Abstract
Complex interactions between symbiotic bacteria and insects ultimately result in equilibrium in all aspects of life in natural insect populations. In this study, abundance of principal symbiotic bacteria was estimated using qPCR in 1553 individuals of aphids, Aphis gossypii. Aphids were sampled from primary and secondary host plants—hibiscus and cotton. Hibiscus aphids were collected from 24 different locations in April, September, and November, whereas cotton aphids were collected between 2015 and 2017 from areas with wide variations in climatic conditions. About 30%–45% aphids were recorded with the most dominant symbiont, Arsenophonus. The other symbionts were in low frequency, and about 7% of aphids were noted with Hamiltonella, Acinetobacter, and Microbacterium, and 3% of aphids were verified with Serratia and Pseudomonas. Aphids infected with Hamiltonella, Arsenophonus, and Serratia can influence Buchnera densities. Hamiltonella has positive interaction with densities of Arsenophonus and Serratia. Almost 100% coinfection of Hamiltonella and Arsenophonus was detected in Xinxiang aphids and 50% coinfection was reported in aphids from North China, while no coinfection was detected in Hainan aphids. These findings describe the prevalence pattern and richness of core community of symbiotic bacteria in naturally occurring populations of A. gossypii and provide new insights for the study of symbiotic bacteria.
Keywords: abundance, cotton aphid, geographical distribution, infection frequencies, symbiotic bacteria
About 30%–45% and 10% aphids were found infected with Arsenophonus and Hamiltonella, respectively. Aphids infected with Hamiltonella, Arsenophonus, and Serratia can influence Buchnera densities with different variations. Host plant may play important role than biotypes on bacterial symbiont infection frequencies and densities.

1. INTRODUCTION
The cotton aphid, Aphis gossypii Glover, is an important global pest that sucks sap and transmits viral diseases to host plants, causing serious economic losses in agriculture (Figure 1) (Blackman & Eastop, 2000). It colonizes more than 600 plant species, many of which are important crops (Ebert & Cartwright, 1997). Plant transfer experiments and genetic diversity analysis have highlighted the existence of A. gossypii host biotypes (Carletto et al., 2009; Wang et al., 2016; Zhang et al., 2018). A. gossypii is distributed on a large geographical scale with a wide range of host plants, which provides a basis for a highly variable life cycle, with a holocyclic pattern in cold winters and anholocyclic forms in warm regions (Margaritopoulos et al., 2006).
FIGURE 1.

The mutualistic interaction between Aphis gossypii and ants on cotton. The cotton aphid, A. gossypii Glover, is an important global pest that sucks sap and transmits viral diseases to host plants, causing serious economic losses in agriculture. Due to the difference in the life cycle and other factors, such as the mutualistic interaction between cotton aphids and ants, the synergy between symbiotic bacteria and aphids is quite different within aphid species or different in populations of same species. In this study, seven bacterial abundances in 1553 A. gossypii were estimated using quantitative PCR, providing novel information about bacterial community related to biotypes, host plants, prevalence time, and geography
Aphis gossypii usually migrates between the primary and the secondary hosts throughout the year (Kwon & Kim, 2017; Xia et al., 1999). In many areas, A. gossypii uses hibiscus plants as the primary host, between April and mid‐May; then migrate to secondary host plants; and again return to feed on hibiscus from October to November, whereas some aphids eat hibiscus all the year long (Charaabi et al., 2008; Zhang & Zhong, 1990).
Symbiotic bacteria exist in many insects, known as primary symbionts (obligate symbionts), secondary symbionts, and facultative symbionts (Bright & Bulgheresi, 2010). Symbiotic bacteria have conditionally positive effects on the physical condition of the host insects. During host adaptation, bacterial symbionts suppress plant resistance (Su et al., 2015) or increases the expression of detoxifying enzymes (Singh et al., 2020) that efficiently enhance the host adaptation. Recent studies showed that symbiotic bacteria contribute to host variation, heat tolerance (Zhang, Leonard, et al., 2019), and temperature preference (Hague et al., 2020). Meanwhile the host should also bear the metabolic and fitness‐dependent cost to symbiont bacterial presence (Engl et al., 2020; Oliver et al., 2008); bacterial symbionts even can shape their host evolution (Coffman & Burke, 2020). Therefore, it is very important and necessary for insects to maintain a balanced population of symbiotic bacteria.
In primary symbionts, Buchnera is essential for sap‐sucking aphids—reported in almost all aphid species, and responsible for essential amino acids and other nutrients for growth, reproduction (van Ham et al., 2003), and to improve heat tolerance (Zhang, Leonard, et al., 2019). The interaction of many facultative symbionts within aphids were well‐studied, such as the bacterium Hamiltonella increases resistance to its parasitoid via toxin protein (Brandt et al., 2017) and Rickettsiella changes body color from red to green in its natural populations (Tsuchida et al., 2010).
Due to the difference in the life cycle, host type, pesticide selection pressure, dispersal, and migration, the synergy between symbiotic bacteria and aphids is quite different within aphid species or different in populations of same species. Some studies have been carried out in bacterial communities of A. gossypii based on 16S rRNA gene sequencing on Illumina platforms (Gallo‐Franco et al., 2019; Xu et al., 2020; Zhang, Luo, Wang, et al., 2019; Zhao et al., 2016), quantitative PCR, and normalized host genes (Ayoubi et al., 2020; Chong & Moran, 2016; Zhang, Cao, et al., 2016). These findings reflect that symbiotic bacteria play an important role in A. gossypii, but the information about absolute quantity of symbiotic bacteria in natural populations and their changing trends within seasons, geographic areas, and hosts is still lacking. In this study, seven bacterial abundance in 1553 A. gossypii were estimated using quantitative PCR (qPCR), providing novel information about bacterial community related to biotypes, host plants, prevalence time, and geography.
2. MATERIAL AND METHODS
2.1. Field sampling and DNA extracting
For Illumina MiSeq DNA sequence analysis, wingless A. gossypii were collected from cotton fields located in Anyang (Henan Province) and Shihezi (Xinjiang Province) in late June 2016. Different instars of wingless aphids were picked up from cotton and then put in nuclease‐free Eppendorf tubes. In order to make the sample data representative, only one aphid per cotton plant in each sampling field was collected. In this way, more than 50 aphids from 50 different fields were collected, mixed, and considered as one sample; 10 samples from each city were collected and immediately immersed in liquid nitrogen and stored at −80°C for further study.
For qPCR analysis, A. gossypii were collected from cotton and hibiscus plants, each adult wingless aphid was put in a nuclease‐free Eppendorf tube. (a) From cotton, A. gossypii were collected from Xinjiang (2015) and Henan (2015–2017) in June and from Hainan (2015) Province in February. Only one wingless adult aphid per plant from 8 to 48 cotton plants per site was collected to avoid sampling from the offspring of a single female (Zhang et al., 2018); aphids were immersed in liquid nitrogen and stored at −80°C for further study. (b) From hibiscus, A. gossypii were collected from ornamental hibiscus located in urban areas from each location; the locations are shown in Appendix S1. Aphids were collected in April, September, and November 2016. In order to avoid the confounding of the area caused by the transplanting of seedlings, we choose hibiscus which has been transplanted more than two years. Only one aphid per plant was sampled, and the next sample was collected from plants more than 10 meters away from the previous plants from where the sample was drawn. Sampled aphids were placed in 95% ethanol and stored in room temperature for further study.
