Abstract
Background
Emerging studies have shown that long noncoding RNAs (lncRNAs) predominantly function in the carcinogenesis of multiple developing human tumors. The current study aimed to investigate the underlying mechanisms of LINC00337 in lung adenocarcinoma.
Methods
We analyzed TCGA and GTEx datasets and chose LINC00337 as the research object. Cell proliferation, cell apoptosis, cell cycle, migration, and invasion were detected in the gain and loss experiments of LINC00337 both in vitro and in vivo. Moreover, RNA pull-down, luciferase reporter assays, western blotting analysis, and rescue experiments were performed to investigate the underlying molecular mechanisms of LINC00337 function.
Results
LINC00337 expression was remarkably upregulated in lung adenocarcinoma. In addition, LINC00337 knockdown was shown to repress cell migration, invasion, and proliferation, as well as the cell cycle, and gear up apoptosis in lung adenocarcinoma in vitro and in vivo. With respect to the mechanism, LINC00337 knockdown boosted miR-1285-3p expression and then restrained YTHDF1 expression post-transcriptionally. Crucially, both miR-1285-3p decrement and YTHDF1 overexpression successfully reversed the influence on cell proliferation, migration, invasion, and apoptosis caused by LINC00337 shRNA.
Conclusions
These results suggest that LINC00337 acts as an oncogenic lncRNA, targeting miR-1285-3p and regulating YTHDF1 expression, to promote the progression of lung adenocarcinoma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-021-02253-8.
Keywords: Lung adenocarcinoma, lncRNA, LINC00337, Cell invasion, Cell proliferation
Background
As a frequently seen malignant tumor, lung cancer is the chief cause of cancer-related deaths worldwide [1, 2]. Lung cancer is histologically classified into large cell carcinoma, squamous cell carcinoma, adenocarcinoma, and bronchoalveolar carcinoma [3]. Lung cancer patients have a low overall 5-year survival rate (approximately 18.1%). Lung adenocarcinoma accounts for nearly 40% of lung cancer cases [4, 5]. Therefore, it is of great significance to identify novel biomarkers and targets for the early diagnosis and treatment of lung cancer.
Human genomic sequencing uncovers the active transcription of more than 90% of genomes, among which 2% is the RNA that encodes proteins, and the remaining is the RNA unable to encode proteins [6, 7]. Long noncoding RNAs (lncRNAs) are ncRNAs longer than 200 nt [8, 9], many of which show cell type-specific expression [10–12] and had specific subcellular compartment locations [13–15]. Additionally, the expression of numerous lncRNAs has been demonstrated to be related to the progression of diverse cancers, which are able to modulate cancer cell proliferation and apoptosis [16–19]. According to reports, LINC00337 is a pro-tumor factor in gastric cancer [20] and esophageal cancer [21], but its function in lung adenocarcinoma remains elusive.
In the current study, we analyzed TCGA and GTEx datasets and found that LINC00337 was dramatically higher in lung adenocarcinoma tissues than in para-tumor tissues. In the TCGA dataset, high LINC00337 levels indicated a shorter overall survival. We also examined samples surgically resected from 46 lung adenocarcinoma cases at our institution to determine differences in the expression level of LINC00337 between lung adenocarcinoma tissues and normal tissues, and the results were consistent with the analyses of TCGA and GTEx datasets. We then conducted a series of experiments to explore whether LINC00337 participates in the onset and development of lung adenocarcinoma and the mechanism of its function.
Materials and methods
Collection of clinical samples
From 2017 to 2019, 46 paired fresh lung adenocarcinomas and para-tumor tissues were harvested at our hospital and snap-frozen at − 80 °C. The patients did not receive preoperative chemotherapy or radiotherapy. All included subjects provided informed consent, and the study was approved by the Institutional Review Board of Xinxiang Central Hospital. The detailed clinicopathological characteristics of the patients with lung adenocarcinoma are summarized in Table 1.
Table 1.
The correlation of LINC00337 expression with clinical parameters in patients with lung adenocarcinoma
| Clinicopathological features | Number of cases | LINC00337 expression | P value | |
|---|---|---|---|---|
| High (n=23) | Low (n=23) | |||
| Gender | 0.2362 | |||
| Male | 25 | 10 | 15 | |
| Female | 21 | 13 | 8 | |
| Age | 0.7683 | |||
| < 60 | 24 | 13 | 11 | |
| ≥ 60 | 22 | 10 | 12 | |
| Tumor size | 0.0377* | |||
| ≤ 5 | 24 | 8 | 16 | |
| > 5 | 22 | 15 | 7 | |
| TNM stages | 0.0058* | |||
| I/II | 28 | 9 | 19 | |
| III/IV | 18 | 14 | 4 | |
| Lymph node metastasis | 0.0072* | |||
| Present | 24 | 17 | 7 | |
| Absent | 22 | 6 | 16 | |
Total data from 46 lung adenocarcinoma patients were analyzed. For the expression of LINC00337 was assayed by qRT-PCR, the median expression level was used as the cutoff. Data were analyzed by chi-squared or Fisher’s exact test. P-value in bold indicates statistically significant
Cell culture
Lung adenocarcinoma cell lines (PC-9, H1373, HCC827, and A549) and a normal human lung epithelial cell line (BEAS2B) were collected from the Cell Resource of CAMS (Beijing, China), cultured in RPMI-1640 medium with 10% FBS (Gibco, CA, USA), and preserved at 37 °C with 5% CO2.
shRNAs and anti-miRNA inhibitors
Overall, shRNAs targeting LINC00337 (shRNA#1, 2), shRNA targeting YTHDF1 (sh-YTHDF1), and negative control shRNA(sh-NC) with no definite target were adopted and synthesized by Genechem (Shanghai, China). Anti-miR-1285-3p inhibitor (anti-miR-1285-3p) and anti-miR negative control (anti-miR-NC) were purchased from RiboBio Co. (Guangzhou, China). PC-9 or A549 cells were seeded in 6-well plates 24 h prior to transfection with 40–60% confluence, and then transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Transfected cells were harvested 48 h after transfection. Stable cell lines were selected by treatment with neomycin (500 µg/mL) for 4 weeks.
