Abstract
We analyse the time-series evolution of the cumulative number of confirmed cases of COVID-19, the novel coronavirus disease, for some African countries. We propose a mathematical model, incorporating non-pharmaceutical interventions to unravel the disease transmission dynamics. Analysis of the stability of the model’s steady states was carried out, and the reproduction number , a vital key for flattening the time-evolution of COVID-19 cases, was obtained by means of the next generation matrix technique. By dividing the time evolution of the pandemic for the cumulative number of confirmed infected cases into different regimes or intervals, hereafter referred to as phases, numerical simulations were performed to fit the proposed model to the cumulative number of confirmed infections for different phases of COVID-19 during its first wave. The estimated declined from 2.452–9.179 during the first phase of the infection to 1.374–2.417 in the last phase. Using the Atangana–Baleanu fractional derivative, a fractional COVID-19 model is proposed and numerical simulations performed to establish the dependence of the disease dynamics on the order of the fractional derivatives. An elasticity and sensitivity analysis of was carried out to determine the most significant parameters for combating the disease outbreak. These were found to be the effective disease transmission rate, the disease diagnosis or case detection rate, the proportion of susceptible individuals taking precautions, and the disease infection rate. Our results show that if the disease infection rate is less than 0.082/day, then is always less than 1; and if at least 55.29% of the susceptible population take precautions such as regular hand washing with soap, use of sanitizers, and the wearing of face masks, then the reproduction number remains below unity irrespective of the disease infection rate. Keeping values below unity leads to a decrease in COVID-19 prevalence.
Keywords: COVID-19, Africa, Mathematical modelling, Stability analysis, Fractional derivatives
1. Introduction
Towards the end of December 2019, the infectious Coronavirus disease known as COVID-19 was first detected in Wuhan, the capital city of the Hubei province in China. Caused by the severe acute respiratory syndrome coronavirus SARS-CoV-2 [1], COVID-19 has caused a global health emergency. The World Health Organization (WHO) declared it to be a public health emergency of international concern on 30 January 2020 [2], and as a pandemic on 11 March 2020 [3]. By 15 June 2020, the outbreak had infected around 7.8 million people globally with total fatalities of around 430,000 people. Following Africa’s first case recorded in Egypt on 14 February 2020, there had been over 246,636 confirmed cases with over 6571 deaths by 16 June 2020. COVID-19 is a rapidly spreading contagious zoonotic disease with symptoms that manifest after an incubation period of approximately 5 days following infection. Symptoms are highly variable, but range from fever, dry cough, and fatigue to less common ones like aches, sore throat, conjunctivitis, diarrhoea, and loss of smell and taste. Because efficient vaccination is not yet widely available, and there are few validated medications for treatment, COVID-19 control strategies employed by government agencies are still largely dominated by non-pharmaceutical interventions such as social distancing, wearing of face masks, regular washing of hands with soap, and use of hand sanitizer. However, the efficacy of these control strategies are not yet well-quantified, and their effectiveness is likely to change as new COVID-19 mutants take the stage.
Infectious disease modelling is a very active scientific research field. The activity is motivated, in part, by the need to gain deeper insight into disease dynamics in order to predict the trend of an epidemic outbreak, through being able to validate and test the effectiveness of control measures proposed to check the spread of the disease [4]. In recent years, mathematical modelling has been playing a key role in understanding the dynamics of infectious diseases and their control measures. It has recently been applied to study, for example Ebola [5], [6], [7], [8], [9], Dengue fever [10], [11], [12], [13], [14], Zika virus [15], [16], [17], [18], [19], [20], and Tuberculosis [21], [22], [23], [24]. Research on comprehending and predicting the trend of COVID-19 has been focused mainly on Europe [25], [26], [27], [28], [29], [30], Asia [31], [32], [33], [34], [35], [36], [37], and the Americas [38], [39] due partly to the degree of spread and impact on these continents. However, a few research papers have appeared on cases in Africa, and in Nigeria in particular [40], [41], [42], [43], while earlier, Gilbert et al. [44] evaluated the vulnerability and preparedness of the African continent against the risk of importing the disease. The importance of investigating the dynamics of COVID-19 in Africa can hardly be overemphasized, not only for the sake of Africa itself, but also because the people of the African continent are constantly visiting or migrating to other continents in the pursuit of further education, business, or other bilateral purposes; thereby creating a high risk of spreading the disease, as already witnessed and reported. Moreover, the investigation of COVID-19 dynamics will provide reliable information to decision-makers on the implementation of possible strategies and control measures aimed at stemming the spread of the pandemic [42], [43]. Recently, Manchein et al. [45] analysed the growth of the cumulative number of confirmed infected cases of COVID-19 up to March 27, 2020, from countries of Asia, Europe, North America, and South America using the power-law: , where is a deviation accounting for the uncertainty in the observed values. They found values of , and for nine countries of Asia, Europe, North America, and South America and employed a distance correlation to show that the power-law curves between the countries are statistically highly correlated [45]; but African countries were not considered.
Inspired by the work of Manchein et al. [45], this present paper analyses the time-series evolution of COVID-19 for 6 African countries: Egypt, Ethiopia, Kenya, Nigeria, Senegal and South Africa. A modified SEIR model with integer order derivatives is proposed, incorporating some non-pharmaceutical interventions, to estimate the reproduction number of the infection in the various countries, and also to highlight the effectiveness of the interventions in flattening the time-evolution of new COVID-19 cases. In addition, a fractional order equivalent is considered using the Atangana–Baleanu derivative [46]. This derivative is to be preferred over other fractional derivatives, such as the Caputo and Caputo–Fabrizio, because of its non-singular and non-local kernel. It has already found numerous applications in diverse models arising in science, engineering and medicine [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58].
The paper is organized as follows: in Section 2, we carry out an analysis of data collected for the countries to be studied. In Section 3, the model and its basic dynamical properties are presented. Section 4 provides a qualitative analysis of the model, including a determination of the stability properties of the equilibrium points based on the next generation matrix method [59], [60] employed in obtaining the reproduction number. Numerical simulations, elasticity and sensitivity analysis, and fractional numerical simulations, are presented in Sections 5, 6, 7, respectively. The work is summarized and conclusions drawn in Section 8. Appendix providing details of the numerical scheme used for solution of the fractional derivative model.
2. Data analysis
The data for the countries to be analysed were collected from the daily situation reports published by the World Health Organization (WHO) [61]. The choice of countries to be analysed was made so as to ensure geographical spread, as well as relatively high population densities. Nigeria and Senegal were chosen in the West Africa zone, Ethiopia and Kenya in East Africa, Egypt in North Africa and South Africa in Southern Africa (Fig. 1). Fig. 2 shows the cumulative number of confirmed infected COVID-19 cases as a function of days since first case was recorded up till 20th June 2020 for the selected countries. Note that the values on the horizontal and vertical axes are different for each country due to the different levels of disease progression and different inception dates. The black-continuous and the blue-dashed curves in Fig. 2 represent respectively the cubic function, and the power law function, that was fitted to the time-series. Numerical values of the fitted parameters , , for the cubic and power law equation for each country are given in Table 1. It is clear that the cubic equation fits best to the actual data for the cumulative number of confirmed cases in comparison with the power law equation of Manchein et al. [45]. However, the results of the cubic and power law fitting were comparable during the early stages of disease progression for all the countries. It is noteworthy that the fitted cubic equation for each country is such that there exists no maximum point for the curve beyond days. Such a maximum point would correspond to a time when a maximum in the cumulative number of infections is being approached so that the curve begins to flatten. The fact that no maximum value is approached reduces the usefulness of the cubic equation for investigating future dynamics of the pandemic and possible actions needed to flatten the curve of disease progression. Nevertheless, it can be adopted as a tool for forecasting the expected number of new infections.
