Abstract
Background
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that leads to a breakdown of tolerance to self-antigens resulting in inflammation and organ damage. The anti-inflammatory activity of CD73-derived adenosine is well documented, however, its role in SLE pathogenesis is unknown.
Methods
Human peripheral blood immune cells were obtained from adult SLE patients (SLE) and healthy controls (HC). Expression and activity of purinergic ectoenzymes were assessed by qRT-PCR, flow cytometry and HPLC. Genes encoding purinergic ectoenzymes in SLE patients were analysed with targeted DNA sequencing.
Findings
Among circulating immune cells (both in HC and SLE), CD73 was most highly expressed on B cells, which was mirrored by high enzymatic activity only in HC. CD73 protein molecular weight was unchanged in SLE, however, the enzymatic activity of CD73 on SLE B cells was almost fully abolished. Accordingly, AMP accumulated in cultured SLE B cells. A similar discrepancy between protein expression and enzymatic activity was observed for NAD-degrading CD38 on SLE B cells. No differences were found in the rate of extracellular ATP degradation and expression of CD39, CD203a/c, and CD157. DNA sequencing identified no coding variants in CD73 in SLE patients.
Interpretation
We describe a new pathomechanism for SLE, by which inactivation of CD73 on B cells produces less anti-inflammatory adenosine, resulting in immune cell activation. CD73 inactivation was not due to genetic variation but may be related to posttranslational modification.
Funding
The German Research Council, Medical Faculty of the Heinrich-Heine-University Duesseldorf, Hiller Research Foundation, and Cardiovascular Research Institute Duesseldorf.
Keywords: Purines, Metabolism, Autoimmune disease, CD38, CD39
Research in context.
Evidence before this study
SLE patients show multiple B cell abnormalities including an increase in plasmablasts, decreased B cell regulatory activity and increased immunoglobulin production so that modulation of B cell function has been generally viewed as promising therapeutic target. Human circulating B cells are known to express the ectonucleotidases CD39 and CD73, which form anti-inflammatory adenosine. Whether there are changes in purinergic metabolism in SLE patients has not been explored.
Added value of this study
B cells showed the highest CD73 surface expression among human circulating immune cells. In SLE patients the activity of CD73 and CD38 was found to be selectively silenced in B cells. Since CD73 is the bottleneck of extracellular nucleotide degradation to anti-inflammatory adenosine, this pathway is likely to be a crucial step in the pathophysiology of SLE involving B cell - immune cell interactions.
Implications of all the available evidence
While CD73 is highly expressed in human B cells, it was found to be enzymatically inactive on SLE B cells. Since CD73 expression is negligible in mice B cells, this pathway cannot adequately be studied in mice models of SLE. Substitution therapy with soluble human CD73 may constitute a new therapeutic approach for the treatment of SLE.
Alt-text: Unlabelled box
1. Introduction
Systemic Lupus Erythematosus (SLE) is an autoimmune disease that can involve many organs and typically affects women between puberty and menopause [1]. Both genetic and environmental factors influence the development of SLE. In most patients, SLE is a quantitative trait with several genes contributing to the risk of developing the disease [1].
T cells and B cells are important contributors to SLE pathogenesis. Alterations in T cell signalling, in the production of cytokines, in proliferation, and in regulatory functions have been documented in patients with SLE [2]. Impaired B cell regulation in SLE contributes to the production of autoantibodies, cytokines and augmented presentation of antigen to T cells. Autoantibodies, like double stranded DNA antibodies (anti-dsDNA Ab), are traditionally viewed as essential mediators of pathology in SLE [1]. It is now widely accepted that breach of B cell tolerance and abnormal activation represent critical steps in the initiation of the pathogenic cascade leading to clinical disease [3]. The detailed mechanisms by which the abnormal interplay between innate and adaptive immunity is initiated, finally resulting in widespread tissue and organ inflammation and insult to multiple organ systems, are presently not fully understood.
Several lines of evidence highlight the importance of adenosine as a crucial regulatory autocrine and paracrine factor that modulates tissue inflammation [4]. The concentrations of this nucleoside, normally present at low nanomolar levels in the interstitial fluid of unstressed tissues, can rapidly rise to micromolar concentrations in response to stress and in response to pathophysiological conditions, such as hypoxia, ischemia, inflammation or trauma [5]. Thus, adenosine behaves as an ‘alarm’ or danger signal to activate adenosine receptors on target cells, which generate various cellular responses, e.g. by modulating cAMP, that aim to restore tissue homeostasis. Adenosine has been shown to generally have protective effects, which shield cells and tissues from an excessive inflammatory response and immune-mediated damage.
Adenosine elicits its responses by binding to and activating one or more of the four transmembrane adenosine receptors, denoted A1R, A2AR, A2BR and A3R [5]. The activation of A1R and A3R leads to decreased intracellular cyclic adenosine monophosphate (cAMP) levels by Gi-coupled signal transduction. A2AR are GαS- or Gαolf-linked receptors that activate adenylyl cyclase, increase cAMP, and activate protein kinase A (PKA) and Epac1/2. A2BR can signal through both GαS and Gq proteins. Particularly lymphocyte function is potently regulated by A2AR, suggesting that the anti-inflammatory effects of A2AR agonists in animal models of autoimmunity and ischemia are mediated, in part, by targeting lymphocytes [5].
There are two principle sites of adenosine production in almost every cell: intracellular and extracellular [6]. While intracellular adenosine formation is strictly oxygen dependent, the extracellular production of adenosine by the ecto-5’-nucleotidase CD73 is influenced by the cellular activation state. Activated cells release ATP which subsequently is degraded to AMP by CD39 or pyrophosphatases CD203a, CD203c [7]. Similarly, extracellular NAD can serve as substrate for the formation of AMP by action of CD38 and CD203a, CD203c [7]. Human peripheral B cells, expressing CD39 and CD73, were already shown to be susceptible for adenosine-mediated suppression of B cell proliferation and cytokine expression [8]. Furthermore, CD73 expression and activity on B cells has been reported to play a critical regulatory role in B cell – T cell interactions [8].
In this study we explored the metabolism of extracellular purines in B cells und demonstrate, that in SLE-patients the CD73 protein on peripheral B cells is enzymatically inactive despite unaltered abundance. This suggests that lack of B cell-derived anti-inflammatory adenosine is importantly involved in the pathogenesis of SLE.
2. Methods
2.1. Patients
Blood samples were obtained from a pool of 68 adult SLE patients attending the outpatients’ Rheumatology department of University Hospital Düsseldorf. All patients fulfilled the criteria of the Systemic Lupus Collaborating Clinics (SLICC) for SLE [9]. B cell-depleting immunosuppression or cyclophosphamide therapy led to exclusion from the study. None of the patients was in clinically active states as defined by the clinical assessment of a specialist rheumatologist, which was verified in the patients’ clinical documentation. Additionally, SLEDAI score was four or below in most patients, which is defined as mild disease [10]. The main immunosuppressive medication consisted of prednisolone in half of the patients in a dose range of 1-10 mg/day and hydroxychloroquine in two thirds of the patients. Some patients (<10%) were also treated with mycophenolate mofetil or azathioprine. The mean SLEDAI in the patient pool was 3·0±3·0, CRP 0·3±0·3 mg/dl, and anti-dsDNA antibodies 230·4±219·0 IU/ml (Supplementary Tab. 1).
