Abstract
Helicobacter spp., except for Helicobacter cinaedi, have only rarely been reported in cases of septicemia. A patient with X-linked (Bruton’s) agammaglobulinemia was found to have persistent sepsis with a Helicobacter-like organism despite multiple courses of antibiotics. His periods of sepsis were associated with leg swelling thought to be consistent with cellulitis. The organism was fastidious and required a microaerophilic environment containing H2 for growth. Optimal growth was observed at 35 to 37°C on sheep blood, CDC anaerobe, and Bordet-Gengou agars. Serial subcultures every 4 to 5 days were required to maintain viability. The organism was strongly urease positive and showed highest relatedness to Helicobacter-like organisms with the vernacular name “Flexispira rappini” by 16S rRNA gene sequence analysis. Genomic DNA hybridization studies, however, found 24 to 37% relatedness to “F. rappini” and even less to other Helicobacter spp. Although the organism phenotypically resembles “Flexispira” and Helicobacter, it is thought to represent a new taxon. The patient’s infection was eventually cleared with a prolonged (5-month) course of intravenous imipenem and gentamicin.
“Flexispira rappini” is a urease-producing, fusiform gram-negative organism which has been shown to be closely related to Helicobacter spp., based on its 16S rRNA gene sequence as well as growth and morphologic characteristics. It has been isolated from aborted sheep fetuses (4), mouse intestinal mucosa (19), and stools from humans with mild chronic diarrheal disease (17). Recently, an isolate thought to be “F. rappini” on the basis of 16S rRNA gene sequencing was isolated from blood of a child with pneumonia, although it was not clear that the organism was actually the cause of the pneumonia (25). This organism tested negative for urease production, which is an atypical reaction for “F. rappini”.
Helicobacter-like organisms are occasional causes of septicemia occurring in AIDS patients. Helicobacter cinaedi is the organism most often identified, but several Helicobacter-like organisms have been implicated as well, such as Helicobacter westmeadii (26) and Helicobacter strain Mainz (8, 13). The involvement of Helicobacter and “Flexispira” organisms in cases of sepsis may be more common than previously thought, given the evidence to be presented that it is difficult to detect, grow, and identify these organisms.
We report here an unusual case of recurrent bacteremia with an organism that by 16S rRNA gene sequencing most closely aligns with “F. rappini” but by whole-cell DNA-DNA hybridization has been found to represent a new taxon of organism that is closely related to “Flexispira” and Helicobacter (22). The patient was an immunocompromised host with X-linked (Bruton’s) agammaglobulinemia who had recurrent sepsis with this organism over a 7-month period until it was successfully eradicated with intravenous (i.v.) antibiotics.
CASE REPORT
The patient is a 36-year-old black male with X-linked (Bruton’s) agammaglobulinemia, which was diagnosed at age 4. He was treated with intramuscular gamma globulin until age 32, when he was switched to i.v. gamma globulin. He reports that as a child he had chronic swelling in his left leg, but this resolved as he got older. In 1994 (age 32) he noted edema of the right ankle which progressed to involve his foot and calf. He was presumed to have cellulitis and was placed on antibiotics, with some improvement. In late 1995 he had recurrent episodes of leg swelling and was again placed on antibiotics. By January of 1996, he had worsening of the leg swelling and onset of various systemic symptoms, including fevers, night sweats, and decreased appetite. In March 1996 he had persistent left leg swelling, and aerobic blood cultures drawn during this hospitalization grew a fastidious gram-negative bacillus that proved difficult to grow when subcultured to agar plates. He was treated with i.v. ampicillin-sulbactam for 4 weeks, followed by a 2-week course of amoxicillin-clavulanic acid and trimethoprim-sulfamethoxazole. Subsequent blood cultures were still positive with the same organism, and he was placed on oral ciprofloxacin and clarithromycin. He remained on these drugs until late July, when blood cultures were again positive for the same fastidious gram-negative bacillus. At this point he was treated with i.v. ampicillin-sulbactam for 2 weeks, but because of i.v. access problems he was placed on oral ciprofloxacin and clarithromycin; nevertheless, his problems persisted. When the patient was evaluated at the National Institutes of Health (NIH) in August 1996, blood cultures were positive for the same organism, which was then subjected to antibiotic susceptibility testing. On the basis of this testing, the patient was treated initially with oral antibiotics (doxycycline and metronidazole) and had initial improvement in leg swelling; blood cultures remained positive, and treatment was changed to oral amoxicillin-clavulanic acid, minocycline, and rifampin. Again, initial improvement was followed by symptom recurrence, and in January 1997 i.v. gentamicin and imipenem were initiated and continued for 5 months (until June 1997). This resulted in clearing of both local and systemic symptoms as well as in negative follow-up blood cultures.