Fig. 1.
Map of Africa showing the locations of the countries studied and the corresponding numbers of confirmed cases (NOC) and numbers of deaths (NOD) due to COVID-19 [61].
Fig. 2.
Cumulative number of confirmed infected cases by COVID-19 as a function of time from inception (first case) for South Africa, Egypt, Nigeria, Senegal, Ethiopia and Kenya. The black-continuous and the blue-dashed curves represent respectively the functions and that fit the actual time-series represented by the red dots. The parameters , , for each country are described in Table 1.
Table 1.
Parameters of the cubic and power-law fitting curves.
| Country | ||||||
|---|---|---|---|---|---|---|
| South Africa | −4223.492 | 671.706 | −21.445 | 0.214 | 0.072 | 2.875 |
| Egypt | −1911.406 | 258.244 | −7.608 | 0.070 | 0.001 | 3.522 |
| Nigeria | 186.931 | −14.660 | −0.324 | 0.017 | 0.002 | 3.260 |
| Senegal | 226.162 | −28.806 | 0.739 | −0.000196 | 0.062 | 2.365 |
| Ethiopia | −299.237 | 51.666 | −1.771 | 0.017 | 0.092 | 2.046 |
| Kenya | −155.907 | 28.809 | −0.903 | 0.011 | 0.133 | 2.142 |
The cubic equation was used to predict the expected cumulative number of confirmed infected cases from June 21 to June 30 with Root Mean Square Error (RMSE) and Mean Percentage Error (MPE) used as performance indicators for the prediction (Table 2):
where . and . are, respectively, the observed and estimated values of the cumulative number of confirmed infected cases and is the number of observations used. In general, the lower the RMSE and MPE, the better the model. A positive MPE value indicates overestimation in calculated values, while a negative MPE value indicate underestimation. From Table 2, we observe that the lowest and highest RMSE were obtained for Ethiopia and South Africa respectively and the MPEs obtained were of magnitude between of the actual cumulative number of confirmed infected cases.
Table 2.
Performance indicator for the cubic equation.
| South Africa |
Egypt |
Nigeria |
|||
|---|---|---|---|---|---|
| RMSE | MPE | RMSE | MPE | RMSE | MPE |
| 6488.75 | −3.005 | 2616.93 | 2.716 | 335.16 | 1.022 |
| Senegal |
Ethiopia |
Kenya |
|||
| RMSE | MPE | RMSE | MPE | RMSE | MPE |
| 279.72 | 3.047 | 276.00 | 2.806 | 324.42 | 4.290 |
3. Model formulation
Here, we propose a new epidemiological model for the COVID-19 epidemic. The proposed model is an extended form of the well-known Susceptible Exposed Infected Recovered (SEIR) compartmental model that takes into account some features such as quarantine, isolation and asymptomatic infections, commonly employed in epidemiological studies of communicable diseases such as, Ebola, Zika, COVID-19, etc [62], [63], [64]. An asymptomatic transmission refers to transmission of the virus through a person, who does not develop any symptoms despite having been infected. The model contains seven epidemiological compartments namely: Susceptible , Exposed , Infected , Asymptomatic , Quarantined , Hospitalized and Recovered . The complete flow chart of the interactions between different classes of the proposed model is shown in Fig. 3. The susceptible population represents the totality of the entire population that is at risk of being infected with the virus. This population is assumed to be increasing at a constant rate . The increase is not a net increase because is the natural death rate common to all the classes of the population. Exposure and transmission of the virus to the susceptible population involves the action of individuals in the infected class and the asymptomatic class . We assume that the infected class consists of people that develop symptoms while the asymptomatic class involves people that are without symptoms and therefore unaware of their positive COVID-19 status. denotes the rate of disease transmission with representing a measure of the relative (reduced) effectiveness of individuals in the asymptomatic class as disease spreaders. The spread of the disease to the susceptible population can be controlled by several precautionary measures such as use of soap and sanitizers, lockdown, social distancing and use of face masks and other Personal Protective Equipment (PPE). We assume that represents the portion of the population that maintains these precautions with the disease only transmitted to portion of the susceptible population. is the infection rate for the model with and the portions of the exposed class that go into the infected class and asymptomatic class , respectively. The quarantine class involves the quarantining of exposed individuals usually through contact tracing, at a rate and people who develop symptoms in quarantine are also hospitalized at a rate . As the current procedure in most African countries is to limit testing mostly to people who develop COVID-19 symptoms, we assume that the infectious and symptomatic class are tested at a rate (diagnosis or detection rate of infected symptomatic individuals) and moved into the hospitalized class . Also, quarantined individuals who develop symptoms are moved to the hospitalized class at a rate while those who do not develop symptoms after days are back into the susceptible population. Let and be respectively, the recovery rate of the isolated/hospitalized infected population and untreated asymptomatic population into the recovered population . and denote the COVID-19 induced death from the hospitalized and asymptomatic class, respectively with usually very small compared to . is the natural death rate common to all the classes of the population. We assume, based on current scientific evidence, that the COVID-19 deceased are not infectious, and that individuals develop antibodies and become immune to the disease once they are recovered.
Fig. 3.
Flowchart of the COVID-19 Africa model (1).
On the basis of the above assumptions, the nonlinear system of differential equations describing the COVID-19 model used to analysed data from African countries can be written mathematically as:
| (1) |
with the initial conditions , , , , , and . At every instant of time, the quantity represents the number of deaths caused by the disease at time while represents the total number of confirmed COVID-19 cases at time . Furthermore, let the total size of the population be . Definitions of the model parameters (1) are presented in Table 3.
Table 3.
Description and unit of model parameters.
| Parameter | Description | Unit |
|---|---|---|
| Constant population growth rate | Persons day−1 | |
| Natural population death rate | day−1 | |
| Effective disease transmission rate | Persons−1 day−1 | |
| Relative infectiousness of class with respect to | dimensionless | |
| Portion of taking precautionary measures | dimensionless | |
| Infection rate (1/Incubation Period) | day−1 | |
| Proportion of symptomatic infections | dimensionless | |
| Quarantine rate of exposed individuals | day−1 | |
| Diagnosis or case detection rate | day−1 | |
| Recovery rate of isolated/hospitalized individuals | ||
| Recovery rate of the untreated asymptomatic | ||
| Isolation rate of individuals from class to class | ||
| Transition rate from class to class | ||
| , | COVID-19 death rate |
4. Qualitative analysis of the system
4.1. Positivity of the solutions
Theorem 1
If,,,,,and, then the solutions of system (1) ; , , , , , and are positive for all .
Proof
From the first equation of system (1), we have
(2) where . From Eq. (2), we have
(3) Eq. (3) means that the solution of system (1) for is positive. Similar expression for , , , , and can be obtained from system (1) as
(4)
(5)
(6)
(7)
(8)
(9) where , and . Therefore, we can say that , , , , , and for all . This completes the proof.
4.2. Boundedness of the system
Theorem 2
All solutions of system (1) that initiate in are bounded uniformly in the region .
Proof
Let be any solution of system (1) with any given non-negative initial condition. Also, let . Then
(10) Thus, by the differential inequality theory [65], we obtain
It thus follows that, for ,
(11) This implies that, is positively invariant so that all solutions of (1) with initial conditions in are confined in .
Furthermore, the interacting functions of the system (1) are continuous and have continuous partial derivatives on . Hence, they are Lipschitzian on . Additionally, Theorem 2 implies that the solutions of Eq. (1) with initial conditions in are uniformly bounded. Therefore, the initial value problem (IVP) is well posed.