Buffy coats from healthy control (HC) donors (random anonymous blood donors, n = 36) were purchased from the Transfusion Unit of the University Hospital Düsseldorf. Blood samples and buffy coats, respectively, were processed as described below and, whenever possible, used for multiple assays. The number of samples analysed within each assay of individual HC and SLE donors are given in the Figure Legends. For CD73 enzymatic activity measurements – central to this study – B cells from 15 individuals of the SLE patient pool were used and their detailed patient characteristics are shown in Tab. 1.
Table 1.
Characteristics of SLE donors for B cells used for CD73 enzymatic activity measurements.
| Patient characteristics [units, normal range] | mean | (SD) | ||
|---|---|---|---|---|
| number of patients [n] | 15 | |||
| age [yrs] | 46.33 | (18.63) | ||
| sex (%) | female | 12 | (80.0) | |
| male | 3 | (20.0) | ||
| disease duration [yrs] | 16.60 | (10.36) | ||
| SLE Disease Activity Index 2000 (SLEDAI-2K) | 4.15 | (4.51) | ||
| SLE Questionnaire for Population Studies (SLAQ) | 5.00 | (4.58) | ||
| BMI [kg/m2] | 24.17 | (2.30) | ||
| CRP [mg/dl, < 0.5] | 0.32 | (0.35) | ||
| LDH [U/l, < 247] | 216.93 | (43.58) | ||
| AST [U/l, < 31] | 25.00 | (3.82) | ||
| ALT [U/l, < 35] | 21.13 | (5.97) | ||
| hemoglobin [mg/dl, 11.9 – 14.6] | 13.20 | (1.12) | ||
| leukocytes [x1000/µl, 4.5 – 12.7] | 6.85 | (1.94) | ||
| C3 [mg/dl, 90 - 180] | 98.67 | (30.16) | ||
| C4 [mg/dl, 10 - 40] | 16.87 | (12.03) | ||
| IgG [mg/dl, 700 - 1600] | 1242.47 | (287.63) | ||
| IgA [mg/dl, 70 - 500] | 235.53 | (59.90) | ||
| IgM [mg/dl, 40 - 280] | 133.67 | (97.63) | ||
| anti-dsDNA antibodies [IU/ml, < 80] | 315.80 | (343.87) | ||
| creatinin [mg/dl, < 0.9] | 0.85 | (0.22) | ||
| GFR [ml/min, 90 - 140] | 90.13 | (21.95) | ||
| Urin analysis: | ||||
| -erythrocytes [ /µl, < 23] | 18.33 | (12.43) | ||
| -leukocytes [ /µl, < 20] | 54.33 | (102.20) | ||
| -protein [mg/gCreatinin, < 150] | 119.45 | (183.23) |
2.2. Ethics
All samples were obtained in compliance with the Declaration of Helsinki. All subjects provided informed consent, and the study was approved by the local Medical Ethics Committee of the Heinrich-Heine-University Düsseldorf (# 5714R).
2.3. B cell isolation
After preparation of peripheral blood mononuclear cells via Histopaque-1077/1119 (Sigma-Aldrich, St. Louis, MO, US) density centrifugation, total B cells were purified by magnetic cell sorting using Pan B Cell Isolation Kit (Miltenyi, Bergisch Gladbach, Germany), respectively, according to the manufacturer's instructions. Shortly, non-B cells were depleted by magnetically labelling of CD2, CD3, CD4, CD14, CD15, CD16, CD34, CD56, CD61, CD235a and FceRIa.
2.4. Western Blot analysis
Isolated pan-B cells were washed with ice-cold PBS and total protein was extracted using RIPA buffer (Tris-HCl 50 mM, pH 8·0, NaCl 150 mM, EDTA 2 mM, NP-40 1%, SDS 0·1%, Natriumdesoxycholat 1%, Natriumorthovanadat 1 mM, PMSF 1 mM, Natriumfluorid 50 mM) with Protease-Inhibitor-Cocktail. After snap freezing, the lysates were centrifuged at 10,000x g for 10 min at 4°C and supernatants were mixed with 4x Laemmli buffer (Bio-Rad, Hercules, CA, US) with 10% 2-mercaptoethanol, boiled 5 min at 95°C and resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis gels. Protein bands were transferred to a nitrocellulose membrane, which was blocked (Tris buffer, 5% non-fat dry milk, 0·05% Tween20) and probed with the primary antibody for CD73 (ab133582, Abcam, Cambridge, UK; dilution: 1:1,000). The blot was developed with an HRP-conjugated anti-rabbit IgG secondary antibody (Dako, Agilent, Santa Clara, CA, US; dilution: 1:200). Visualization was performed by ECL solution (Luminol, Para-Hydroxycoumarinsäure, DMSO and H2O2) and ChemiDoc Touch Imaging System (Bio-RAD, Hemel Hempstead, UK). Re-probing was performed with housekeeper GAPDH (#2118, Cell Signalling Technology, Danvers, MA, US; dilution: 1:2,000).
2.5. Flow cytometry
After erythrocyte lysis, 1×106 leukocytes were stained with CD4-APC/Cy7 (RRID:AB_314085), CD8a-AF647 (RRID:AB_2564166), CD19-PE (RRID:AB_2564142), CD38-PerCP/Cy5.5 (RRID:AB_893316), CD56-Bv421 (RRID:AB_11218798), and CD73-PE/Cy7 (RRID:AB_2561541) antibodies (Biolegend, San Diego, CA, US) for 30 min at 4°C after Fc receptor blocking using TruStain FcX (Biolegend). Flow cytometric analysis was performed at a FACS Canto II cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and analysed using DIVA 6 software (BD Biosciences). After exclusion of cell doublets via size (forward scatter-H/forward scatter-A plot), populations of granulocytes, monocytes and lymphocytes were identified according to their forward scatter / side scatter characteristics. In the lymphocyte population, CD19+ B cells, CD4+ or CD8+ T cells and CD56+ NK cells were gated.
Flow cytometry of B cell subpopulations and detection of ectoenzymes of the adenosine pathway was performed using a MACS Quant flow cytometer (Miltenyi) on isolated pan-B cells. Doublets were excluded via size and a restrictive forward and side scatter gate; dead cells were excluded using PI and AnnexinV. Before staining with B cell subpopulation surface markers, cells were pre-incubated with FcR Blocking Reagent (Miltenyi) for 10 minutes. Results were expressed as percent of gated cells using FlowJo (V.10 Becton Dickinson, Ashland, OR, US) software. The following anti-human monoclonal antibodies and respective isotype controls from Miltenyi Biotec were used: CD138 (clone 44F9) conjugated to VioBlue (RRID:AB_2751882), CD19 (clone LT-19) conjugated to VioBlue (RRID:AB_2661288), CD21 (clone HB5) conjugated to PerCP-Vio700 (RRID:AB_2656291), CD27 (clone M-T271) conjugated to APC (RRID:AB_2751154), CD38 (clone REA671) conjugated to PE (RRID:AB_2733811), CD73 (clone AD2) conjugated to PE-Vio770 (RRID:AB_2752181), IgD (clone IgD26) conjugated to FITC (RRID:AB_2819581), CD39 (MZ18-23C8) conjugated to VioBlue (RRID:AB_2893087), CD203c (FR3-16A11) conjugated to APC (RRID:AB_2811555), CD157 (REA465) conjugated to APC-Vio770 (RRID:AB_2655318), Mouse IgG1 (clone IS5-21F5) conjugated to VioBlue (RRID:AB_2733969), Mouse IgG1 (clone IS5-21F5) conjugated to APC (RRID:AB_2733440), Mouse IgG1 (clone IS5-21F5) conjugated to FITC (RRID:AB_2733683), Mouse IgG1 (clone IS5-21F5) conjugated to PE-Vio770 (RRID:AB_2733275), Mouse IgG2a (clone S43.10) conjugated to PerCP-Vio700 (RRID:AB_2733064), REA Control (S) (clone REA293) conjugated to PE (RRID:AB_2733893) and APC-Vio770 (RRID:AB_2733167). Further antibodies used: CD203a (NovusBio, polyclonal) conjugated to FITC (RRID:AB_2893088), sheep IgG (R&D Systems, IC016G) conjugated to FITC (RRID:AB_10891881).