Each sample was washed with 70% ethanol and rinsed three times with nuclease‐free water, and total DNA from individual aphids or mixed samples was extracted using the TIANamp Genomic DNA Kit (TIANGEN Biotech (Beijing) LTD., China) according to the manufacturer's instruction. In order to break gram‐positive bacterial cells, additional lysozyme (50 mg/ml) was added at the incubation step. Elution buffer, 30 μl, was added at the last step, and re‐elution was done for one more time. Negative DNA extraction (control) includes DNA extractions of the nuclease‐free water. The quantity and quality of the DNA were measured with a NanoDrop 2000C spectrophotometer (Thermo Scientific). The purified DNA samples were stored at −20°C, and the samples having lower concentration, <25 ng/μl, were excluded from the study.
2.2. 16S rRNA gene amplification and sequencing
The V3‐V4 hypervariable regions of the bacterial 16S rRNA gene were amplified using the primers 338F (5′‐ACTCCTACGGGAGGCAGCAG ‐3′) and 806R (5′‐GGACTACHVGGGTWTCTAAT‐3′). DNA from mixed aphid samples was used as template DNA. Amplicon generation of PCR products, quantification and qualification, PCR product mixing and purification, library preparation, and sequencing were carried out on an Illumina MiSeq platform at Shanghai Major Bio‐pharm Technology Co., Ltd. Bioinformatics. Sequences with ≥97% similarity were assigned to the same OTUs.
2.3. Quantification of symbiotic bacteria
qPCR was used to determine copies of 16S rRNA genes of dominant bacteria, and entire DNA from individual aphids—diluted 10 times—was used as template DNA. Bacterial special primers (Appendix S2) were designed according 16S rRNA genes using BEACON DESIGNER 7.6 (PREMIER Biosoft International, CA, USA). Primer PCR efficiencies were tested using series‐diluted templates (Appendix S2). The standard template was prepared, as previously reported (Zhang, Luo, Jiang, et al., 2019), and the general step was cloning the target sequence in a plasmid of pEASY‐T3 cloning vector (TransGen Biotech, China). Escherichia coli DH5α was used as a host for plasmid propagation, and the target sequence in the plasmid vectors was confirmed by sequencing. Each PCR reaction contained 5 μl 2×TransStart Green qPCR SuperMix (TransGen Biotech, China), 0.2 μl each of 10 mM forward and reverse primers, 2.0 μl template DNA (2.0 μl negative DNA considered as negative controls), and 0.2 μl 50 × ROX; nuclease‐free water was added to make up to 10 μl. The Step OnePlus™ Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) was used to perform PCR, according to cycling conditions of 95°C for 3 min followed by 40 cycles of a two‐step PCR (95°C for 5 s, 60°C for 30 s). All qPCR reactions were done in triplicate for each individual aphid DNA, and each reaction plate generated a corresponding standard curve. To make sure the primer specificity validated for each primer, checking steps were carried out, as previously reported (Zhang et al., 2019).
2.4. Statistical analysis
Bioinformatics of 250‐bp paired‐end reads were conducted, as previously reported (Zhang, Luo, Jiang, et al., 2019). Principal component analysis (PCA) based on the Bray–Curtis method was performed to study alpha diversity (ACE and Chao1 estimators, Good's coverage estimates, and Shannon and Simpson diversity indices) among observed species. Alpha diversity and beta diversity analyses were executed based on normalized output data by random selected sequences per sample according to the sequence number of the sample with the least sequences, and the statistical analyses were carried out by using the independent two‐sample t test to identify the differences between two groups, at p < .05, which is considered significant. All analyses and estimates were carried out on the freely accessible online Majorbio Cloud Platform (https://cloud.majorbio.com).
The abundance of symbiotic bacteria in one microliter of DNA solution was estimated by measuring the 16S rRNA gene of each bacterium. Standard curves were generated using series dilutions of the standard plasmid vector, and 6 proportional dilutions of the plasmid were prepared with the lowest concentration, composed of nearly 100 copies. Due to addition of lowest concentration, the standard curve cannot be constructed well. Other 5 dilutions of standard plasmid vectors were used as a result of aphid's abundance lower than the lowest point of the standard curve and considered as undetectable bacterial individuals.
The analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) revealed that the most important biological factor (hosts, seasons, and geography) effected on symbiont and secondary symbiont community structures. ANOSIM and PERMANOVA were performed using Adonis and Anosim functions in R vegan package (version 2.5‐7, https://cran.r‐project.org/web/packages/vegan/), respectively, based on the Bray–Curtis community dissimilarity index with 999 permutations. SPSS 20.0 was used to evaluate the differences between infection frequency and abundance. For comparison of infection frequency between two samples, Pearson's chi‐squared test was used when n ≥ 40, and Fisher's exact test was used when n < 40. Group comparisons of symbiotic bacteria and abundance were evaluated with the Mann–Whitney U test (group number > 2) and Kruskal–Wallis test (n > 2). Correlation between different bacteria was carried out using the Pearson correlation of the means of these parameters.
3. RESULT
3.1. Overview of the bacterial diversity from Henan and Xinjiang
The V3‐V4 region of 16S rRNA gene was amplified from A. gossypii samples collected from Henan and Xinjiang provinces, and the Illumina MiSeq PE300 platform was used to generate raw reads. There were 78206–140328 raw reads from Henan samples, and 86988–145634 raw reads from Xinjiang‐collected samples. Good's coverage estimates of sequencing data were noted with maximum coverage in all samples, remained more than 99% (Appendix S3). The quality filtering sequences for aphids from Henan and Xinjiang was assigned 274 and 453 OTUs containing 170 co‐existed OTUs, respectively.
Xinjiang‐collected aphids were found with higher bacterial biodiversity estimates (ACE and Chao1) than the aphids collected from Henan. Aphids from Xinjiang were also found with higher diversity of bacterial species (Figure 2a,b). The PCA‐based studies of the 20 samples showed all clusters together, except two Xinjiang samples (Figure 2c). The most abundant symbiotic bacterial phylum was Proteobacteria, which accounted for 99.76% and 96.04% in Henan and Xinjiang samples, respectively. The most abundant genus was the primary symbiont Buchnera, having a relative abundance of 83.78% in aphids from Henan and 74.93% in aphids from Xinjiang. Generally, Arsenophonus, Acinetobacter, Serratia, Brevundimonas, and Pseudoxanthomonas were the top 5 most abundant facultative symbionts at the genus level in aphids from Henan, which accounted for 15.65% of the total. The bacterial genera Arsenophonus, Hamiltonella, Exiguobacterium, and Kosakonia including unclassified genus of the family Enterobacteriaceae were the 5 most abundant facultative symbionts in aphids from Xinjiang and accounted for 20.87% of the total (Figure 2d).
FIGURE 2.