4.3. Basic reproduction number
Here, we employ the next generation matrix technique to determine the basic reproduction number , representing the number of secondary infections caused by a single infected individual in the entire duration of their infection [59], [60]. The classes which are directly involved in the spread of disease are , , and . Therefore, from the system equation (1), we obtain the reduced system:
| (12) |
In compact matrix form, system (1) can be written as:
| (13) |
where ,
and
In epidemiology, the matrix is referred to as the matrix of new infections and is the transfer matrix of individuals between compartments. The transition matrices and are obtained from the partial derivatives of and with respect to , , and , evaluated at the disease-free equilibrium .
We define the next generation matrix by , where the reproduction number is given by the spectral radius of [60].
| (14) |
The reproduction number obtained for the model is the sum of two other reproduction numbers and , where
| (15) |
represents the number of secondary infections caused by an infected individual during their time spent in the infected population. It is a measure of the number of the susceptible population that are infected by people in the infected group with a bilinear transmission rate , with being the time an infected individual remains in the infected group and being the time an individual remains in the exposed group. represents the number of secondary infections because of an asymptomatic individual during their time spent in the asymptotic group. It represents the number of the susceptible population that are infected by people in the asymptomatic group with an enhanced transmission rate with being the time an individual remains in the asymptomatic group.
4.4. Equilibria of the system
The equilibrium points of the model system (1) are obtained by setting the interacting functions . The disease-free equilibrium is given by , while the endemic equilibrium is given by , where
with and . We find that the disease-free equilibrium always exists, whereas, the endemic equilibrium is only feasible if and .
4.5. Local stability analysis
Here, we discuss the local asymptotic stability criteria of the equilibria of system (1) by evaluating the Jacobian or community matrix and the resulting characteristic equation. We then examine the signs of the eigenvalues based on the Routh–Hurwitz conditions and/or Descartes rule of sign. It is easy to show that the Jacobian of system (1) is given as:
| (16) |
where , , , , , and .
Theorem 3
The disease-free equilibrium is locally asymptotically stable if and unstable if .
Proof
The Jacobian matrix of system (1) evaluated at the disease-free equilibrium, is given by
(17) where, , , , , , , , , , , , , , , , and . The characteristic equation of model system (1) evaluated at the disease free equilibrium point, , is given by
(18) where, , and . It is clear from Eq. (18) that the four eigenvalues, , , and have negative values and the remaining eigenvalues can be easily obtained by finding the roots of the cubic polynomial in Eq. (18). Applying the Routh–Hurwitz criteria on the cubic polynomial in (18) requires that , and for the other three eigenvalues to be negative or have negative real parts.
Hence, the Routh–Hurwitz criterion is satisfied if, and we may conclude that the COVID-19 model (1) is locally asymptotically stable at the free equilibrium point, .
Theorem 4
The disease-endemic equilibrium is locally asymptotically stable, if and and unstable otherwise.
Proof
The Jacobian matrix of system (1) evaluated at the endemic equilibrium, is given by
(19) where,
The characteristic equation of system (1) evaluated at the disease-endemic equilibrium, , is given by
(20) where
It is clear from Eq. (20) that the two eigenvalues, and are negative, and that the remaining five eigenvalues are roots of the quintic polynomial in (20). Using , and guaranteed by the feasibility condition of the endemic equilibrium, we observe that . By employing the Descartes’ rule of signs, we find that the number of positive eigenvalues of the quintic polynomial given in Eq. (20) is equal to the number of sign-changes from to , which equals zero. Hence, the system (1) is locally asymptotically stable if and .
4.6. Global stability analysis
Theorem 5
The disease-free equilibrium is globally asymptotically stable if and unstable if .
Proof
Consider the Lyapunov function , where and are non-negative parameters. It is easy to see that . Also, and it is positive definite .
(21) Replacing and , from system (1) into Eq. (21) yields
(22) Choosing , and plugging and , we have
(23) Since follows from , therefore, it is clear that , when and also , if . Hence, by LaSalle’s Invariance principle [66], [67], the disease-free equilibrium is globally asymptotically stable.
Theorem 6
If , then there exist a disease-endemic equilibrium and it is globally asymptotically stable in the interior of .
Proof
Given that , then the existence and local asymptotic stability of the disease-endemic equilibrium is guaranteed. Consider the Lyapunov function
(24) The time derivative of is given by
(25) However, at the endemic state, we have
(26) Then using Eq. (26) in Eq. (25), we have
(27) Since , , , and , then Eq. (27) becomes
(28) It follows from arithmetic–geometric inequality that
(29) Therefore, and also , only if , , , and . Hence, by LaSalle’s Invariance principle [66], [67], the endemic equilibrium is globally asymptotically stable.
5. Numerical simulations
We now carry out numerical simulations to compare our proposed COVID-19 Africa model (1) to the data for cumulative number of confirmed infected cases obtained from the World Health Organization (WHO) [61] for South Africa, Egypt, Nigeria, Senegal, Ethiopia and Kenya. The starting point of our simulation will be a day before the index case was recorded in each country. The demographical parameters and are estimated from the total population size () and life expectancy (L.E.) data obtained from the 2018 United Nations data bank [68]. For example, Nigeria has a life expectancy of 54.332 years with population size estimate of . Hence, the average death rate, (), used for the simulation will be /day with constant population growth rate, . We assume that the proportion of new infections that are symptomatic, (), is 60% and the portion of the susceptible population taking precautionary measures, (), is 30% for all the countries except 40% used for South Africa and Egypt [69], [70]. The relative infectiousness of the asymptomatic class, (), is set to 0.5 [40], [71]. It is assumed that individuals in quarantine, either develop symptoms after an average of 7 days and are moved to isolation/hospitalization at a rate /day or are released from quarantine, after an average of 14 days without developing symptoms, into the susceptible population at a rate /day. The average remission time is set to 14 days and 7 days for individuals in the hospitalized and asymptomatic classes respectively. Hence, the recovery rates /day and /day were used in the model simulations. The COVID-19-induced death rates (, ) are estimated from the percentage of case fatalities recorded. The remaining parameters, the effective disease transmission rate (), infection rate (), quarantine rate of exposed individuals () and diagnosis/case detection rate () are obtained from fitting the model to the data using the NonlinearModelFit function in Mathematica. Table 4 gives the values of the parameters used in the simulations. The following values of the initial conditions were also used for all the simulated countries: , , , , , and . Moreover, the simulations and parameter estimations were performed such that new initial conditions and new values of fitting parameters were obtained whenever the percentage daily increase in cases was more than 30%. This approach divided the time evolution of the pandemic for the cumulative number of confirmed infected cases into different regimes or intervals, which we refer to as phases. (Note that this definition of phase is quite distinct from an interval of constant conditions determined by e.g. a particular “Tier” or level of lockdown restrictions as used in the UK.) South Africa, Egypt, Nigeria and Kenya all have two phases of the infection, and Senegal has three phases, while Ethiopia has only one phase of the infection. Table 5 gives the estimated values of the parameters , , and , as well as the calculated reproduction number for the different phases of the infection for all the countries. Fig. 4 gives the results of the numerical simulations for the different phases of the infection for all the countries. From Fig. 4, we observe that our model (1) was well-fitted to the actual data for cumulative number of confirmed infected cases for all the countries, with values of between 1.374 and 9.179 and highest during the first phase of the infection for all the countries. The observed decrease in the values beyond the first phase may be due to the impact of the strict lockdown measure and other preventive policies enforced by the authorities.
Table 4.