According to the expression of IgD, CD21, CD27, CD38 and CD138, the following B cell subsets were identified: CD21+CD27−CD38+ transitional B cells, CD21+CD27−CD38−CD138− mature naïve B cells, IgD+CD27+ non-switched memory B cells, IgD−CD27+ switched memory B cells, IgD−CD21+CD27+CD38− resting memory B cells, IgD−CD27−CD21−CD138− exhausted (double-negative) B cells, IgD-CD21+CD27+CD38high early plasmablasts, IgD−CD21−CD27+CD38high late plasmablasts, CD21+/−CD27+CD38high plasmablasts (all) and CD38highCD138+ plasma cells. In addition, percentage of B cells positive for ectoenzymes of the adenosine pathway (CD39, CD203a, CD203c, CD157) were determined on pan B cells.
2.6. Quantitative Real-Time PCR (qRT-PCR)
RNA was purified from isolated B cells with the RNA Mini Kit (Qiagen, Hilden, Germany). Quantification of RNA was performed with a NanoPhotometer (Implen, Munich, Germany). cDNA was produced by reverse transcription using the Quantitect Reverse Transcription Kit (Qiagen). For quantification of mRNA expression, we used predesigned and validated primers from Qiagen for human CD38 (#QT00073192), CD39 (#QT00081473), CD73 (#QT00027279), CD203a (#QT00094787), CD203c (#QT00086744), and CD157 (#QT00033880). The expression levels of all target genes were normalized against beta-2-microglobulin (B2m forward: 5´-GGCTCACACTGAATTCACCC-3`, B2m backward: 5`-GTCTCGATCCCAGTAGACGG-3`), generating a Δ cycle threshold (Ct) value. Relative expression values were then calculated by the ΔΔCt method, with the mean of the control group as calibrator.
2.7. High pressure liquid chromatography (HPLC)
For analysis of extracellular purine metabolism, B cells were isolated as described above. T cells were purified using the Pan T Cell Isolation Kit (Miltenyi) according to the manufacturer's instructions. 1×105 cells / time point were incubated with100 µl of 20 µM AMP, NAD or ATP (Sigma-Aldrich) in HBSS for 10 min, 20 min, and 40 min at 37°C. For analysis of the endogenous extracellular purine metabolism, 1×105 cells were incubated in 100 µl HBSS only for 10 min at 37°C.
In separate experiments, we explored the role of unspecific phosphatases in the degradation of AMP by B cells, by measuring AMPase activity in the presence of the CD73-specific inhibitor α,β-methylene-ADP (AOPCP). We found that AOPCP inhibited AMPase activity on HC B cells by 91·0±2·8% (20 µM AMP, 50 µM AOPCP, incubation for 40 min, n=5 donors), indicating that the role of other phosphatases on B cells is negligible relative to CD73.
To analyse purine concentrations in serum, 4 ml blood from healthy individuals and SLE patients was immediately mixed 1:1 with a stopping solution adapted from Gorman et al [11]. to inhibit purine metabolism and uptake. The stopping solution contained 118 mM NaCl, 5 mM KCl, 40 mM tricine buffer, 4·15 mM EDTA, 100 µM NBMPR, 40 µM dipyridamole, 100 µM 5-iodotubericidin and 100 µM EHNA at pH 7·4. The blood mix was transferred to a precooled glass tube and centrifuged for 8 min at 800 g at 4°C. The supernatant was deproteinized by addition of 1 volume 80% acetonitrile / 1% trifluoroacetic acid / 19% ddH2O. After 10 min incubation at 4°C, the sample was centrifuged for 10 min at 12,000 g at 4°C. Solvents were evaporated from the supernatant by lyophilization and the precipitate was dissolved in 50 µl HBSS.
Cell supernatants (50 µl injection volume) and serum preparations (35 µl injection volume) were applied to an ACQUITY UPLC Bio H-Class System equipped with a Cortecs C18+ UPLC column (3·0×150 mm, particle size 1·6 µm) (Waters, Milford, MA, US). Purine separation was performed as previously described [12], using a liner gradient of buffer A (200 mM KH2PO4 / 200 mM KCl, pH 6) and buffer B (200 mM KH2PO4 / 200 mM KCl / 7·5% acetonitrile, pH 6). Absorbance was measured at 254 nm.
2.8. Next Generation Sequencing (NGS) of target genes
Genomic DNA was isolated from 1 ml whole blood with EDTA as anticoagulant using the FlexiGene DNA Kit (Qiagen) according to the manufacturer's instructions. Tagmentation-based DNA library preparation and target gene enrichment were performed with the SureSelect QXT reagent kit (Agilent) and an Agilent SureSelect XT custom targets library (Agilent) according to the manufacturer's instructions. Paired-end short-read next generation sequencing was performed on an Illumina MiSeq DNA Sequencer Instrument using the Illumina MiSeq Reagent Kit v2. The bioinformatic analysis followed established best-practices using the Genome Analysis Toolkit (GATK) v.4.1.3.0. Briefly, this comprised sequence read adapter trimming (Cutadapt v.2.5, https://github.com/marcelm/cutadapt); sequence read alignment to the human reference genome version GRCh38 (bwa 0.7.17, http://bio-bwa.sourceforge.net); alignment processing and calling of sequence variants (GATK); and variant annotation (annovar v.2018-04-16, https://annovar.openbioinformatics.org).
2.9. Statistics
Data are presented as means ± SD; n indicates the number of biological replicates (blood samples or cell isolates from different healthy individuals or SLE patients). Data were statistically analysed using GraphPad Prism with multiple t tests (comparison of HC group and SLE group in different cell populations / time points / analytes) or t test with Welch's correction (comparison of HC group and SLE group for expression of individual target genes or proteins) as stated in the figure legends. Correction for multiple testing was performed using the Holm-Sidak method with alpha = 0·05. The threshold for statistical significance was set at p<0·05.
2.10. Role of funders
Funders had no role in study design, data collection, data analyses, interpretation, or writing of the manuscript.