MATERIALS AND METHODS
Blood cultures.
The organism was first grown in aerobic pediatric BacTAlert (BTA) (Organon Teknika Corp., Durham, N.C.) bottles at the Medical College of Georgia. Although gram-negative rods were seen on Gram staining, they failed to grow on subculture to a variety of enriched media, under different temperature and atmospheric conditions, both at the Medical College of Georgia and at NIH, where a positive blood culture bottle had been sent. However, another positive culture bottle, sent to the Bacteriology Laboratory at the Georgia Department of Human Resources (DHR), was successfully subcultured by using microaerophilic conditions that included H2. Upon admission of the patient to NIH, each blood culture set performed for this patient consisted of one aerobic and one anaerobic BTA bottle and four blood agar plates that had been inoculated with Isolator (Wampole Labs, Cranbury, N.J.) concentrate and incubated under microaerophilic conditions with H2. If flagged as positive, or after 4 days of incubation, BTA cultures were stained with Gram stain and acridine orange (AO) stain and then subcultured under microaerophilic conditions with H2. Isolator plates were incubated for 2 weeks.
Growth characteristics.
Optimum conditions for growth of the organism were determined by comparing growth aerobically in ambient air, aerobically in 6% CO2, anaerobically (85%N2, 10% CO2, and 5%H2), and under the following microaerophilic conditions: CampyGen (a CO2 generator from Oxoid, Basingstoke, Hampshire, England), Campy Gas (10% CO2, 5%O2, and 85% N2) (Columbia Diagnostics, Springfield, Va.), and BBL CampyPak Plus (a CO2-H2 generator from Becton Dickinson, Cockeysville, Md.). Optimum plating media were chosen by comparing growth on horse and sheep blood agars, brucella agar, CDCA (anaerobic blood agar), Bordet-Gengou agar with methicillin, cefoperozone-vancomycin agar (Remel Labs, Lenexa, Kans.), chocolate agar, and charcoal-yeast extract agar incubated at 25, 35, and 42°C. Permanent stocks of the organism were made by freezing early-stationary-phase growth (either suspended in defibrinated rabbit blood and frozen with liquid nitrogen or quick frozen in Trypticase soy broth with glycerol, stored at −70°C).
Biochemical and morphologic evaluation.
Extensive biochemical evaluation of the organism was performed at the Georgia DHR Bacteriology Laboratory, the Microbiology Service of the Clinical Center of NIH, and the Special Bacteriology Reference Laboratory of the Centers for Disease Control and Prevention (CDC) (not all tests were done by all laboratories). The following biochemical characteristics were assessed by using inocula from cultures incubated under microaerophilic conditions at 35°C or tubes or plates incubated under those conditions as appropriate for the test: production of alkaline phosphatase, catalase, H2S, oxidase, and urease; reduction of nitrate; hydrolysis of hippurate and indoxyl acetate; growth on MacConkey agar; motility; and susceptibility to cephalothin, nalidixic acid, and metronidazole. Additional biochemical tests performed at the CDC laboratory included Simmons citrate, nitrite reduction, indole, methyl red, Voges-Proskauer, triple sugar iron agar, esculin hydrolysis, gelatin hydrolysis, growth in nutrient broth with and without 6.5% NaCl, litmus milk, and acid production from d-glucose, d-xylose, d-mannitol, lactose, sucrose, and maltose. The CDC tests were performed by previously described methods (29). The acid production tests were done in enteric base supplemented with 15% rabbit serum, and the urease test was performed with a heavy inoculum to detect preformed enzyme activity. Cell wall fatty acid analyses were performed by both the Georgia DHR and CDC laboratories. Organism morphology was determined by AO staining, dark-field examination, Ryu flagellum staining (Remel Labs), and electron microscopy (EM).
EM.
Organisms grown for 4 days on blood agar were washed with phosphate-buffered saline, suspended in 4% formaldehyde, and processed for transmission EM. The microscopy was performed by the Laboratory of Cell and Molecular Structure (Science Applications INternational Corp., Frederick, Md.).
CFA analysis.
Cells were grown for 3 to 5 days on heart infusion agar with 5% rabbit blood at 35°C in an H2-enriched atmosphere as described above. The cellular fatty acid (CFA) compositions were determined as described previously (29) with a commercially available software package (MIDI, Newark, Del.).
Antibiotic susceptibility testing.