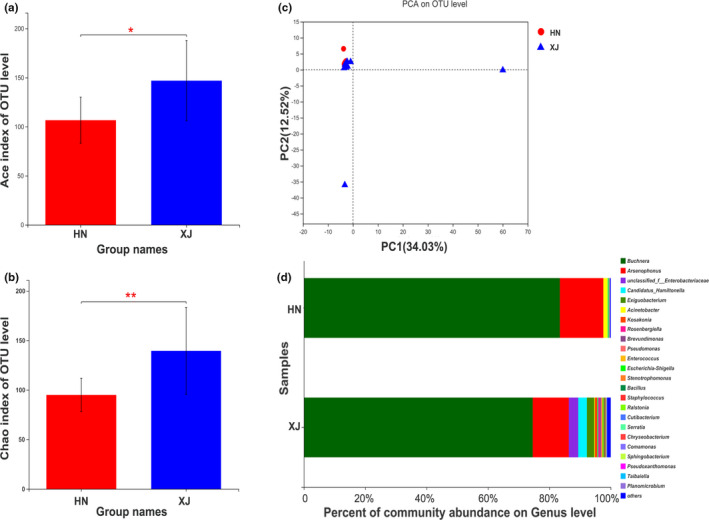
Bacterial diversity of Aphis gossypii from Henan (HN) and Xinjiang (XJ) provinces by high‐throughput sequencing approaches. Community richness and diversity measured by Ace (a) and Chao1 (b), Student’s t test was used for analysis significant differences of group mean value, *p < .05 and **p < .01. Principal component analysis based on the 20 samples (c) and symbiotic bacteria relative abundance of the bacteria in A. gossypii on genus level (d)
3.2. The relationship between symbiotic bacteria
The infection frequency and abundance of 7 main symbiotic bacteria in single A. gossypii were analyzed by qPCR. From 1533 A. gossypii collected from North China—Xinjiang and Hainan provinces—100% individuals were found with primary symbiont Buchnera, 31.51% individuals were noted by at least one facultative symbiont, and 11.87% individuals were observed with two or more symbionts. Arsenophonus was the most prevalent facultative symbiont, and 29.94% individuals were perceived with it, followed by Acinetobacter (7.31%), Hamiltonella (7.11%), Microbacterium (6.91%), Serratia (3.26%), and Pseudomonas (2.22%). During this study, it was noticed that 56.62% individuals were not found by any facultative symbiont.
The density of Buchnera was influenced by some facultative symbionts. Both Hamiltonella‐ and Arsenophonus‐infected aphids have higher density of Buchnera than the uninfected ones (Mann–Whitney‐U tests, p < .001) (Figure 3a,b). In Hamiltonella‐infected aphids, Buchnera density was negatively correlated with the Hamiltonella density (r = −.4855, p < .001) (Figure 3d), and Buchnera density was positively correlated with Arsenophonus density in Arsenophonus infected aphids (r = .5876, p < .001) (Figure 3e). Interestingly, Buchnera density has no significant difference between Serratia‐infected aphids and Serratia‐uninfected aphids (Mann–Whitney‐U tests, p = .611) (Figure 3c), but when aphids were infected with Serratia, Buchnera density was clearly correlated with Serratia density (r = .2934, p = .0387) (Figure 3f). Buchnera density was not remained significant, between Acinetobacter‐infected aphid and ‐uninfected aphids (Mann–Whitney‐U tests, p = .890), among Microbacterium‐infected aphids and uninfected aphids (Mann–Whitney‐U tests, p = .264), and between Pseudomonas‐infected aphids and uninfected aphids (Mann–Whitney‐U tests, p = .650). However, the densities of Acinetobacter (r = .1624, p = .0870), Microbacterium (r = −.1589, p = .1038), and Pseudomonas (r = −.3273, p = .0589) were not found with any impact on the density of Buchnera bacterium (Appendix S4).
FIGURE 3.
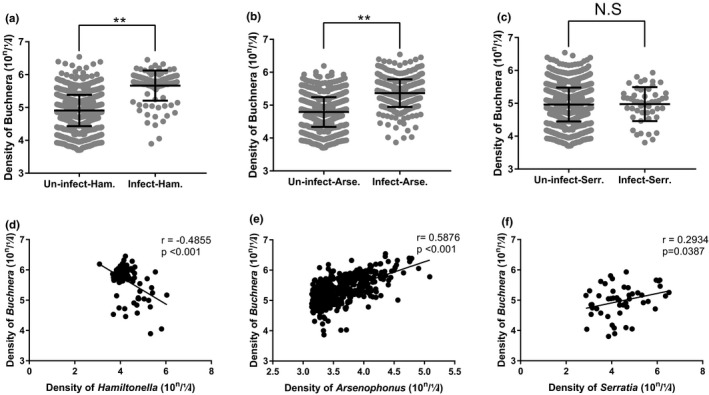
The density of Buchnera and its relationship with facultative symbionts. The Buchnera copy numbers in one microliter of aphid DNA were show in a. (Hamiltonella‐infected aphids (Infect‐Ham.) vs. uninfected aphids (Un‐infect‐Ham.)), b. (Arsenophonus‐infected individuals (Infect‐Arse.) vs. uninfected individuals (Un‐infect‐Arse.)), c. (Serratia‐infected individuals (Infect‐Serr.) vs. uninfected individuals (Uninfect‐Serr.)), scatter dot plot with mean ± standard deviation. **p < .01 and N.S = no significant difference, data were analysis by Mann–Whitney U test. The correlation between copy numbers of Buchnera and Hamiltonella (d), Arsenophonus (e), and Serratia (f) were test according to bacteria copy numbers in one microliter of aphid DNA
Coinfection with facultative symbionts was common in A. gossypii. It was found that 10.44% and 1.37% aphids were recorded with two or three facultative symbionts simultaneously. Only one aphid was coinfected with four facultative symbionts. The maximum coinfection type was of Arsenophonus and Hamiltonella, accounted for 4.44%, followed by coinfection of Arsenophonus and Microbacterium (2.15%) and coinfection of Arsenophonus and Acinetobacter (1.44%).
The Hamiltonella‐infected aphids have recorded with higher coinfection frequency with other facultative symbionts (96.33%), followed by Pseudomonas‐infected aphids (55.38%). The density of Hamiltonella detected with obvious positive correlation with Arsenophonus density (r = .2227, p = .0457) and Serratia (r = .8604, p < .001) in A. gossypii (Figure 4a,d). The Arsenophonus‐infected aphids have recorded with a higher Arsenophonus titer level when observed with Hamiltonella (Mann–Whitney‐U tests, p < .001), and Hamiltonella‐infected aphids have lower Hamiltonella titer level when detected with Hamiltonella (Mann–Whitney‐U tests, p < .001) but has a higher Hamiltonella titer level when noticed with Serratia (Figure 4b–e). The aphids observed with Hamiltonella were all coinfected with Arsenophonus in Xinjiang population except one aphid, 50% aphids, collected from North China, coinfected with Arsenophonus, whereas coinfection was not recorded in Hainan‐collected aphid samples.
FIGURE 4.