3. Results
3.1. Similar CD73 and CD38 expression on peripheral blood leukocytes from SLE patients and healthy individuals
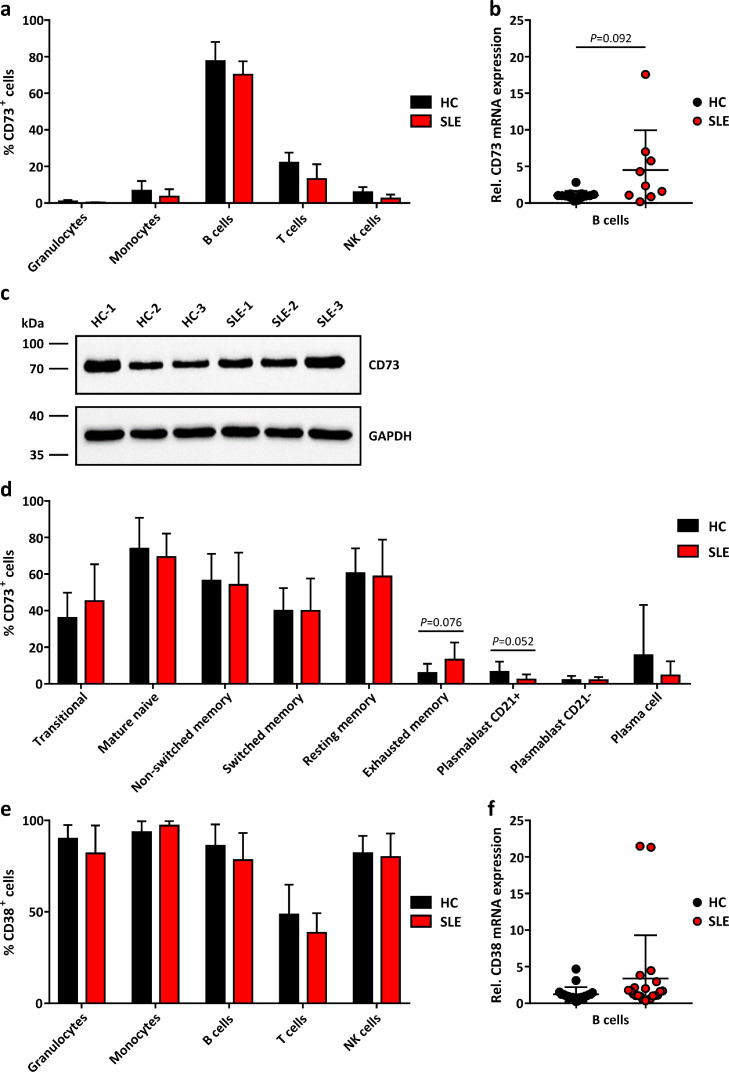
Comparative protein expression analysis in peripheral blood leukocytes (granulocytes, monocytes, B cells, T cells, NK cells) by flow cytometry revealed that the frequency of cells positive for CD73 (AMP → adenosine) in SLE patients was similar to healthy controls (HC) (Fig. 1a). Similarly, the mean expression level of CD73 per cell was unaltered (Supplementary Fig. 1a).
Fig. 1.
Expression of CD73 and CD38 on peripheral blood leukocytes.
a Proportion of CD73+ cells in peripheral blood leukocyte populations from healthy controls (HC) and SLE patients (SLE) analysed by flow cytometry (HC n=5, SLE n=4 donors). Means ± SD. P>0·05 [Multiple t tests]. b Gene expression of CD73 in B cells isolated from HC and SLE peripheral blood analysed by qRT-PCR (HC n=13, SLE n=9 donors). P=0·092 [Welch's t test]. c Western Blot analysis of CD73 in B cells from HC and SLE patients (n=3 each, full blots can be found in Supplementary Data File 1). d CD73+ cells among B cell subsets from HC and SLE peripheral blood analysed by flow cytometry (HC n=15, SLE n=23 donors). Means ± SD. P>0·05 [Multiple t tests]. e CD38+ cells in peripheral blood leukocytes analysed by flow cytometry (HC n=5, SLE n=4 donors). Means ± SD. P>0·05 [Multiple t tests]. f CD38 gene expression in peripheral blood B cells analysed by qRT-PCR (HC n=21, SLE n=22 donors). *P<0·05 [Welch's t test].
As to be expected [13], B cells showed the highest frequency of CD73+ cells which by far exceeded that of T cells (Fig. 1a). Unchanged expression of CD73 on B cells of SLE patients was confirmed by Western Blot (Fig. 1c), while gene expression of CD73 tended to be slightly higher in SLE patients (Fig. 1b).
B cell subset analysis revealed that CD73 expression was prevalent in early B cell stages (Fig. 1d). The relative distribution of B cell subsets showed that transitional B cells, plasma cells and plasmablasts tended to be enriched in the SLE B cell pool compared to HC B cells (Supplementary Fig. 2), which is consistent with previous studies [14].
We also measured surface expression of CD38 (NAD → → AMP). As shown in Fig. 1e, CD38 protein was abundant on peripheral leucocytes. In B cells, neither the mean CD38 protein expression level per cell (Supplementary Fig. 1b) nor CD38 gene expression (Fig. 1F) was different between SLE patients and HC. Only granulocytes from SLE patients showed a minor increase in mean CD38 expression level per cell (Supplementary Fig. 1b).
3.2. Severely impaired AMP- and NAD-degrading activity on the surface of B cells from SLE patients
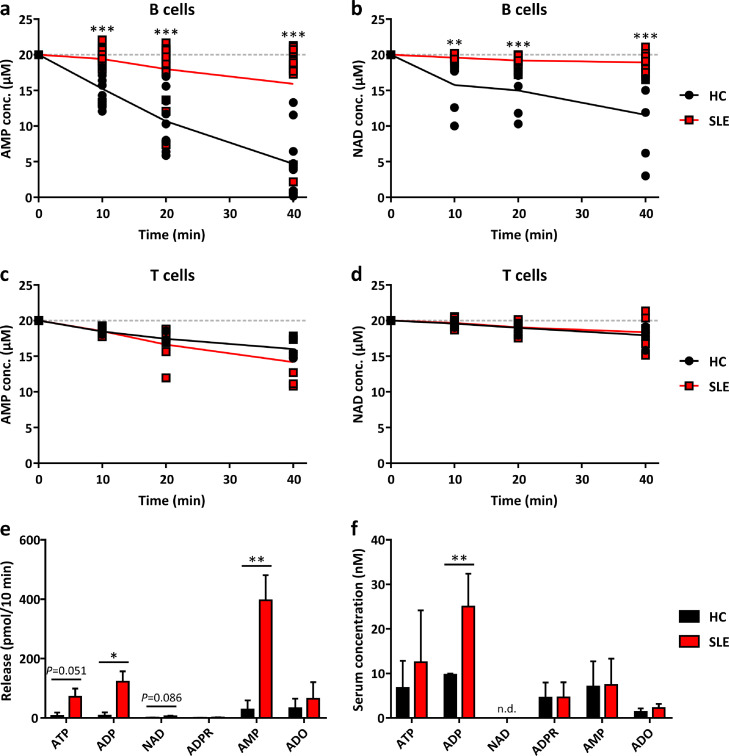
Next, we studied whether the similar protein expression of CD73 and CD38 as measured by flow cytometry translates in similar enzymatic activities in HC and SLE B cells. To this end, we measured the extracellular degradation of AMP and NAD over time when starting the enzyme kinetics with a saturating substrate concentration of 20 µM. While B cell CD73 in HC rapidly degraded AMP, this effect was almost fully abolished in 12 of the 15 analysed B cell samples from SLE patients (Fig. 2a), showing a clear dissociation between abundant surface expression (Fig. 1a) and enzymatic activity. Surprisingly, a similar dissociation, though less striking, was observed for NAD: eight of 13 analysed B cell samples of SLE patients were unable to degrade NAD as compared to healthy controls (Fig. 2b). The observed effect was B cell-specific, since the rate of AMP and NAD degradation in T cells was not different between SLE patients and HC (Fig. 2c+d) The lower rate of AMP degradation in T cells compared to B cells in healthy controls is consistent with the lower abundance of CD73+ cells in T cells (Fig. 1a).