The fastidious nature of the organism precluded the use of traditional disk or microdilution methods. Instead, in vitro antibiotic susceptibility testing was performed with the use of E-test strips (AB Biodisk, Piscataway, N.J.) placed on heavily inoculated sheep blood agar and incubated in a microaerophilic atmosphere with H2. The E-test strips were generally read after 2 to 3 days of incubation. Along with the patient’s organism, control organisms for which the MICs were known were tested with the same conditions and medium to validate the expected activities of the antibiotics. Although there are no National Committee for Clinical Laboratory Standards recommended breakpoints for this organism, susceptibility or resistance for each antibiotic was estimated based on MIC breakpoints used for other organisms. The following agents were tested: amoxicillin-clavulanate, ampicillin, azithromycin, ceftriaxone, ciprofloxacin, clindamycin, chloramphenicol, doxycycline, gentamicin, imipenem, minocycline, and metronidazole. Beta-lactamase production was tested for by using DrySlide nitrocefin (Difco, Detroit, Mich.). Susceptibilities were determined with the first blood isolate recovered at NIH in August 1996; repeat testing against selected agents was done on the recurrent blood isolates (1 and 4 months later) to look for the development of resistance to the agents being used.
16S rRNA gene sequencing (NIH).
Crude DNA extracts were prepared by using DNA extraction reagent (Perkin-Elmer, Norwalk, Conn.). PCR of the 16S rRNA gene was performed with the primers pH156 (5′-AGA-GTT-TGA-TCC-TGG-CT-3′) and p1394R (5′-ATG-GTG-TGA-CGG-GCG-G-3′), which were designed based on an alignment of Helicobacter and Campylobacter sequences obtained from GenBank. PCR products were cloned into pCR 2.1 sequencing plasmids (Invitrogen, Carlsbad, Calif.) and transformed into Escherichia coli INVαF′. Plasmids were extracted, purified, and sequenced with M13 sequencing primers. The sequencing reactions were performed with the ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer) according to the manufacturer’s instructions. Sequences were assembled and aligned by using GeneWorks 2.4 software and compared to sequences in the GenBank and EMBL databases. A phylogenetic tree (not shown) was constructed by maximum-parsimony methods (GeneWorks software).
16S rRNA gene sequencing (CDC).
Purified genomic DNA was used to amplify the 16S rRNA gene with the primers fD1 and rD1, which were originally described for amplifying the 16S rRNA genes of many eubacteria (28). The linker sequences were omitted from the primers, as previously described (5). The PCR product was checked by agarose gel electrophoresis, purified, and then sequenced. The sequencing reactions were performed with the ABI PRISM kit (Perkin-Elmer). The primer set used for sequencing was derived from those designed by Stackebrandt and Charfreitag (21). The sequencing reaction products were resolved on a 5% acrylamide–8 M urea gel electrophoresed on an ABI 373S automated sequencer (Perkin-Elmer). The sequence data was edited and compiled by using the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.). The resulting sequence was 1,459 bases long and corresponded to bases 37 to 1537 in E. coli (3). The 16S rRNA gene sequence was aligned with four “F. rappini” sequences, eight Helicobacter sequences, and Wolinella succinogenes as the outgroup by using the Genetics Computer Group program PILEUP (Table 1). The multiple-sequence alignment was edited to remove the 5′ and 3′ hypervariable regions (11), the intervening sequence found in some of the genes, and any other ambiguous regions. The final alignment was 1,381 nucleotides in length. The edited alignment was used in PHYLIP (version 3.5; J. Felsenstein, University of Washington, Seattle) to derive a phylogenetic dendrogram with the nucleotide substitution model of Jukes and Cantor (14) and the neighbor-joining method of Saitou and Nei (18).
TABLE 1.
Ribosomal sequences used for alignment of the NIH isolate
| Species | Strain | GenBank accession no. |
|---|---|---|
| “F. rappini” | NADC 1893 | M88137 |
| NADC 1937 | M88138 | |
| ATCC 49317 | AF047851 | |
| FH 9702248 | AF034135 | |
| H. cinaedi | CCUG 18818 | M88150 |
| Helicobacter sp. | No designation | U44756 |
| H. bilis | Hb1 | U18766 |
| H. canis | NCTC 12739 | L13464 |
| H. hepaticus | Fred1 | L39122 |
| H. trogontum | LRB 8581 | U65103 |
| Helicobacter sp. | Dog-4, Eaton 1302 | U51874 |
| H. cholecystus | Hkb1 | U46129 |
| W. succinogenes | ATCC 29543 | M88159 |
DNA-DNA hybridization studies (CDC).
“F. rappini” ATCC 43966, H. cinaedi ATCC 35683T, and Helicobacter muridarum ATCC 49282T were obtained from the American Type Culture Collection and cultured on heart infusion agar with 5% rabbit blood (Becton Dickinson). DNA relatedness among these organisms was determined by the hydroxyapatite method as described previously (2). Isolated DNAs from the NIH patient isolate and “F. rappini” ATCC 43966 were labeled with [32P]dCTP by using a nick translation kit (Gibco BRL, Gaithersburg, Md.) as described by the manufacturer.