Parameters used for numerical simulation.
| Parameters | South Africa | Egypt | Nigeria | Senegal | Ethiopia | Kenya | Status |
|---|---|---|---|---|---|---|---|
| L.E. | 63.857 | 71.825 | 54.332 | 67.665 | 66.240 | 66.342 | [68] |
| [68] | |||||||
| 2512.389 | 3829.249 | 10128.565 | 659.833 | 4635.643 | 2171.148 | ||
| 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | Assumed | |
| 0.4 | 0.4 | 0.3 | 0.3 | 0.3 | 0.3 | Assumed | |
| 0.6 | 0.6 | 0.6 | 0.6 | 0.6 | 0.6 | Assumed | |
| 0.0714 | 0.0714 | 0.0714 | 0.0714 | 0.0714 | 0.0714 | ||
| 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | ||
| 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | ||
| 0.0714 | 0.0714 | 0.0714 | 0.0714 | 0.0714 | 0.0714 | ||
| 0.02 | 0.039 | 0.021 | 0.014 | 0.016 | 0.027 | Assumed | |
| 0.005 | 0.0098 | 0.005 | 0.004 | 0.004 | 0.007 | Assumed |
Table 5.
Fitted parameters for different phases of the infection.
| Countries | Phase | |||||
|---|---|---|---|---|---|---|
| South Africa | 1 | 0.226 | 0.487 | 0.072 | 8.290 | |
| 2 | 0.0095 | 0.035 | 0.073 | 3.794 | ||
| Egypt | 1 | 0.243 | 0.073 | 0.079 | 5.906 | |
| 2 | 0.246 | 0.059 | 0.083 | 1.825 | ||
| Nigeria | 1 | 0.264 | 0.058 | 0.153 | 3.162 | |
| 2 | 0.438 | 0.124 | 0.165 | 1.374 | ||
| Senegal | 1 | 0.252 | 0.200 | 0.078 | 7.409 | |
| 2 | 0.362 | 0.329 | 0.107 | 1.453 | ||
| 3 | 0.234 | 0.478 | 0.071 | 1.623 | ||
| Ethiopia | 1 | 0.258 | 0.538 | 0.074 | 2.452 | |
| 2 | 0.130 | 0.350 | 0.287 | 1.559 | ||
| Kenya | 1 | 0.240 | 0.106 | 0.084 | 9.179 | |
| 2 | 0.064 | 0.028 | 0.074 | 2.417 | ||
Fig. 4.
Cumulative number of confirmed infected cases by COVID-19 as a function of time for South Africa, Egypt, Nigeria, Senegal, Ethiopia and Kenya. The black dots corresponds to the real data while the blue continuous line represents the simulation from model (1).
6. Elasticity and sensitivity analysis of
Here, we analyse the elasticity and sensitivity of the reproduction number, . Sensitivity analysis is a well known technique for identifying the critical parameters or inputs of a model and quantifying their importance relative to one another [58]. For the purpose of elasticity and sensitivity analysis, we used data from Nigeria as a case study. The baseline values and ranges of the system parameters used here are given in Table 6.
Table 6.
Baseline values and ranges of the system parameters.
| Parameter | Baseline value | Range |
|---|---|---|
| 10128.565 | ||
| 0.5 | ||
| 0.3 | ||
| 0.438 | ||
| 0.6 | ||
| 0.124 | ||
| 0.165 | ||
| 0.143 | ||
| 0.005 |
In order to perform the elasticity analysis of , we first calculated the normalized forward sensitivity index. In general, the elasticity (normalized forward sensitivity index) of a variable, , that depends differentiably on a parameter, , is given as [72]:
| (30) |
A negative (or positive) sensitivity index indicates a decrease (or an increase) in the value of the parameter resulting in a decrease (or an increase) in the value of . For, , we obtain the following:
The elasticity or sensitivity index shows the influence of change in one parameter while keeping all other parameters constant. Note that the sensitivity indices of and do not depend on any parameter value. Interestingly, the reproduction number does not depend on the recovery rate of the hospitalized individuals (), or on the isolation rate of quarantined individuals (), or on the transition rate from quarantine class to susceptible class after quarantine (), or on the COVID-19-induced death rate of the hospitalized cases (). We proceed to evaluate the above sensitivity indices using the baseline parameter values in Table 6. A plot showing the sensitivity indices for with respect to its constituent parameters is presented in Fig. 5, from which we can state that increases whenever , , , or increase. On the other hand, whenever , , , , or increase, then decreases. For example, implies that increasing by 10% will increase by 2.71%. Hence, a similar interpretation can be inferred for the remaining parameters in Table 6. Following the elasticity analysis, the most positive sensitivity index was obtained for with a 10% increase in leading to the same proportional increase in . Note that the same value of positive sensitivity index as was obtained for . However, is a demographic variable that cannot be changed in the field. The controllable parameter with the most negative sensitivity index was . With a 10% increase in , a decrease of 7.29% in was found. We observe that the most significant parameters are the effective disease transmission rate (), the disease diagnosis or case detection rate (), and the proportion of susceptible individuals taking precautions (). Therefore, we can conclude that efforts at controlling the disease should concentrate on decreasing the transmission rate through contact reduction via lockdown and social distancing measures. In addition, can be increased by encouraging social campaigns aimed at increasing the number of individuals taking precautionary measures such as regular hand washing with soap and use of sanitizers, use of face mask and other PPEs.
Fig. 5.
Plot showing sensitivity indices of as a function of its constituent parameters.
For the sensitivity analysis of the reproduction number, , the Latin Hypercube Sampling (LHS) technique [73] was implemented for the parameters of the model. The correlation between and the input parameter values obtained from 1000 simulations was quantified using Partial Rank Correlation Coefficients (PRCC). The sensitivity analysis results of , shown in Fig. 6 demonstrate that the parameters with the greatest influence on are , , , , and in order of decreasing sensitivity. Clearly, the sensitivity analysis results are similar to those of the elasticity analysis, as the parameters , , and , identified via sensitivity indices as being the most significant parameters, are also the most sensitive parameters. It is noteworthy that, though the demographic parameters, and had sensitivity indices of and , respectively, the results of the sensitivity analysis shows that they have insignificant influence on due to their very low PRCC values.
Fig. 6.
Plot of partial rank correlation coefficient showing the influence of input parameters on .
