Abstract
Purpose
To examine the performance of radiologists in differentiating COVID-19 from non-COVID-19 atypical pneumonia and to perform an analysis of CT patterns in a study cohort including viral, fungal and atypical bacterial pathogens.
Methods
Patients with positive RT-PCR tests for COVID-19 pneumonia (n = 90) and non-COVID-19 atypical pneumonia (n = 294) were retrospectively included. Five radiologists, blinded to the pathogen test results, assessed the CT scans and classified them as COVID-19 or non-COVID-19 pneumonia. For both groups specific CT features were recorded and a multivariate logistic regression model was used to calculate their ability to predict COVID-19 pneumonia.
Results
The radiologists differentiated between COVID-19 and non-COVID-19 pneumonia with an overall accuracy, sensitivity, and specificity of 88% ± 4 (SD), 79% ± 6 (SD), and 90% ± 6 (SD), respectively. The percentage of correct ratings was lower in the early and late stage of COVID-19 pneumonia compared to the progressive and peak stage (68 and 71% vs 85 and 89%). The variables associated with the most increased risk of COVID-19 pneumonia were band like subpleural opacities (OR 5.55, p < 0.001), vascular enlargement (OR 2.63, p = 0.071), and subpleural curvilinear lines (OR 2.52, p = 0.021). Bronchial wall thickening and centrilobular nodules were associated with decreased risk of COVID-19 pneumonia with OR of 0.30 (p = 0.013) and 0.10 (p < 0.001), respectively.
Conclusions
Radiologists can differentiate between COVID-19 and non-COVID-19 atypical pneumonias at chest CT with high overall accuracy, although a lower performance was observed in the early and late stage of COVID 19 pneumonia. Specific CT features might help to make the correct diagnosis.
Abbreviations: 95% CI, 95% confidence interval; AIC, Akaike information criterion; chi2, Pearson’s chi-squared test; CMV, Cytomegalovirus; COVID 19, Coronavirus disease 2019; GGO, Ground glass opacity; HSV1, Herpes Simplex Virus 1 (HSV1); OR, Odds ratio; PJP, Pneumocystis jiroveci; RSV, Respiratory Syncytial Virus; RT-PCR, Reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, Standard deviation; tt2, Welch’s two-sample t-test
Keywords: CT, COVID-19, Atypical, Viral, Bacteria, Fungal
1. Introduction
Nucleic acid tests, most commonly via reverse transcription polymerase chain reaction (RT-PCR) assay, represent the standardized test for the detection of SARS-CoV-2 RNA from respiratory clinical specimens with a specificity reaching 100% [1], [2]. Besides RT-PCR, chest CT has turned out to be a helpful and fast tool in diagnosing COVID-19 pneumonia, with a moderate to high overall sensitivity of 75–88% [3], [4], [5].
However, compared to the highly specific RT-PCR, the specificity of chest CT in diagnosing COVID-19 is lower, with a reported overall specificity of 46–80% (95% CI: 29–63%) [3], [5]. This can be explained by the fact, that typical signs of COVID-19 pneumonia partially overlap with that of other acute and chronic pulmonary conditions. Some of the findings frequently encountered in COVID-19 pneumonia are: ground glass opacities (GGO), consolidation, crazy paving and enlargement of subsegmental vessels (diameter greater than 3 mm) in areas of GGO [6], [7], [8], [9], [10]. A peripheral and posterior distribution of abnormalities is commonly present [6], [7], [8], [9], [10]. The time course of these findings was investigated by Pan et al. and four stages of COVID-19 pneumonia were reported: early stage (0–4 days after symptom onset) with GGO as main finding, progressive stage (5–8 days after symptom onset) with diffuse GGO, crazy-paving pattern, and consolidation, peak stage (9–13 days after symptom onset) with consolidation becoming more prevalent, and late stage (≥14 days after symptom onset) with gradually absorption of the abnormalities [11]. However, atypical pneumonias other than COVID-19 may have similar patterns. Recent studies have compared the CT findings of COVID-19 pneumonia to that of other viral pneumonias [12], [13], as well as specifically to that of influenza A pneumonia [14], [15]. One of these studies also examined the performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia, revealing an accuracy between 60 and 83% [12].
The aim of this study was to investigate the diagnostic performance of radiologists with different level of experience in differentiating COVID-19 pneumonia from other atypical bacterial, fungal and viral pneumonias. Furthermore, the ability of radiologists to correctly classify infiltrates as COVID-19 pneumonia was tested for every one of the described CT stages of the disease. In addition, this study contains a detailed analysis of infiltrate patterns of all pneumonias included, aiming at identifying those atypical pneumonias most similar to COVID-19 pneumonia and defining imaging markers that might help distinguish COVID-19 pneumonia from its top differential diagnoses.
2. Material and methods
Approval of the institutional ethics commission was obtained for this study (S-293/2020).
2.1. Patients
The patient registry of the department of infectious diseases was searched for the patients with positive RT-PCR for SARS-CoV-2 between January and June 2020 or one of the most common pathogens causing atypical pneumonia between January 2016 and June 2020. Subsequently, the set of identified, positive tested patients was searched for those patients, who in addition underwent a chest CT scan two weeks prior or after the test. Non-COVID-19 pathogens included in this study were: influenza virus, parainfluenza virus, respiratory syncytial virus (RSV), cytomegalovirus (CMV), herpes simplex virus type 1 (HSV1), Mycoplasma pneumoniae, Legionella pneumophila, and Pneumocystis jiroveci (PJP).
For both groups, the identified CT scans were reviewed by a consultant radiologist with three years of experience in thoracic imaging, and a radiology resident in the fourth year of training. Excluded were scans showing a) no acute pulmonary disease, b) acute pneumonia and a positive test result for more than one pathogen, c) acute pulmonary disease other than pneumonia (e.g., pulmonary oedema), d) extensive acute or chronic thoracic pathologies not allowing a certain confirmation of acute pneumonia (e.g., lobar atelectasis). Examples of excluded CT scans are presented in Supplementary Fig. 1.
