Abstract
Campylobacter jejuni is a Gram-negative, pathogenic bacterium which causes campylobacteriosis, a form of gastroenteritis. C. jejuni is the most frequent cause of food borne illness in the world, surpassing Salmonella and E. coli. Coating the surface of C. jejuni is a layer of sugar molecules known as the capsular polysaccharide that, in C. jejuni NCTC 11168, is composed of a repeating unit of d-glycero-l-gluco-heptose, d-glucuronic acid, d-N-acetyl-galactosamine, and d-ribose. The d-glucuronic acid moiety is further amidated with either serinol or ethanolamine. It is unknown how these modifications are synthesized and attached to the polysaccharide. Here we report the catalytic activities of two previously uncharacterized, PLP-dependent enzymes, Cj1436 and Cj1437, from C. jejuni NCTC 11168. Using a combination of mass spectrometry and nuclear magnetic resonance, we determined that Cj1436 catalyzes the decarboxylation of l-serine phosphate to ethanolamine phosphate. Cj1437 was shown to catalyze the transamination of dihydroxyacetone phosphate to (S)-serinol phosphate in the presence of l-glutamate. The probable routes to the ultimate formation of the glucuronamide substructures in the capsular polysaccharides of C. jejuni are discussed.
Graphical Abstract
Introduction
Campylobacter jejuni is a Gram-negative, zoonotic organism which causes campylobacteriosis, the leading food-borne illness in the world.1 In the United States, campylobacteriosis accounts for 1.3 million cases of gastroenteritis each year.2 C. jejuni is a commensal organism in chickens that can easily spread to other farm animals and pets through unsanitary drinking water, meat processing facilities, unpasteurized milk, and pet feces.3 Symptoms of campylobacteriosis include nausea, cramps, fever, and bloody diarrhea.4 Additionally, Campylobacter infections precede ~40% of all Guillain-Barré syndrome diagnoses.5, 6 It is estimated that Campylobacter causes over 25,000 deaths annually in children under 5 years of age, primarily affecting Asia and Africa.7 There is no known FDA approved vaccine for Campylobacter infection making it a significant public health concern. The best vaccine candidates to date have been glycoconjugate vaccines, which are based on the surface-exposed sugars of C. jejuni.7
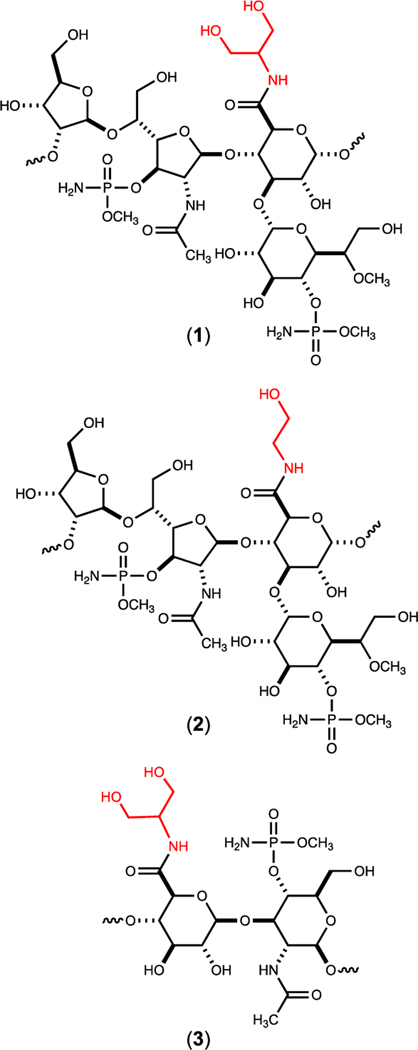
Coating the exterior surface of C. jejuni is a polymeric layer of carbohydrates known as the capsular polysaccharide (CPS). This polysaccharide is quite diverse among different strains of C. jejuni and is used to serotype ~60 different variants of C. jejuni.8 The CPS is known to aid in evasion of the host immune system and adherence to epithelial tissue.9 The CPS of C. jejuni NCTC 11168 (HS:2) is composed of a repeating unit of d-glycero-l-gluco-heptose, d-glucuronic acid, d-N-acetyl-galactosamine, and d-ribose.10, 11 The capsule is further modified by methylation, phosphoramidylation, and amidation. The glucuronic acid moiety of the CPS from C. jejuni serotype HS:2 is found amidated with either ethanolamine or serinol. In the HS:19 serotype the glucuronate is amidated only with serinol.12 The biosynthetic pathways for the formation of these two modifications to the glucuronate moiety are currently unknown. The structures of the capsules found in serotypes HS:2 and HS:19 are provided in Figure 1.
Figure 1.
Structures of the capsular polysaccharides found in C. jejuni NCTC 11168 (HS:2) (1 and 2) and C. jejuni strain RM1285 (HS:19) (3). The amide moiety in each structure is highlighted in red.
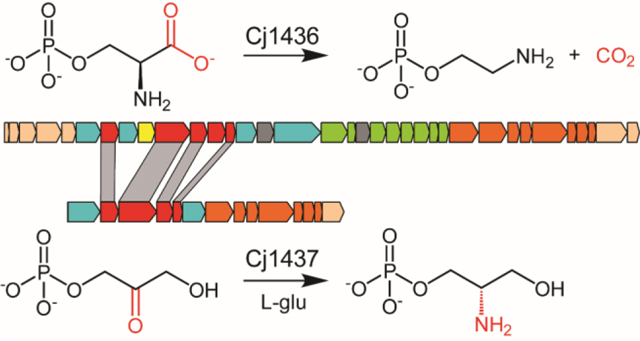
The DNA cluster responsible for the formation of the CPS in C. jejuni NCTC 11168 is composed of 35 genes as illustrated in Figure S1.13 Previous investigations have functionally characterized the genes necessary for the biosynthesis of the d-glycero-l-gluco-heptose moiety and modification (Cj1152, Cj1423, Cj1424, Cj1425, Cj1427, Cj1428, and Cj1430.14–19 In addition, the enzymes Cj1415, Cj1416, Cj1417, and Cj1418 have been shown to be required for biosynthesis of the phosphoramidate modification () and gene knockout experiments have identified the enzymes (Cj1422 and C1421) required for the transfer of the phosphoramidate modifications to specific sugar receptors.8, 20–25 More recently, we have shown that Cj1441 is required for the conversion of UDP-glucose to UDP-glucuronate and that this enzyme is unable to catalyze the formation of an amide bond via the attack of an amine substrate with the putative thioester intermediate formed during the oxidation of UDP-glucose.26 The enzymes required for the biosynthesis of the two amines (serinol and ethanolamine) found in the HS:2 capsule are currently unknown.