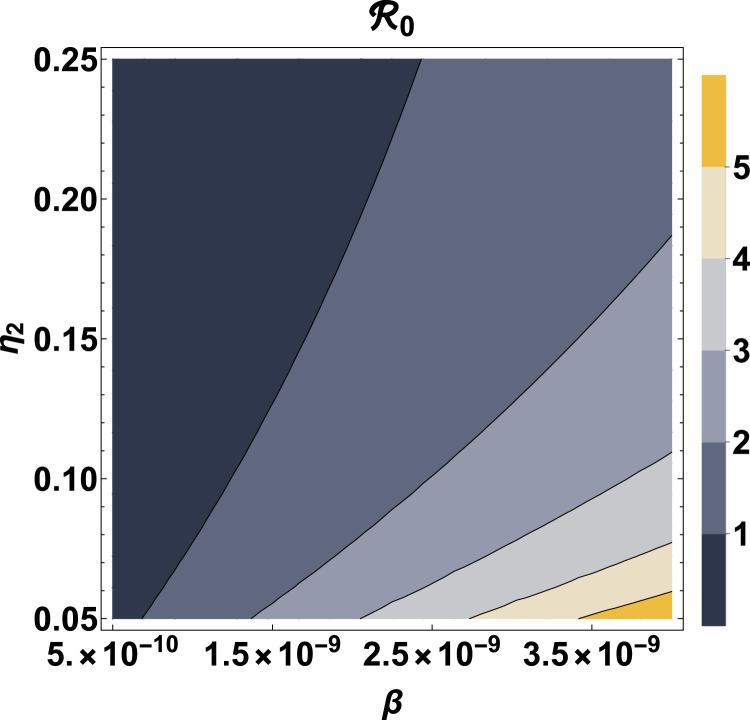
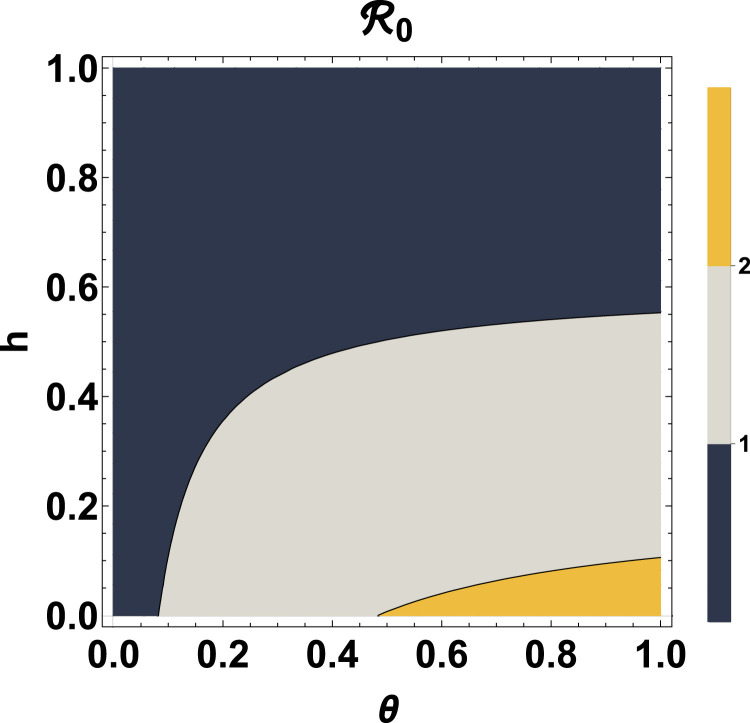
Fig. 7, Fig. 8 provide contour plots of the reproduction number as a function of the parameter pairs and , respectively. From Fig. 7, we observe that for an effective disease transmission rate, , is always less than 1, indicating that, eventually the disease will die out in the population regardless of the value of the diagnosic or case detection rate (). For increase in the value of , can be kept under 1 by also increasing . If and /day, then . Also, if and /day, then . Fig. 8 shows that if the disease infection rate, is less than 0.082/day, then is always less than regardless of the proportion of susceptible individuals taking precautions (). Another remarkable observation from Fig. 8 is that, if at least 55.29% of the susceptible population adheres to the prescribed precautions, such as regular hand washing with the use of soap, use of sanitizers and face masks, then the reproduction number can be kept below unity regardless of the value of the disease infection rate. This result is in agreement with previous studies carried out for Lagos, Nigeria, where at least 55% of the population effectively making use of face masks while in public was recommended for the reproduction number of the disease to be brought below 1 [40]. Moreover, if less than 10.58% of the susceptible population take the prescribed precautions with a disease infection rate greater than 0.483, then .
Fig. 7.
Contour plot of as a function of and .
Fig. 8.
Contour plot of as a function of and .
Fig. 9 shows the influence of the variation in the model parameters on the progression of the number of active cases. The plots in Fig. 9 were obtained by simulating the model (1) numerically using different values of the system parameters, , , and , while other parameters were kept constant. We observe that variations in the parameters have significant influence on the maximum of the infection and the number of days taken to reach this maximum. Notably, for an increase in the values of and , there is a corresponding increase in the maximum infection in addition to this value being attained on a later day. For infection rates of 0.4, 0.5, 0.6 and 0.7, maxima of about 850,000, 750,000, 610,000 and 460,000 may be reached after about , , and days, respectively. For diagnostic or case detection rates of 0.10, 0.13, 0.16 and 0.18, maxima of about 1.5 million, million, 590,000 and 370,000 may be reached after about , , and days, respectively. If 15% of the susceptible population takes precautions, then the projection in Fig. 9 shows that the number of active cases may attain about 1.2 million by around the 240th day of the infection. Alternatively, with 35% of the population taking precautionary measures, then the number of active cases may reach around 320,000 by around the 510th day of the infection.
Fig. 9.
Figure showing the effect of the model parameters (, , , ) on the number of active cases.
7. Fractional model with Atangana–Balenau derivative
Fractional models with the Atangana–Balenau fractional derivative provide more efficient results than the ordinary derivative models [46], [47], [51], [57], [74], [75], [76], [77], [78]. We begin here by defining the Atangana–Balenau fractional derivative and its integral.
Definition 7.1
Let , , ,. The Atangana–Balenau fractional derivative is then defined [46] as:
(31) and the associated fractional integral of the Atangana–Balenau derivative is defined as [46]:
(32) where is the normalization function satisfying , and is the Mittag-Leffer function with one parameter.
Definition 7.2
The Mittag-Leffler function with one parameter is defined [46] as
(33)
We generalize the model (1) by applying the Atangana–Baleanu derivative and thus obtain the following fractional COVID-19 model for Africa:
| (34) |
where is the fractional derivative and represents the fractional order parameter.
The numerical results for the fractional model (34) were obtained by following the procedure described in the Appendix in which the modified Adams–Bashforth scheme developed by Toufik and Atangana [78] was adopted. Readers are referred to some very recent applications of the modified Adams–Bashforth scheme [47], [48], [49], [79]. Fig. 10 shows the dependence of the number of active cases, infectious and symptomatic class, asymptomatic class and the hospitalized class from the fractional model (36) on the magnitude of the order of the fractional derivative, . Fig. 10 was obtained by using the initial conditions , , , , , and and performing numerical simulation of the fractional model (36) using the modified Adams–Bashforth scheme (39)–(45) for , 0.95 and 0.9. The result shows that the magnitude of has a marked impact on the day the maximum is reached, with a right shift in the time at which this happens as decreases from 1.0. However, the order of the fractional derivative () has only an insignificant effect on the projected peak numbers of active cases. Specifically, for , 0.95 and 0.9, the peak numbers of active cases were approximately 590,000, 570,000, 550,000 by about the 370th, 440th and 540th day after the first case of the infection was recorded, respectively. Hence, the order of the fractional derivative () can be used as an effective delay variable for the peak of the infection.
Fig. 10.
The dynamics of the active cases, infectious and symptomatic class , asymptomatic class and hospitalized class of the fractional model (36) for different values of the order of the fractional derivative .
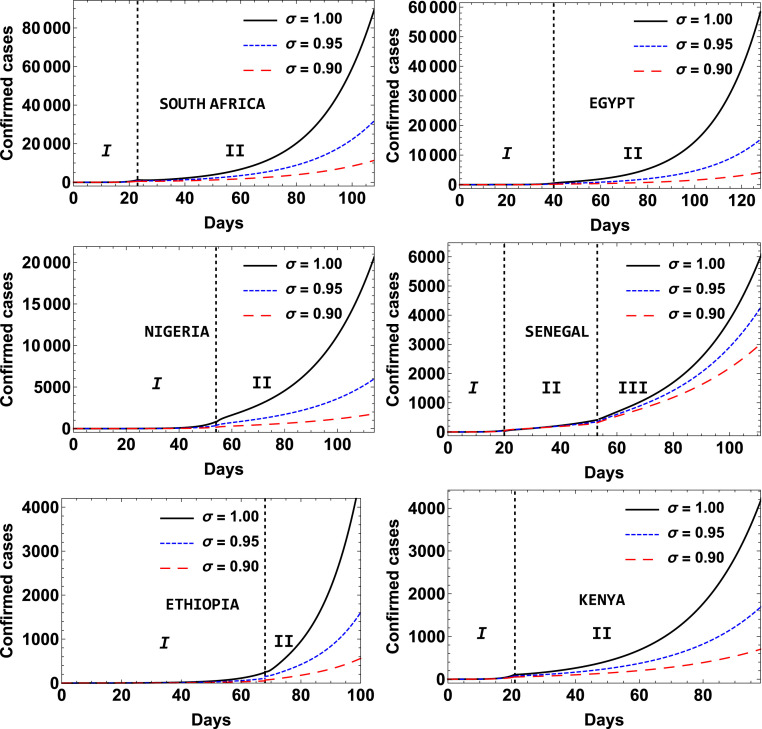
The results of the numerical simulations showing the effect of order of the fractional derivative on the cumulative number of confirmed infected cases by COVID-19 for the different phases of the infection for all the countries is depicted in Fig. 11.