Fig. 2.
Enzymatic activity of CD73 and CD38 on peripheral blood B cells.
a-e B cells (a-b, e) and T cells (c-d) from the peripheral blood of healthy controls (HC) and SLE patients (SLE) were isolated. Purine concentrations in cell supernatants (1×105 cells / sample) were assessed by HPLC after addition of 20 µM AMP (a, c) or 20 µM NAD (b, d) at the indicated time points or without addition of exogenous purines after incubation for 10 min (e). f Purine concentrations in blood serum of HC and SLE patients analysed by HPLC. Means ± SD (a: HC n=9, SLE n=15; b: HC n=6, SLE n=13; c: HC n=5, SLE n=6; d: HC n=5, SLE n=6; e: HC n=3, SLE n=4; f: HC n=5, SLE n=5 donors). *P<0·05, **P<0·01, ***P<0·001 [Multiple t tests]. n.d., not detected.
We then measured in isolated HC and SLE B cells the endogenous release of various purine compounds into the supernatant by HPLC. As shown in Fig 2e, SLE B cells accumulated significantly more AMP compared to B cells from HC, which is consistent with the observed dysfunction in AMP degradation of SLE B cells (Fig. 2a). The accumulated AMP likely derived from the ATP-ADP-AMP degradation pathway, since NAD degradation was found to be impaired (Fig. 2b) and the intermediate ADP was significantly increased in the SLE B cell supernatant (Fig. 2e).
We also explored, whether differences in B cell-associated CD73 activity translates into differences in AMP and adenosine plasma levels in HC and SLE patients. To this end we analysed purine levels in blood serum, using a stopping solution to inhibit purine degradation in freshly drawn blood (see Methods). As shown in Fig. 2f, ADP plasma levels were elevated in SLE patients but there were no differences in the levels of AMP and adenosine. This suggests that the inability of SLE B cells to convert AMP to adenosine does not translate into systemic changes but is likely restricted to a rather local mechanism.
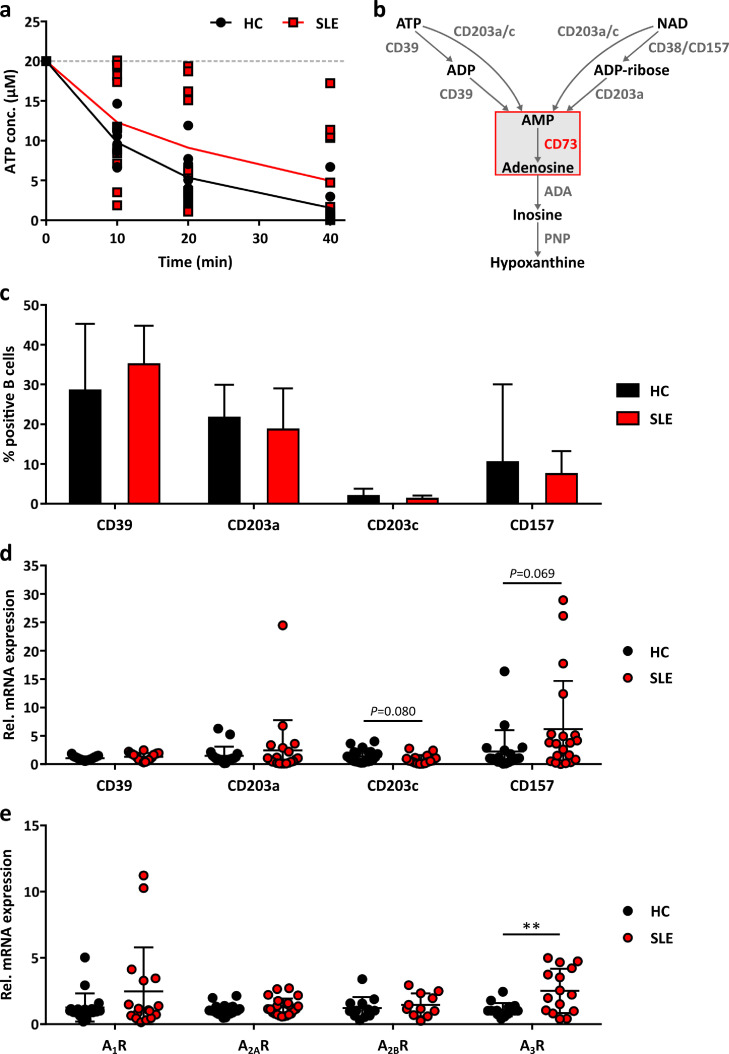
3.3. Unaltered ATP-degrading activity on the surface of SLE B cells
To explore whether there are differences in the extracellular metabolism of ATP, we have additionally measured the kinetics of B cell-related degradation of ATP. As shown in Fig. 3a, degradation of ATP was generally more rapid as compared to AMP (Fig. 2a), but was similar in SLE patients and HC (Fig. 3a). This went parallel with no significant change in the proportion of B cells expressing ATP-degrading CD39 in SLE patients (Fig. 3c). Other ectoenzymes involved in the direct conversion of ATP and NAD to AMP (CD203a, CD203c: two pyrophosphatases; CD157: paralogue of CD38; Fig. 3b) showed also no significant differences in surface distribution (Fig. 3c) and gene expression (Fig. 3d). Analysis of the four adenosine receptors revealed that A3R was significantly higher expressed in SLE patients (Fig. 3e).
Fig. 3.
Expression of other purinergic ectoenzymes in peripheral blood B cells.
a ATP concentrations in the supernatant of B cells isolated from the peripheral blood of healthy controls (HC) and SLE patients (SLE) after addition of 20 µM ATP measured by HPLC (HC n=9, SLE n=12 donors). Means ± SD. P>0·05 [Multiple t tests]. b Scheme of the ectoenzyme cascade facilitating extracellular purine degradation. ADA, adenosine deaminase. PNP, purine nucleoside phosphorylase. c Proportion of CD39+, CD203a+, CD203c+ and CD157+ cells in peripheral blood B cells from HC and SLE patients analysed by flow cytometry (HC n=15, SLE n=24 donors). Means ± SD. P>0·05 [Welch's t test]. d Gene expression of CD39 (HC n=21, SLE n=20 donors), CD203a (HC n=19, SLE n=21 donors), CD203c (HC n=21, SLE n=16 donors) and CD157 (HC n=19, SLE n=20 donors) in HC and SLE B cells analysed by qRT-PCR. P>0·05 [Welch's t test]. e Gene expression of the adenosine receptors A1R (HC n=19, SLE n=17 donors), A2AR (HC n=21, SLE n=22), A2BR (HC n=12, SLE n=11 donors) and A3R (HC n=15, SLE n=15 donors) in HC and SLE B cells analysed by qRT-PCR. **P<0·01 [Welch's t test].