The electronic medical records were reviewed to collect information about the symptom onset and the type of symptoms of the included patients.
2.2. Clinical specimens and pathogen testing
In both groups, nasopharyngeal and oropharyngeal swabs were the most commonly collected clinical specimens. Further clinical specimens were sputum, tracheal aspirate, bronchial secretion and bronchoalveolar lavage (BAL). PCR represented the diagnostic test used in every patient, both in the COVID-19 and the non-COVID-19 group. Details about the pathogen testing are shown in Supplementary Table 1.
2.3. CT imaging
Imaging of COVID-19 patients was performed using 16- to 256-detector row CT scanners (Siemens Healthcare, Philips Medical Systems). Intravenous contrast agent was used in 13 patients (14%), mainly because of suspected pulmonary embolism.
Non-COVID-19 patients were examined using 6- to 256-detector row CT scanners (Siemens Healthcare, Philips Medical Systems). The CT examinations in this group were performed when pneumonia was suspected or as part of examinations in another acute or non-acute setting. 92 (31%) CT scans were performed after application of intravenous contrast agent. For details regarding imaging see Supplementary Table 2.
2.4. Reader study
CT scans were anonymised and all relevant information, including scan-date, was removed from the images. Four board certified radiologists (R1, R2, R3 and R4 with 33, 28, 10 and 4 years of experience in thoracic imaging, respectively) and a radiology resident in the last year of training (R5) reviewed the CT scans independently and blinded to the pathogen test results. The CT scans had to be classified as COVID-19 or non-COVID-19 pneumonia.
2.5. Pattern analysis
Two other board-certified radiologists with two and six years of experience in thoracic imaging, blinded to the results of pathogen tests, performed a consensus reading of the entire cohort, in order to record specific CT features. The CT features were defined based on the latest glossary of terms of the Fleischner society [16].
2.6. Statistical analysis
Means and standard deviations (SD), as well as absolute and relative frequencies were calculated for patients’ characteristics. Continuous and categorical variables were then compared by using 2-sided student-t tests (tt2) and chi-squared tests (chi2), respectively. Diagnostic performance was analyzed by calculating sensitivity, specificity, accuracy, positive and negative predictive value with associated 95% CI for each reader. Furthermore, mean sensitivity, specificity and accuracy with associated SD for the five readers are presented. For each atypical pathogen in the non-COVID-19 group the percentage of correct ratings as “non-COVID-19 pneumonia” was calculated and the average percentage of correct ratings is presented as mean with associated SD.
Interrater agreement was described by calculating the percentage of agreement as a raw measure and Krippendorff’s alpha coefficient as an adjusted measure. The K-alpha values were interpreted as: 0.00–0.20 = slight agreement, 0.21–0.40 = fair agreement, 0.41–0.60 = moderate agreement, 0.61–0.80 = substantial agreement and 0.81–1.00 = almost perfect agreement [17], [18].
All ratings in the COVID-19 group were stratified by early, progressive, peak and late CT stage of COVID-19 pneumonia [11]. For each disease stage, the percentage of correct ratings with associated (binomial) 95% CI, as well as the associated chi-squared p-value, were calculated.
A multivariate logistic regression model was used to model the probability of having COVID-19 pneumonia, including the covariables: axial distribution, craniocaudal distribution, rounded GGO, band like subpleural opacities, subpleural curvilinear lines, reversed halo sign, perilobular pattern, centrilobular nodules, vascular enlargement, and bronchial wall thickening. A combination of forward and backward variable selection procedure (the FAMoS algorithm) based on the Akaike information criterion (AIC) was used to select the most relevant of the above-mentioned variables starting with the model including all preselected variables [19]. Here, odds ratios with associated 95% CI are given. Furthermore, we conducted a cluster analysis (by Gower’s distance) including the same variables in order to identify the atypical pneumonias which demonstrate CT features most similar/dissimilar to COVID-19 pneumonia [20]. The quality of clustering is given by the silhouette width (sw, interpretation: 0 < sw ≤ 0.25 no structure, 0.25 < sw ≤ 0.5 weak, 0.5 < sw ≤ 0.75 middle, 0.75 < sw ≤ 1 strong). We define observations within one cluster more “similar” to each other that observations between clusters (“dissimilar”).
The statistical analysis was performed by an independent statistician using the statistical programming language R [21]. All p values are to be interpreted in a descriptive manner.
3. Results
3.1. Demographic and clinical data
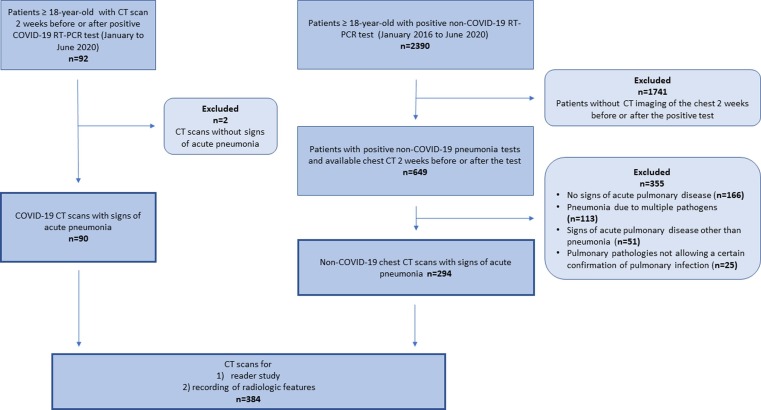
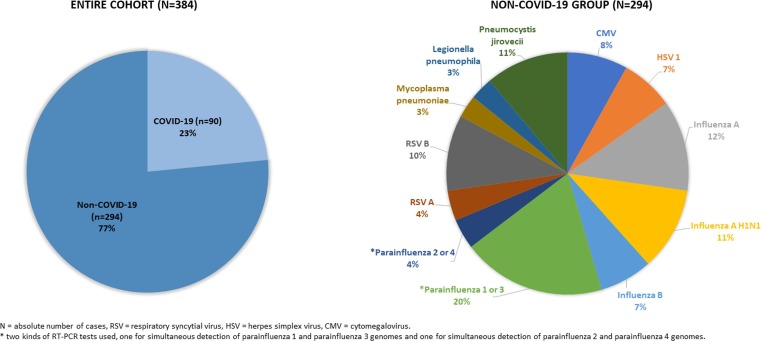
92 consecutive patients tested positive for SARS-CoV-2 and 2390 consecutive patients tested positive for atypical pathogens other than SARS-CoV-2 were identified. As demonstrated in the flowchart of patient enrolment (Fig. 1 ), a total of 90 patients with COVID-19 pneumonia and 294 patients with non-COVID-19 pneumonia were included in the study, resulting in the final cohort of 384 patients. Pie charts demonstrating the size of the COVID-19 and non-COVID-19 group, as well as the distribution of the different pathogens in the non-COVID-19 group are shown in Fig. 2 .