RNA isolation and quantitative real-time PCR (qRT-PCR)
As per the manufacturer’s instructions, total RNA segregation was implemented using TRIzol from Invitrogen, and it was then synthesized into cDNA using stochastic primers with a PrimeScript RT reagent Kit from Takara (Dalian, China) or an miRNA reverse transcription PCR kit commercially offered by RiboBio. qRT-PCR analysis was performed using the SYBR Premix Ex Taq kit (Takara). The following primers were used: LNC00337, 5′-CCAGACTGGAGAACCACAGC-3′ (forward) and 5′-CTGTGTCTATGTGCAGCCCT-3′ (reverse); miR-1285-3p, 5′-TCTGGGCAACAAAGTGAG-3′ (forward) and 5′-CTCAACTGGTGTCGTGGA-3′ (reverse); and YTHDF1, 5′-ACCTGTCCAGCTATTACCCG-3′ (forward) and 5′-TGGTGAGGTATGGAATCGGAG-3′ (reverse). Bulge-Loop miRNA qPCR Primers were obtained from RiboBio, and data were processed using the StepOnePlus Real-Time PCR System obtained from Applied Biosystems (Shanghai, China), whose results were evaluated with GAPDH or U6 expression as a standard.
Western blot analysis
RIPA extraction liquid from Beyotime (Jiangsu, China), in the presence of protease inhibitor cocktail and PMSF (Roche, Shanghai, China), was used to lyse the assembled cells. Following the determination of the protein sample concentration using the BCA Protein Assay Kit from Beyotime, the harvested proteins were separated by SDS-PAGE (10% gel) and transferred to PVDF membranes, which underwent 1-h sealing with Tris-buffered saline (5% defatted milk) and 12-h primary antibody incubation at 4 °C. Next, the optical density method was used to quantitate autoradiographs using Quantity One software (Bio-Rad) with GAPDH (#2118; CST, Shanghai, China) as a reference. Anti-YTHDF1 (#86463), anti-E-cadherin (#3195), and anti-Vimentin (#5741) antibodies were obtained from CST.
Immunohistochemistry
Nude mouse tumor tissues implanted in paraffin were immunostained, and the expression level and position of target proteins were determined using the avidin–biotin–peroxidase method. Next, primary antibodies against E-cadherin and vimentin were diluted 1:200 for later application. Tumor apoptosis and proliferation were evaluated by independently probing for Ki-67 and Bax. Finally, slice visualization was achieved using a microscope from Olympus (Japan).
5-ethynyl-20-deoxyuridine assay (EdU) Assay
Cell proliferation was determined by the ethynyl-2-deoxyuridine incorporation assay using an EdU Apollo DNA in vitro kit (RIBOBIO, Guangzhou, China) according to the manufacturer’s instructions. Briefly, after transfection with the corresponding vector, cells were incubated for 2 h at 37 °C, with 100 µL of 50 µM EdU/well. The cells were identified using fluorescence microscopy. Each experiment was carried out three times.
Cell Counting Kit-8 assay
Cell Counting Kit-8 (Beyotime Inst Biotech, China) was used to determine cell proliferation. Briefly, 5 × 103 cells/well underwent 1-day raising in a 96-well broad-bottomed plate at 37 °C, followed by transfection with the corresponding vectors. Finally, using a microplate reader from Bio-Rad (Shanghai, China), the absorbance was measured at 450 nm, and each experiment was performed three times.
Apoptosis and cell cycle experiments
As per the manufacturer’s instruction, apoptosis determination was implemented via FACS using a PE-Annexin V apoptosis detection kit from BD Pharmingen (Shanghai, China) after 48-h transfection, and the cell cycle was assessed utilizing PI cell cycle assessment kits (BD Pharmingen). Each assay was performed in triplicate.
Wound-healing assays
We cultured different groups of lung adenocarcinoma cells (1 × 106 cells/well) up to 90% confluency. Then, we scratched the monolayer of cells with a sterile pipette tip (100 µL) in each well. After washing three times with phosphate-buffered saline (PBS; Thermo Fisher Scientific, Inc.), the cells were incubated with serum-free PRIM-1640 medium for 24 h at 37 °C in an incubator containing 5% CO2. The plates were viewed under a light microscope (DFC500; Leica, Wetzlar, Germany) at different time points and monitored using AxioVision version 4.7 software (Carl Zeiss Meditec, Dublin, CA, USA).
Transwell assay
Transwell chambers were used to observe the invasion of lung adenocarcinoma cells. We seeded cancer cells in the upper chamber precoated with Matrigel (Corning, USA, dilution ratio: 1:6) at a density of 105 cells per well and supplemented with DMEM containing 1% FBS. We filled 600 µL DMEM with 10% FBS into the lower chamber. The cells were then incubated at 37 °C for 24 h. We fixed cells with 4% methanol and stained them with crystal violet. Then, we counted them in five random 200⋅ microscopic fields after the cells invaded the lower surface of the membrane. Each assay was performed in triplicate.
Dual-Luciferase reporter assay
Genechem designed and synthesized a YTHDF1 full-length promoter reporter vector. A human YTHDF1 3′-untranslated region (UTR) fragment with the supposed binding sites for the miR-1285-3p reporter vector was provided by RiboBio. After transfection for 48 h, the Dual-Luciferase Reporter Assay System from Promega was used for luciferase activity determination according to the manufacturer’s instructions, and the luciferase activity ratio (Firefly/Renilla) was ascertained. Each assay was performed in triplicate.
RNA pull-down assay
The DNA fragment with the full-length LINC00337 or NC sequence was amplified using a primer with T7 and cloned into GV394 from Genechem (Shanghai, China). The restriction enzyme XhoI was used for the linearization of DNA. Next, T7 RNA polymerase (Takara) and Biotin RNA Labeling Mix (Roche, China) were used for reverse transcription of biotin-labeled RNAs that underwent reverse transcription. Thereafter, the products received DNase I (RNase-free, Roche) treatment and purification using the RNeasy Mini Kit (Qiagen, USA), and the extracted RNAs were used for qRT-PCR assessment.
In vivo tumorigenesis and metastasis assays
Six-week-old nude mice were randomly divided into two groups (4 mice/group) and raised with unlimited food and water in sterile conditions without pathogens. To establish a lung adenocarcinoma xenograft model, we subcutaneously injected A549 cells into nude mice. Tumor growth was monitored weekly and calculated as follows: volume = (length) × (width)2/2. Tail intravenous injection models were established for the lung metastasis assays. After 6 weeks, the nude mice were euthanized, and metastatic nodules in each lung were analyzed. Animal assays were performed in the SPF Animal Laboratory at Xinxiang Medical University, and experiments were performed according to the NIH guidelines on animal welfare.