Nucleotide sequence.
The 1,375-bp sequence derived at NIH and the 1,459-bp sequence derived at CDC were aligned, analyzed, and edited based on a global alignment with related species used in the phylogenetic dendrogram. The two sequences shared a 1,346-base overlap. The consensus sequence was 1,488 bases in length.
Nucleotide sequence accession number.
The consensus sequence was submitted to GenBank and assigned accession no. AF118807.
RESULTS
Blood cultures.
The organism grew only in the aerobic BTA bottles (pediatric and standard bottles). Some of these bottles were flagged as positive after 2 to 4 days of incubation. Other aerobic bottles that were read as negative by the BTA automated detection system at 4 days were positive by AO staining and by subculture, so automated detection could not be relied on. By Gram staining the organisms were difficult to see, whereas by AO staining numerous organisms were readily apparent. All anaerobic BTA bottles and Isolator plates were negative.
Growth characteristics.
There was no growth on blood agar plates incubated aerobically (with or without 6% CO2), anaerobically, or under microaerophilic conditions without H2 (such as with CampyGen or CampyGas). Even with enriched blood agar and optimal microaerophilic conditions, growth required 4 to 5 days of incubation. CampyPak Plus generates CO2 and H2 and was adequate for growth, but the best growth was obtained by using a combination of CampyGen and CampyPak Plus. This combination of generators was used for medium comparisons, temperature studies, biochemical tests, and antibiotic susceptibility testing performed at NIH. The organism grew best at 35 to 37°C and did not grow at 25 or 42°C, even after prolonged incubation. Of the variety of agars tested, the best growth was achieved on sheep blood agar, CDCA, and Bordet-Gengou agar. Poorer growth occurred on cefoperozone-vancomycin, brucella, chocolate, and horse blood agars, while no growth occurred on buffered charcoal-yeast extract agar. Growth on blood agar appeared as a thin, spreading, gray film, which was often difficult to discern. The organism became nonviable easily and had to be subcultured every 4 to 5 days to retain viability.
Morphologic and biochemical evaluation.
Organisms grown in blood culture bottles and stained with AO most often showed a long, thin, straight or slightly curved fusiform appearance. However, less frequently, a long, loosely coiled form was also seen. Active motility was seen by dark-field microscopy, while flagellum stains showed the cells to have bipolar flagella (amphitrichous), averaging four to six flagella at each end.
Significant biochemical reactions are shown in Table 2, along with the expected reactions of “F. rappini” and of other Helicobacter spp. either isolated from blood (H. cinaedi Mainz, H. westmeadii, and H. fennelliae) or known to have periplasmic fibers (H. bilis, H. felis, H. trogontum, and H. muridarum). The isolate failed to grow in most of the biochemical test media, including Simmons citrate, nitrate and nitrite reduction, indole, methyl red, Voges-Proskauer, triple sugar iron agar, gelatin, esculin, and litmus milk. In the carbohydrate acidification tests, the isolate grew but produced no detectable acid. All of the biochemical tests described above but not shown in Table 2 were negative. The present isolate had characteristics most similar to those of “F. rappini,” H. bilis, and H. muridarum, all of which produce urease and possess periplasmic fibers.
TABLE 2.
Comparison of the patient’s isolate with other species of Helicobacter isolated from blood (“F. rappini,” H. cinaedi Mainz, H. westmeadii, and H. fennelliae) and those with periplasmic fibersa
| Test or characteristic | Patient isolate | “F. rappini” | H. cinaedi | Strain Mainzc | H. westmeadiid | H. fennelliae | H. bilise | H. felis | H. trogontumf | H. muriduram |
|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | + | vb | + | + | + | + | + | + | + | + |
| Oxidase | + | + | + | + | + | + | + | + | + | + |
| Nitrate reduction | − | − | + | − | + | − | + | + | + | − |
| Urease | + | + | − | − | − | − | + | + | + | + |
| Alkaline phosphatase | + | − | − | − | + | + | NAg | + | —h | + |
| Indole acetate hydrolysis | − | − | − | − | − | + | − | − | NA | + |
| Hippurate | − | − | − | − | + | − | − | − | − | − |
| Growth at: | ||||||||||
| 25°C | − | − | − | NA | − | − | − | − | − | − |
| 37°C | + | + | + | + | + | + | + | + | + | + |
| 42°C | − | + | −i | − | − | − | + | + | + | − |
| Resistancej to: | ||||||||||
| Nalidixic acid | R | R | S | R | S | S | R | R | R | R |
| Cephalothin | R | R | I | S | R | S | R | S | R | R |
| Metronidozole | S/R | R | NA | NA | NA | NA | S | S | S | NA |
| Periplasmic fibers | + | + | − | NA | − | − | + | + | + | + |
| No. of flagella/cell | 8 to 12 | 10 to 20 | 1 to 2 | NA | 1 | 2 | 3 to 14 | 14 to 20 | 5 to 7 | 10 to 14 |
| Distribution | Bipolar | Bipolar | Bipolar | NA | Polar | Bipolar | Bipolar | Bipolar | Bipolar | Bipolar |
Reactions are from reference 7 unless otherwise specified.