3.4. No variants in the CD73-encoding gene sequence in SLE patients
To finally investigate, whether the profound inhibition of CD73 enzymatic activity in SLE B cells might be related to germline DNA sequence variations, DNA sequencing analysis was performed in eight SLE patients with confirmed lack of AMP degradation (Supplementary Fig. 3a). Aside from the gene encoding CD73 (NT5E) also further genes encoding enzymes with potential involvement in the observed phenotype were studied, including ectoenzymes CD38 (CD38), CD157 (BST1), CD203a (ENNP1), CD203c (ENPP3), CD39 (ENTPD1), adenosine-degrading adenosine deaminase (ADA), and ADP-ribosyltransferase 1 (ART1). As summarized in Tab. 2, several uncommon heterozygous sequence variants (< 1 % allele frequency in the GnomAD general population database [15]) that alter the protein-coding sequences of the genes were found in five of the eight sequenced samples. The locations of the identified uncommon variants are displayed in Supplementary Fig. 3b. Each of these variants has been observed before in the general population, including the stop gain variant in ART1 (123 heterozygous variant carriers among 140,762 individuals). Importantly, the CD73-encoding gene NT5E was unaltered, suggesting that the observed enzymatic dysfunction was not due to changes in CD73 protein sequence.
Table 2.
DNA sequencing of genes encoding purinergic ectoenzymes in SLE B cells.
Genes encoding purinergic ectoenzymes in B cells from SLE patients (No. 1-8) were analysed with targeted high-throughput DNA sequencing. Identified variants in genes of the extracellular purinergic metabolism shown together with dbSNP (https://www.ncbi.nlm.nih.gov/snp) identifiers, allele frequencies in exome sequencing population data (GnomAD v. 2.1.1, https://gnomad.broadinstitute.org), available ClinVar information (https://www.ncbi.nlm.nih.gov/clinvar) and in silico impact predictions of missense variants taken from dbNSFP [34]. HGNC gene symbols and HGVS nomenclature are shown. het: heterozygous, N: neutral effect on protein structure or function; D: possible/potential alteration of protein structure or function.
| Missense impact prediction |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein name | Gene symbol | HGVS description | Zygosity | dbSNP identifier | GnomAD exomes allele frequency | ClinVar | CADD | SIFT | Polyphen2 | Mutation Taster | Provean | In patient No. |
| CD203a | ENPP1 | NM_006208.2:c.523G>A; p.(Asp175Asn) | het | rs142001296 | 0.000111 | . | 23.3 | N | D | D | D | 6 |
| CD203a | ENPP1 | NM_006208.2:c.2657G>C; p.(Arg886Thr) | het | rs8192683 | 0.004085 | benign | 21.2 | N | N | N | N | 7 |
| CD203c | ENPP3 | NM_005021.4:c.130G>A; p.(Gly44Arg) | het | rs76529435 | 0.002635 | . | 23.5 | N | D | D | N | 6 |
| CD203c | ENPP3 | NM_005021.4:c.458C>T; p.(Pro153Leu) | het | rs61741318 | 0.002629 | . | 23.5 | D | D | D | D | 6 |
| CD203c | ENPP3 | NM_005021.4:c.1498G>A; p.(Gly500Arg) | het | rs373462240 | 0.000004 | . | 29.6 | D | D | D | D | 8 |
| A3R | ADORA3 | NM_000677.3:c.749A>G; p.(Asn250Ser) | het | rs375139289 | 0.000107 | . | 23.9 | D | D | D | D | 2 |
| CD296 | ART1 | NM_004314.2:c.734C>G; p.(Pro245Arg) | het | rs150574054 | 0.005759 | . | 25 | D | D | D | D | 1 |
| CD296 | ART1 | NM_004314.2:c.225T>G; p.(Tyr75*) | het | rs141826088 | 0.000920 | . | 32 | . | . | . | . | 7 |
4. Discussion
Among the human immune cells analysed, B cells showed the highest expression of CD73, already pointing to an important role of purinergic signalling in B cell biology. Here we report that CD73 on human peripheral B cells from SLE patients is functionally inactive, despite unaltered expression of this ectoenzyme as assessed by flow cytometric and Western Blot analysis. This effect was not observed on T cells, suggesting a B cell-specific defect. Obviously, peripheral B cells from SLE patients lacked the ability to fully hydrolyse extracellular adenine nucleotides to generate adenosine, a nucleoside well known for its anti-inflammatory activity e.g. on neutrophils, macrophages and T cells [4]. The capacity to degrade extracellular ATP, however, remained unaltered in SLE B cells, so that as a consequence of the CD73-related metabolic defect, AMP accumulated extracellularly. Lack of B cell CD73 activity was found in 19 of 22 patients analysed which is surprising in view of the clinical heterogeneity of SLE [1].
Extracellular degradation of NAD by CD38 or CD203a/c is an alternative pathway for adenosine formation [7]. Both, ATP and NAD degradation converge into the formation of AMP, the substrate of CD73. We found ATP degradation to be more rapid on B cells as compared to NAD degradation. Importantly, NAD degradation was reduced in SLE B cells, which could result in the accumulation of extracellular NAD. However, under in vitro conditions the release of NAD from B cells was only small and not significantly increased in SLE B cells. Interestingly, daratumumab, a human monoclonal antibody that targets CD38, was recently reported to result in substantial clinical improvement in two patients with live-threatening SLE [16], most likely by depleting plasma cells. The role of the NAD pathway (NAD → AMP → adenosine) in the pathogenesis of SLE remains to be investigated.
B cells in SLE are able to stimulate T cells via up-regulation of costimulatory molecules, and reciprocally activated T cells provide substantial help to autoreactive B cells, thus driving autoantibody production [17]. It is also well established that adenosine generated by human regulatory T cells initiates immune suppression [18], which is mediated by A2AR activation. Furthermore, activated human B cells were already reported to accumulate AMP and in coculture downregulated T cell proliferation [8]. Thus, alteration in extracellular AMP and adenosine in SLE B cells is likely to have important paracrine signalling consequences on T cells, known to be critically involved in SLE pathogenesis [1].
There still may be another purine-related mechanism involved in SLE pathogenesis. As a consequence of inactive CD73, not only is there less adenosine formed, but AMP accumulates extracellularly. It has recently been described that both AMP and adenosine act as ligands for A2BR signalling [19]. This opens the interesting possibility that AMP accumulating around SLE B cells may be a locally acting A2B agonist, which is linked to the formation of IL-6 [20]. IL-6 is a proinflammatory cytokine that is well known to promote the maturation of naïve B cells into memory or plasma cells, differentiation of follicular T helper cells, formation of germinal centres, and production of autoantibodies. Blockade of IL-6 was already proposed as therapy in SLE [21].