Fig. 1.
Flowchart of patient enrolment.
Fig. 2.
Pie charts demonstrating the size of the COVID-19 and non-COVID-19 group, as well as the distribution of the different pathogens in the non-COVID-19 group.
There was no significant difference in patient age between the COVID-19 and non-COVID-19 group (mean age 62 years in both groups, p = 0.894tt2). The COVID-19 group comprised slightly more men (67% vs 59%; p = 0.228chi2). In terms of the time of CT-imaging in relation to the symptom onset, patients in the COVID-19 group were examined later compared to the patients in the non-COVID-19 group (mean of 9.3 vs 7.1 days, p = 0.011tt2). Regarding the time interval between pathogen testing and CT imaging, on average patients in the COVID-19 group were examined two days after pathogen testing, whereas patients in the non-COVID-19 group 0,8 days prior to the test (p < 0.001tt2). The demographics and clinical symptoms of all patients, as well as the time of the CT-imaging in relation to symptom onset and pathogen testing are listed in Table 1 .
Table 1.
Patient characteristics and information about the time of the CT-imaging in relation to symptom onset and pathogen testing.
| Variable | COVID 19 | Non-COVID-19 | P value |
|---|---|---|---|
| N | 90 | 294 | |
| Age, mean (SD), years | 62 (14) | 62 (13) | 0.894tt2 |
| Sex, n (%) | 0.228chi2 | ||
| Female | 30 (33) | 121 (41) | |
| Male | 60 (67) | 173 (59) | |
| Time from symptom onset to CT, mean (SD), days* | 9,3 (6) | 7,1 (9,7) | 0.011tt2 |
| Time CT after test, mean (SD), days | 2 (4.3) | −0.81 (5.3) | <0.001tt2 |
| Symptoms, n (%) ** | |||
| Fever | 70 (78) | 181 (62) | 0.007chi2 |
| Cough | 68 (76) | 131 (45) | <0.001chi2 |
| Shortness of breath | 62 (69) | 133 (45) | <0.001chi2 |
| Other | 58 (64) | 85 (29) | <0.001chi2 |
N = absolute number of cases, SD = standard deviation, chi2 = Pearson’s chi-squared test, tt2 = Welch’s two-sample t-test.
*Information available for all the patients in the COVID-19 group (n = 90) and for 79% of the patients in the non-COVID-19 group (n = 232).
** Information available for 99% of the patients in the COVID-19 group (n = 89) and for 92% of the patients in the non-COVID-19 group (n = 271).
3.2. Diagnostic performance of radiologists and interrater agreement
The readerś diagnostic accuracy in differentiating COVID-19 pneumonia from non-COVID-19 pneumonia was 92% (R5), 90% (R2 and R4), 85% (R1), and 82% (R3). Sensitivity ranged from 72 to 87% and specificity from 80 to 95% (Table 2 ). The average diagnostic accuracy, sensitivity, and specificity was 88% ± 4 (SD), 79% ± 6 (SD), and 90% ± 6 (SD), respectively.
Table 2.
Years of experience and performance of radiologist in differentiating COVID-19 from non-COVID-19 pneumonia.
| Reader | Experience in Thoracic Radiology, years | Rated as COVID-19 pneumonia, n / Rated as non-COVID-19 pneumonia, n | Accuracy, % (95% CI) | Sensitivity, % (95% CI) | Specificity, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|---|---|---|---|---|
| R1 | 33 | 101/283 | 85 (81–89) | 74 (64–83) | 88 (84–92) | 66 (56–75) | 92 (88–95) |
| R2 | 28 | 93/291 | 90 (86–93) | 80 (70–88) | 93 (89–96) | 77 (68–85) | 94 (90–96) |
| R3 | 10 | 136/248 | 82 (78–86) | 87 (78–93) | 80 (75–85) | 57 (49–66) | 95 (92–97) |
| R4 | 4 | 80/304 | 90 (86–92) | 72 (62–81) | 95 (92–97) | 81 (71–89) | 92 (88–95) |
| R5 | Radiology resident in last year of training | 88/296 | 92 (89–95) | 82 (73–89) | 95 (92–97) | 84 (75–91) | 95 (91–97) |
N = absolute number of cases, 95% CI = 95% confidence intervals.
A moderate interrater agreement was observed in classifying the CT scans as COVID-19 or non-COVID-19 pneumonia (percent agreement = 0.84, Krippendorff's α = 0.58, 95% CI: 0.52–0.65).
PJP, Influenza A H1N1, CMV, and HSV1 were the atypical pneumonias most often misdiagnosed as COVID-19 with an average percentage of false ratings of 19%, 15%, 15% and 13% respectively (Table 3 ).
Table 3.
