ:λ) ratio to 44.75. hematological studies did not reveal any toxic effect on hematopoiesis associated with the therapy (HGB 137 g/L, HCT 42%, WBC 7.57 × 109/l, RBC 4.81 × 1012/l, PLT 312 × 109/L and MCV 86.4 fl). Bone marrow karyotype was normal – 46,XY and IGH (14q32) rearrangements (14q32 break probe), 1q+/1p- anomalies and p53 deletion were not detected. With the exception of single hypolobulated eosinophilic megakaryocytes, no signs of dysplasia in bone marrow where found, which confirms that bortezomib has a good effect on low-risk MDS [7].
Fig. 1.
A) Partial G-banding karyotype of homologues №20 showing del(20) (q11.2q13.1). B) FISH examination of bone marrow cells – (a) granulocyte with one red (PTPRT gene) and one green signal (MAPRE1 gene)(clone №2); (b) mononuclear cell with one red and three green signals (clone №3); (c) mononuclear cell with one red and two green signals (clone №1); (d) mononuclear cell with normal FISH pattern – two red and two green signals. C) FISH examination of purified CD138+ bone marrow cells showing one red and three green signals (clone №3).
3. Discussion
The deletion of 20q occurs mainly in myeloid malignances, but rarely it is also observed in lymphoproliferative disorders including multiple myeloma [2]. Significant proportion of MM cases with 20q- has been associated with a myeloproliferative diseases and/or dysplastic changes of the hematopoietic tissue and the anomaly in most of these cases was found in the non-plasma cells [4]. Nilsson et al., [6] described 8 MM cases with 20q- as the sole anomaly, without any evidence of MDS/AML. Moreover, in a case with isolated 20q- and monoclonal gammopathy of undetermined significance, the authors found that the 20q- anomaly was presented in the myeloid cells (CD15+), B-cells (CD19+), progenitor cells (CD34+CD38+) and hematopoietic stem cells (CD34+CD38-) and suggest that the 20q- might arise at a multipotent progenitor cell level.
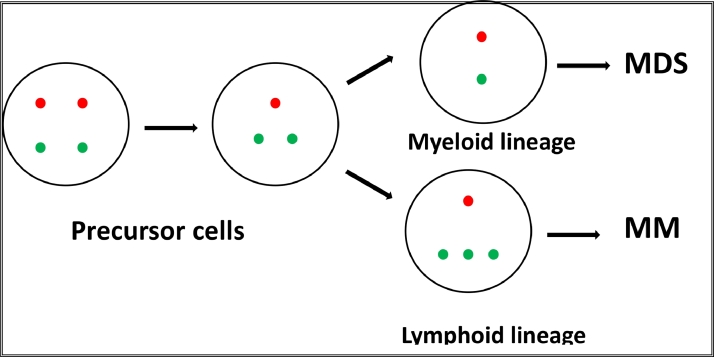
In the presented case three clones were observed with three different variants of the 20q abnormality. Clone №1 was with deletion of the PTPRT gene, clone №2 with deletion of both genes PTPRT and MAPRE1, and clone №3 with deletion of the PTPRT gene and duplication of the MAPRE1 gene. The anomaly of the first clone was presented only in mononuclear cells, the anomaly of the second clone in mononuclear cells and granulocytes and the anomaly of the third clone only in plasma cells. These data indicate that in the described case 20q- occurs also at the level of precursor cells because both myeloid and lymphoid lines are affected. It is logical to suppose that the anomaly of the precursor cells is the deletion only of PTPRT/MAFB gene locus (clone №1) and that during clonal evolution this stemline generates two sidelines – one with deletion of the gene loci of PTPRT, MAFB and MAPRE1 found in the myeloid line (clone №2) and another with deletion of PTPRT and MAFB gene loci in combination with duplication of MAPRE1 gene locus (clone №3) found in the myeloma cells. In other words, during clonal evolution in myeloid line MAPRE1 is deleted, while in myeloma cells it is duplicated (Fig. 2). Since the myeloid clone (clone №2) and myeloma clone (clone №3) have different genome alterations, it can be assumed that two different molecular mechanisms are responsible for both diseases MDS and MM.
Fig. 2.
Schematic representation of the possible clonal evolution of the patient's bone marrow cells leading to the two diseases - MDS and MM (red signal PTPRT gene; green signal MAPRE1 gene).