Together it appears that the metabolic consequences of downregulated B cell CD73 activity on purinergic signalling may be twofold: The reduced adenosine formation by B cells sends a proinflammatory signal to T cells. At the same time the accumulating AMP may send a pro-inflammatory signal to B cells via A2BR, which is coupled to IL-6 production. Therefore, the balance between pro- and anti-inflammatory activity in purinergic signalling may be strongly tilted towards inflammation in SLE. The patients in our study received anti-inflammatory medication and therefore their disease activity was well controlled, despite dysfunction of B cell CD73. In view of the well-known immune suppressive role of adenosine in adaptive immunity, this does not argue against a role of B cell CD73 in disease pathogenesis. Rather adequate immunosuppressive treatment can successfully counteract the pro-inflammatory consequences of diminished adenosine production. The detailed mechanism of B cell CD73 dysfunction in SLE pathogenesis needs to be addressed in future studies, e.g. by investigating the local B cell – T cell interactions in human lymph node germinal centres.
Genetic factors play an important role in the pathogenesis of SLE and many SLE susceptibility genes have been identified [1]. A missense single-nucleotide polymorphism in the gene encoding purine nucleoside phosphorylase (PNP: inosine → hypoxanthine) was reported to lead to a loss-of-function PNP variant, associated with cell cycle abnormalities and interferon pathway activation in B cells [22]. CD73 is upstream of PNP (Fig. 3b) and, as we have shown, functionally inactive on SLE B cells. Therefore, one would expect a reduced hypoxanthine production which in a similar way as a PNP polymorphism may be pathophysiologically relevant.
Our DNA sequencing analyses of genes involved in extracellular ATP and NAD metabolism revealed no uncommon genetic alterations in the coding sequence of the CD73-encoding gene NT5E in SLE patients. On the post-transcriptional level, a CD73 splice variant lacking 50 amino acids (∼5 kDa) was reported, that was expressed at low abundance in human tissues and was catalytically inactive [23]. Since Western Blot analysis of B cell CD73 did not show differences in molecular weight, aberrant splicing of CD73 transcripts in SLE B cells due to genetic alterations not evident in the sequencing approach seems to be unlikely. Together this suggests that the observed enzymatic dysfunction was not due to changes in CD73 protein sequence but rather involved post-translational modifications (PTM). Very recently, extracellular NAD+ was shown to reduce the enzymatic activity of CD73 in mice by ecto-ADP-ribosyltransferase (ARTC)2.2-mediated mono-ADP-Ribosylation [24]. This may well be the mechanism involved in the depressed CD73 activity in B cells of SLE patients. However, there are major species differences: While mice express three ARTC family members (ARTC1, ARTC2.1, ARTC2.2), mono-ADP-ribosylation at the human cell surface is exclusively dependent on ARTC1 [25] and it is presently unknown whether human CD73 activity is also controlled by ADP-ribosylation. Demonstration of ARTC1-dependent mono-ADP-ribosylation in SLE patient B cells will be technically demanding. Despite highly sensitive mass spectrometry techniques, identification of the mouse T cell ADP-ribosylome required the isolation of T cells from seven spleens [24] (corresponding to about 2·3×108 T cells), while from a 20 ml sample of human peripheral blood only about 1·4-10·6×106 B cells can be isolated. It also should be kept in mind, that aside of ADP-ribosylation PTM include acetylation, lipidation, persulfidation, AMPylation, disulfide formation, phosphorylation, methylation, nitration, and ubiquitin-like modifications [26].
That adenosine may play a critical role in the pathogenesis of SLE is also reflected by the finding that several drugs which interfere with adenosine metabolism have shown clinically favourable effects in some studies. This includes methotrexate (MTX) which elevates adenosine by blocking adenosine deaminase by the accumulation of 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) [27]. Similarly, dipyridamole elevates extracellular adenosine by blocking its cellular reuptake [28]. Note, however, that MTX and dipyridamole are not commonly used agents for SLE treatment. Furthermore, serum adenosine deaminase activity in SLE patients was found to be significantly increased compared with that in HC [29], again suggesting a reduction in biological active adenosine in SLE patients. Finally, mesenchymal stem cells highly expressing CD39 and CD73 were recently shown to be therapeutically beneficial in lupus nephritis [30].
There appear to be major species differences in B cell purinergic signalling. While human B cells showed the highest fraction of CD73+ cells within all immune cells analysed, B cells of mice only show negligible CD73 expression [31]. Because of these profound differences in extracellular nucleotide degradation, the pathomechanism suggested by us cannot be adequately studied in mice. This raises the important question how well the different lupus experimental models used in the past mimic the true clinical situation. Note that each model only shares specific subsets of attributes of SLE observed in humans, so that identification of therapeutic targets and screening for treatments may be misleading.
Autoreactive B cells are key drivers in the pathogenesis of SLE and a wide range of biologics targeting B cells have been evaluated for therapeutic use. Despite numerous clinical trials addressing various B cell-related targets, belimumab, a humanized monoclonal antibody specifically inhibiting B cell activating factor (BAFF), was the only drug since the year 1957 that has been approved as a treatment of adult SLE patients [32]. Our findings, that B cells of SLE patients are unable to form anti-inflammatory adenosine because of inactive CD73 and at the same time accumulate AMP as potential pro-inflammatory A2BR ligand, suggest novel treatment options. Systemic administration of soluble CD73 could substitute for the inactive B cell CD73 to lower extracellular AMP and shift to anti-inflammatory adenosine. Whether specific adenosine receptor agonists/antagonists [33] may be therapeutically beneficial remains to be explored.
In summary, the present study uncovered a severe metabolic defect in adenosine generation by B cell CD73 in SLE patients. The associated alterations in purinergic signalling are likely to influence B cell to immune cell crosstalk that is critical in the pathophysiology of SLE. These new insights provide new options for the treatment of SLE.
Contributors
JS and GP conceived, supervised the study, interpreted results, and wrote the manuscript. JH analysed data, finalized figures and tables, and wrote the manuscript. JS, GP, and JH have verified the underlying data. MSH acquired patients for the study, analysed data, and designed experiments. BS performed the HPLC measurements. CA participated in data analysis and interpretation. MeS, NH, MLS, MW, RB performed experiments and preanalysed data. MatS acquired patients for the study and revised the manuscript. HS and DW performed DNA sequencing and data interpretation. All authors read and approved the final version of the manuscript.
Declaration of competing interest
None.
Acknowledgments
Acknowledgments
JS was supported by a grant of the German Research Council (DFG project identifier: 236177352) and the Cardiovascular Research Institute Duesseldorf (CARID). JH was supported by the Research Committee of the Medical Faculty of the Heinrich-Heine-University Düsseldorf (project identifier: 2018-12). GP was supported by a grant of the German Research Council (DFG project identifier: 323627809) and an unlimited grant from the Hiller Research Foundation.
We thank Birgit Opgenoorth, the Team of the Rhinevit-Biobank and all participating physicians in the outpatient clinic for their excellent technical and logistic support.
Data Sharing Statement
Source data are available upon request to the corresponding author.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103616.
Contributor Information
Jürgen Schrader, Email: schrader@uni-duesseldorf.de.
Georg Pongratz, Email: georg.pongratz@med.uni-duesseldorf.de.