Data are from reference 13.
Data are from reference 26.
Data are from reference 10.
Data are from reference 16.
NA, not available.
One of six strains was positive.
Kiehlbauch et al. 15 reported that 100% of isolates grew poorly at 42°C under microaerophilic conditions.
R, resistant; S, susceptible.
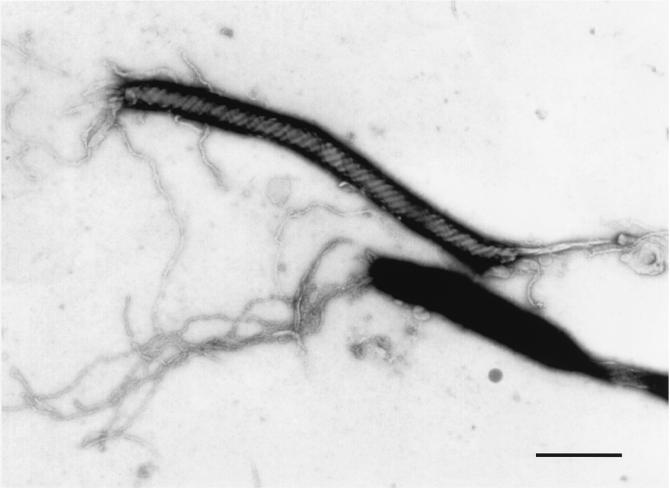
As shown in Fig. 1, EM verified the presence of four to six bipolar flagella and the slightly tapered morphology of the organism. Periplasmic fibers, a significant taxonomic feature for this group of bacteria, were clearly visible. The organism measured 0.2 to 0.3 μm wide by 3.7 to 5.5 μm long.
FIG. 1.
Transmission electron micrograph of the NIH isolate, showing the presence of periplasmic fibers. Bar, 1.0 μm.
The CFA compositions of saponified whole cells of the NIH patient isolate and “F. rappini” ATCC 43966 are given in Table 3. Both of these organisms are characterized by large amounts of 16:0 (43%) and 18:1ω7c (22%), moderate amounts of 12:0 (16%) and 14:0 (7%), and smaller amounts of 3-OH-14:0 (4%), 16:1ω7c (1%), 18:2 (2%), 18:1ω9c (1%), and 18:0 (4%). This profile was not observed with any of the other CFA groups in the CDC library.
TABLE 3.
CFA compositions of the NIH patient isolate and “F. rappini” ATCC 43966
| Fatty acida | % of total fatty acids (arithmetic mean) in:
|
|
|---|---|---|
| NIH patient isolate | “F. rappini” ATCC 43966 | |
| 12:0 | 16 | 6 |
| 14:0 | 7 | 7 |
| 3-OH-14:0 | 4 | 4 |
| 16:1ω7c | 1 | 1 |
| 16:0 | 43 | 44 |
| 18:2 | 2 | 5 |
| 18:1ω9c | 1 | 3 |
| 18:1ω7c | 22 | 23 |
| 18:0 | 4 | 7 |
The number before the colon indicates the number of carbons, the number after the colon is the number of double bonds, ω indicates the position of the double bond counting from the hydrocarbon end of the carbon chain, and OH is a hydroxy group at the 3(β) position from the carboxyl end. c, cis isomer.
Antibiotic susceptibility testing.
Results of the antibiotic testing are shown in Table 4. Upon admission of the patient to NIH, the organism showed in vitro resistance to ciprofloxacin and intermediate susceptibility to amoxicillin-clavulanate. Both of these antibiotics had been used for treatment early in the patient’s course. Despite ampicillin resistance, the organism did not produce detectable beta-lactamase. The in vitro multiresistance of this isolate to a variety of antibiotics narrowed therapeutic options to imipenem, gentamicin, minocycline, and metronidazole. On subsequent testing of later isolates, the organism remained susceptible to imipenem, gentamicin, and minocycline but had become resistant to metronidazole (>32 μg/ml). The patient had received all of these antimicrobial agents.