Statistical analysis
Differences in data in terms of normal distribution and equal variance were processed using a two-tailed Student’s test (two-group comparisons) or ANOVA, and the post-hoc Bonferroni test (multigroup comparisons) was implemented as appropriate. Differences in the data of non-normal distribution or unequal variance were processed by a nonparametric Mann-Whitney U test (two‐group comparisons) or the Kruskal-Wallis test followed by the post-hoc Bonferroni test (multigroup comparisons). P < 0.05 was considered as statistically significant. All tests were performed using SPSS (version 22.0; SPSS, Chicago, IL, USA).
Results
LINC00337 is elevated in lung adenocarcinoma tissues and cells, and is predominantly localized in the cell cytoplasm
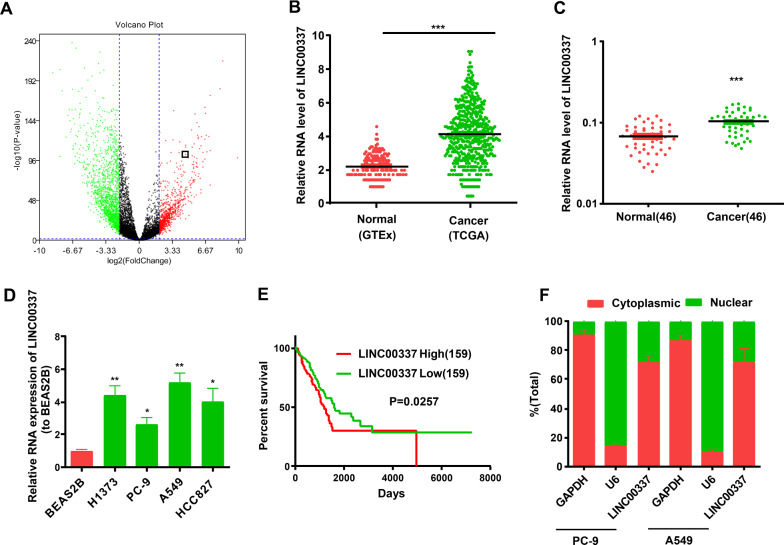
Through the analysis of TCGA and GTEx databases, we found that LINC00337 was significantly increased in lung adenocarcinoma tissues (from TCGA database) relative to normal tissues (from GTEx database) (Fig. 1A, B). We then verified 46 lung adenocarcinoma tissues and adjacent non-tumorous tissues by qRT-PCR assays, and the results were consistent with previous analyses of TCGA and GTEx databases (Fig. 1C). Similarly, higher LINC00337 levels were observed in lung adenocarcinoma cells (PC-9, H1373, HCC827, and A549) than in the normal human lung epithelial cell line BEAS2B (Fig. 1D). Additionally, PC-9 and A549 cells were selected for the subsequent assays. In addition, LINC00337 expression levels in lung adenocarcinoma were evidently interrelated to high-grade cancer, lymph node metastasis, and tumor size, as opposed to other parameters, such as age or sex (Table 1). TCGA database showed that the overall survival rate of patients with low LINC00337 levels was higher than that of patients with high LINC00337 levels (Fig. 1E). Next, we examined the subcellular localization of LINC00337 and found that most of the LINC00337 were present in the cytoplasm of lung adenocarcinoma cells (Fig. 1 F).
Fig. 1.
LINC00337 is raised in lung adenocarcinoma tissues and cells, and is significantly present in the cell cytoplasm. A Volcano plot of TCGA + GTEx database, and LINC00337 is increased in lung adenocarcinoma. B The RNA level of LINC00337 in lung adenocarcinoma samples (from TCGA database) and normal samples (from GTEx database). C LINC00337 expression in 46 pairs of lung adenocarcinoma tissues and para-tumor tissues from our data. D LINC00337 expression in lung adenocarcinoma PC-9, H1373, HCC827, and A549 cell lines and normal human lung epithelial BEAS2B cell line. E Kaplan–Meier curves of overall survival from TCGA database. The median expression level of LINC00337 was used as the cutoff. F Localization of LINC00337 by nucleocytoplasmic separation experiment. Data represent the mean ± SD; *P < 0.05, ** P < 0.01, ***P < 0.001
Knockdown of LINC00337 curbs the cell cycle, as well as proliferation, and invasion, and increases apoptosis of lung adenocarcinoma cells
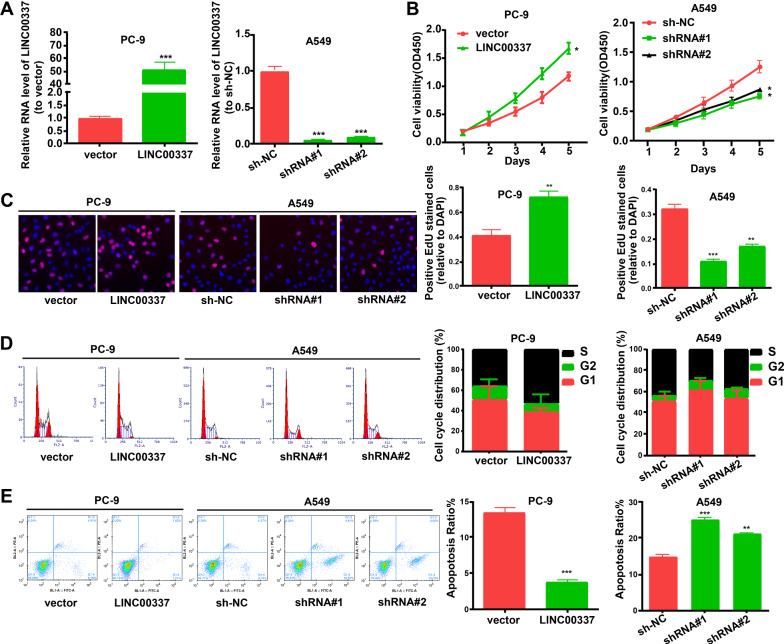