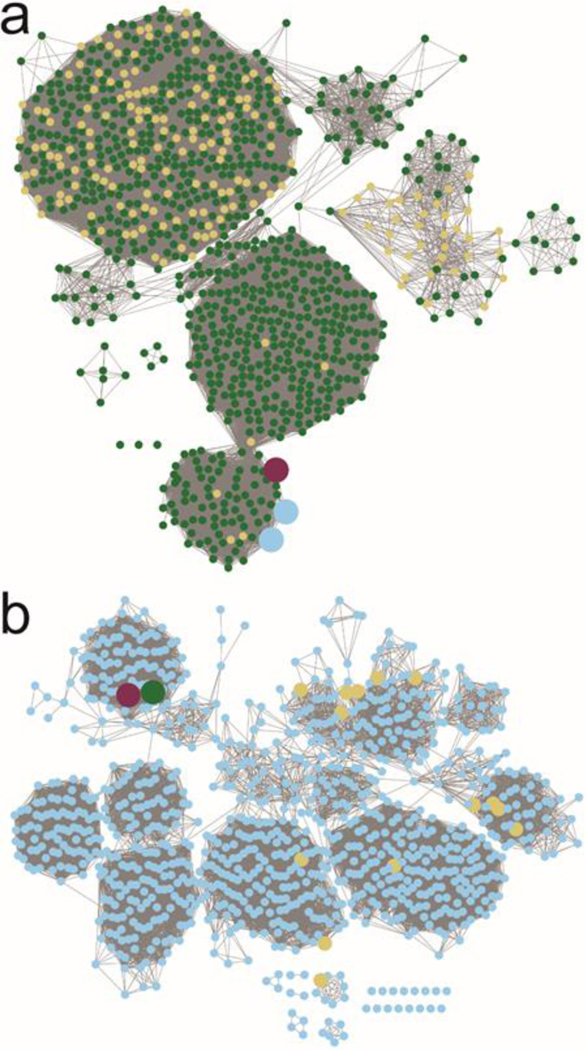
The three most likely enzymes required for the biosynthesis of serinol and ethanolamine in the HS:2 capsule of C. jejuni are Cj1435, Cj1436, and Cj1437. Cj1435 is predicted to be a HAD phosphatase, while Cj1436 and Cj1437 are anticipated to be pyridoxal phosphate (PLP) dependent enzymes. Close homologs to Cj1435 and Cj1437 are found in the gene cluster for the HS:19 serotype, but none to Cj1436. A sequence similarity network (SSN) for the closest 1000 protein sequences to Cj1436 is presented in Figure 2a at a threshold of 39% sequence identity.27, 28 The nearest functionally characterized enzyme is l-serine phosphate decarboxylase (SMUL_1544) with an overall sequence identity of 46%. This enzyme has been shown to catalyze the formation of ethanolamine phosphate during the biosynthesis of norcobamides.29, 30 The SSN with Cj1437 for the nearest 1000 sequences is presented in Figure 2b at a sequence identity of 41%. The nearest functionally characterized enzyme to Cj1437 is histidinol phosphate aminotransferase, which catalyzes the transamination of imidazole acetol phosphate, in the presence of l-glutamate, during the biosynthesis of l-histidine. Here we have purified Cj1436 and Cj1437 and have functionally characterized these two enzymes. Cj1436 is found to catalyze the decarboxylation of l-serine phosphate to generate ethanolamine phosphate and CO2, while Cj1437 catalyzes the transamination of dihydroxyacetone phosphate to (S)-serinol phosphate in the presence of l-glutamate.
Figure 2.
Sequence similarity networks for Cj1436 and Cj1437 using the 1000 nearest protein sequences. (a) The SSN for Cj1436 clustered at a sequence identity of 39%. Maroon node is Cj1436. Blue nodes are enzymes sharing 99% sequence identity to the functionally characterized l-serine phosphate decarboxylase (SMUL_1544 from Sulfurospirillum multivorans). Yellow nodes are annotated as l-threonine phosphate decarboxylases. Green nodes are described as aminotransferases. (b) SSN for Cj1437 clustered at a sequence identity of 41%. The maroon node is Cj1437 while the green node is HS19.10. Yellow nodes are proteins with a Swissprot designation as histidinol phosphate aminotransferases. Blue nodes do not have a Swissprot annotation, but are generically described as aminotransferases.
Materials and Methods
Cloning, Expression, and Purification of Cj1436 and Cj1437.
The genes for Cj1436 (UniProt id: Q0P8H8) and Cj1437 (UniProt id: Q0P8H7) from serotype HS:2 were cloned from the genomic DNA of C. jejuni NCTC 11168 obtained from ATCC. The genomic DNA served as the starting template for PCR using Phusion DNA polymerase (New England Biolabs). Primers were designed that incorporated NdeI and XhoI restriction sites and the resulting fragments were digested at 37 °C and ligated into a pET31b expression vector, which carried a C-terminal hexahistidine tag. The gene encoding the homolog to Cj1437 (HS19.10) from the HS:19 serotype in C. jejuni strain RM1285 (Uniprot id: Q5M6M3) was codon optimized for expression in E. coli and ordered from Twist Biosciences in a pET28a expression vector with an N-terminal hexahistidine tag.
The pET31b vector was used to transform BL21 E. coli cells (Novagen) via electroporation. Cells harboring the pET31b-Cj1436, pET31b-Cj1437 and pET28a-HS19.10 plasmids were cultured in lysogeny broth with 100 mg/L ampicillin for the pET31b constructs and 50 mg/L kanamycin for the pET28a construct. The cells were grown at 37 °C with shaking and induced with 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the optical density reached 0.8 at 600 nm. The cells were allowed to express protein at 21 °C for 18 h after induction and then harvested by centrifugation at 15,000 rcf at 4 °C. The cell pellet was resuspended in loading buffer (50 mM HEPES/K+, 300 mM KCl, 20 mM imidazole, pH 8.0) and lysed with sonication. The sonicated cells were centrifuged at 25,000 rcf at 4 °C before the lysate was passed through a 0.45 μm filter (Whatman). The sample was loaded onto a prepacked 5-mL HisTrap (GE Healthcare) nickel affinity column. The protein was eluted from the column using 50 mM HEPES/K+, pH 8.0, 300 mM KCl, and 250 mM imidazole over a gradient of 25 column volumes. The protein was pooled and dialyzed against 10 mM HEPES/K+ pH 8.0, and 200 mM KCl. Cj1436 was concentrated to 10 mg/mL and flash frozen using liquid nitrogen before being stored at −80 °C. Approximately 5 mg of protein was obtained per liter of cell culture. Cj1437 (~3 mg of protein per liter of cell culture) was concentrated to 3.5 mg/mL and flash frozen before being stored at −80 °C. HS19.10 (~10 mg of protein per liter of cell culture) was concentrated to 6 mg/mL and flash frozen before being stored at −80 °C.