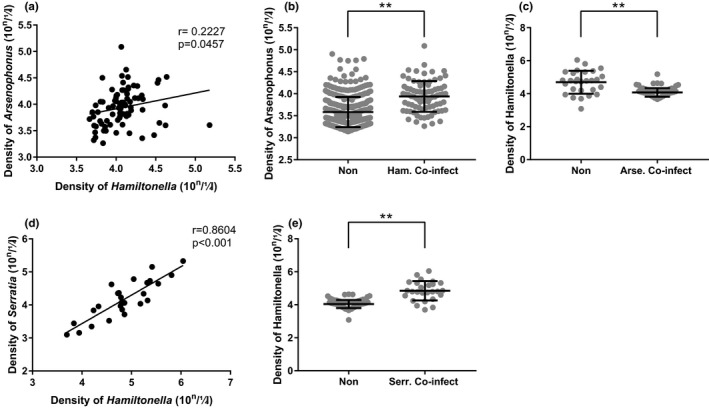
The relationship between facultative symbionts. The correlation between Arsenophonus and Hamiltonella copy numbers (a), Arsenophonus copy numbers in aphid co‐infection with Hamiltonella (Ham.Co‐infect) and aphids no co‐infection with Hamiltonella (Non) (b), Hamiltonella copy numbers in aphid co‐infection with Arsenophonus (Arse.Co‐infect) and aphids no co‐infection with Arsenophonus (Non) (c). The correlation between Serratia and Hamiltonella copy numbers (d), Hamiltonella copy numbers in aphid co‐infection with Serratia (Serr.Co‐infect) and aphids no co‐infection with (Non) (e). Scatter dot plot with mean ± standard deviation. **p < .01, data were analysis by Mann–Whitney U test
3.3. Assessment of infection frequency in population of aphids of Henan, Hainan and Xinjiang
3.3.1. Aphid samples collected from cotton plant
Multivariate analysis showed that there were significant differences in bacterial community compositions among geographic populations feeding on cotton (ADONIS: F 2,433 = 37.587, R 2 = .148, p = .001; ANOSIM: R = .022, p = .001). There were significant differences in infection frequency of Hamiltonella and Arsenophonus belonging to the three geographic populations collected from cotton plants (χ2 = 91.909, p < .001 and χ2 = 72.767, p < .001, respectively). The Hamiltonella infection frequency was higher in Xinjiang (36.96%, n = 184) and Hainan (22.86%, n = 35), although very low infection frequency was recorded in Henan (0.46%, n = 217). Arsenophonus infection frequency was higher in Xinjiang (75.54%) and Henan (46.08%), whereas lower infection frequency was observed in Hainan (5.71%). Microbacterium infection frequency was higher in Henan (29.95%). Hainan populations have higher Serratia and Pseudomonas infection frequencies among these three geographic populations, 31.43% and 34.29%, respectively, whereas in other two geographic populations, the infection frequencies were less than 6.60%, even have not recorded any infection of some symbiotic bacterial species in some aphid populations.
The abundance of Buchnera and four facultative symbionts were significantly dissimilar in different geographical aphid populations. Buchnera has highest abundance in Xinjiang populations, which was 1.70‐fold and 5.04‐fold more than Henan (Mann–Whitney‐U tests, p < .001) and Hainan populations (Mann–Whitney‐U tests, p < .001), respectively. Hamiltonella has a higher titer level in the Hainan population (n = 8), which was 23.84‐fold greater than in Xinjiang (n = 68). The titer level of Serratia and Pseudomonas was maximum in Hainan (n = 11 and n = 12, respectively) than that in Henan populations (n = 6 and n = 8, respectively), which were 6.59‐fold and 1.79‐fold larger, respectively. The abundance of Arsenophonus was 1.96‐fold greater in the Xinjiang population than that in Henan (Mann–Whitney‐U tests, p < .001) (Figure 5).
FIGURE 5.
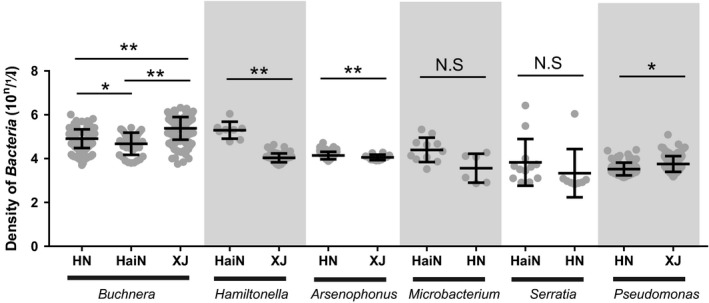
The densities of symbiotic bacteria in different Aphis gossypii geographic populations collected on cotton. The symbiotic bacteria copy numbers in Henan province (HN), Hainan province (HaiN), and Xinjiang province (XJ) were showed, scatter dot plot with mean ± standard deviation. *p < .05, **p < .01, N.S = no significant difference, data were analysis by Mann–Whitney U test
3.3.2. Aphid samples collected from hibiscus plants
Hibiscus is the main primary host of cotton aphid. During the occurrence, cotton aphid was collected from 24 geographic populations from hibiscus plants grown in North China. Samples from all the geographic populations contained infected aphids with Buchnera and Arsenophonus. Fifteen of 24 samples collected from various geographic populations contained aphids infected with Microbacterium. The other four facultative symbionts detected ranged from 33.33% to 45.83% in 24 geographic studied populations (Appendix S5). Multivariate analysis showed that there were significant differences in bacterial community compositions among geographic populations feeding on hibiscus (ADONIS: F 23,1,073 = 7.675, R 2 = .141, p = .001; ANOSIM: R = .083, p = .001).
Generally, the aphid's infection frequency with facultative symbionts were at a low level in each geographic population. Average aphids (18.97%) were recorded with Arsenophonus, with highest infection frequency (44.90%), although lowest infection frequency remained at 4.88%. About 4.87% aphids were infected with Microbacterium, 6.68% were observed with Serratia and 2.97% aphids recorded with Pseudomonas. In Hamiltonella‐infected 10 populations, 1.52%–13.98% aphids were infected with Hamiltonella. All the 7 populations collected in Henan Province were infected with Acinetobacter, and 3 10 populations collected in Shandong Province were infected with Acinetobacter, but the infection frequency in each population was lower than that in the Henan population (Appendix S5).
The aphids collected from Handan city (Hebei Province) had a higher abundance of Buchnera, 9.47‐fold higher than Baoding city population, whereas the Baoding city population has the lowest abundance of Buchnera among 24 geographic populations. Interestingly, three populations recorded with maximum Buchnera copies/abundance were from adjacent regions, Handan city, Xingtai city (Hebei Province), and Anyang city (Henan Province), with a significantly higher Buchnera abundance than Baoding city, Hebei Province (Mann–Whitney‐U tests, p < .001); Jining city (Mann–Whitney‐U tests, p < .001); and Kaifeng city, Henan Province (Mann–Whitney‐U tests, p < .001), and no significant difference with other cities (Figure 6).
FIGURE 6.