Appendix. Supplementary materials
References
- 1.Kaul A., Gordon C., Crow M.K. Systemic lupus erythematosus. Nat Rev Dis Primer. 2016;2:1–21. doi: 10.1038/nrdp.2016.39. [DOI] [PubMed] [Google Scholar]
- 2.Crispín J.C., Kyttaris V.C., Terhorst C., Tsokos G.C. T cells as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6:317–325. doi: 10.1038/nrrheum.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tipton C.M., Hom J.R., Fucile C.F., Rosenberg A.F., Sanz I. Understanding B-cell activation and autoantibody repertoire selection in systemic lupus erythematosus: a B-cell immunomics approach. Immunol Rev. 2018;284:120–131. doi: 10.1111/imr.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haskó G., Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. doi: 10.3389/fimmu.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haskó G., Linden J., Cronstein B., Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesse J., Alter C., Schrader J. In: The Adenosine Receptors. Borea PA, Varani K, Gessi S, Merighi S, Vincenzi F, editors. Springer International Publishing: Cham; 2018. Adenosine Signalling in the injured heart; pp. 439–460. (eds) [Google Scholar]
- 7.Hesse J., Leberling S., Boden E. CD73-derived adenosine and tenascin-C control cytokine production by epicardium-derived cells formed after myocardial infarction. FASEB J. 2017;31:3040–3053. doi: 10.1096/fj.201601307R. [DOI] [PubMed] [Google Scholar]
- 8.Saze Z., Schuler P.J., Hong C.-S., Cheng D., Jackson E.K., Whiteside T.L. Adenosine production by human B cells and B cell-mediated suppression of activated T cells. Blood. 2013;122:9. doi: 10.1182/blood-2013-02-482406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petri M., Orbai A.-M., Alarcón G.S. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–2686. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bombardier C., Gladman D.D., Urowitz M.B., Caron D., Chang C.H. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 11.Gorman M.W., Marble D.R., Ogimoto K., Feigl E.O. Measurement of adenine nucleotides in plasma. Luminescence. 2003;18:173–181. doi: 10.1002/bio.721. [DOI] [PubMed] [Google Scholar]
- 12.Smolenski R.T., Lachno D.R., Ledingham S.J., Yacoub M.H. Determination of sixteen nucleotides, nucleosides and bases using high-performance liquid chromatography and its application to the study of purine metabolism in hearts for transplantation. J Chromatogr. 1990;527:414–420. doi: 10.1016/s0378-4347(00)82125-8. [DOI] [PubMed] [Google Scholar]
- 13.Thomson L.F., Ruedi J.M., Glass A. Production and characterization of monoclonal antibodies to the glycosyl phosphatidylinositol-anchored lymphocyte differentiation antigen ecto-5′-nucleotidase (CD73) Tissue Antigens. 1990;35:9–19. doi: 10.1111/j.1399-0039.1990.tb01750.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu C., Gershwin M.E., Chang C. Diagnostic criteria for systemic lupus erythematosus: a critical review. J Autoimmun. 2014:10–13. doi: 10.1016/j.jaut.2014.01.004. 48-49. [DOI] [PubMed] [Google Scholar]
- 15.Karczewski K.J., Francioli L.C., Tiao G. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostendorf L., Burns M., Durek P. Targeting CD38 with Daratumumab in Refractory Systemic Lupus Erythematosus. N Engl J Med. 2020;383:1149–1155. doi: 10.1056/NEJMoa2023325. [DOI] [PubMed] [Google Scholar]
- 17.Moulton V.R., Tsokos G.C. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J Clin Invest. 2015;125:2220–2227. doi: 10.1172/JCI78087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deaglio S., Dwyer K.M., Gao W. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holien J.K., Seibt B., Roberts V. AMP and adenosine are both ligands for adenosine 2B receptor signaling. Bioorg Med Chem Lett. 2018;28:202–206. doi: 10.1016/j.bmcl.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Feoktistov I., Biaggioni I. Role of adenosine A2B receptors in inflammation. Adv Pharmacol San Diego Calif. 2011;61:115–144. doi: 10.1016/B978-0-12-385526-8.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tackey E., Lipsky P., Illei G. Rationale for interleukin-6 blockade in systemic lupus erythematosus. Lupus. 2004;13:339–343. doi: 10.1191/0961203304lu1023oa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghodke-Puranik Y., Dorschner J.M., Vsetecka D.M. Lupus-associated functional polymorphism in PNP causes cell cycle abnormalities and interferon pathway activation in human immune cells. Arthritis Rheumatol. 2017;69:2328–2337. doi: 10.1002/art.40304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snider N.T., Altshuler P.J., Wan S., Welling T.H., Cavalcoli J., Omary M.B. Alternative splicing of human NT5E in cirrhosis and hepatocellular carcinoma produces a negative regulator of ecto-5′-nucleotidase (CD73) Mol Biol Cell. 2014;25:4024–4033. doi: 10.1091/mbc.E14-06-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leutert M., Duan Y., Winzer R. Identification of the Mouse T Cell ADP-Ribosylome Uncovers ARTC2.2 mediated regulation of CD73 by ADP-Ribosylation. Front Immunol. 2021;12:3328. doi: 10.3389/fimmu.2021.703719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Girolamo M., Fabrizio G. Overview of the mammalian ADP-ribosyl-transferases clostridia toxin-like (ARTCs) family. Biochem Pharmacol. 2019;167:86–96. doi: 10.1016/j.bcp.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Lin H., Caroll K.S. Introduction: posttranslational protein modification. Chem Rev. 2018;118:887–888. doi: 10.1021/acs.chemrev.7b00756. [DOI] [PubMed] [Google Scholar]
- 27.Friedman B., Cronstein B. Methotrexate mechanism in treatment of rheumatoid arthritis. Joint Bone Spine. 2019;86:301–307. doi: 10.1016/j.jbspin.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan M.K.X., Heng T.Y.J., Mak A. The potential use of metformin, dipyridamole, N-acetylcysteine and statins as adjunctive therapy for systemic lupus erythematosus. Cells. 2019;8:323. doi: 10.3390/cells8040323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z., Zhao G., Zhang Z. Serum adenosine deaminase activity is increased in systemic lupus erythematosus patients and correlated with disease activity. Immunol Res. 2018;66:299–304. doi: 10.1007/s12026-018-8984-9. [DOI] [PubMed] [Google Scholar]
- 30.Dang J., Xu Z., Xu A. Human gingiva-derived mesenchymal stem cells are therapeutic in lupus nephritis through targeting of CD39−CD73 signaling pathway. J Autoimmun. 2020;113 doi: 10.1016/j.jaut.2020.102491. [DOI] [PubMed] [Google Scholar]
- 31.Bönner F., Borg N., Burghoff S., Schrader J. Resident cardiac immune cells and expression of the ectonucleotidase Enzymes CD39 and CD73 after ischemic injury. PLOS ONE. 2012;7:e34730. doi: 10.1371/journal.pone.0034730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stohl W. Inhibition of B cell activating factor (BAFF) in the management of systemic lupus erythematosus (SLE) Expert Rev Clin Immunol. 2017;13:623–633. doi: 10.1080/1744666X.2017.1291343. [DOI] [PubMed] [Google Scholar]
- 33.Borea P.A., Gessi S., Merighi S., Vincenzi F., Varani K. Pharmacology of adenosine receptors: the state of the art. Physiol Rev. 2018;98:1591–1625. doi: 10.1152/physrev.00049.2017. [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Li C., Mou C., Dong Y., Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103. doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.