TABLE 4.
Antibiotic susceptibility determined by the E test on sheep blood agar after 48 h of incubation
| Antimicrobial agent | MIC (μg/ml)a for blood isolate obtained in (mo/yr):
|
||
|---|---|---|---|
| 8/96 | 9/96 | 1/97 | |
| Amoxicillin-clavulanate | 16-8 (I) | 12-6 (I) | NTb |
| Ampicillin | >256 (R) | NT | NT |
| Azithromycin | >256 (R) | NT | NT |
| Ceftriaxone | >32 (R) | NT | NT |
| Chloramphenicol | >256 (R) | NT | NT |
| Ciprofloxacin | >32 (R) | NT | NT |
| Clindamycin | >256 (R) | NT | NT |
| Doxycycline | 8 (I) | 6 (I) | 2 (S) |
| Gentamicin | 1 (S) | 1.5 (S) | 0.38 (S) |
| Imipenem | 0.05 (S) | 0.03 (S) | 0.02 (S) |
| Metronidazole | 4 (S) | >32 (R) | >32 (R) |
| Minocycline | 2 (S) | 0.75 (S) | 1.0 (S) |
| Rifampin | NT | >32 (R) | NT |
R, resistant; I, intermediate; S, susceptible.
NT, not tested.
16S rRNA gene sequence analyses.
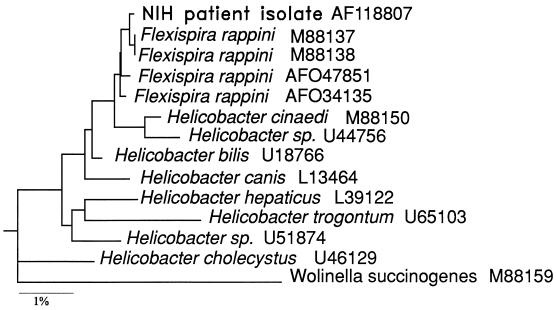
Results of two independent sequence analyses with two different primer sets for the 16S rRNA gene showed excellent concordance over the 1,346-base overlap. A 1,488-bp consensus sequence was derived. The consensus sequence was aligned with several closely related sequences in GenBank and was found to be most similar to the 16S rRNA gene sequences of “F. rappini” and several Helicobacter spp. As shown in Fig. 2, a phylogenetic dendrogram generated by using PHYLIP computer software in conjunction with the 16S rRNA sequence alignment placed our isolate in a group with two “F. rappini” sequences (NADC 1893 and NADC 1937). This small cluster was closely associated with two other “F. rappini” sequences (ATCC 49317 and strain FH 9702248). The similarity matrix derived from the sequence alignment indicates that the NIH isolate 16S rRNA gene sequence has 99.85% similarity to the 16S rRNA gene sequence of “F. rappini” NADC 1937 (GenBank accession number M88138) and 99.78, 99.56, and 99.64% similarity to those of “F. rappini” NADC 1893, ATCC 49317, and FH 9702248, respectively. The H. bilis Hb1 sequence was 99.06% similar to that of the NIH isolate, while H. cinaedi was 98.91% similar.
FIG. 2.
Phylogenetic tree based on 1,381 nucleotides of the 16S rRNA gene, showing the relationship of the NIH isolate to “F. rappini” and Helicobacter sequences. W. succinogenes was used as the outgroup. The bar represents a 1% difference in gene sequence.
DNA-DNA hybridization.
Results of a DNA-DNA hybridization analysis which included the NIH patient isolate and reference strains of “F. rappini,” H. cinaedi, H. pylori, and H. muridarum are given in Table 5. DNA from the NIH isolate showed a low level of binding to DNAs of all of the reference strains tested, with relative binding ratios ranging from 24% against “F. rappini” (ATCC 43966) to less than 1% against H. pylori. In order to confirm the relationship between the patient isolate and “F. rappini,” DNA from strain ATCC 43966 was labeled and hybridized with DNA from the patient isolate. A similar relative binding ratio (37%) was obtained.
TABLE 5.