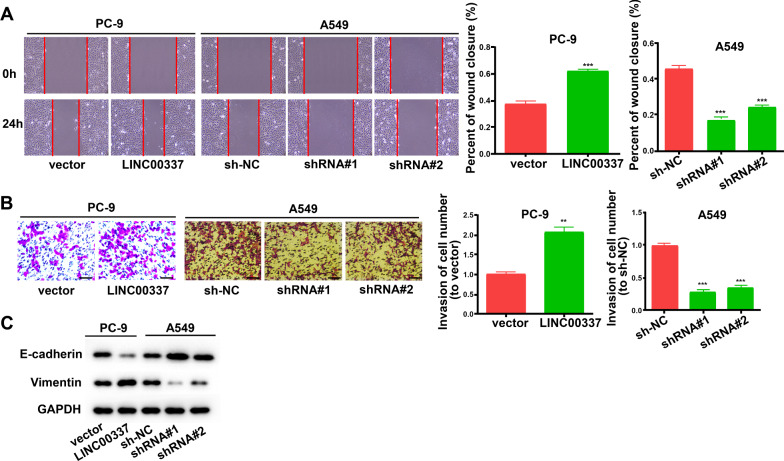
To determine whether LINC00337 functions in lung adenocarcinoma cells, we performed a variety of in vitro assays to assess the impact of shRNA knockdown LINC00337 and overexpression of LINC00337 on cell functions, including proliferation, apoptosis, and invasion. PC-9 cells were transfected with LINC00337 overexpression vector, and A549 cells were transfected with sh-LINC00337 (Fig. 2A). CCK-8 and EdU assays showed that overexpression of LINC00337 promoted the proliferation of PC-9 cells, and sh-LINC00337 attenuated the proliferation of A549 cells (Fig. 2B, C). LINC00337 knockdown reduced cell cycle arrest at the S phase in A549 cells compared with the negative control, and overexpression of LINC00337 resulted in cell cycle arrest at the S phase in PC-9 cells (Fig. 2D). As shown in Fig. 2E, an evidently elevated apoptotic cell ratio was observed in the sh-LINC00337 group relative to sh-NC cells, and reduced the proportion of apoptotic cells in LINC00337-transfected cells relative to vector-transfected cells. Meanwhile, the migration and invasion of cells were considerably elevated by overexpression of LINC00337 and decreased by sh-LINC00337 (Fig. 3A, B). Western blot assays indicated that overexpression of LINC00337 promoted the expression of Vimentin and inhibited the expression of E-cadherin (Fig. 3C). All the above-mentioned data confirmed that knockdown of LINC00337 curbs the cell cycle, as well as proliferation, and invasion, and increases apoptosis of lung adenocarcinoma cells.
Fig. 2.
Knock-down of LINC00337 curbs proliferation, and increases apoptosis of lung adenocarcinoma cells. A Transfection efficiency of sh-LINC00337#1(shRNA#1), sh-LINC00337 #2(shRNA#2), and LINC00337-overexpressing vectors (LINC00337). B CCK-8 and C EdU assays were performed to test cell proliferation. D The influence of LINC00337 on the cell cycle. E The influence of LINC00337 on cell apoptosis. Data represent the mean ± SD of three independent experiments; *P < 0.05, ** P < 0.01, ***P < 0.001
Fig. 3.
Knock-down of LINC00337 inhibits cell migration and invasion of lung adenocarcinoma cells. A Cell migration was determined using wound-healing assays. B Cell invasion was determined using Transwell assays. C Western blot was performed to assess the protein level of E-cadherin and Vimentin in vitro. Data represent the mean ± SD of three independent experiments; ** P < 0.01, ***P < 0.001
LINC00337 interplays with miR-1285-3p in a direct manner
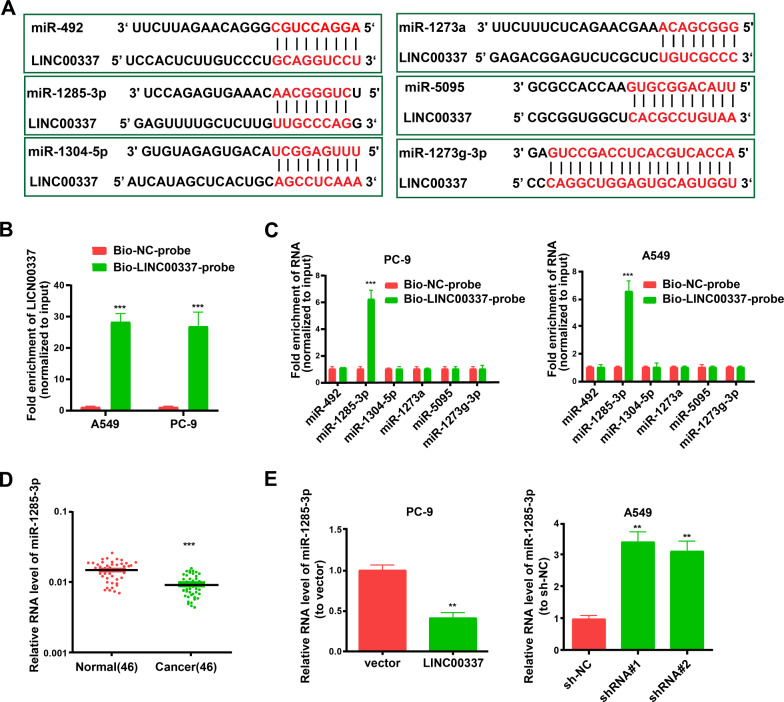
LncRNA is a newly discovered regulatory mechanism affecting post-transcriptional control, disturbing miRNA pathways, and acting as a natural miRNA sponge to reduce the binding of endogenous miRNAs to target genes [22–25]. By searching an online bioinformatics database (RegRNA 2.0, http://regrna2.mbc.nctu.edu.tw/), we observed that six miRNAs (hsa-miR-492, hsa-miR-1285-3p, hsa-miR-1304-5p, hsa-miR-1273a, hsa-miR-5095, and hsa-miR-1273 g-3p) possessed putative binding sites for LINC00337 (Fig. 4A).
Fig. 4.