Bioinformatic Analysis of Cj1436 and Cj1437.
The amino acid sequences for Cj1436 and Cj1437 were obtained from the Uniprot database. Individually, the protein sequences for the two genes were input as the search queries for the BLAST section of the EFI-EST webtool to generate the sequence similarity networks (SSNs).27, 28 The parameters were set such that the BLAST search would retrieve the 1000 closest sequences to the search query. Sequence identities for Cj1436 and Cj1437 were set at 39% and 41%, respectively. The networks were created using the yFile organic layout and all images were created using Cytoscape version 3.7.2.
Determination of Kinetic Constants for Cj1437 and HS19.10.
The kinetic constants for Cj1437 and HS19.10 were determined using an l-glutamate dehydrogenase coupled assay to monitor the formation of α-ketoglutarate. The reaction mixtures contained 150 nM enzyme, 250 μM PLP, 250 μM NADPH, 15 mM ammonium chloride, 2.0 mM l-glutamate, 5.0 units of l-glutamate dehydrogenase, varying levels of DHAP (0.005 mM – 2.0 mM), and 50 mM HEPES/K+ buffer at pH 8.0. The reaction was monitored spectrophotometrically at 340 nm and 25 °C with a Spectramax340 UV-visible spectrophotometer. The kinetic parameters were determined by fitting the initial rates to eqn. 1 using GraFit 5, where ν is the initial velocity of the reaction, Et is the enzyme concentration, kcat is the turnover number, [A] is the substrate concentration, and Km is the Michaelis constant.
| (1) |
Determination of Reaction Products for Cj1436.
The reaction catalyzed by Cj1436 was determined by following the decarboxylation of l-serine phosphate via 1H and 31P NMR spectroscopy. The reaction was conducted in 50 mM phosphate buffer, pD 8.0, in D2O and initiated by the addition of 1.0 μM Cj1436 with a fixed level of PLP (250 μM) and a fixed concentration of l-serine phosphate (5.0 mM). The catalytic activity of Cj1436 was also tested with l-serine, l-threonine phosphate, l-aspartate, and l-glutamate at a fixed concentration of 5.0 mM. The reaction was also monitored using 31P NMR spectroscopy by monitoring the phosphorus resonance for the product (ethanolamine phosphate) relative to the 31P resonance for substrate (serine phosphate) as a function of time. The reaction was conducted in 50 mM HEPES/K+ at pH 8.0 containing 250 μM PLP, 5.0 mM l-serine and initiated by the addition of 1.0 μM enzyme.
Membrane Inlet Mass Spectrometry (MIMS) for the Determination of the Kinetic Constants for Cj1436.
Steady-state kinetic constants for Cj1436 were determined by following the decarboxylation of l-serine phosphate using MIMS.31 – 33. In these experiments, assay mixtures contained 0.1, 0.5, 1.0, 5.0, and 10 mM l-serine phosphate dissolved in 100 mM HEPES/K+ buffer, pH 8.0. Reactions were initiated at 25 oC by the addition of an enzyme/PLP mixture in 100 mM HEPES/K+ buffer, pH 8.0, to give final concentrations of 1.0 μM Cj1436 and 250 μM PLP (total volume 2.0 mL). The catalytic activity of Cj1436 was also tested with l-serine at a fixed concentration of 5.0 mM.
Reaction Product Confirmation Using Mass Spectrometry.
Cj1436 (1.0 μM was incubated with 5.0 mM l-serine phosphate, 250 μM PLP, and 50 mM ammonium bicarbonate at pH 8.0. The reaction was monitored using 31P NMR spectroscopy until the reaction was complete. The solution was filtered through a GE Healthcare Vivaspin 500 10 kDA MWCO filter and 100 μL of the flow-through was submitted for mass spectrometry analysis using a Thermo Scientific Q Exactive Focus mass spectrometer. Cj1437 (1.0 μM) was incubated with 5.0 mM DHAP, 15 mM l-glutamate, and 250 μM PLP in 50 mM ammonium bicarbonate buffer at pH 8.0 until the reaction reached equilibrium as determined by 31P NMR spectroscopy. The solution was filtered through a GE Healthcare Vivaspin 500 10 kDa MWCO filter and 100 μL of the flow-through was analyzed using a Thermo Scientific Q Exactive Focus mass spectrometer.
Chemical Synthesis of (R/S)-Serinol phosphate and (S)-Serinol Phosphate.
The synthesis of racemic and (S)-serinol phosphate is described in the Supplementary Information.
Stereochemical Analysis of Serinol Phosphate Produced by Cj1437 and HS19.10.
The stereochemistry of the chiral center at C2 of the serinol phosphate produced by Cj1437 and HS19.10 was determined by monitoring the exchange of the hydrogen at C2 of (R/S)-serinol phosphate and (S)-serinol phosphate with solvent deuterium. Cj1437 or HS19.10, at a concentration of 3.0 μM, was allowed to incubate with 3.0 mM (R/S)-serinol in 50 mM phosphate buffer (D2O) at pH 8.0. The exchange of the hydrogen at C2 of serinol phosphate with deuterium was monitored using 1H NMR spectroscopy (Avance III 400 MHz). A similar experiment was conducted with the (S)-serinol phosphate enantiomer. The exchange reactions were also conducted in the presence of 250 μM PLP and 250 μM α-ketoglutarate using the HS19.10 enzyme.
Results
Determination of the Reaction Catalyzed by Cj1436.