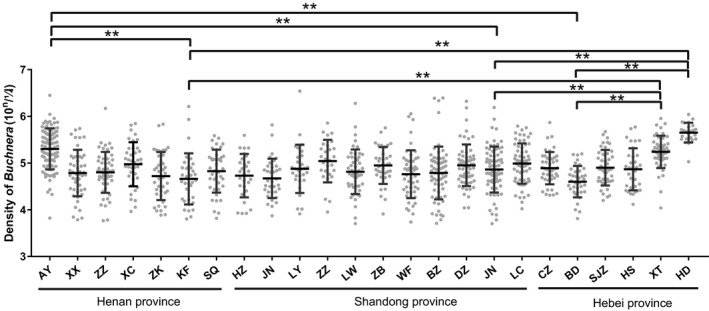
The densities of Buchnera in different Aphis gossypii geographic populations collected on hibiscus. The Buchnera copy numbers in Anyang city (AY), Xinxiang city (XX), Zhengzhou city (ZZ), Xuchang city (XC), Zhoukou city (ZK), Kaifeng city (KF), Shangqiu city (SQ), Heze city (HZ), Jining city (JN), Linyi city (LY), Zaozhuang city (ZZH), Laiwu city (LW), Zibo city (ZB), Weifang city (WF), Binzhou city (BZ), Dezhou city (DZ), Jinan city (JN), Liaocheng city (LC), Cangzhou city (CZ), Baoding city (BD), Shijiazhuang city (SJZ), Hengshui city (HS), Xingtai city (XT), and Handan city (HD) were showed, scatter dot plot with mean±standard deviation. **p < .01, others showed no significant difference, data were analysis by Mann–Whitney U test
3.4. Assessment of infection frequency within geographic populations of aphids collected during different time periods
3.4.1. Three‐year infection comparison among geographic populations from Anyang, Henan
The infection frequency of facultative symbionts in cotton aphids has significant difference from 2015 to 2017 from Anyang, Henan (ADONIS: F 2,214 = 21.197, R 2 = .165, p = .001; ANOSIM: R = .226, p = .001). About 50% aphids were found infected with Arsenophonus both in 2015 and 2016, but only 13.33% individuals in 2017 (Fisher's exact test, both p < .001). There were 41.83% aphids infected with Microbacterium in 2015, but almost none of infected‐aphid was detected in 2016 and 2017 (Appendix S5). The abundance of Buchnera has no significant difference between 2015 and 2016 (Mann–Whitney‐U tests, p = .071), although 3.13‐fold (Mann–Whitney‐U tests, p < .001) and 4.62‐fold (Mann–Whitney‐U tests, p < .001) higher than in 2017, respectively (Figure 7a).
FIGURE 7.
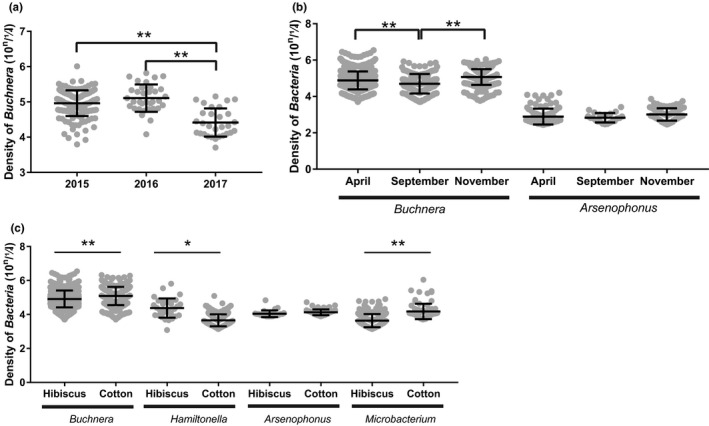
The densities of symbiotic bacteria in Aphis gossypii collected at different times and on different hosts. The copy numbers of Buchnera in A. gossypii from Anyang city during 2015–2017 (a), the copy numbers of Buchnera and Acinetobacter in A. gossypii collected on hibiscus in April September November (b), the copy numbers of Buchnera, Hamiltonella, Arsenophonus, and Microbacterium in A. gossypii collected on hibiscus and cotton (c) were showed, scatter dot plot with mean ± standard deviation. *p < .05, **p < .01, others showed no significant difference, data were analysis by Mann–Whitney U test
3.4.2. Infection comparison among geographic populations from North China
Aphis gossypii were collected from hibiscus plants in three varying time periods in 2016. Multivariate analysis showed that there were significant differences in bacterial community compositions among populations feeding on hibiscus at three periods (ADONIS: F 2,1094 = 24.490, R 2 = .043, p = .001; ANOSIM: R = .062, p = .001). About 7 bacteria were detected in sampled aphids, except Hamiltonella in September and Acinetobacter in November. There were about 15% aphids infected with Arsenophonus in April and September, lower than the infection frequency in November, 30.82% (χ2 = 11.844, p = .001 and χ2 = 29.124, p < .001, respectively). There were 16.00% aphids infected with Acinetobacter in April, infection frequency dropped up to 1.41% in September, and no infected aphid was detected in November. The highest infection frequency with Microbacterium was detected in November, 6.56%; meanwhile, 1.08% and 0.70% aphids were found infected with Microbacterium in April and September, respectively. Serratia has highest infection frequency in September, 6.34%, and 1.54% and 3.93% individuals were infected with Microbacterium in April and November, respectively.
Both Buchnera and Arsenophonus abundance were lowest in September. The Buchnera abundance in April and November was 1.56‐fold (Mann–Whitney‐U tests, p < .001) and 1.76‐fold (Mann–Whitney‐U tests, p < .001) higher than that in September, respectively. The Arsenophonus abundance in April and November was 1.94‐fold (Mann–Whitney‐U tests, p = .811) and 1.71‐fold (Mann–Whitney‐U tests, p = .030) higher than that in September, respectively (Figure 7b).
3.5. Assessment of infection frequency among aphid populations collected from various host plants
All bacteria were detected in A. gossypii feeding on cotton and hibiscus. Multivariate analysis showed that there were significant differences in bacterial community compositions between aphids feeding on cotton and hibiscus (ADONIS: F 1,1531 = 33.916, R 2 = .022, p = .001; ANOSIM: R = .062, p = .001). The frequency of four bacterial infection was higher in aphids feeding on cotton than those feeding on hibiscus. These infection frequencies were Hamiltonella (17.66% in aphid on cotton vs. 2.92% in aphids on hibiscus, χ2 = 102.683, p < .001), Arsenophonus (55.28% vs. 19.87%, χ2 = 186.421, p <.001), Microbacterium (17.89% vs. 2.55%, χ2 = 114.740, p < .001), and Pseudomonas (4.59% vs. 1.28%, χ2 = 15.771, p < .001). The infection frequency of Acinetobacter was higher in aphids feeding on hibiscus than aphids feeding on cotton (9.66% vs. 1.38%, χ2 = 31.635, p < .001), and Serratia infection frequency has no significant difference between aphids feeding on hibiscus and cotton (2.83% vs. 4.36%, χ2 = 2.321, p = .128).