DNA relatedness of the NIH patient isolate, a reference strain of “F. rappini,” and type strains of three Helicobacter species
| Source of unlabeled DNA | Binding (%) with labeled DNA from NIH patient isolate at 50°C |
|---|---|
| NIH patient isolate | 100 |
| “F. rappini” ATCC 43966 | 24 |
| H. cinaedi ATCC 35683 | 7 |
| H. pylori ATCC 43504 | 1 |
| H. muridarum ATCC 49282 | 7 |
DISCUSSION
The genus Helicobacter became prominent following the recognition that H. pylori colonization of the stomach wall is a significant cause of chronic gastritis and peptic ulcer disease and may be associated with the development of gastric cancers. Subsequently, other Helicobacter-like organisms have been found to cause infection in humans, largely in patients with AIDS. These infections are most commonly attributed to two species, H. cinaedi and H. fennelliae, which cause asymptomatic colonization as well as serious infections, including septicemia. In recent years, infections with other Helicobacter spp. have been described, predominantly from other mammals such as rats, mice, ferrets, dogs, cats, swine, cheetahs, and nonhuman primates, as well as two avian species. Some of the infections in animal species are characterized by gastritis, while other infections may involve other areas of the gastrointestinal tract. Concomitantly, there has been an increase in case reports of human infections caused either by animal Helicobacter spp. or by new, previously undescribed species resembling Helicobacter. These infections in humans include septicemia in AIDS patients caused by H. westmeadii (26), septicemia and septic arthritis caused by Helicobacter sp. strain Mainz (8, 13), gastroenteritis caused by Helicobacter pullorum (23, 24), and chronic gastritis caused by Helicobacter heilmanii (6, 12, 20).
As noted above, the majority of reported cases of Helicobacter sepsis have occurred in AIDS patients, and its occurrence in our patient with X-linked (Bruton’s) agammaglobulinemia is unusual. Whether such patients have an immune defect that predisposes them to Helicobacter sepsis remains to be determined. It should be noted that the patient described here had prolonged sepsis despite several courses of antimicrobial therapy. This suggests that the organism may have developed resistance to the antimicrobial agents used or perhaps had developed a sequestered focus of infection that was difficult to clear with oral therapy. The latter possibility is suggested by the fact that the patient noted associated leg swelling with each of his recurrent episodes of sepsis and had been treated for “cellulitis.” In vitro susceptibility tests were used to help select antibiotics to effectively eradicate the organism after several courses oral and i.v. therapies had failed. The spectrum of antibiotic resistance that this organism demonstrated suggests that susceptibility studies should be attempted for serious infections with Helicobacter, particularly when the infection fails to be cleared.
The present case demonstrates the difficulty involved in the detection, isolation, and identification of very fastidious Helicobacter spp. Their growth is not reliably picked up by automated blood culture systems, and Gram stains of positive bottles may appear negative since the faintly staining organisms are difficult to distinguish from background debris. On this basis, regular use of AO staining of bottles that have negative Gram stains but positive growth indices is recommended to detect Helicobacter as well as other fastidious organisms. Our patient’s organism did not grow in anaerobic blood culture bottles and failed to grow on Isolator plates incubated under optimal growth conditions for a prolonged period of time. Isolation of this organism as well as some of the other fastidious Helicobacter spp. requires the use of microaerophilic conditions with H2, which is not a feature of all microaerophilic systems used by clinical laboratories. The lack of H2 in the microaerophilic atmospheres routinely used by two of our laboratories resulted in their initial failure to subculture the organism from the blood culture bottles.
Once a Helicobacter-like organism is isolated, unambiguous identification by traditional biochemical methods is limited due to the paucity of useful tests. H. pylori, a strongly urease-positive organism isolated from gastric sources, is often identified on the basis of these criteria coupled with appropriate Gram stain morphology. Other Helicobacter spp. are isolated from a variety of sources, including blood, and can be either misidentified as Campylobacter spp. or misidentified as to the species of Helicobacter. While there are at least 20 described human and animal Helicobacter-like species, there are only a few biochemical traits that are useful for species identification. Although our isolate’s biochemical profile was similar to that of “F. rappini,” it was not consistent with those of previously described strains due to a positive alkaline phosphatase reaction and lack of growth at 42°C. Based on its biochemical evaluation, this organism was considered unidentified. However, the NIH patient isolate and the reference strain of “F. rappini” shared a unique CFA profile which differed from profiles of all of the other CFA groups in the CDC library, including most Campylobacter and Helicobacter species. This suggested that the NIH patient isolate and “F. rappini” represent either a unique taxon or closely related taxa.
In recent years, most published work on Helicobacter isolates has relied on 16S rRNA gene sequencing data to identify the species, as was originally attempted in this case. The high degree of homology (>99%) found with the GenBank strain of “F. rappini” suggested that this was the most likely identification. However, in DNA-DNA hybridization studies our isolate showed only 24 to 37% relatedness to the reference strain of “F. rappini” and substantially lower relatedness to the three Helicobacter spp. tested. Using the criteria of Wayne et al. (27), which indicate that a relative binding ratio of at least 70% is required for species-level relatedness, the NIH patient isolate is not related at the species level to “F. rappini” or to any of the Helicobacter strains tested. These findings reinforce the idea that close identity of 16S rRNA gene sequences is not always synonymous with species identity (9, 22). Despite low relatedness to the “Flexispira” and the three Helicobacter spp. tested and the lack of conclusive information from biochemical and CFA analyses, the morphology of the organism, including the presence of periplasmic fibers, strongly suggests a relationship to these species (Table 2). We suggest that the organism belongs to a taxon closely related to but separate from either “Flexispira” or Helicobacter. At this time, since our findings are based on the study of only one isolate, we believe that the proposal of a specific epithet for this organism is premature. In any case, greater proficiency at detection, growth, and identification of these fastidious organisms will help us to better understand the type of localized and systemic infections caused by Helicobacter spp. and the underlying host factors leading to these infections.