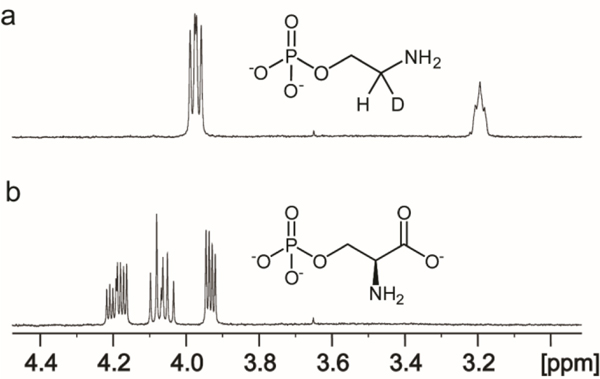
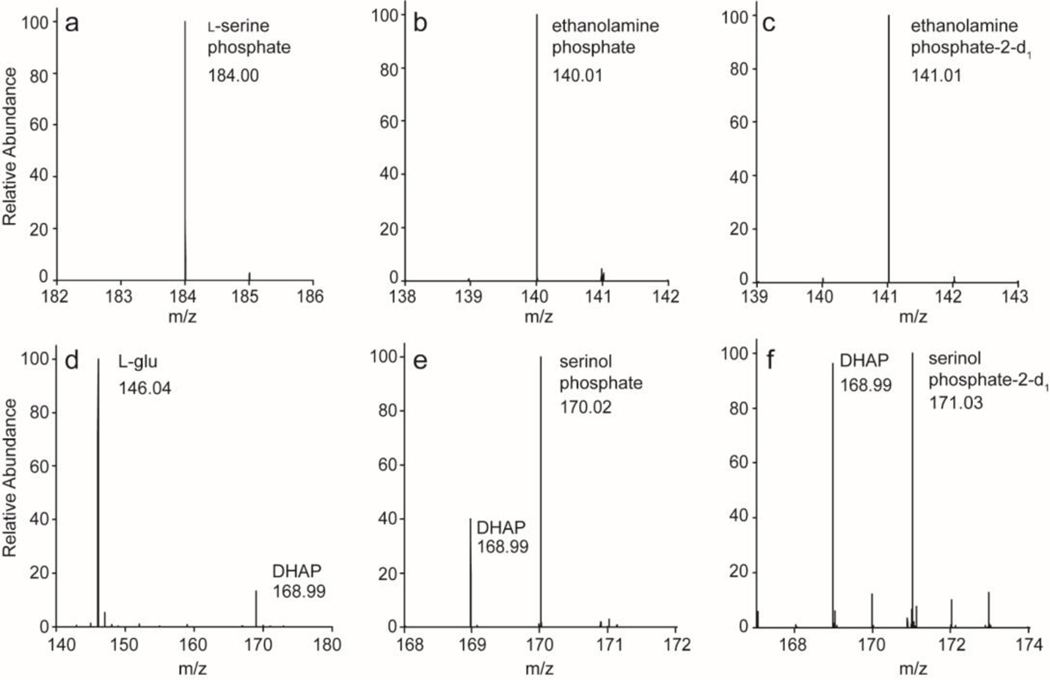
The closest functionally characterized enzyme to Cj1436 is SMUL_1544 from S. multivorans and this enzyme has been shown to catalyze the decarboxylation of l-serine phosphate to ethanolamine phosphate and CO2 (29, 30). 1H NMR spectroscopy was used to monitor the reaction catalyzed by Cj1436 in D2O with l-serine phosphate and the results are presented in Figure 3 showing that the product is ethanolamine phosphate. The two hydrogens from C2 resonate at 3.96 ppm and appear as a doublet of doublets due to coupling with the adjacent phosphate and the single proton from C1 with coupling constants of 1.89 and 12.14 Hz, respectively (Figure 3a). The single proton from C1 appears as a broadened triplet at 3.19 ppm. The 1H NMR spectrum of the substrate prior to the addition of enzyme is provided in Figure 3b. In the 31P NMR spectrum, the substrate exhibits a single resonance at 3.89 ppm (Figure 4b) whereas the product resonates at 3.82 ppm (Figure 4a). The formation of ethanolamine phosphate was confirmed using ESI negative mode mass spectrometry. The substrate of the reaction, l-serine phosphate is present at an m/z of 184.0 for the M-H ion (Figure 5a). The product of the reaction, ethanolamine phosphate, is found at an m/z of 140.01 when the reaction is conducted in H2O (Figure 5b) and 141.0 when the reaction is conducted in D2O (Figure 5c).
Figure 3.
1H NMR spectrum of the reaction catalyzed by Cj1436 in D2O. (a) The product of the reaction catalyzed by Cj1436 in the presence of l-serine phosphate is ethanolamine phosphate. (b) The substrate of the reaction, l-serine phosphate, prior to the addition of enzyme. Additional details are provided in the text.
Figure 4.
31P NMR spectrum of the reaction catalyzed by Cj1436. (a) Ethanolamine phosphate resonates at 3.82 ppm. A small amount of remaining substrate appears at 3.89 ppm. (b) Control reaction in the absence of enzyme showing the substrate, l-serine phosphate at 3.89 ppm. Additional details are provided in the text.
Figure 5.
Mass spectrometry data for Cj1436 and Cj1437. (a) l-Serine phosphate, the substrate for Cj1436 appears in a no-enzyme control at an m/z of 184.00 for the M-H ion. (b) Product of the reaction catalyzed by Cj1436. Ethanolamine phosphate appears at an m/z of 140.01 for the M-H ion. (c) Product of the reaction catalyzed by Cj1436 in the presence of D2O. The deuterated product, (2-d1)-ethanolamine phosphate appears at an m/z of 141.01 for the M-H ion. (d) l-glutamate and DHAP, substrates for the reaction catalyzed by Cj1437 appear in a no-enzyme control at m/z of 146.04 and 168.99 respectively for the M-H ions. (e) Product of the reaction catalyzed by Cj1437. The product serinol phosphate appears at an m/z equal to 170.02 while leftover DHAP substrate appears at 168.99 for the M-H ions. (f) The product of the reaction catalyzed by Cj1437 in the presence of D2O. (2-d1)-Serinol phosphate appears at an m/z of 171.03 while leftover DHAP substrate appears at 168.99 for the M-H ions.
Determination of Reaction Catalyzed by Cj1437.