LINC00337 interplays with miR-1285-3p directly. A The binding sequence of LINC00337 with miR-1285-3p, miR-1304-5p, miR-492, miR-1273a, miR-5095, and miR-1273 g-3p, as predicted by RegRNA 2.0. B Detection of LINC00337 using qRT-PCR in samples pulled down by biotinylated LINC00337 and a negative control probe. C Examination of miR-492, miR-1285-3p, miR-1304-5p, miR-1273a, miR-5095, and miR-1273 g-3p in the same sample was reduced by biotinylation of LINC00337 and the NC probe. D The level of miR-1285-3p in 46 pairs of lung adenocarcinoma and para-tumor tissues. E The expression levels of miR-1285-3p in cells transfected with either shRNA or LINC00337 were evaluated by qRT-PCR. Data represent the mean ± SD; ** P < 0.01, ***P < 0.001
Later, we utilized a biotin pull-down system to continuously probe miRNAs that directly interplay with LINC00337. We unraveled a significant amount of miR-1285-3p in the LINC00337 pull-down pellet relative to the control group, as examined by qRT-PCR, but the proportions of miR-492, miR-1273a, miR-5095, miR-1273 g-3p, and miR-1304-5p in the LINC00337 pull-down pellet displayed a clear elevation relative to the control group (Fig. 4B, C). Moreover, miR-1285-3p was expressed in lung adenocarcinoma samples at a lower level than in normal samples (Fig. 4D). Overexpression of LINC00337 decreased miR-1285-3p expression levels as determined by qRT-PCR (Fig. 4E). All the above-mentioned data confirmed that LINC00337 could directly sponge miR-1285-3p in a highly specific manner.
LINC00337 regulates the miR-1285-3p target gene YTHDF1
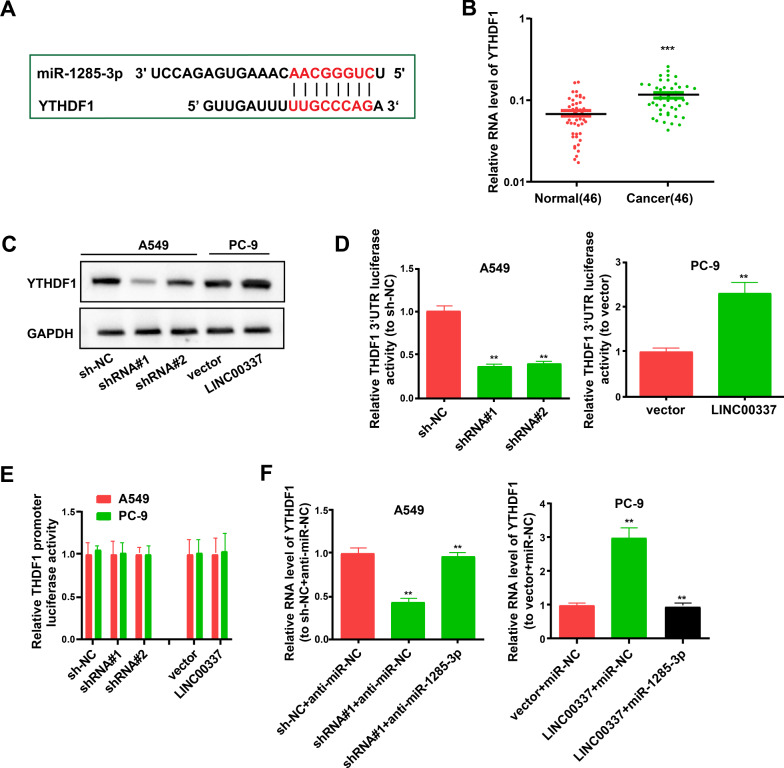
By searching miRDB, we observed seven target genes of miR-1285-3p, with scores >95 (http://mirdb.org/): AHI1, DAB2IP, BTRC, YTHDF1, TMEM41B, SIKE1, and FMO5 (Additional file 1: Table S1). After reviewing the literature, we found that only YTHDF1 was bound to lung adenocarcinoma [26–28]. Therefore, we surmised that LINC00337 functions by influencing YTHDF1 expression in lung adenocarcinoma. The binding sites of YTHDF1 and miR-1285-3p are shown in Fig. 5A. Results of qRT-PCR showed that the YTHDF1 levels in cancer tissues were significantly increased (Fig. 5B), and western blotting showed that downregulation of LINC00337 decreased the protein level of YTHDF1 (Fig. 5C).
Fig. 5.
LINC00337 controls the miR-1285-3p target gene YTHDF1. A Proposed binding sequence of miR-1285-3p with YTHDF1. B YTHDF1 levels in 46 paired lung adenocarcinoma and para-tumor tissues. C The protein levels of YTHDF1 in cells undergoing shRNA or LINC00337 transfection were evaluated by western blot assay, with GAPDH as a standard. D Relative luciferase activities of YTHDF1 3′-UTR reporter in stable A549 and PC-9 cells undergoing shRNA or LINC00337 transfection were examined using the Dual-Luciferase Reporter Assay System. E Relative luciferase activities of YTHDF1 promoter reporter in stable A549 and PC-9 cells receiving shRNA or LINC00337 transfection. F YTHDF1 expression level in stable A549 and PC-9 cells after transfection. Data represent the mean ± SD; ** P < 0.01, ***P < 0.001
Dual-luciferase reporter assays were implemented using a human YTHDF1 3′-UTR fragment with supposed binding sites of miR-1285-3p and the YTHDF1 promoter reporter vector for the notarizing effect of miR-1285-3p on YTHDF1. Cells transfected with stable sh-LINC00337 exhibited a dramatically decreased relative luciferase activity of YTHDF1-3′-UTR (Fig. 5D). Overexpression of LINC00337 increased luciferase activity in cells transfected with the stable LINC00337 overexpression vector (Fig. 5D). However, transfection of the sh-LINC00337 or LINC00337 overexpression vector did not alter the promoter activity of YTHDF1 in PC-9 and A549 cells (Fig. 5E). Furthermore, decrement of miR-1285-3p with anti-miR-1285-3p successfully hindered the decrease in YTHDF1 protein levels induced by LINC00337 shRNA (Fig. 5F). It could be inferred that LINC00337 controlled YTHDF1 expression at the miR-1285-3p-adjusted posttranscriptional level. The transfection efficiency of the miR-1285-3p mimics and anti-miR-1285-3p is shown in Additional file 2: Figure S1A.