Percentage and number of correct ratings as non-COVID-19 pneumonia for every pathogen in the non-COVID-19 group.
| Pathogen, n | Cases correctly rated as non-COVID-19 Pneumonia, % (n) |
Mean percentage of correct rating for all readers, % (SD) | ||||
|---|---|---|---|---|---|---|
| Reader 1 | Reader 2 | Reader 3 | Reader 4 | Reader 5 | ||
| Influenza virus (all included types and subtypes), 90 | 91 (82) | 90 (81) | 83 (75) | 93 (84) | 92 (83) | 90 (4) |
| Influenza A (except H1N1), 36 | 94 (34) | 92 (33) | 89 (32) | 92 (33) | 89 (32) | 91 (2) |
| Influenza A H1N1, 33 | 85 (28) | 82 (27) | 73 (24) | 90 (30) | 94 (31) | 85 (8) |
| Influenza B, 21 | 95 (20) | 100 (21) | 90 (19) | 100 (21) | 95 (20) | 96 (4) |
| Parainfluenza virus (all included types), 68* | 96 (65) | 96 (65) | 94 (64) | 96 (67) | 93 (63) | 95 (2) |
| Parainfluenza 1 or 3, 57 | 97 (55) | 97 (55) | 93 (53) | 98 (56) | 93 (53) | 95 (2) |
| Parainfluenza 2 or 4, 11 | 91 (10) | 91 (10) | 100 (11) | 100 (11) | 91 (10) | 95 (5) |
| RSV (all included types), 42 | 93 (39) | 88 (37) | 88 (37) | 100 (42) | 98 (41) | 93 (5) |
| RSV A, 12 | 100 (12) | 83 (10) | 92 (11) | 100 (12) | 100 (12) | 95 (7) |
| RSV B, 30 | 90 (27) | 90 (27) | 87 (26) | 100 (30) | 97 (29) | 93 (5) |
| CMV, 23 | 70 (16) | 96 (22) | 70 (16) | 96 (22) | 96 (22) | 85 (14) |
| HSV 1, 21 | 81 (17) | 95 (20) | 67 (14) | 90 (19) | 100 (21) | 87 (13) |
| Pneumocystis jiroveci, 32 | 78 (25) | 94 (30) | 47 (15) | 84 (27) | 100 (32) | 81 (21) |
| Legionella pneumophila, 9 | 89 (8) | 100 (9) | 67 (6) | 100 (9) | 100 (9) | 91 (14) |
| Mycoplasma pneumoniae, 9 | 89 (8) | 100 (9) | 100 (9) | 100 (9) | 100 (9) | 98 (5) |
N = absolute number of cases, SD = standard deviation, SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2, RSV = respiratory syncytial virus, CMV = Cytomegalovirus, HSV 1 = herpes simplex virus 1. * two kinds of RT-PCR tests used, one for simultaneous detection of parainfluenza 1 and parainfluenza 3 genomes and one for simultaneous detection of parainfluenza 2 and parainfluenza 4 genomes.
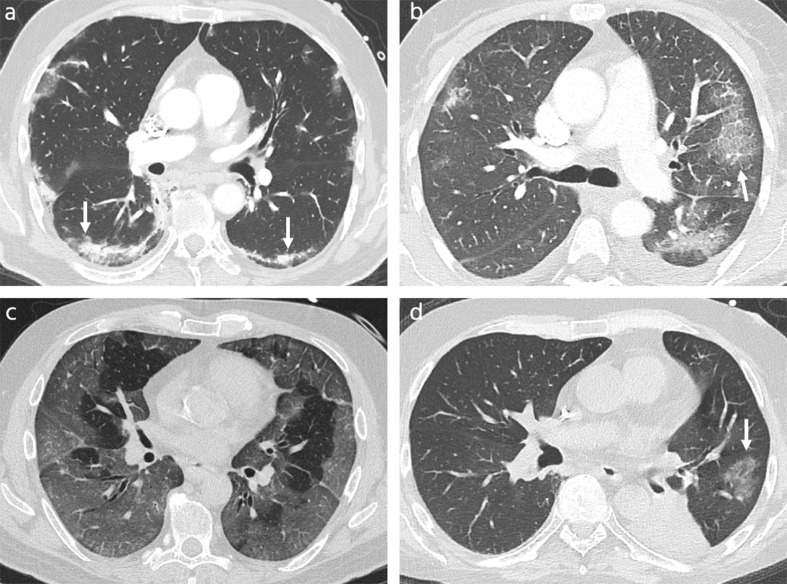
Exemplary cases, which all radiologists or the majority of them misdiagnosed as COVID-19 or non-COVID-19 pneumonia are shown in Fig. 3, Fig. 4 .
Fig. 3.
CT scans of non-COVID-19 atypical pneumonia misinterpreted as COVID-19 pneumonia (a) Only CT scan misdiagnosed by all five radiologists: 52-year-old man with influenza A pneumonia, 7 days after symptom onset, showing subpleural curvilinear lines (arrows). (b, c) Examples misdiagnosed by four of five radiologists: (b) 66-year-old woman with CMV-pneumonia, 3 days after symptom onset, with ground glass opacities and peripheral crazy paving (arrow), (c) 57-year-old man with influenza A H1N1 pneumonia, 20 days after symptom onset, demonstrating extensive, mostly subpleural ground glass opacities. (d) Example misdiagnosed from three of five radiologists: 50-year-old-man with RSV B pneumonia, 2 days after symptom onset, with solitary rounded ground glass opacity in left upper lobe (arrow). Note the complete atelectasis of the left lower lobe.
Fig. 4.
CT scans of COVID-19 pneumonia mistaken for non-COVID-19 pneumonia (a) Only case misdiagnosed by all five radiologists: 64-year-old woman with COVID-19 pneumonia, 14 days after symptom onset, showing nodules of ground glass opacity (arrows). (b-d) Examples misdiagnosed by four of five radiologists: (b) 79-year-old man, 2 days after symptom onset, demonstrating diffuse ground glass opacities, (c) 67-year-old woman, 20 days after symptom onset, demonstrating consolidation in the right lower lobe (arrow). (d) Example misdiagnosed by three of five radiologists: 70-year-old man, 4 days after symptom onset, demonstrating a solitary rounded ground glass opacity in the left upper lobe (arrow).