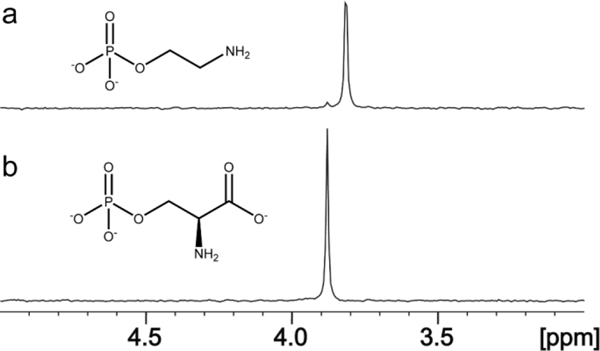
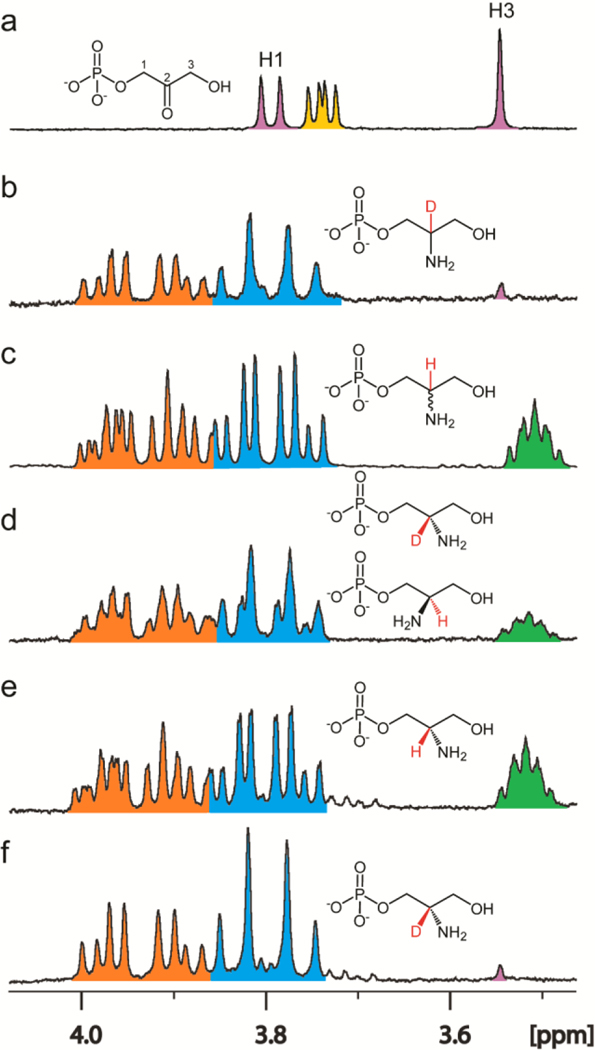
The sequence similarity network for Cj1437 indicates that this enzyme will most likely catalyze a transamination reaction since the closest functionally characterized enzyme is histidinol phosphate aminotransferase. Since the pathway for the biosynthesis of the CPS in serotype HS:2 must eventually make serinol, the most probable reaction catalyzed by Cj1437 is the transamination of DHAP with an amino donor such as l-glutamate to make serinol phosphate. When tested with dihydroxy acetone (DHA) as a potential substrate, the rate was less than 3% of the rate with DHAP. The reaction catalyzed by Cj1437 was initially characterized using 1H NMR spectroscopy using 3.0 mM DHAP and 15 mM l-glutamate. A portion of the 1H NMR spectrum of the mixture of DHAP and l-glutamate in D2O is presented in Figure 6a. After the addition of Cj1437, deuterated serinol phosphate is formed as indicated by the pair of doublet of doublets for the two protons at C1 centered at 3.95 ppm and the doublet of doublets for the two protons at C3 centered at 3.80 ppm (Figure 6b). The NMR spectrum of the chemically synthesized racemic serinol phosphate is shown for comparison in Figure 6c, where the hydrogen at C2 resonates at 3.51 ppm thus confirming the formation of serinol phosphate.
Figure 6.
1H NMR spectra of reactions catalyzed by Cj1437. (a) Control spectrum of a mixture of DHAP (pink) and l-glutamate (yellow). The resonance for the H1 hydrogen of DHAP is centered at 3.79 ppm, while the H3 hydrogen is centered at 3.55 ppm. The Cα of l-glutamate appears as a doublet of doublets centered at 3.74. (b) Product of the reaction catalyzed by Cj1437. The H1 hydrogens (orange) are centered at 3.94 ppm. The C3 hydrogens (blue) are centered at 3.80 ppm. A small amount of DHAP is left over as shown by the pink singlet at 3.55 ppm. (c) Chemically synthesized (R/S)-serinol phosphate. The H1 protons are centered at 3.93 ppm and the H2 hydrogen (green) is centered at 3.51 ppm. (d) Incubation of Cj1437 with (R/S)-serinol phosphate with 50 μM αKG in D2O for 2 h. (e) Chemically synthesized (S)-serinol phosphate. (f) Incubation of Cj1437 with (S)-serinol phosphate and 250 μM α-KG for 2 h. Additional details are provided in the text.
The product of the reaction catalyzed by Cj1437 has a chiral center at C2. To determine the stereochemistry at C2, the enzyme was incubated with racemic serinol phosphate (3.0 mM) and α-ketoglutarate (250 μM) and the exchange of the hydrogen at C2 with solvent deuterium was monitored by 1H NMR spectroscopy. Prior to the addition of enzyme (3.0 μM) the integrated ratio of the resonances for the hydrogens at C1 and C3, relative to the hydrogen at C2 was 4.0:1.04. After the addition of enzyme, the ratio gradually changed to 4.0:0.55 (Figure 6d), where the mixture of the serinol phosphate with hydrogen and deuterium at C2 is now clearly visible. When the same reaction was conducted using chemically synthesized (S)-serinol phosphate (Figure 6e) the hydrogen at C2 completed exchanged with solvent deuterium after 2 h (Figure 6f) thus demonstrating that Cj1437 catalyzes the synthesis of (S)-serinol phosphate from DHAP and l-glutamate.
The reaction products formed after the incubation of Cj1437 with DHAP and l-glutamate were confirmed to be serinol phosphate and α-ketoglutarate by ESI mass spectrometry (negative mode). In the control experiment in the absence of enzyme the substrates, DHAP and l-glutamate are identified at m/z of 146.05 and 168.99, respectively for the M-H ions (Figure 5d). The products, serinol phosphate is found at 170.02 (Figure 5e). Furthermore, when the reaction is conducted in D2O the serinol phosphate product incorporates a single deuterium as demonstrated by the appearance of the signal at an m/z of 171.03 (Figure 5f).
Kinetic Analysis of the Reaction Catalyzed by Cj1436.
Steady-state kinetic parameters for Cj1436 were obtained by monitoring the rate of decarboxylation at various concentrations of l-serine phosphate using membrane inlet mass spectrometry. These experiments provided values for kcat, kcat/Km, and Km of 0.21 ± 0.02 s−1, 350 ± 120 M−1 s−1 and 0.6 ± 0.2 mM, respectively. The value of kcat/Km for Cj1436 is 3–4 fold lower than that determined for Cj1437 (see below). No activity (<0.01 s−1) was observed when l-serine replaced l-serine phosphate in the assay.
Kinetic Analysis of the Reaction Catalyzed by Cj1437.
The reaction catalyzed by Cj1437 and HS19.10 was followed using a glutamate dehydrogenase coupled assay to measure the rate of formation of α-ketoglutarate. Using DHAP and l-glutamate as substrates, the values of kcat, kcat/Km, and Km with Cj1437 were determined to be 0.50 ± 0.02 s−1, 1.2 (± 0.2) x 104 M−1 s−1 and 42 ± 6 μM. Using the homolog to Cj1437 from the HS:19 serotype (HS19.10), the values of kcat, kcat/Km, and Km were determined to be 1.10 ± 0.01 s−1, of 2.7 (± 0.1) x 104 M−1 s−1 and 41 ± 2 μM, respectively. The turnover using l-aspartate (15 mM) was less than 50% of the rate exhibited by l-glutamate (15 mM) at a fixed concentration of DHAP (3.0 mM) as monitored by 1H NMR spectroscopy. Essentially no activity could be detected using l-alanine.