The abundance of Buchnera in aphid populations feeding on cotton was 1.48‐fold higher than aphid populations feeding on hibiscus (Mann–Whitney‐U tests, p < .001). Microbacterium abundance was 1.12‐fold higher in the population feeding on cotton than aphid populations feeding on hibiscus (Mann–Whitney‐U tests, p = .002). Aphids feeding on hibiscus have 1.47‐fold higher Hamiltonella abundance than aphids feeding on cotton (Mann–Whitney‐U tests, p = .029). There was no significant difference between Arsenophonus abundance between aphids feeding on hibiscus and aphids feeding on cotton (Mann–Whitney‐U tests, p = .277) (Figure 7c).
4. DISCUSSION
Symbiotic bacteria are necessary for the survival, reproduction, host adaption, and resistance to biotic or abiotic stresses of most insects, especially for the sap‐sucking insect‐like aphids (Hosokawa et al., 2010; Ley et al., 2008; Moran, 2007; Teixeira et al., 2008). First, we used the 16S rRNA gene sequencing on Illumina platforms to compare the differences of cotton aphid symbiotic bacteria between Xinjiang and Henan populations and identify the dominant symbiotic bacteria. There is no doubt that the primary symbiont Buchnera exists in the cotton aphids in a dominant proportion. In this study, the relative abundance of Buchnera was more than 74% in the aphid population from both locations Henan and Xinjiang; however, the linear distance between the two places exceeds 2,500 km. Aphids from Henan Province have a higher proportion of facultative symbionts than aphids collected from Xinjiang Province, and both richness estimates and biodiversity of bacteria were higher in Xinjiang. PCA studies showed some aphid population in Xinjiang has large variations. Henan is warm temperate‐subtropical area, whereas Xinjiang has wide range of temperature differences between days and nights. Varying environmental conditions may be the cause of these differences during the spread of cotton aphid populations (Dunbar et al., 2007).
Symbiont abundance were previously measured in whole insects as the number of symbiotic bacteria per aphid genome using qPCR and normalized by single‐copy gene abundance (Chong & Moran, 2016). In this study, adult aphids were used, during the early stage of adult aphid, and the number of symbiotic bacteria was basically stable (Ayoubi et al., 2020), so the age of adult aphids has little effect on the community of symbiotic bacteria. When aphids reproduce via viviparous parthenogenesis, ovaries occupy a large proportion of the female body, such as the last stage maturing embryo length was longer than 0.8 mm compare with the mother's body size 4.0–5.0 mm of Acyrthosiphon pisum (Rabatel et al., 2013). During development, the embryo of A. pisum just receives a small proportion of Buchnera from the mother at the beginning, immediately following rapid multiplication of the bacterium (Wilkinson et al., 2003). This causes the uneven distribution of symbiotic bacteria among the adult aphids, so we used the abundance of symbiotic bacteria in a single aphid to measure its richness. The DNA of each adult aphid for qPCR analysis was extracted according to a consistent method to ensure that the same DNA yields for each individual. To ensure the consistency of extraction, samples with lower concentrations were discarded. Aphid samples having DNA concentrations near 30–40 ng/μl were used for qPCR studies.
So far, the infection of symbiotic bacteria in the natural population of cotton aphids is still poorly understood. In this study, the abundance of 7 bacteria in individual aphids were appraised by using qPCR. The seven bacteria were Buchnera, Arsenophonus, Acinetobacter, Hamiltonella, Serratia, Microbacterium, and Pseudomonas. Arsenophonus, Acinetobacter, Hamiltonella, and Serratia were the top genera, recorded in aphids, collected from Xinjiang and Henan. Comparable studies also show a similar trend in relative abundance of the bacteria in cotton aphid (Ayoubi et al., 2020; Tian et al., 2019; Xu et al., 2020; Zhang, Pan, et al., 2016; Zhao et al., 2016). The abundance of the infrequent bacterium Pseudomonas was higher in some cotton aphids (Gallo‐Franco et al., 2019). Pseudomonas is considered as a widespread aphid pathogen, and A. pisum can reduce the infection by avoiding the highly virulent strain of Pseudomonas (Hendry et al., 2018). Microbacterium belongs to Actinobacteria, which is widely present in air, soil, water, and plants, as well exists in guts of larvae and adult insects (Dantur et al., 2015), and has not influenced by spirotetramat insecticide used against cotton aphids (Zhang, Pan, et al., 2016). In the current study, there was a group of symbiotic bacteria, which accounts for a large proportion in cotton aphids, identified to family level, Enterobacteriaceae, and excluded due to ineffective detection by specific primers, utilized against Exiguobacterium.
The infection frequency of symbiotic bacteria in cotton aphids draws different conclusions in different research methods. Through a wide range of single‐individual qPCR studies, the presence of the main symbiotic bacteria of A. gossypii was basically outlined. Using the diagnostic PCR method, about 44.58% (n = 1,200) aphids were infected with Arsenophonus in cotton aphids from the Nanjing population (Tian et al., 2019), which is significantly higher than infection frequency in this study (29.94%, n = 1533) (χ2 = 62.356, p < .001). High‐throughput 16S rRNA sequencing showed 5.45% (n = 110) A. gossypii were infected with Hamiltonella, and most of them were collected from Beijing, North China, and Xinjiang (Xu et al., 2020). The Hamiltonella infection frequency was not significant in our study (χ2 = 0.432, p = .511), indicating that Hamiltonella infection frequency in the natural population of cotton aphids was low. Other studies showed that North American A. pisum populations have higher infection frequencies of Hamiltonella (32.39%, n = 318); it may reflect those aphid populations have different needs, depending on the benefits provided by Hamiltonella, which needs a balance with fitness costs of the bacterium (Hafer‐Hahmann & Vorburger, 2020; Vorburger & Gouskov, 2011). More than 36% Serratia infection frequency has reported in pea aphids (Parker et al., 2017), but in current studies related to the natural population of cotton aphids, Serratia frequency was low, significantly lower than previous study (χ2 = 144.405, p < .001) (Xu et al., 2020). Different studies showed different infection frequencies of Serratia and Arsenophonus in cotton aphids but not Hamiltonella; the reason for this was the infected aphids have low abundance of Arsenophonus and Serratia, existing among the aphid samples. In this study, DNA was first diluted to 10‐fold for qPCR. Aphids having bacterial abundance lower than the lowest point of the standard curve were considered not infected with this bacterial, resulting in a relatively lower infection frequency than diagnostic PCR direct use original DNA solution. Although Hamiltonella‐infected individuals always have a high abundance in natural cotton aphid populations, few aphids were considered undetected individuals in data processing.
Based on resources, survival niches, and interaction between aphids and bacterial symbionts, there are complex relationships among them, known as metabolic tug of war (Smith & Moran, 2020). Not all facultative symbionts can affect the abundance of Buchnera in cotton aphids. Subsequently, in aphids infected with Hamiltonella, the abundance of primary Buchnera is increased, but in infected aphids, there was a negative correlation of Hamiltonella on the abundance of Buchnera in cotton aphids (Xu et al., 2020; Zhang, Luo, Wang, et al., 2019). The infection of Arsenophonus in cotton aphids showed different results, for example, when infected with Arsenophonus, the abundance of Buchnera reported to be increased (Tian et al., 2019), and continuously increases with the increase in Arsenophonus abundance. The opposite coinfection relationship was showed by using high‐throughput 16S rRNA sequencing (Xu et al., 2020) as the use of relative abundance was easy in incorrectness due to the inconsistency of the total abundance. In several aphid species, loss of genes in Buchnera complements in the Serratia genome, and Buchnera and Serratia reported to be evolved together during the biosynthesis of tryptophan (Lamelas et al., 2011; Monnin et al., 2020). Serratia has a low infection level in the natural cotton populations, indicating that it is not necessary for the survival of cotton aphids. But Serratia infection can influence the Buchnera densities in cotton aphids; this implies that there may be a close interaction between them.