The diagnostic performance of the radiologists was analysed for each one of the four described CT stages of COVID-19 pneumonia [11]. The average percentage of correct ratings was 68% (95% CI: 58–77) in the early, 85% (95% CI: 79–90) in the progressive, 89% (95% CI: 82–95) in the peak, and 71% (95% CI: 63–78) in the late stage. For details see Supplementary Table 3.
3.3. Analysis of CT features
All recorded CT-features both for the COVID-19 and the non-COVID-19 group are illustrated in detail in Table 4 .
Table 4.
Recorded CT features of the entire cohort.
| Parameter | COVID-19 | Non-COVID-19 | p value |
|---|---|---|---|
| N | 90 | 294 | |
| Laterality, n (%) | 0.391chi2 | ||
| Unilateral | 3 (3) | 19 (6) | |
| Bilateral | 87 (97) | 275 (94) | |
| Axial direction, n (%) | <0.001chi2 | ||
| Central | 2 (2) | 13 (4) | |
| Peripheral | 40 (44) | 28 (10) | |
| No predilection | 48 (53) | 253 (86) | |
| Craniocaudal direction, n (%) | <0.001chi2 | ||
| Upper | 7 (8) | 35 (12) | |
| Lower | 20 (22) | 132 (45) | |
| No predilection | 63 (70) | 127 (43) | |
| Anteroposterior direction, n (%) | 0.806chi2 | ||
| Anterior | 1 (1) | 3 (1) | |
| Posterior | 27 (30) | 78 (27) | |
| No predilection | 62 (69) | 213 (72) | |
| Subpleural sparing, n (%) | 6 (7) | 20 (7) | >0.999chi2 |
| Extent, n (%) * | <0.001chi2 | ||
| Mild | 13 (14) | 107 (36) | |
| Moderate | 46 (51) | 107 (36) | |
| Severe | 31 (34) | 80 (27) | |
| Number of pulmonary segments involved, mean (SD) | 13 (4.6) | 11 (5.2) | <0.001tt2 |
| Decreased attenuation pattern, n (%) | <0.001chi2 | ||
| GGO | 39 (43) | 121 (41) | |
| Consolidation | 3 (3) | 26 (9) | |
| Crazy paving | 2 (2) | 1 (0) | |
| GGO and consolidation | 29 (32) | 83 (28) | |
| GGO and crazy paving | 5 (6) | 10 (3) | |
| Consolidation and crazy paving | 0 (0) | 1 (0) | |
| GGO, consolidation and crazy paving | 11 (12) | 11 (4) | |
| Rounded GGO, n (%) | 37 (41) | 33 (11) | <0.001chi2 |
| Band like subpleural opacities, n (%) | 57 (63) | 27 (9) | <0.001chi2 |
| Reversed halo sign, n (%) | 22 (24) | 10 (3) | <0.001chi2 |
| Perilobular pattern, n (%) | 17 (19) | 16 (5) | <0.001chi2 |
| Halo sign, n (%) | 3 (3) | 13 (4) | 0.880chi2 |
| Solid and/or GGO centrilobular nodules, n (%) | 4 (4) | 172 (59) | <0.001chi2 |
| Tree-in-bud, n (%) | 0 (0) | 12 (4) | 0.109chi2 |
| Subpleural curvilinear lines, n (%) | 40 (44) | 33 (11) | <0.001chi2 |
| Septal thickening, n (%) | 11 (12) | 101 (34) | <0.001chi2 |
| Irregular reticulation, n (%) | 4 (4) | 8 (3) | 0.621chi2 |
| Air bronchogram, n (%) | 14 (16) | 58 (20) | 0.464chi2 |
| Bronchial wall thickening, n (%) | 8 (9) | 143 (49) | <0.001chi2 |
| Vascular thickening, n (%) ** | 29 (32) | 13 (4) | <0.001chi2 |
| Bronchiectasis/Bronchiolectasis, n (%) | 9 (10) | 20 (7) | 0.437chi2 |
| Pleural effusion, n (%) | 18 (20) | 104 (35) | 0.009chi2 |
| Enlarged mediastinal lymph nodes, n (%) | 9 (10) | 16 (5) | 0.197chi2 |
N = absolute number of cases, SD = standard deviation, chi2 = Pearson’s chi-squared test, tt2 = Welch’s two-sample t-test, GGO = ground glass opacity.
Based on the percentage of affected lung parenchyma: mild < 25%, moderate 25 – 50%, severe >50%.
Enlargement of subsegmental vessels (diameter greater than 3 mm) in areas of lung abnormalities.
Based on the AIC, the variables reversed halo sign and perilobular pattern were removed from the final multivariate analysis. A peripheral distribution of pathologic findings was associated with a 2.13-fold risk of COVID-19 pneumonia, when compared to a distribution with no zonal predilection (p = 0.116). The presence of rounded GGO, band like subpleural opacities, and subpleural curvilinear lines was associated with a 1.96-, 5.55-, and 2.52-fold risk of COVID-19 pneumonia, respectively (p = 0.099, p < 0.001, and p = 0.021). Supplementary Fig. 2 illustrates the difference between band like subpleural opacities and subpleural curvilinear lines. Enlargement of subsegmental vessels (diameter greater than 3 mm) in areas of lung abnormalities was associated with a 2.63-fold risk of COVID-19 pneumonia (p = 0.072).