Discussion
The HS:2 serotype of Campylobacter jejuni is surrounded by a capsular polysaccharide consisting of a repeating unit of d-glycero-l-gluco-heptose, d-glucuronic acid, d-N-acetyl-galactosamine, and d-ribose.10, 11 The glucuronic acid is amidated with either serinol or ethanolamine. This same modification can be found within the CPS of the C. jejuni serotype HS:19 except that the amide bond is formed only with serinol. The exact role of the amide bond containing molecule within the CPS is unknown. Other modifications to the CPS are critical for the evasion of the immune system and adherence to epithelial tissue.9, 34 Additionally, the phase variable phosphoramidate modification is a target for recognition by bacteriophage F336.35 Phase variation may play a role in the biosynthesis of glucuronamide. The gene which encodes for Cj1437 appears to have a poly-G tract of 9 nucleotides (877–885). Poly-G tracts are common to C. jejuni and are known to introduce phase variation which creates phenotypic heterogeneity in populations of C. jejuni. This allows C. jejuni to avoid the immune system as well as phage attack.36
To form the amide bond found in the CPS of C. jejuni HS:2, the bacterium must synthesize two primary amines, ethanolamine and serinol. Here we have shown that the first step in the biosynthesis of the ethanolamine product is the decarboxylation of l-serine phosphate to ethanolamine phosphate catalyzed by Cj1436. This result is consistent with the prior identification of an l-serine phosphate decarboxylase (SMUL_1544), which was shown previously to catalyze the decarboxylation of l-serine phosphate.29 SMUL_1544 is the only other functionally characterized l-serine phosphate decarboxylase reported to date and is used in the biosynthesis of norpseudovitamin B12 in S. multivorans.29, 30 l-Threonine phosphate is also a weaker substrate, but no activity could be detected with l-serine, l-threonine, l-aspartate, or l-glutamate. The phosphate group of the substrate is thus necessary for substrate recognition and is required for catalytic activity. The homologous three-dimensional structures of SMUL_1544 and CobD of S. enterica have identified the residues used to bind the phosphate group of the substrate.29, 37 In SMUL_1544 these residues are Arg-356, Arg-368 and Ser-25. A sequence alignment of Cj1436 with SMUL_1544 reveals conservation of those residues required to bind the phosphate group of the substrate (Figure S2).
Cj1437 participates in the first step in serinol formation where it catalyzes the transamination of DHAP to serinol phosphate using l-glutamate as the amine donor. The stereochemistry of the reaction was shown to produce the (S)-enantiomer of serinol phosphate using solvent deuterium exchange with chemically synthesized (S)-serinol phosphate. This stereochemical outcome is consistent with the known product formation of l-histidinol phosphate in the reaction catalyzed by histidinol phosphate transaminase.38 Serinol phosphate has been shown to be an important precursor for serinol production.39 Serinol is an intermediate in the formation of other chemicals including the anti-cancer drugs N-palmitoyl-2-amino-1,3-propanediol, and the first orally administered treatment for multiple sclerosis, Gilenya® (Novartis).40–42
The precise sequence of the enzymatic transformations required for the biosynthesis of the glucuronamide moiety in the capsular polysaccharide of C. jejuni HS:2 has not yet been elucidated. A previous bioinformatic investigation identified a probable candidate enzyme, Cj1438, which contains an N-terminal glycosyltransferase domain as well as a C-terminal ATP-grasp domain that could be used for amide bond formation. However, at this point it is unclear whether the ATP-grasp domain would utilize serinol phosphate or serinol as the amine substrate or whether the acceptor carboxylate would be contributed by UDP-glucuronate or the growing polysaccharide chain. In either case the putative phosphatase (Cj1435) will ultimately have to be used to remove the phosphoryl group from either serinol phosphate or the phosphorylated glucuronamide moiety. Efforts to unravel these possible scenarios are in progress.
Glucuronamides are found within the lipopolysaccharide O-antigen and CPS of many bacteria including Acinetobacte baumannii G7, E. coli O143, E. coli L-19, Vibrio cholerae H11 (non-O1), Shigella boydii type 8, Pseudomonas aeruginosa, Proteus mirabilis O27, V Vibrio vulnificus strain 6353, Bacillu halodurans, Providencia stuartii O25 and Proteus. penneri strain 14.43–53 In A. baumannii G7 E. coli O143, E. coli L-19, S. boydii type 8, and B. halodurans there is an ATP-grasp enzyme that is located in the O-antigen or CPS biosynthetic gene cluster. In addition to the organisms listed, C. jejuni HS:19 contains a glucuronamide as well as an ATP-grasp containing enzyme. An analysis of other C. jejuni genomes revealed that C. jejuni HS:22 also contains the genes necessary for biosynthesis of a CPS modified with a glucuronamide. A comparison of the capsular polysaccharide biosynthetic gene clusters for HS:2, HS:19, and HS:22 is presented in Figure S1.
Conclusion
Here we report the functional characterization of two enzymes necessary for the formation of the C. jejuni HS:2 capsular polysaccharide, Cj1436 and Cj1437. Cj1436 is a PLP-dependent decarboxylase, which catalyzes the decarboxylation of l-serine phosphate to ethanolamine phosphate. The products were confirmed by mass spectrometry and 1H NMR spectroscopy. The rate and catalytic constants were measured using NMR spectroscopy and membrane inlet mass spectrometry. Cj1437 is a PLP-dependent transaminase, which catalyzes the transamination of DHAP using l-glutamate to form (S)-serinol phosphate. The product of the reaction was characterized using mass spectrometry and 1H NMR spectroscopy. The rate and catalytic constants were determined spectrophotometrically using a glutamate dehydrogenase coupled assay.
Supplementary Material
Acknowledgments
Funding
This research was supported by the National Institutes of Health (GM 139428 and GM 122825).
Footnotes
ASSOCIATED CONTENT
Support Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional synthetic methods and protein sequence comparisons.
The authors have no competing financial interests.