The LINC00337/miR-1285-3p/YTHDF1 axis regulates the behavior of lung adenocarcinoma cells
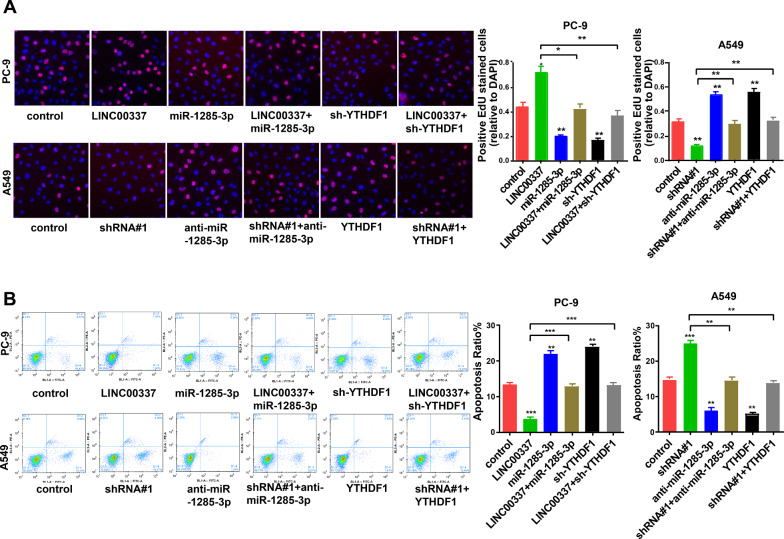
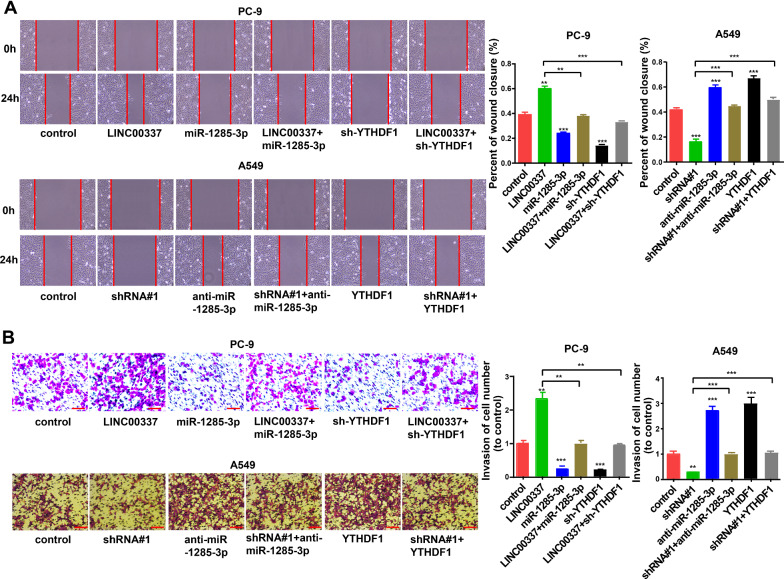
Subsequently, we explored the effect of LINC00337 on the miR-1285-3p/YTHDF1 axis in lung adenocarcinoma. We transfected sh-YTHDF1 in PC-9 cells and transfected the YTHDF1 overexpression vector into A549 cells (Additional file 2: Figure S1B). As shown in Fig. 6A, knockdown of miR-1285-3p and upregulation of YTHDF1 reversed the cell proliferation that was reduced by LINC00337 shRNA. Knockdown of YTHDF1 and upregulation of miR-1285-3p reversed the cell proliferation induced by LINC00337 overexpression. Cell apoptosis, migration, and invasion assays showed a similar phenomenon: both knockdown of YTHDF1 and upregulation of miR-1285-3p reversed the effects caused by overexpression of LINC00337 on cell apoptosis (Fig. 6B), migration, and invasion (Fig. 7A, B). These data suggest that LINC00337 modulates lung adenocarcinoma in vitro through the miR-1285-3p / YTHDF1 axis.
Fig. 6.
The LINC00337/miR-1285-3p/YTHDF1 axis regulates the behavior of lung adenocarcinoma cells. A Using the EdU assay to detected the cell proliferation of A549 and PC-9 cells after transfection. B Apoptosis of A549 and PC-9 cells after transfection was detected. Data represent the mean ± SD of three respective experiments; *P < 0.05, ** P < 0.01, ***P < 0.001
Fig. 7.
The LINC00337/miR-1285-3p/YTHDF1 axis controls behaviors of lung adenocarcinoma cells. A Cell migration of A549 and PC-9 cells after transfection. B Cell invasion of A549 and PC-9 cells after transfection. Data represent the mean ± SD of three respective experiments; ** P < 0.01, ***P < 0.001
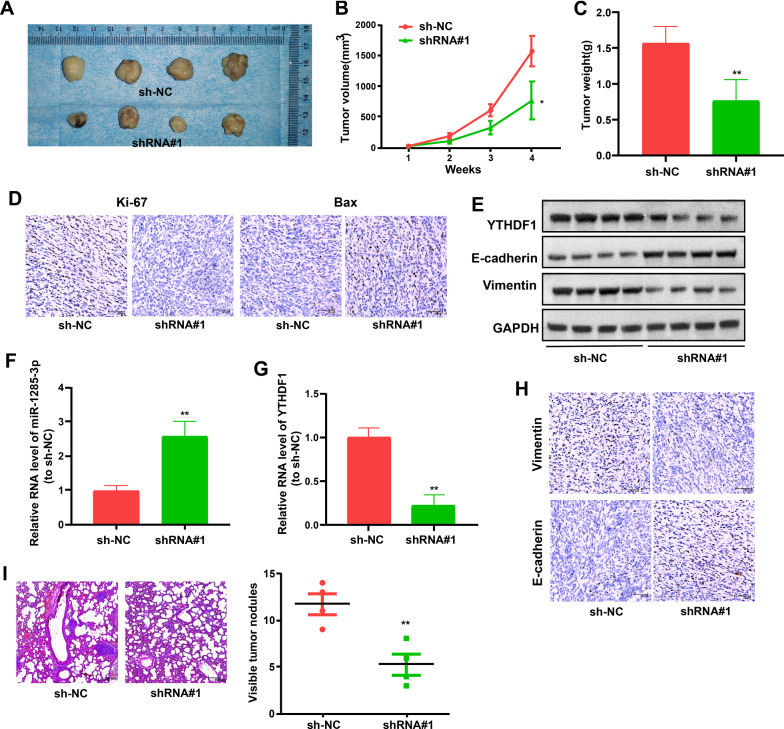
Inhibition of LINC00337 suppresses lung adenocarcinoma tumor growth and metastasize in vivo
To further determine the anti-tumorigenesis potential of LINC00337 inhibition in vivo, stable A549 cells transfected with sh-NC or sh-LINC00337 were inoculated into nude mice. Mice in the sh-LINC00337 group had decreased tumor volume and weight after the assay relative to the sh-NC group (Fig. 8A–C). Furthermore, LINC00337 knockdown inhibited tumor proliferation and boosted cell apoptosis (Fig. 8D). Western blot assay, qRT-PCR, and histological results of excised tumor tissues implied a positive correlation between LINC00337 expression and YTHDF1 and Vimentin, as well as an inverse relationship with miR-1285-3p and E-cadherin, in LINC00337 repression and control groups (Fig. 8E–H). Furthermore, HE staining of mouse lung slices revealed that suppressing LINC00337 reduced the number of metastatic nodules in the lung relative to the sh-NC group (Fig. 8I). The above-mentioned findings uncovered the potential of LINC00337 in terms of tumor metastasis and proliferation, and offered more support for treatments targeting LINC00337 in lung adenocarcinoma.