When compared to a distribution of pathologic findings with no zonal predilection, a lower- or an upper-zone preference was associated with decreased risk of COVID-19 pneumonia with odds ratio (OR) of 0.26 and 0.35, respectively (p = 0.054 and p < 0.01). The presence of signs primarily indicating airway disease, namely bronchial wall thickening and centrilobular nodules, was also associated with decreased risk of COVID-19 pneumonia with OR of 0.30 and 0.10 respectively (p = 0.013 and p < 0.001,). The results of the multivariate analysis are demonstrated in Table 5 .
Table 5.
Results of the multivariate analysis.
| Variable | OR (95% CI) | p value |
|---|---|---|
| Axial direction (“no predilection” as reference category) | ||
| Peripheral distribution | 2.13 (0.83–5.49) | 0.116 |
| Craniocaudal direction (“no predilection” as reference category) | ||
| Upper distribution | 0.35 (0.11–0.97) | 0.054 |
| Lower distribution | 0.26 (0.11–0.61) | 0.003 |
| Rounded GGO | 1.96 (0.88–4.33) | 0.099 |
| Band like subpleural opacities | 5.55 (2.55–12.42) | < 0.001 |
| Subpleural curvilinear lines | 2.52 (1.15–5.54) | 0.021 |
| Centrilobular nodules | 0.10 (0.03–0.28) | < 0.001 |
| Bronchial wall thickening | 0.30 (0.11–0.74) | 0.013 |
| Vascular enlargement | 2.63 (0.94–7.73) | 0.072 |
95% CI = 95% confidence interval, OR = odds ratio, GGO = ground glass opacity.
When identifying two clusters in the data (silhouette width = 0.30), most of the pneumonias in Cluster 1 (“COVID-19-cluster”) were cases of COVID-19 pneumonia (86/90 = 96%). Furthermore, PJP, HSV1 and CMV pneumonia were often assigned to this cluster (86%, 81% and 74%, respectively). On the other hand, Mycoplasma and Influenza B pneumonia were most often assigned to cluster 2 (the “non-COVID-19-cluster”, 100% and 71%, respectively). For details about the cluster analysis see Supplementary Table 4.
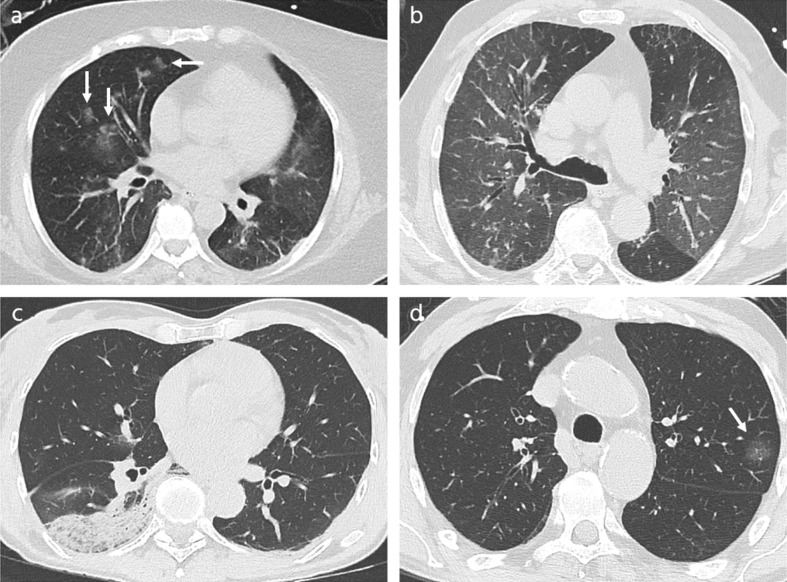
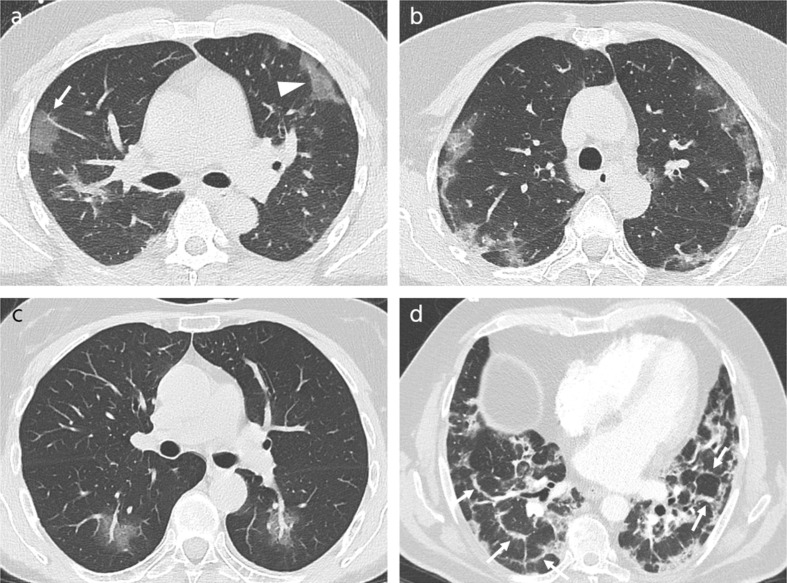
CT-scans of COVID-19 patients, demonstrating signs suggestive of COVID-19 pneumonia are illustrated in Fig. 5 .
Fig. 5.
CT-scans of COVID-19 patients, demonstrating signs suggestive of COVID-19 pneumonia (a) 39-year-old man, 6 days after symptom onset, showing rounded (arrow) and band-like (arrowhead) ground glass opacities in the periphery of the lung. (b) 70-year-old woman, 8 days after symptom onset, showing subpleural curvilinear lines in both lungs. (c) 54-year-old woman, 2 days after symptom onset, demonstrating enlargement of segmental and subsegmental vessels inside areas of ground glass opacity in both lower lobes. (d) 59-year-old man, 22 days after symptom onset, demonstrating extended perilobular opacities in both lungs (arrows).