References
- 1).Epps SV, Harvey RB, Hume ME, Phillips TD, Anderson RC, and Nisbet DJ (2013) Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. International Journal of Environmental Research and Public Health, 10, 6292–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, and Griffin PM (2011) Foodborne illness acquired in the United States--major pathogens. Emerging Infect. Dis 17, 7–15, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Burnham PM and Hendrixson DR (2018) Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol 16, 551–565, [DOI] [PubMed] [Google Scholar]
- 4).Allos BM (2001) Campylobacter jejuni Infections: update on emerging issues and trends. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 32, 1201–1206. [DOI] [PubMed] [Google Scholar]
- 5).Nachamkin I, Allos BM, and Ho T. (1998) Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev 11, 555–567, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Yuki N. (1997) Molecular mimicry between gangliosides and lipopolysaccharides of Campylobacter jejuni isolated from patients with Guillain-Barré Syndrome and Miller Fisher Syndrome. J. Infect. Dis 176, S150–S153. [DOI] [PubMed] [Google Scholar]
- 7).Poly F, Noll AJ, Riddle MS, and Porter CK (2019) Update on Campylobacter vaccine development. Human Vaccines & Immunotherapeutics, 15, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).McNally DJ, Lamoureux MP, Karlyshev AV, Fiori LM, Li J, Thacker G, Coleman RA, Khieu NH, Wren BW, Brisson J-R, Jarrell HC, and Szymanski CM (2007) Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem 282, 28566–28576, [DOI] [PubMed] [Google Scholar]
- 9).van Alphen LB, Wenzel CQ, Richards MR, Fodor C, Ashmus RA, Stahl M, Karlyshev AV, Wren BW, Stintzi A, Miller WG, Lowary TL, and Szymanski CM (2014) Biological roles of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. PLoS One 9, e87051, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).St. Michael F, Szymanski CM, Chan Li, J., Khieu KH Larocque NHS, Wakarchuk WW, Brisson JR, and Monteiro MA. (2002) The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem 269, 5119–5136, [DOI] [PubMed] [Google Scholar]
- 11).Young KT, Davis LM, and Dirita VJ (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol 5, 665–679, [DOI] [PubMed] [Google Scholar]
- 12).McNally DJ, Jarrell HC, Khieu NH, Li J, Vinogradov E, Whitfield DM, Szymanski CM, and Brisson JR (2006) The HS:19 serostrain of Campylobacter jejuni has a hyaluronic acid-type capsular polysaccharide with a nonstoichiometric sorbose branch and O-methyl phosphoramidate group. FEBS J. 273, 3975–3989. [DOI] [PubMed] [Google Scholar]
- 13).Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, and Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668, [DOI] [PubMed] [Google Scholar]
- 14).Huddleston JP, Anderson TK, Spencer KD, Thoden JB, Raushel FM, and Holden HM (2020) Structural analysis of Cj1427, an essential NAD-dependent dehydrogenase for the biosynthesis of the heptose residues in the capsular polysaccharides of Campylobacter jejuni. Biochemistry 59, 1314–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Huddleston JP and Raushel FM (2020) Functional characterization of Cj1427, a unique ping-pong dehydrogenase responsible for the oxidation of GDP-d-glycero-α-d-manno-heptose in Campylobacter jejuni. Biochemistry 59, 1328–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Huddleston JP and Raushel FM (2019) Biosynthesis of GDP-d-glycero-α-d-manno-heptose for the capsular polysaccharide of Campylobacter jejuni. Biochemistry 58, 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).McCallum M, Shaw GS, and Creuzenet C. (2013) Comparison of predicted epimerases and reductases of the Campylobacter jejuni d-altro- and l-gluco-heptose synthesis pathways. J. Biol. Chem 288, 19569–19580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Barnawi H, Woodward L, Fava N, Roubakha M, Shaw SD, Kubinec C, Naismith JH, and Creuzenet C. (2021) Structure-function studies of the C3/C5 epimerases and C4 reductases of the Campylobacter jejuni capsular heptose modification pathways. The Journal of Biological Chemistry, 296, 100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Huddleston JP, Anderson TK, Girardi NM, Thoden JB, Taylor Z, Holden HM, & Raushel FM (2021) Biosynthesis of d-glycero-l-gluco-Heptose in the Capsular Polysaccharides of Campylobacter jejuni. Biochemistry, 60, 1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Taylor ZW, Brown HA, Holden HM, and Raushel FM (2017) Biosynthesis of nucleoside diphosphoramidates in Campylobacter jejuni. Biochemistry 56, 6079–6082. jejuni. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Taylor ZW, Chamberlain AR, and Raushel FM (2018) Substrate specificity and chemical mechanism for the reaction catalyzed by glutamine kinase. Biochemistry 57, 5447–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Taylor ZW and Raushel FM (2019) Manganese-induced substrate promiscuity in the reaction catalyzed by phosphoglutamine cytidylyltransferase from Campylobacter jejuni. Biochemistry 58, 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Taylor ZW and Raushel FM (2018) Cytidine diphosphoramidate kinase: an enzyme required for the biosynthesis of the O-methyl phosphoramidate modification in the capsular polysaccharides of Campylobacter jejuni. Biochemistry 57, 2238–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Taylor ZW, Brown HA, Narindoshvili T, Wenzel CQ, Szymanski CM, Holden HM, and Raushel FM (2017) Discovery of a glutamine kinase required for the biosynthesis of the O-methyl phosphoramidate modifications found in the capsular polysaccharides of Campylobacter jejuni. J. Am. Chem. Soc 139, 9463–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Sternberg MJ, Tamaddoni-Nezhad A, Lesk VI, Kay E, Hitchen PG, Cootes A, van Alphen LB, Lamoureux MP, Jarrell HC, Rawlings CJ, Soo EC, Szymanski CM, Dell A, Wren BW, and Muggleton SH (2013) Gene function hypotheses for the Campylobacter jejuni glycome generated by a logic-based approach. J. Mol. Biol 425, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Riegert AS, and Raushel FM (2021) Functional and Structural Characterization of the UDP-Glucose Dehydrogenase Involved in Capsular Polysaccharide Biosynthesis from Campylobacter jejuni. Biochemistry, 60, 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Atkinson HJ, Morris JH, Ferrin TE, and Babbitt PC (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Keller S, Wetterhorn KM, Vecellio A, Seeger M, Rayment I, and Schubert T. (2019) Structural and functional analysis of an l-serine O-phosphate decarboxylase involved in norcobamide biosynthesis. FEBS letters, 593, 3040–3053. [DOI] [PubMed] [Google Scholar]