ACKNOWLEDGMENTS
We acknowledge the careful work carried out by the technologists at each of the laboratories working on cultures from this patient. We particularly thank Carolyn Dorworth-Fukuda for maintaining the viability of this fragile organism and for performing the antibiotic susceptibility tests.
REFERENCES
- 1.Archer J R, Romero S, Ritchie A E, Hamacher M E, Steiner J H, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner D J, McWhorter A C, Leete Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Biochemistry. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryner J H, Ritchie A E, Pollet L, Kirkbride C A. Experimental infection and abortion of pregnant guinea pigs with a unique spirillum-like bacterium isolated from aborted ovine fetuses. Am J Vet Res. 1987;48:91–95. [PubMed] [Google Scholar]
- 5.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O’Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dieterich C, Wiesel P, Neiger R, Blum A, Corthesy-Theulaz I. Presence of multiple “Helicobacter helimannii” strains in an individual suffering from ulcers and in his two cats. J Clin Microbiol. 1998;36:1366–1370. doi: 10.1128/jcm.36.5.1366-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton K A, Dewhirst F E, Radin M J, Fox J G, Paster B J, Krakowka S, Morgan D R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 8.Fleish F, Burnens A, Weber R, Zbinden R. Helicobacter species strain Mainz isolated from cultures of blood from two patients with AIDS. Clin Infect Dis. 1998;26:526–527. doi: 10.1086/517110. [DOI] [PubMed] [Google Scholar]
- 9.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 10.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray M W, Sankoff D, Cedergren R J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilzenrat N, Lamoureux E, Weintrub I, Alpert E, Lichter M, Alpert L. Helicobacter heilmannii-like spiral bacteria in gastric mucosal biopsies. Arch Pathol Lab Med. 1995;119:1149–1153. [PubMed] [Google Scholar]
- 13.Husmann M, Gries C, Jehnichen P, Woelfel T, Gerken G, Ludwig W, Bhakdi S. Helicobacter sp. strain Mainz isolated from an AIDS patient with septic arthritis: case report and nonradioactive analysis of 16S rRNA sequence. J Clin Microbiol. 1994;32:3037–3039. doi: 10.1128/jcm.32.12.3037-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 15.Kiehlbauch J A, Brenner D J, Cameron D N, Steigerwalt A G, Makowski J M, Baker C N, Patton C M, Wachsmuth I K. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J Clin Microbiol. 1995;33:2940–2947. doi: 10.1128/jcm.33.11.2940-2947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes E N, Queiroz D M M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 17.Romero S, Archer J R, Hamacher M E, Bologna S M, Schell R F. Case report of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:142–143. doi: 10.1128/jcm.26.1.142-143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Schauer D B, Ghori N, Falkow S. Isolation and characterization of “Flexispira rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solnick J V, O’Rourke J, Lee A, Paster B J, Dewhirst F E, Tompkins L S. An uncultured spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990;136:37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 23.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 24.Steinbrueckner B, Haerter G, Pelz K, Weiner S, Rump J A, Deissler W, Bereswill S, Kist M. Isolation of Helicobacter pullorum from patients with enteritis. Scand J Infect Dis. 1997;29:315–318. doi: 10.3109/00365549709019053. [DOI] [PubMed] [Google Scholar]
- 25.Tee W, Leder K, Karroum E A, Dyall-Smith M. “Flexispira rappini” bacteremia in a child with pneumonia. J Clin Microbiol. 1998;36:1679–1682. doi: 10.1128/jcm.36.6.1679-1682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trivett-Moore N L, Rawlinson W D, Yuen M, Gilbert G L. Helicobacter westmeadii sp. nov., a new species isolated from blood cultures of two AIDS patients. J Clin Microbiol. 1997;35:1144–1150. doi: 10.1128/jcm.35.5.1144-1150.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 28.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jordan J J, Cook E C, Danshvar M I. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria. 2nd ed. Baltimore, Md: The William & Wilkins Co.; 1996. [Google Scholar]