4. Discussion
Our study proved that the radiologists were able to differentiate COVID-19 from non-COVID-19 pneumonia with high overall accuracy (88% ± 4 SD) and specificity (90% ± 6 SD), but somewhat lower sensitivity (79% ± 6 SD). The radiologistś performance did not directly correlate with their level of experience. With this study, we were able to identify those atypical pneumonias that look most similar to COVID-19 pneumonia in CT images and are therefore most frequently misdiagnosed as COVID-19 pneumonia. Furthermore, correlation between the CT stage of COVID-19 pneumonia and the readerś diagnostic performance was analysed, revealing that in the early and late stage the number of correct ratings was lower. Our analysis indicated that there are patterns suggestive of COVID-19 pneumonia (e.g., subpleural curvilinear lines, band like subpleural opacities, vascular enlargement). Additionally, our study suggested that the presence of signs primarily indicating airway disease (bronchial wall thickening and centrilobular nodules) was associated with non-COVID-19 pneumonia.
The diagnostic accuracy of radiologists in our study was higher compared to that reported by Bai et al. [12]. This is, most likely, the result of the continuous growing experience of radiologists with the imaging findings of COVID-19 pneumonia since the detection of SARS-CoV-2 in December 2019. Most commonly mistaken for COVID-19 pneumonia were PJP and viral pneumonias due to influenza A virus H1N1, CMV, and HSV1. These findings are in line with the results of the cluster analysis, showing that CT patterns of PJP, HSV1 and CMV pneumonia are most similar to those of COVID-19 pneumonia. This can be explained by the fact that GGO represents the main pattern of the above-mentioned atypical pneumonias, similarly to COVID-19 pneumonia [22]. COVID-19 pneumonia was more frequently misdiagnosed as non-COVID-19 pneumonia in the early and late CT stage of the disease. This possibly reflects the fact that GGO is the main pathologic finding in the early stage, and that the absorption of the typical findings of COVID-19 pneumonia in the late phase is commonly accompanied by signs of organising pneumonia or early signs of fibrosis. These features can be encountered in multiple infectious diseases of the lung, making the CT appearance somewhat unspecific [22], [23].
The chest CT findings of COVID-19 pneumonia in our study were very similar to previous studies [6], [7], [8], [9], [10], [11]. In contrast to previous studies, which compared the CT findings between COVID-19 and viral pneumonias [12], [13], our study included well balanced subgroups of viral as well as fungal and atypical bacterial pneumonias. Including convincing case numbers of influenza pneumonia in particular, is crucial, as influenza pneumonia represents the main differential diagnosis of COVID-19 pneumonia during the cold months in temperate regions. Moreover, the number of COVID-19 patients was intentionally chosen to be comparable to that of the subgroups of atypical pneumonias. This was done to create a more realistic setting, possibly simulating the incidence and prevalence of COVID-19 pneumonia after the end of its pandemic phase. To our knowledge, this study is the first to examine the radiologists’ performance in relation to the stage of the COVID-19 pneumonia and the first to search for the atypical pneumonias most often misdiagnosed as COVID-19. Furthermore, as far as we know, our study is the first to work out pattern-based top differential diagnoses for COVID-19 pneumonia.
The results of the presented work need to be seen in light of the study design and its limitations. The readers had to assess the CT images without any additional information about the patientś symptoms or medical background. This might be a potential bias, as precise clinical information usually helps the radiologist make the correct diagnosis [24]. This study showed that even less experienced radiologists in training are able to reliably differentiate COVID-19 from other atypical pneumonias. As only one of the five readers was a radiologist in training, the performance of this reader might not be representative and therefore should not be generalized. However, the authors of this study strongly believe that due to the tremendous research activities as well as the educational activities focusing on diagnosing COVID-19, a notable increase in knowledge was obtained within the last year, independent of the level of experience in radiology. Due to its retrospective character, the CT protocols used in this study were not standardized. This can be seen as a flaw in study design. However, it reflects the daily routine when, for example, pulmonary embolism is suspected, but pneumonic infiltrates are diagnosed [25]. A selection bias, also associated with the retrospective character of this study, represents the fact that, in comparison to the suspected cases of COVID-19 pneumonia, the CT scans of atypical pneumonias were not ordered according to fixed criteria. One of the initial steps in this study was to identify different pathogens forming the group of non-COVID-19 pneumonias. As inclusion of all known atypical pneumonias is not possible, the choice had to be narrowed down, resulting in a possible bias. The selection process was performed taking into account the characteristic patterns of COVID-19 pneumonia, and aimed at including atypical pneumonias most similar to it. This is why, for example, comparatively more common fungal pneumonias that look quite different from COVID-19 pneumonia on chest CT, such as aspergillosis, were not included in this study. The multivariate regression analysis revealed highly clinically relevant results regarding the CT features suggestive of COVID-19 pneumonia, although not all results were statistically significant (interpreted as “p ≤ 0.05”). The statistical significance of the variables “peripheral distribution of abnormalities” (OR = 2.13, p = 0.116), “rounded GGO” (OR = 1.96, p = 0.099) and “vascular enlargement in areas of GGO” (OR = 2.63, p = 0.072) might be shown in future confirmatory clinical studies with a greater number of patients, providing for higher statistical power to detect the relevant effects.
In conclusion, this study showed that radiologists are capable of differentiating between COVID-19 and non-COVID-19 atypical pneumonia on chest CT. The fact that reliable and confident differentiation between COVID-19 and other atypical pneumonias is possible, apparently independent from the level of experience, might serve as an indicator for successful education during the pandemic. Our work reveals the most important COVID-19 differential diagnoses within the studied patient cohort. This indicates atypical pneumonias a radiologist should take into account when being asked to assess a suspected COVID-19 case. Last but not least, the presented study is meant to provide assistance to radiologists, focusing on pattern analysis of atypical pneumonias most similar to COVID-19 pneumonia, presenting pitfalls in diagnosing atypical pneumonias and developing reliable distinguishing criteria.