Fig. 11.
Effect of order of the fractional derivative on the cumulative number of confirmed infected cases by COVID-19 as a function of time for South Africa, Egypt, Nigeria, Senegal, Ethiopia and Kenya.
The effective reproduction number , defined as the actual average number of secondary cases per primary case at time (for ) is a useful time-varying threshold in epidemiology for measuring the trajectory and rate of spread of the disease at any point in time during the course of the epidemic. The effective reproduction number for Eq. (1) is given by
| (35) |
Generally, the number of disease cases rises when , attains a peak when , and declines when . For more details on the effective reproduction number, see Refs. [80], [81], [82]. Fig. 12, Fig. 13 gives an illustration of the effective reproduction number for the selected countries for different values of the order of the fractional derivative . As seen in Fig. 12, Fig. 13, the COVID-19 epidemic had an effective reproduction number that generally decreases with time for the different phases of the infection for all the countries. Also, the effective reproduction number falls more rapidly with time for and least for . This observation is consistent with Fig. 10 which shows that the peak numbers of active cases was reached earliest for .
Fig. 12.
Evolution of the effective reproduction number with time for South Africa, Egypt and Nigeria for different values of the order of the fractional derivative .
Fig. 13.
Evolution of the effective reproduction number with time for Ethiopia, Kenya and Senegal for different values of the order of the fractional derivative .
8. Conclusion
In this paper, a mathematical model for the novel coronavirus (COVID-19) disease which incorporates some non-pharmaceutical interventions was proposed and used to investigate the transmission dynamics in selected African countries, namely, South Africa, Egypt, Nigeria, Senegal, Ethiopia and Kenya. The model contains seven epidemiological compartments namely: Susceptible , Exposed , Infected , Asymptomatic , Quarantine , Hospitalized and Recovered and also takes into account some specific features of the COVID-19 epidemic such as quarantine, isolation and asymptomatic infections. We obtained the critical points, identified the disease-free states and the endemic states. The basic reproduction number, was computed using the next generation matrix approach. Numerical simulations to fit the proposed model to the actual data for cumulative number of confirmed infected cases was performed for the different phases of the infection for all the countries, with values of between 1.311 and 9.179 obtained. As expected, we observe that the condition is necessary for the stability (or instability) of the disease-free(or endemic state). A fractional version of the model was introduced using the Atangana–Baleanu derivative and numerical simulations were performed for better understanding of the dependence of the dynamics of the disease on the order of the fractional derivative, . The result shows that the magnitude of has a pronounced effect on the day the maximum is reached with a right shift observed in the time taken for the maximum to be attained as decreases from 1.0. However, the order of the fractional derivative has insignificant effect on the projected peak number of active cases. Hence, can be used as an effective delay variable for the peak of the infection. Elasticity and sensitivity analyses show that the most significant parameters are the effective disease transmission rate (), disease diagnosis or case detection rate (), proportion of susceptible taking precautions () and the disease infection rate (). If the disease infection rate, is less than 0.082/day, then is always less than regardless of the proportion of susceptible taking precautions (). Another remarkable inference from the study is that, if at least 55.29% of the susceptible population take precautions such regular hand washing with the use of soap, use of sanitizers and wearing of face masks, then, the reproduction number can be kept below irrespective of the value of the disease infection rate. The most important practical conclusion is that efforts to control the disease should concentrate on decreasing the transmission rate by contact reduction, via lockdown and social distancing measures. In addition, can be increased by encouraging social campaigns aimed at increasing the number of individuals taking prescribed precautionary measures.
CRediT authorship contribution statement
O.T. Kolebaje: Methodology, Investigation, Writing – original draft. O.R. Vincent: Conceptualization, Software, Formal analysis. U.E. Vincent: Conceptualization, Validation, Writing – review & editing. P.V.E. McClintock: Supervision, Validation, review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
All data used for this study were obtained from the daily situation reports published by the World Health Organization (WHO) and are available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. The codes used are available on request to replicate the results in this paper. The Appendix also provides information on our numerical scheme.
Acknowledgements
We are grateful for support from the Engineering and Physical Sciences Research Council (United Kingdom) under research grants Nos. EP/D000610/1 and EP/M015831/1.
Appendix. Numerical scheme
We now describe the numerical procedure used for solution of the fractional model (34) by adopting the modified Adams–Bashforth scheme developed by Toufik and Atangana [78]. Some recent applications of the modified Adams–Bashforth scheme, include [47], [48], [49], [79]. Before applying the procedure in Toufik and Atangana [78], we write the fractional COVID-19 model (34) in the following form:
| (36) |
Following the procedure in Toufik and Atangana [78], the fractional model can take the form:
| (37) |
Using , , in (37), we obtain:
| (38) |
Employing the two-step Lagrange-polynomial approximation for the integral in (38), we obtain the following the numerical scheme for the fractional model (34):
| (39) |
| (40) |
| (41) |
| (42) |
| (43) |
| (44) |
| (45) |
where .
References
- 1.WHO . World Health Organization; 2020. Naming the coronavirus disease (COVID-19) and the virus that causes it. [Google Scholar]
- 2.WHO . World Health Organization; 2020. Statement on the second meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-ncov) [Google Scholar]
- 3.WHO . Speeches. World Health Organization; 2020. Director-general’s opening remarks at the media briefing on COVID-19 -11 March 2020. [Google Scholar]
- 4.Rahman B, Aziz IA, Khdhr FW, Mahmood DFD. Preliminary estimation of the basic reproduction number of SARS-CoV-2 in the Middle East, Bull. World Health Organ. Preprint.