Fig. 8.
Inhibition of LINC00337 suppresses lung adenocarcinoma tumor growth and metastasize in vivo. A Images of xenografts. B Tumor volume and C weight of xenografts. D Ki-67 was used to assess proliferation via immunohistochemical staining, and Bax was utilized for apoptosis assay. E Western blotting was used to assess the protein level of YTHDF1, E-cadherin, and Vimentin in tumors. F MiR-1285-3p and G YTHDF1 levels in tumors were detected. H Vimentin and E-cadherin from tumors were assessed via immunohistochemistry. I HE stained lung sections. Data represent the mean ± SD; *P < 0.05, **P < 0.01
Discussion
In this study, we analyzed TCGA and GTEx datasets and chose LINC00337 as the research object, which was expressed at a notably higher level in lung adenocarcinoma tissues and paired para-tumor tissues. In addition, LINC00337 knockdown significantly curbed the ability of lung adenocarcinoma cells to proliferate and invade, as well as arrest the cell cycle, but increased apoptosis in vitro and in vivo.
Like proteins, the function of lncRNAs depends on their subcellular localization [29]. Cytoplasmic lncRNAs that mostly localize and function in the cytoplasm can influence gene regulation by acting as decoys for miRNAs and proteins [30, 31]. Several studies have revealed that lncRNAs are sponges of many miRNAs, exerting the same function as ceRNAs in tumorigenesis[32, 33].
Through subcellular localization experiments, we found that most of the LINC00337 was present in the cytoplasm of lung adenocarcinoma cells, suggesting that LINC00337 might function at the post-transcriptional level. In this case, LINC00337 may act as a ceRNA to disturb miRNA pathways and control the suppression of miRNA targets. Therefore, we predicted the miRNA and its downstream targets that may be associated with LINC00337 by searching in RegRNA 2.0, miRDB. RNA pull-down assay, dual-luciferase reporter assay, qRT-PCR, and western blotting were performed to confirm the association. The results showed that LINC00337 functions as a miR-1285-3p sponge to control YTHDF1 in a positive manner. Subsequent cell function tests confirmed that both knockdown of YTHDF1 and upregulation of miR-1285-3p reversed the influence caused by overexpression of LINC00337 on cell invasion, proliferation, and apoptosis.
This study had several limitations. First, a larger sample size is required to verify the clinical value of LINC00337. Second, more lncRNAs should be explored in the pathogenesis of lung adenocarcinoma. We will further explore these in future studies.
Conclusions
In conclusion, we identified that LINC00337 was upregulated in lung adenocarcinoma and correlated with poor survival outcomes in patients with lung adenocarcinoma. LINC00337 acts as an oncogenic lncRNA, targeting miR-1285-3p and regulating YTHDF1 expression, to promote the progression of lung adenocarcinoma.
Supplementary Information
Additional file 1: TableS1. The 7 target genes of miR-1285-3p which score >95 in miRDB.
Additional file 2: FigureS1. The transfection efficiency in PC-9 and A549 cells.
Acknowledgements
Not applicable.
Authors’ contributions
RZ designed and conducted the majority of the experiments and manuscript writing. DW assisted with the results collection and processing. LW and GG instructed data analysis and figure production. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The present research gained approval from the Ethics Committee of Xinxiang Central Hospital and Written informed consent from patients was obtained. Animal experiments took place in SPF Animal Laboratory at Xinxiang Medical University. All animal assays were implemented as per the Guide for the Care and Use of Laboratory Animals by NIH.