The low infection frequency may indicate that cotton aphids were more likely to acquire symbiont bacteria through horizontal transfer because of more complicated coinfection of secondary bacterial symbionts. Nearly 100% coinfection of Hamiltonella and Arsenophonus was recorded in Xinjiang population, where cotton aphids have to deal with more complex biological and abiotic factors. A previous study showed that Arsenophonus together with Hamiltonella contributed to the fitness of A. gossypii by enhancing its performance (Ayoubi et al., 2020). Recent research shows Hamiltonella also can have an effect on insecticide resistance of aphids (Li et al., 2020) or can alter the dynamics of host metabolic interactions with co‐occurring microorganisms (Blow et al., 2020). Costs and benefits of coinfection have influenced within‐host interactions between these symbionts, resulting in coinfection dynamic changes within natural populations (Russell et al., 2013).
Geographical distribution affects the common bacterial symbiont in insects (De Cock et al., 2020; Pan et al., 2012). Cotton is widely grown in China and serves as the main secondary host of cotton aphids. Hibiscus is the main primary host. A. gossypii were collected from cotton from three regions with obvious climate differences. In the studied regions, cotton aphids have a holocyclic life cycle, that is, in Henan and Xinjiang and reproduce continuously by apomictic parthenogenesis in Hainan, recorded with significant difference in bacterial symbiont infection frequencies and abundance among the three geographic populations of aphids. This situation also arises in hibiscus populations collected at different times and regions. Hamiltonella can confer tolerance to high temperatures in A. pisum (Russell & Moran, 2006), and some Serratia are known to be involved in the detoxification of insecticides (van den Bosch & Welte, 2017), or it could increase insect susceptibility to exposed insecticides (Skaljac et al., 2018). Although A. gossypii were collected from hibiscus, infection frequencies and abundance remained greatly different among various geographic populations. The symbiotic bacteria may help the host organism to improve adaptability, affected by environmental and/or historical factors (Tsuchida et al., 2002).
In this study, A. gossypii were collected from hibiscus in April, September, and November from North China, where eggs of the aphid hatched on the host plant—hibiscus—and later, after the month of April, migrated to the host plant in summer. There were some cotton aphids still hosting hibiscus plants in September, and alate adults return to the hibiscus to mate and oviposit in November (Xia, 1997). The natural population of cotton aphids was suppressed to a small number several times a year because of the natural environment, especially due to rain and or absence of food plants (Hu et al., 2017; Liu et al., 2017), so migration of aphids among host plants occurs several times a year. There were differences in infection frequencies of bacterial symbionts among geographic populations, and most of the facultative symbionts spread more likely through horizontal transfer and can pass through plants (Henry et al., 2013), indicating that A. gossypii migrate in a small area. The infection of facultative symbionts, in the aphids, remained significantly different among different years from the same place with significant fluctuations in abundance. It shows that there is no obvious dominant population in the aphid population, in response to environmental changes; populations and symbiotic bacteria have great variability.
In conclusion, using the 16S rRNA gene sequencing, the dominant symbiotic bacteria were identified, and the abundance of symbiotic bacteria was estimated using qPCR in natural populations of A. gossypii. The low infection frequency of facultative symbionts in the natural population of cotton aphids were found across host plants, geographic area, and time periods. The infection of Serratia, Hamiltonella, and Arsenophonus can affect the density of Buchnera with different modes, and there was also obvious interaction between facultative symbionts. By analysis of the symbiotic bacteria from different geographic areas, we speculate that A. gossypii migrate in a small area.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
AUTHOR CONTRIBUTIONS
Shuai Zhang: Conceptualization (equal); Data curation (equal); Funding acquisition (equal); Methodology (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Honghua Su: Writing‐review & editing (equal). Weili Jiang: Data curation (equal). Daowu Hu: Resources (equal). Intazar Ali: Writing‐review & editing (equal). Tianxing Jing: Methodology (equal). Yizhong Yang: Supervision (equal); Writing‐review & editing (equal). Xiaoyan Ma: Project administration (equal); Resources (equal); Writing‐review & editing (equal).
ETHICS APPROVAL
This study does not contain any studies with human participants or vertebrate performed by any of the authors. Cotton aphids are invertebrate insects and, according to the IUCN criteria, are not considered as endangered or protected species. Cotton aphid samples were collected from farmer's fields and ornamental hibiscus after obtaining their permission.
INFORMED CONSENT
Informed consent was obtained from all individual participants included in the study.
Supporting information
Appendix S1
Appendix S2
Appendix S3
Appendix S4
Appendix S5
ACKNOWLEDGMENTS
This study was supported by a grant from the National Natural Science Foundation of China (31972355).
Zhang, S. , Su, H. , Jiang, W. , Hu, D. , Ali, I. , Jin, T. , Yang, Y. , & Ma, X. (2021). Symbiotic microbial studies in diverse populations of Aphis gossypii, existing on altered host plants in different localities during different times. Ecology and Evolution, 11, 13948–13960. 10.1002/ece3.8100
Contributor Information
Shuai Zhang, Email: shuaizhang@yzu.edu.cn.
Yizhong Yang, Email: yzyang@yzu.edu.cn.
Xiaoyan Ma, Email: maxiaoyan@caas.cn.
DATA AVAILABILITY STATEMENT
All raw sequences were deposited in the NCBI Sequence Read Archive under accession number SRA Accession no. PRJNA725955.