The molecular pathogenesis of MDS with isolated 20q- is thought to be associated with the inactivation of one or more tumor suppressor genes (TSGs) mapped in the “Common Deleted Regions” (CDR) on 20q13.11–q13.12 [8], while the pathogenesis of MM with isolated 20q- is still unknown. The deletion in the myeloid lineage of a large 20q segment (q11.2-q13.1) in the presented case means that the mechanisms related to MDS are complex. Along with the deletions of multiple genes, the deletion of the TSG PTPRT is probably also important, as it is located in the noted CDR. With respect to the MM, a more specific molecular mechanism may be suggested. The deletion of MAFB is possibly not pathogenetically significant, because this gene plays a role in the onset of MM if it is overexpressed as a result of t(14;20)(q32;q12) [9]. Unlike it the deletion of PTPRT and the duplication of MAPRE1 could deregulate signaling pathways that are frequently aberrant in MM. The duplication of MAPRE1 occurs in the “Common Retained Region” mapped at 20q11.2 where in myeloid malignances duplication or amplification has been observed. RT-PCR examinations revealed that there is correlation between amplification and increased expression of some of the genes located in this region [10]. Accordingly, MAPRE1 duplication in myeloma cells will results in elevation of its protein expression (EB1) that could promotes Wnt/Beta-catenin signaling pathway via inactivating tumor suppressor APC (adenomatous polyposis coli), as has been demonstrated in esophageal squamous cell carcinoma and colorectal carcinoma [11,12]. On the other hand it is known that protein tyrosine phosphatases, including PTPRT, are negative regulators of STAT3 [13]. Thus, deletion of PTPRT will diminish its phosphatase activity resulting in STAT3 upregulation. It could be summarized that the described abnormalities in myeloma cells probably lead to deregulation of key signaling pathways, such as Wnt/Beta-catenin and IL-6/STAT3 and, respectively, their downstream targets CCND1, MYC, MCL-1, BCL-XL and BCL2 which is known to be crucial event for the occurrence of MM.
Declaration of Competing Interest
None.
References
- 1.Bilhou-Nabera C. del(20q) in myeloid malignancies. Atlas Genet. Cytogenet. Onco. Haematol. 2001;5(1):33–34. doi: 10.4267/2042/37701. [DOI] [Google Scholar]
- 2.Jawad M.D., Shi M., Oliveira J.L. Clinical course of patients with incidental finding of 20q- in the bone marrow without a morphologic evidence of multiple myeloma. Am. J. Hematol. 2016;91(6):556–559. doi: 10.1002/ajh.24347. [DOI] [PubMed] [Google Scholar]
- 3.Mossafa H., Defasque S., Fourcade C. Coexistance of multiple myeloma and myelodysplastic syndrome or lymphoid diseasesat diagnosis: evidence for two clonal and independent chromosome abnormalities. Blood. 2011;118 doi: 10.1182/blood.V118.21.5063.5063. 5063-5063. [DOI] [Google Scholar]
- 4.White J.S., Zordan A., Campbell L.J. Deletion(20q) as the sole abnormality in plasma cell myeloma is not associated with plasma cells as identified by clg FISH. Cancer Genet. 2012;205(12):644–652. doi: 10.1016/j.cancergen.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Andrea D., Hayden M., Rao K.W. Isolated chromosome 20q deletions in patients with non-myeloid disorders. Blood. 2012;120 doi: 10.1182/blood.V120.21.1442.1442. 1442-1442. [DOI] [Google Scholar]
- 6.Nilsson T., Nilsson L., Lenhoff S. MDS/AML-associated cytogenetic abnormalities in multiple myeloma and monoclonal gammopathy of undetermined significance: evidence for frequent de novo occurrence and multipotent stem cell involvement of del(20q) Genes Chromosom. Cancer. 2004;41(3):223–231. doi: 10.1002/gcc.20078. [DOI] [PubMed] [Google Scholar]
- 7.Terpos E., Verrou E., Banti A. Bortezomib is an effective agent for MDS/MPD syndrome with 5q- anomaly and thrombocytosis. Leuk. Res. 2007;31(4):559–562. doi: 10.1016/j.leukres.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Bacher U., Haferlach T., Schnittger S. Investigation of 305 patients with myelodysplastic syndromes and 20q deletion for associated cytogenetic and molecular genetic lesions and their prognostic impact. Br. J. Haematol. 2014;164(6):822–833. doi: 10.1111/bjh.12710. [DOI] [PubMed] [Google Scholar]
- 9.A. Micale M. t(14;20)(q32;q12) IGH/MAFB in plasma cell myeloma. Atlas Genet. Cytogenet. Oncol. Haematol. 2019;23(8):230–233. doi: 10.4267/2042/70482. [DOI] [Google Scholar]
- 10.Mackinnon N., Selan C., Wall M. The paradox of 20q11.21 amplification in a subset of cases of myeloid malignancy with chromosome 20 deletion. Genes, Chromosom. Cancer. 2010;49(11):988–1013. doi: 10.1002/gcc.20806. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Zhou X., Zhu H. Overexpression of EB1 in human esophageal squamous cell carcinoma (ESCC) may promote cellular growth by activating β-catenin/TCFpathway. Oncogene. 2005;24:6637–6645. doi: 10.1038/sj.onc.1208819. [DOI] [PubMed] [Google Scholar]
- 12.Stypula-Cyrus Y., Mutyal N.N., Dela Cruz M.A. End-binding protein 1 (EB1) up-regulation is an early event in colorectal carcinoma. FEBS Lett. 2014;588(5):828–835. doi: 10.1016/j.febslet.2014.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Guo A., Yu J. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. USA. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]