Consent for publication
All authors involved in the study had given their consent for submitting this article for publication.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: 10.1186/s12935-024-03431-0
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/14/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s12935-024-03431-0
References
- 1.Hao X, Han F, Ma B, Zhang N, Chen H, Jiang X, Yin L, Liu W, Ao L, Cao J, et al. SOX30 is a key regulator of desmosomal gene suppressing tumor growth and metastasis in lung adenocarcinoma. J Exp Clin Cancer Res. 2018;37(1):111. doi: 10.1186/s13046-018-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee G, Park H, Sohn I, Lee SH, Song SH, Kim H, Lee KS, Shim YM, Lee HY. Comprehensive computed tomography radiomics analysis of lung adenocarcinoma for prognostication. Oncologist. 2018;23(7):806–13. doi: 10.1634/theoncologist.2017-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y, Zhao J, Yin X, Yuan X, Guo J, Bi J. miR-297 acts as an oncogene by targeting GPC5 in lung adenocarcinoma. Cell Prolif. 2016;49(5):636–43. doi: 10.1111/cpr.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam H, Li N, Dhar SS, Wu SJ, Lv J, Chen K, Flores ER, Baseler L, Lee MG. HP1gamma promotes lung adenocarcinoma by downregulating the transcription-repressive regulators NCOR2 and ZBTB7A. Cancer Res. 2018;78(14):3834–48. doi: 10.1158/0008-5472.CAN-17-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegab AE, Ozaki M, Kameyama N, Gao J, Kagawa S, Yasuda H, Soejima K, Yin Y, Guzy RD, Nakamura Y, et al. Effect of FGF/FGFR pathway blocking on lung adenocarcinoma and its cancer-associated fibroblasts. J Pathol. 2019;249(2):193–205. doi: 10.1002/path.5290. [DOI] [PubMed] [Google Scholar]
- 6.Tichon A, Perry RB, Stojic L, Ulitsky I. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev. 2018;32(1):70–8. doi: 10.1101/gad.309138.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2018;19(5):930–45. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 8.Shen SN, Li K, Liu Y, Yang CL, He CY, Wang HR. Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol Oncol. 2019;13(12):2554–73. doi: 10.1002/1878-0261.12555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lian Y, Xiong F, Yang L, Bo H, Gong Z, Wang Y, Wei F, Tang Y, Li X, Liao Q, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253. doi: 10.1186/s13046-018-0918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Wang T, Yang Y, Zheng S. Tumor heterogeneity uncovered by dynamic expression of long noncoding RNA at single-cell resolution. Cancer Genet. 2015;208(12):581–6. doi: 10.1016/j.cancergen.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Baskozos G, Dawes JM, Austin JS, Antunes-Martins A, McDermott L, Clark AJ, Trendafilova T, Lees JG, McMahon SB, Mogil JS, et al. Comprehensive analysis of long noncoding RNA expression in dorsal root ganglion reveals cell-type specificity and dysregulation after nerve injury. Pain. 2019;160(2):463–85. doi: 10.1097/j.pain.0000000000001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy AS, O’Brien D, Pisat N, Weichselbaum CT, Sakers K, Lisci M, Dalal JS, Dougherty JD. A comprehensive analysis of cell type-specific nuclear RNA from neurons and glia of the brain. Biol Psychiatry. 2017;81(3):252–64. doi: 10.1016/j.biopsych.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41(9):761–72. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Liu Z, Gong R. Long noncoding RNA: an emerging player in diabetes and diabetic kidney disease. Clin Sci (Lond) 2019;133(12):1321–39. doi: 10.1042/CS20190372. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL, et al. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446:49–61. doi: 10.1016/j.canlet.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 16.Shang AQ, Wang WW, Yang YB, Gu CZ, Ji P, Chen C, Zeng BJ, Wu JL, Lu WY, Sun ZJ, et al. Knockdown of long noncoding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3. Am J Physiol Gastrointest Liver Physiol. 2019;317(2):G222–32. doi: 10.1152/ajpgi.00357.2018. [DOI] [PubMed] [Google Scholar]
- 17.Song F, Li L, Liang D, Zhuo Y, Wang X, Dai H. Knockdown of long noncoding RNA urothelial carcinoma associated 1 inhibits colorectal cancer cell proliferation and promotes apoptosis via modulating autophagy. J Cell Physiol. 2019;234(5):7420–34. doi: 10.1002/jcp.27500. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Zhao L, Li J, Zhong L. Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J Cell Physiol. 2019;234(11):20980–91. doi: 10.1002/jcp.28702. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Yan G, Zhang J, Yu L. Knockdown of long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) inhibits proliferation, migration, and invasion and promotes apoptosis by targeting miR-124 in retinoblastoma. Oncol Res. 2018;26(4):581–91. doi: 10.3727/096504017X14953948675403. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Hu B, Wang X, Li L. Long noncoding RNA LINC00337 promote gastric cancer proliferation through repressing p21 mediated by EZH2. Am J Transl Res. 2019;11(5):3238–45. [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Shen S, Zheng X, Ye K, Ge H, Sun Y, Lu Y. Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. FASEB J. 2020;34(5):6055–69. doi: 10.1096/fj.201900731RR. [DOI] [PubMed] [Google Scholar]
- 22.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425(19):3723–30. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58(5):1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Gao Q, Wang M, Xin H. LncRNA SNHG1 contributes to the regulation of acute myeloid leukemia cell growth by modulating miR-489-3p/SOX12/Wnt/beta-catenin signaling. J Cell Physiol. 2020;236:653–63. doi: 10.1002/jcp.29892. [DOI] [PubMed] [Google Scholar]
- 25.Podralska M, Ciesielska S, Kluiver J, van den Berg A, Dzikiewicz-Krawczyk A, Slezak-Prochazka I. Non-coding RNAs in cancer radiosensitivity: MicroRNAs and lncRNAs as regulators of radiation-induced signaling pathways. Cancers (Basel) 2020;12(6):1662. doi: 10.3390/cancers12061662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Li N, Huang L, Xu S, Zheng X, Hamsath A, Zhang M, Dai L, Zhang H, Wong JJ, et al. Is hydrogen sulfide a concern during treatment of lung adenocarcinoma with ammonium tetrathiomolybdate? Front Oncol. 2020;10:234. doi: 10.3389/fonc.2020.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang Z, Chen L, Mao Y, Zheng Q, Li H, Huang Y, Hu Z, Jin Y. Diagnostic, progressive and prognostic performance of m(6)A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci. 2020;16(11):1785–97. doi: 10.7150/ijbs.39046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10(1):4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23(7):1080–7. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang R, Xing L, Wang M, Chi H, Zhang L, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs/mRNAs and miRNAs with associated ceRNA networks in triple-negative breast cancer. Cell Physiol Biochem. 2018;50(2):473–88. doi: 10.1159/000494162. [DOI] [PubMed] [Google Scholar]
- 31.Tran DDH, Kessler C, Niehus SE, Mahnkopf M, Koch A, Tamura T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene. 2018;37(1):75–85. doi: 10.1038/onc.2017.312. [DOI] [PubMed] [Google Scholar]
- 32.Rui X, Xu Y, Huang Y, Ji L, Jiang X. lncRNA DLG1-AS1 promotes cell proliferation by competitively binding with miR-107 and up-regulating ZHX1 expression in cervical cancer. Cell Physiol Biochem. 2018;49(5):1792–803. doi: 10.1159/000493625. [DOI] [PubMed] [Google Scholar]
- 33.Shen SN, Li K, Liu Y, Yang CL, He CY, Wang HR. Silencing lncRNAs PVT1 upregulates miR-145 and confers inhibitory effects on viability, invasion, and migration in EC. Mol Ther Nucleic Acids. 2020;19:668–82. doi: 10.1016/j.omtn.2019.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: TableS1. The 7 target genes of miR-1285-3p which score >95 in miRDB.
Additional file 2: FigureS1. The transfection efficiency in PC-9 and A549 cells.
Data Availability Statement
Datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.