- 5.Banton S., Roth Z., Pavlovic M. In: 26th southern biomedical engineering conference SBEC 2010, April 30 - May 2, 2010, College Park, Maryland, USA. K. E. Herold W. E. Bentley., editor. Springer; Berlin, Heidelberg: 2010. Mathematical modeling of Ebola Virus dynamics as a step towards rational vaccine design; pp. 196–200. [Google Scholar]
- 6.Chretien J., Riley S., George D.B. Mathematical modeling of the West Africa Ebola epidemic. eLife. 2015;4 doi: 10.7554/eLife.09186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buceta J., Johnson K. Modeling the Ebola zoonotic dynamics: Interplay between enviroclimatic factors and bat ecology. PLOS ONE. 2017;12(6):1–14. doi: 10.1371/journal.pone.0179559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latha V., Fathalla A., Rakkiyappan R., Velmurugan G. A fractional-order model for Ebola Virus infection with delayed immune response on heterogeneous complex networks. J Comput Appl Math. 2018;339:134–146. [Google Scholar]
- 9.Chowell G., Nishiura H. Transmission dynamics and control of Ebola Virus disease (EVD): A review. BMC Med. 2014;12(3):196. doi: 10.1186/s12916-014-0196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derouich M., Boutayeb A. Dengue fever: Mathematical modelling and computer simulation. Appl Math Comput. 2006;177(2):528–544. [Google Scholar]
- 11.Chan M., Johansson M.A. The incubation periods of Dengue viruses. PLOS ONE. 2012;7(11):1–7. doi: 10.1371/journal.pone.0050972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO-VMI Dengue Vaccine Modeling Group Assessing the potential of a candidate Dengue vaccine with mathematical modeling. PLoS Negl. Trop. Dis. 2012;6(3):1–6. doi: 10.1371/journal.pntd.0001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings D.A.T., Iamsirithaworn S., Lessler J.T., McDermott A., Prasanthong R., Nisalak A., Jarman R.G., Burke D.S., Gibbons R.V. The impact of the demographic transition on Dengue in Thailand: Insights from a statistical analysis and mathematical modeling. PLOS Med. 2009;6(9):1–10. doi: 10.1371/journal.pmed.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diethelm K. A fractional calculus based model for the simulation of an outbreak of Dengue fever. Nonlinear Dynam. 2013;71:613–619. [Google Scholar]
- 15.Bonyah E., Okosun K.O. Mathematical modeling of Zika virus. Asian Pacific J Trop Dis. 2016;6(9):673–679. [Google Scholar]
- 16.Padmanabhan P., Seshaiyer P., Castillo-Chavez C. Mathematical modeling, analysis and simulation of the spread of Zika with influence of sexual transmission and preventive measures. Lett Biomath. 2017;4(1):148–166. [Google Scholar]
- 17.Best K., Perelson A.S. Mathematical modeling of within-host Zika virus dynamics. Immun Rev. 2018;285(1):81–96. doi: 10.1111/imr.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiratsudakul A., Suparit P., Modchang C. Dynamics of Zika virus outbreaks: an overview of mathematical modeling approaches. PeerJ. 2018;6 doi: 10.7717/peerj.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goswami N.K., Srivastav A.K., Ghosh M., Shanmukha B. Mathematical modeling of Zika virus disease with nonlinear incidence and optimal control. J Phys Conf Ser. 2018;1000 [Google Scholar]
- 20.Biswas S.K., Ghosh U., Sarkar S. Mathematical model of Zika Virus dynamics with vector control and sensitivity analysis. Inf Dis Model. 2020;5:23–41. doi: 10.1016/j.idm.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Li Y., Zhang X. Mathematical modeling of tuberculosis data of China. J Theor Biol. 2015;365:159–163. doi: 10.1016/j.jtbi.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Goutelle S., Bourguignon L., Jelliffe R.W., Conte J.E., Maire P. Mathematical modeling of pulmonary tuberculosis therapy: Insights from a prototype model with rifampin. J Theor Biol. 2011;282(1):80–92. doi: 10.1016/j.jtbi.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Vynnycky E., Sumner T., Fielding K.L., Lewis J.J., Cox A.P., Hayes R.J., Corbett E.L., Churchyard G.J., Grant A., White R.G. Tuberculosis control in South African gold mines: Mathematical modeling of a trial of community-wide isoniazid preventive therapy. Amer J Epidem. 2015;181(8):619–632. doi: 10.1093/aje/kwu320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avilov K.K., Romanyukha A.A. Mathematical modeling of tuberculosis propagation and patient detection. Autom Remote Control. 2007;68:1604–1617. [Google Scholar]
- 25.Linka K., Peirlinck M., Costabal F.S., Kuhl E. Outbreak dynamics of COVID-19 in Europe and the effect of travel restrictions. Comput Methods Biomech Biomed Eng. 2020:1–8. doi: 10.1080/10255842.2020.1759560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J., Li M., Lv G., Lu Z.K. Monitoring transmissibility and mortality of COVID-19 in Europe. Int J Inf Dis. 2020;95:311–315. doi: 10.1016/j.ijid.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson H.C., Gossner C.M., Colzani E., Kinsman J., Alexakis L., Beauté J., Würz A., Tsolova S., Bundle N., Ekdahl K. Potential scenarios for the progression of a COVID-19 epidemic in the European Union and the European Economic Area, March. Eurosurveillance. 2020;25(9) doi: 10.2807/1560-7917.ES.2020.25.9.2000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picchiotti N., Salvioli M., Zanardini E., Missale F. 2020. Covid-19 pandemic: a mobility-dependent SEIR model with undetected cases in Italy, Europe and US. arXiv:2005.08882. [DOI] [PubMed] [Google Scholar]
- 29.Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alberti T., Faranda D. On the uncertainty of real-time predictions of epidemic growths: A COVID-19 case study for China and Italy. Commun Nonlin Sci Numer Simul. 2020;90 doi: 10.1016/j.cnsns.2020.105372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang K. Mathematical model of infection kinetics and its analysis for COVID-19, SARS and MERS. Infect Genet Evol. 2020;82 doi: 10.1016/j.meegid.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R, Pal S. Trend analysis and forecasting of COVID-19 outbreak in India, medRxiv: 10.1101/2020.03.26.20044511. [DOI]
- 33.Zhou X, Ma X, Hong N, Su L, Ma Y, He J, Jiang H, Liu C, Shan G, Zhu W, Zhang S, Long Y. Forecasting the Worldwide spread of COVID-19 based on logistic model and SEIR Model, medRxiv: 10.1101/2020.03.26.20044289. [DOI]
- 34.Sunhwa C., Moran K. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health. 2020;42 doi: 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhutta ZA, Basnyat B, Saha S, Laxminarayan R. Covid-19 risks and response in South Asia, BMJ 368. [DOI] [PubMed]
- 36.Shearer F, Walker J, Tellioglu N, McCaw J, McVernon J, Black A, Geard N. Assessing the risk of spread of COVID-19 to the Asia Pacific region, medRxiv: 10.1101/2020.04.09.2005725. [DOI]
- 37.Paul A, Chatterjee S, Bairagi N. Prediction on COVID-19 epidemic for different countries: Focusing on South Asia under various precautionary measures, medRxiv: 10.1101/2020.04.08.20055095. [DOI]
- 38.Atkeson A. What will be the economic impact of COVID-19 in the US? Rough estimates of disease scenarios, Working Paper 26867, National Bureau of Economic Research (2020). 10.3386/w26867. [DOI]
- 39.Mizumoto K., Chowell G. Transmission potential of the novel coronavirus (COVID-19) onboard the diamond princess cruises ship. Inf Dis Model. 2020;5:264–270. doi: 10.1016/j.idm.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuonghae D., Omame A. Analysis of a mathematical model for covid-19 population dynamics in Lagos, Nigeria. Chaos, Solit Fract. 2020;139 doi: 10.1016/j.chaos.2020.110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayinde K., Lukman A.F., Rauf R.I., Alabi O.O., Okon C.E., Ayinde O.E. Modeling Nigerian COVID-19 cases: A comparative analysis of models and estimators. Chaos Solit Fract. 2020;138 doi: 10.1016/j.chaos.2020.109911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acheme I.D., Vincent O.R. Data Science for COVID-19. Academic Press; 2021. Machine learning models for predicting survivability in COVID-19 patients; pp. 317–336. [DOI] [Google Scholar]
- 43.Lawal O.M., Vincent O.R. Data Science for COVID-19. Academic Press; 2021. A two-level deterministic reasoning pattern to curb the spread of COVID-19 in africa; pp. 565–581. [DOI] [Google Scholar]