CRediT authorship contribution statement
Athanasios Giannakis: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Dorottya Móré: Data curation, Investigation, Writing – review & editing. Stella Erdmann: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. Laurent Kintzelé: Investigation, Writing – review & editing. Ralph Michael Fischer: Investigation, Writing – review & editing. Monika Nadja Vogel: Investigation, Writing – review & editing. David Lukas Mangold: Investigation, Writing – review & editing. Oyunbileg von Stackelberg: Project administration, Software, Supervision, Writing – review & editing. Paul Schnitzler: Conceptualization, Data curation, Investigation, Validation, Writing – review & editing. Stefan Zimmermann: Conceptualization, Data curation, Investigation, Validation, Writing – review & editing. Claus Peter Heussel: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing. Hans-Ulrich Kauczor: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing. Katharina Hellbach: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejrad.2021.110002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12(546):eabc1931. doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- 2.Böger B., Fachi M.M., Vilhena R.O., Cobre A.F., Tonin F.S., Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021;49(1):21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatami F., Saatchi M., Zadeh S.S.T., Aghamir Z.S., Shabestari A.N., Reis L.O., Aghamir S.M.K. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-80061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waller J.V., Allen I.E., Lin K.K., Diaz M.J., Henry T.S., Hope M.D. The limited sensitivity of chest computed tomography relative to reverse transcription polymerase chain reaction for severe acute respiratory syndrome coronavirus-2 infection: a systematic review on COVID-19 diagnostics. Invest. Radiol. 2020;55(12):754–761. doi: 10.1097/RLI.0000000000000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Islam, S. Ebrahimzadeh, J.-P. Salameh, et al., Thoracic imaging tests for the diagnosis of COVID-19. Cochrane Infectious Diseases Group, ed. Cochrane Database Syst. Rev. doi:10.1002/14651858.CD013639.pub4. [DOI] [PMC free article] [PubMed]
- 6.Bernheim A., Mei X., Huang M., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruso D., Zerunian M., Polici M., Pucciarelli F., Polidori T., Rucci C., Guido G., Bracci B., De Dominicis C., Laghi A. Chest CT Features of COVID-19 in Rome, Italy. Radiology. 2020;296(2):E79–E85. doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Z., Zhang Y., Wang Y.i., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z., Guo D., Li C., Fang Z., Chen L., Yang R., Li X., Zeng W. Coronavirus disease 2019: initial chest CT findings. Eur. Radiol. 2020;30(8):4398–4406. doi: 10.1007/s00330-020-06816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwee T.C., Kwee R.M. Chest CT in COVID-19: what the radiologist needs to know. RadioGraphics. 2020;40(7):1848–1865. doi: 10.1148/rg.2020200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan F., Ye T., Sun P., Gui S., Liang B.o., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.-B., Wang D.-C., Mei J.i., Jiang X.-L., Zeng Q.-H., Egglin T.K., Hu P.-F., Agarwal S., Xie F.-F., Li S., Healey T., Atalay M.K., Liao W.-H. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296(2):E46–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Fang X.u., Bian Y., Lu J. Comparison of chest CT findings between COVID-19 pneumonia and other types of viral pneumonia: a two-center retrospective study. Eur. Radiol. 2020;30(10):5470–5478. doi: 10.1007/s00330-020-06925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Wei R., Rao G., Zhu J., Song B. Characteristic CT findings distinguishing 2019 novel coronavirus disease (COVID-19) from influenza pneumonia. Eur. Radiol. 2020;30(9):4910–4917. doi: 10.1007/s00330-020-06880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Z., Kang Z., Yang D., Ding S., Luo H., Xiao E. A Comparison of clinical and chest CT findings in patients with influenza A (H1N1) virus infection and coronavirus disease (COVID-19) Am. J. Roentgenol. 2020;215(5):1065–1071. doi: 10.2214/AJR.20.23214. [DOI] [PubMed] [Google Scholar]
- 16.Hansell D.M., Bankier A.A., MacMahon H., McLoud T.C., Müller N.L., Remy J. Fleischner society: glossary of terms for thoracic imaging. Radiology. 2008;246(3):697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 17.The IST-3 Collaborative Group, G. Mair, R. von Kummer, et al., Observer reliability of CT angiography in the assessment of acute ischaemic stroke: data from the Third International Stroke Trial, Neuroradiology (2015) doi: 10.1007/s00234-014-1441-0. [DOI] [PMC free article] [PubMed]
- 18.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977 doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 19.M. Gabel, T. Hohl, A. Imle, O.T. Fackler, F. Graw, FAMoS: A Flexible and dynamic Algorithm for Model Selection to analyse complex systems dynamics. Asquith B, ed, PLoS Comput. Biol. (2019) doi: 10.1371/journal.pcbi.1007230. [DOI] [PMC free article] [PubMed]
- 20.Gower J.C. A general coefficient of similarity and some of its properties. Biometrics. 1971 doi: 10.2307/2528823. [DOI] [Google Scholar]
- 21.R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2020, URL https://www.R-project.org/.
- 22.Koo H.J., Lim S., Choe J., Choi S.-H., Sung H., Do K.-H. Radiographic and CT features of viral pneumonia. RadioGraphics. 2018;38(3):719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 23.Parekh M., Donuru A., Balasubramanya R., Kapur S. Review of the chest CT differential diagnosis of ground-glass opacities in the COVID era. Radiology. 2020;297(3):E289–E302. doi: 10.1148/radiol.2020202504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie A., Jones A.J., Goddard P.R. The influence of clinical information on the reporting of CT by radiologists. BJR. 2000;73(874):1052–1055. doi: 10.1259/bjr.73.874.11271897. [DOI] [PubMed] [Google Scholar]
- 25.Green D.B., Raptis C.A., Alvaro Huete Garin I., Bhalla S. Negative computed tomography for acute pulmonary embolism. Radiol. Clin. North Am. 2015;53(4):789–799. doi: 10.1016/j.rcl.2015.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.