- 30).Keller S, Treder A, von Reuss SH, Escalante-Semerena JC, and Schubert T. (2016) The SMUL_1544 gene product governs norcobamide biosynthesis in the tetrachloroethene-respiring bacterium Sulfurospirillum multivorans. Journal of Bacteriology, 198, 2236–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Sheng X, Zhu W, Huddleston J, Xiang DF, Raushel FM, Richards NGJ, and Himo F. (2017) A combined experimental-theoretical study of the LigW-catalyzed decarboxylation of 5-carboxyvanillate in the metabolic pathway for lignin degradation. ACS Catal., 7, 4968–4974. [Google Scholar]
- 32).Zhu W, Easthon LM, Reinhard LA, Tu C-K, Cohen SE, Silverman DN, Allen KN, and Richards NGJ (2016) Substrate binding mode and molecular basis of a specificity switch in oxalate decarboxylase. Biochemistry, 55, 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Moral MEG, Tu C-K, Richards NGJ, and Silverman DN (2011) Membrane inlet for mass spectrometric measurement of catalysis by enzymatic decarboxylases. Analyt. Biochem, 418, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Guerry P, Poly F, Riddle M, Maue AC, Chen YH, and Monteiro MA (2012) Campylobacter polysaccharide capsules: virulence and vaccines. Frontiers in Cellular and Infection Microbiology, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Sørensen MC, van Alphen LB, Harboe A, Li J, Christensen BB, Szymanski CM, and Brøndsted L. (2011) Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. Journal of Bacteriology, 193, 6742–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Cheong CG, Escalante-Semerena JC, & Rayment I. (2002) Structural studies of the l-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes. Biochemistry, 41, 9079–9089. [DOI] [PubMed] [Google Scholar]
- 37).Haruyama K, Nakai T, Miyahara I, Hirotsu K, Mizuguchi H, Hayashi H, and Kagamiyama H. (2001) Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5’-phosphopyridoxyl)-l-glutamate: double substrate recognition of the enzyme. Biochemistry, 40, 4633–4644. [DOI] [PubMed] [Google Scholar]
- 38).Andreessen B, and Steinbüchel A. (2012) Biotechnological conversion of glycerol to 2-amino-1,3-propanediol (serinol) in recombinant Escherichia coli. Applied Microbiology and Biotechnology, 93, 357–365. [DOI] [PubMed] [Google Scholar]
- 39).Bieberich E, Kawaguchi T, and Yu RK (2000) N-acylated serinol is a novel ceramide mimic inducing apoptosis in neuroblastoma cells. The Journal of Biological Chemistry, 275, 177–181. [DOI] [PubMed] [Google Scholar]
- 40).Ueoka R, Fujita T, Iwashita T, van Soest RW, and Matsunaga S. (2008) Inconspicamide, new N-acylated serinol from the marine sponge Stelletta inconspicua. Bioscience, biotechnology, and biochemistry, 72, 3055–3058. [DOI] [PubMed] [Google Scholar]
- 41).Buranachokpaisan T, Dannenfesler RM, and Li P, (2006) Compound formulations of 2-amino-1,3-propanediol compounds. (WO 2006010630 A1) World Intellectual Property Organization (PCT).
- 42).Kenyon JJ, Senchenkova S, Shashkov AS, Shneider MM, Popova AV, Knirel YA, and Hall RM (2020) K17 capsular polysaccharide produced by Acinetobacter baumannii isolate G7 contains an amide of 2-acetamido-2-deoxy-d-galacturonic acid with d-alanine. International Journal of Biological Macromolecules, 144, 857–862. [DOI] [PubMed] [Google Scholar]
- 43).Landersjö C, Weintraub A, and Widmalm G. (1996) Structure determination of the O-antigen polysaccharide from the enteroinvasive Escherichia coli (EIEC) O143 by component analysis and NMR spectroscopy. Carbohydrate research, 291, 209–216. [DOI] [PubMed] [Google Scholar]
- 44).Vinogradov EV, Holst O, Thomas-Oates JE, Broady KW, and Brade H. (1992) The structure of the O-antigenic polysaccharide from lipopolysaccharide of Vibrio cholerae strain H11 (non-O1). European Journal of Biochemistry, 210, 491–498. [DOI] [PubMed] [Google Scholar]
- 45).Vinogradov EV, Knirel YA, Shashkov AS, and Kochetkov NK (1987) Determination of the degree of amidation of 2-deoxy-2-formamido-d-galacturonic acid in O-specific polysaccharides of Pseudomonas aeruginosa O4 and related strains. Carbohydrate Research, 170, c1–c4. [DOI] [PubMed] [Google Scholar]
- 46).Vinogradov EV, Krajewska-Pietrasik D, Kaca W, Shashkov AS, Knirel YA, and Kochetkov NK (1989) Structure of Proteus mirabilis O27 O-specific polysaccharide containing amino acids and phosphoethanolamine. European Journal of Biochemistry, 185, 645–650. [DOI] [PubMed] [Google Scholar]
- 47).Reddy GP, Hayat U, Xu Q, Reddy KV, Wang Y, Chiu KW, Morris JG Jr, and Bush CA. (1998) Structure determination of the capsular polysaccharide from Vibrio vulnificus strain 6353. European Journal of Biochemistry, 255, 279–288. [DOI] [PubMed] [Google Scholar]
- 48).Iyer LM, Abhiman S, Maxwell Burroughs A, and Aravind L. (2009) Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Molecular bioSystems, 5, 1636–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Aono R. (1990) The poly-alpha- and -beta-1,4-glucuronic acid moiety of teichuronopeptide from the cell wall of the alkalophilic Bacillus strain C-125. The Biochemical Journal, 270, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Kocharova NA, Ovchinnikova OG, Bialczak-Kokot M, Shashkov AS, Knirel YA, and Rozalski A. (2011) Structure of the O-polysaccharide of Providencia alcalifaciens O25 containing an amide of d-galacturonic acid with N(ε)-[(R)-1-carboxyethyl]-l-lysine. Biochemistry. Biokhimiia, 76, 707–712 [DOI] [PubMed] [Google Scholar]
- 51).Vinogradov E, Sidorczyk Z, and Knirel YA (2002) Structure of the core part of the lipopolysaccharides from Proteus penneri strains 7, 8, 14, 15, and 21. Carbohydrate Research, 337, 643–649. [DOI] [PubMed] [Google Scholar]
- 52).Zdorovenko EL, Varbanets LD, Liu B, Valueva OA, Wang Q, Shashkov AS, Garkavaya EG, Brovarskaya OS, Wang L, and Knirel YA (2014) Structure and gene cluster of the O antigen of Escherichia coli L-19, a candidate for a new O-serogroup. Microbiology (Reading, England), 160, 2102–2107. [DOI] [PubMed] [Google Scholar]
- 53).Bayliss CD, Bidmos FA, Anjum A, Manchev VT, Richards RL, Grossier JP, Wooldridge KG, Ketley JM, Barrow PA, Jones MA, and Tretyakov MV (2012) Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Research, 40, 5876–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.