- 44.Gilbert M., Pullano G., Pinotti F., Valdano E., Poletto C., Boëlle P.-Y., D’Ortenzio E., Yazdanpanah Y., Eholie S.P., Altmann M., Gutierrez B., Kraemer M.U.G., Colizza V. Preparedness and vulnerability of african countries against importations of COVID-19: a modelling study. Lancet. 2020;395(10227):871–877. doi: 10.1016/S0140-6736(20)30411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manchein C., Brugnago E., da Silva R., Mendes C., Beims M. Strong correlations between power-law growth of COVID-19 in four continents and the inefficiency of soft quarantine strategies. Chaos. 2020;30(4) doi: 10.1063/5.0009454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atangana A., Baleanu D. New fractional derivatives with nonlocal and non-singular kernel: Theory and application to heat transfer model. Therm Sci. 2016;20:763–769. [Google Scholar]
- 47.Khan M.A., Ullah S., Farooq M. A new fractional model for tuberculosis with relapse via Atangana-Baleanu derivative. Chaos Solit Fract. 2018;116:227–238. [Google Scholar]
- 48.Khan M.A., Kolebaje O., Yildirim A., Ullah S., Kumam P., Thounthong P. Fractional investigations of zoonotic visceral leishmaniasis disease with singular and non-singular kernel. Eur Phys J Plus. 2019;134(10):481. [Google Scholar]
- 49.Gomez-Aguilar J.F., Abro K.A., Kolebaje O., Yildirim A. Chaos in a calcium oscillation model via Atangana-Baleanu operator with strong memory. Eur Phys J Plus. 2019;134(10):140. [Google Scholar]
- 50.Owolabi K.M., Atangana A. Springer; Singapore: 2019. Numerical Methods for Fractional Differentiation. [Google Scholar]
- 51.Owolabi K.M., Pindza E. Modeling and simulation of nonlinear dynamical system in the frame of nonlocal and nonsingular derivatives. Chaos Solit Fract. 2019;127:146–157. [Google Scholar]
- 52.Owolabi K.M., Atangana A. Mathematical analysis and computational experiments for an epidemic system with nonlocal and nonsingular derivative. Chaos Solit Fract. 2019;126:41–49. [Google Scholar]
- 53.Naik P.A., Zu J., Owolabi K.M. Modeling the mechanics of viral kinetics under immune control during primary infection of HIV-1 with treatment in fractional order. Physica A. 2019;13 [Google Scholar]
- 54.Shah S.A.A., Khan M.A., Farooq M., Ullah S., Alzahrani E.O. A fractional order model for hepatitis b virus with treatment via Atangana-Baleanu derivative. Physica A. 2019;538 [Google Scholar]
- 55.Jan R., Khan M.A., Kumam P., Thounthong P. Modeling the transmission of dengue infection through fractional derivatives. Chaos Solit Fract. 2019;127:189–216. [Google Scholar]
- 56.Wang W., Khan M.A., Kumam P., Thounthong P. A comparison study of bank data in fractional calculus. Chaos Solit Fract. 2019;126:369–384. [Google Scholar]
- 57.Khan M.A. The dynamics of a new chaotic system through the Caputo–fabrizio and Atanagan-Baleanu fractional operators. Adv Mech Eng. 2019;11(7) [Google Scholar]
- 58.Kolebaje O., Popoola O., Khan M.A., Oyewande O. An epidemiological approach to insurgent population modeling with the Atangana–Baleanu fractional derivative. Chaos Solit Fract. 2020;139 [Google Scholar]
- 59.Diekmann O., Heesterbeek J.A.P., Metz J.A.J. On the definition and the computation of the basic reproduction ratio in models for infectious diseases in heterogeneous populations. J Math Biol. 1990;28:365–382. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- 60.Diekmann O., Heesterbeek J.A.P. Wiley; New York, USA: 2000. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. [Google Scholar]
- 61.WHO . 2020. Coronavirus disease (COVID-2019) situation reports, situation reports. [Google Scholar]
- 62.Tulu T.W., Tian B., Wu Z. Modeling the effect of quarantine and vaccination on Ebola disease. Adv Differential Equations. 2017;178:1–14. [Google Scholar]
- 63.Imran M., Usman M., Malik T., Ansari A.R. Mathematical analysis of the role of hospitalization/isolation in controlling the spread of Zika fever. Virus Res. 2018;255:95–104. doi: 10.1016/j.virusres.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dénes A., Gumel A.B. Modeling the impact of quarantine during an outbreak of Ebola virus disease. Inf Dis Model. 2019;4:12–27. doi: 10.1016/j.idm.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perko L. third ed. Springer; New York, USA: 2001. Differential Equation and Dynamical Systems. [Google Scholar]
- 66.LaSalle J.P. The Stability of Dynamical Systems. vol. 25. SIAM; Philadelphia, USA: 1976. (CBMS-NSF Regional Conference Series in Applied Mathematics). [Google Scholar]
- 67.Tewa J.J., Dimi J.L., Bowong S. Lyapunov functions for a dengue disease transmission model. Chaos Solitons Fractals. 2009;39(2):936–941. [Google Scholar]
- 68.United Nations . United Nations; 2020. United Nations Data Bank: Tech. rep. [Google Scholar]
- 69.Kinoshita R., Anzai A., Jung S.M., Linton N.M., Miyama T., Kobayashi T., Hayashi K., Suzuki A., Yang Y., Akhmetzhanov A.R., Nishiura H. Containment, contact tracing and asymptomatic transmission of novel coronavirus disease (COVID-19): A modelling study. J Clin Med. 2020;9 doi: 10.3390/jcm9103125. 3125(1–9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanes-Lane M., Winters N., Fregonese F., Bastos M., Perlman-Arrow S., Campbell J.R., Menzies D. Proportion of asymptomatic infection among COVID-19 positive persons and their transmission potential: A systematic review and meta-analysis. PLOS ONE. 2020;15(11):1–21. doi: 10.1371/journal.pone.0241536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen T, Rui J, Wang Q, Zhao Z, Cui J, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect. Dis. Pov. 9 (24). [DOI] [PMC free article] [PubMed]
- 72.Chitnis N., Hyman J.M., Cushing J.M. Determining important parameters in the spread of malaria through the sensitivity analysis of a mathematical model. Bull Math Biol. 2008;70(5):1272–1296. doi: 10.1007/s11538-008-9299-0. [DOI] [PubMed] [Google Scholar]
- 73.Blower S.M., Dowlatabadi H. Sensitivity and uncertainty analysis of complex models of disease transmission: an HIV model, as an example. Int Stat Rev. 1994;62:229–243. [Google Scholar]
- 74.Solis-Perez J., Gomez-Aguilar J., Escobar-Jimenez R., Reyes-Reyes J. Blood vessel detection based on fractional hessian matrix with non-singular Mittag–Leffler Gaussian kernel. Biomed Signal Process Control. 2019;54 [Google Scholar]
- 75.Avalos-Ruiz L., Zuniga-Aguilar C., Gomez-Aguilar J., Escobar-Jimenez R., Romero-Ugalde H. FPGA implementation and control of chaotic systems involving the variable-order fractional operator with Mittag–Leffler law. Chaos Solit Fract. 2018;115:117–189. [Google Scholar]
- 76.Atangana A., Gomez-Aguilar J. Decolonisation of fractional calculus rules: breaking commutativity and associativity to capture more natural phenomena. Eur Phys J Plus. 2018;133(4):166. [Google Scholar]
- 77.Coronel-Escamilla A., Lavin-Delgado J., Gomez-Aguilar J., Torres L. Fractional dynamics and synchronization of kuramoto oscillators with nonlocal, nonsingular and strong memory. Alexandria Eng J. 2019;59:1941–1952. [Google Scholar]
- 78.Toufik M., Atangana A. New numerical approximation of fractional derivative with non-local and non-singular kernel: application to chaotic models. Eur Phys J Plus. 2017;132(10):444. [Google Scholar]
- 79.Alzahrani E.O., Khan M.A. Modeling the dynamics of Hepatitis E with optimal control. Chaos Solit Fract. 2018;116:287–301. [Google Scholar]
- 80.Nishiura H., Chowell G. Mathematical and statistical estimation approaches in epidemiology. 2009. The effective reproduction number as a prelude to statistical estimation of time-dependent epidemic trends; pp. 103–121. [Google Scholar]
- 81.Ngonghala C.N., Iboi E., Gumel A.B. Could masks curtail the post-lockdown resurgence of covid-19 in the US? Math Biosci. 2020 doi: 10.1016/j.mbs.2020.108452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gumel A.B., Iboi E., Ngonghala C.N., Elbasha E.H. A primer on using mathematics to understand COVID-19 dynamics: Modeling, analysis and simulations. Inf. Dis. Model. 2021;6:148–168. doi: 10.1016/j.idm.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]