REFERENCES
- Ayoubi, A. , Talebi, A. A. , Fathipour, Y. , & Mehrabadi, M. (2020). Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Science, 27, 86–98. [DOI] [PubMed] [Google Scholar]
- Blackman, R. L. , & Eastop, V. F. (2000). Aphids on the world's crops. An identification guide. Wiley‐Interscience. [Google Scholar]
- Blow, F. , Ankrah, N. Y. D. , Clark, N. , Koo, I. , Allman, E. L. , Liu, Q. , Anitha, M. , Patterson, A. D. , & Douglas, A. E. (2020). Impact of facultative bacteria on the metabolic function of an obligate insect‐bacterial symbiosis. Mbio, 11, e00402‐20. 10.1128/mBio.00402-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, J. W. , Chevignon, G. , Oliver, K. M. , & Strand, M. R. (2017). Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proceedings of the Royal Society B: Biological Sciences, 284(1866), 20171925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright, M. , & Bulgheresi, S. (2010). A complex journey: Transmission of microbial symbionts. Nature Reviews. Microbiology, 8, 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletto, J. , Lombaert, E. , Chavigny, P. , Brevault, T. , Lapchin, L. , & Vanlerberghe‐Masutti, F. (2009). Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Molecular Ecology, 18, 2198–2212. [DOI] [PubMed] [Google Scholar]
- Charaabi, K. , Carletto, J. , Chavigny, P. , Marrakchi, M. , Makni, M. , & Vanlerberghe‐Masutti, F. (2008). Genotypic diversity of the cotton‐melon aphid Aphis gossypii (Glover) in Tunisia is structured by host plants. Bulletin of Entomological Research, 98, 333–341. [DOI] [PubMed] [Google Scholar]
- Chong, R. A. , & Moran, N. A. (2016). Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proceedings of the National Academy of Sciences of the United States of America, 113, 13114–13119. 10.1073/pnas.1610749113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman, K. A. , & Burke, G. R. (2020). Genomic analysis reveals an exogenous viral symbiont with dual functionality in parasitoid wasps and their hosts. PLoS Pathogens, 16, e1009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantur, K. I. , Enrique, R. , Welin, B. , & Castagnaro, A. P. (2015). Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express, 5, 15. 10.1186/s13568-015-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cock, M. , Virgilio, M. , Vandamme, P. , Bourtzis, K. , De Meyer, M. , & Willems, A. (2020). Comparative microbiomics of Tephritid frugivorous pests (Diptera: Tephritidae) from the field: A tale of high variability across and within species. Frontiers in Microbiology, 11, 1890. 10.3389/fmicb.2020.01890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, H. E. , Wilson, A. C. C. , Ferguson, N. R. , & Moran, N. A. (2007). Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLOS Biology, 5, e96. 10.1371/journal.pbio.0050096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, T. A. , & Cartwright, B. (1997). Biology and ecology of Aphis gossypii glover (Homoptera: Aphididae). Southwestern Entomologist, 22, 116. [Google Scholar]
- Engl, T. , Schmidt, T. H. P. , Kanyile, S. N. , & Klebsch, D. (2020). Metabolic cost of a nutritional symbiont manifests in delayed reproduction in a grain pest beetle. Insects, 11(10), 717. 10.3390/insects11100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo‐Franco, J. J. , Duque‐Gamboa, D. N. , & Toro‐Perea, N. (2019). Bacterial communities of Aphis gossypii and Myzus persicae (Hemiptera: Aphididae) from pepper crops (Capsicum sp.). Scientific Reports, 9, 12. 10.1038/s41598-019-42232-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafer‐Hahmann, N. , & Vorburger, C. (2020). Parasitoids as drivers of symbiont diversity in an insect host. Ecology Letters, 23, 1232–1241. 10.1111/ele.13526 [DOI] [PubMed] [Google Scholar]
- Hague, M. T. J. , Caldwell, C. N. , & Cooper, B. S. (2020). Pervasive effects of Wolbachia on Host temperature preference. mBio, 11(5), e01768‐20. 10.1128/mbio.01768-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry, T. A. , Ligon, R. A. , Besler, K. R. , Fay, R. L. , & Smee, M. R. (2018). Visual detection and avoidance of pathogenic bacteria by aphids. Current Biology, 28, 3158–3164.e4. 10.1016/j.cub.2018.07.073 [DOI] [PubMed] [Google Scholar]
- Henry, L. M. , Peccoud, J. , Simon, J.‐C. , Hadfield, J. D. , Maiden, M. J. C. , Ferrari, J. , & Godfray, H. C. J. (2013). Horizontally transmitted symbionts and host colonization of ecological niches. Current Biology, 23, 1713–1717. 10.1016/j.cub.2013.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, T. , Koga, R. , Kikuchi, Y. , Meng, X.‐Y. , & Fukatsu, T. (2010). Wolbachia as a bacteriocyte‐associated nutritional mutualist. Proceedings of the National Academy of Sciences of the United States of America, 107, 769–774. 10.1073/pnas.0911476107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D. W. , Zhang, S. , Luo, J. Y. , Lu, L. M. , Cui, J. J. , & Zhang, X. (2017). An example of host plant expansion of host‐specialized Aphis gossypii Glover in the field. PLoS One, 12, e0177981. 10.1371/journal.pone.0177981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. H. , & Kim, D. S. (2017). Effects of temperature and photoperiod on the production of sexual morphs of Aphis gossypii (Hemiptera: Aphididae) in Jeju, Korea. Journal of Asia‐Pacific Entomology, 20, 53–56. 10.1016/j.aspen.2016.11.006 [DOI] [Google Scholar]
- Lamelas, A. , Gosalbes, M. J. , Manzano‐Marín, A. , Peretó, J. , Moya, A. , & Latorre, A. (2011). Serratia symbiotica from the aphid Cinara cedri: A missing link from facultative to obligate insect endosymbiont. PLoS Genetics, 7, e1002357. 10.1371/journal.pgen.1002357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R. E. , Lozupone, C. A. , Hamady, M. , Knight, R. , & Gordon, J. I. (2008). Worlds within worlds: Evolution of the vertebrate gut microbiota. Nature Reviews. Microbiology, 6, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Sun, J. , Qin, Y. , Fan, J. , Zhang, Y. , Tan, X. , Hou, M. , & Chen, J. (2021). Reduced insecticide susceptibility of the wheat aphid Sitobion miscanthi after infection by the secondary bacterial symbiont Hamiltonella defensa . Pest Management Science, 77, 1936–1944. 10.1002/ps.6221 [DOI] [PubMed] [Google Scholar]
- Liu, X. D. , Xu, T. T. , & Lei, H. X. (2017). Refuges and host shift pathways of host‐specialized aphids Aphis gossypii . Scientific Reports, 7, 9. 10.1038/s41598-017-02248-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritopoulos, J. T. , Tzortzi, M. , Zarpas, K. D. , Tsitsipis, J. A. , & Blackman, R. L. (2006). Morphological discrimination of Aphis gossypii (Hemiptera: Aphididae) populations feeding on Compositae. Bulletin of Entomological Research, 96, 153–165. [DOI] [PubMed] [Google Scholar]
- Monnin, D. , Jackson, R. , Kiers, E. T. , Bunker, M. , Ellers, J. , & Henry, L. M. (2020). Parallel evolution in the integration of a co‐obligate Aphid symbiosis. Current Biology, 30, 1949–1957.e6. 10.1016/j.cub.2020.03.011 [DOI] [PubMed] [Google Scholar]
- Moran, N. A. (2007). Symbiosis as an adaptive process and source of phenotypic complexity. Proceedings of the National Academy of Sciences of the United States of America, 104(Suppl. 1), 8627–8633. 10.1073/pnas.0611659104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K. M. , Campos, J. , Moran, N. A. , & Hunter, M. S. (2008). Population dynamics of defensive symbionts in aphids. Proceedings of the Royal Society B: Biological Sciences, 275, 293–299. 10.1098/rspb.2007.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, H. , Li, X. , Ge, D. , Wang, S. , Wu, Q. , Xie, W. , Jiao, X. , Chu, D. , Liu, B. , Xu, B. , & Zhang, Y. (2012). Factors affecting population dynamics of maternally transmitted endosymbionts in Bemisia tabaci . PLoS One, 7, e30760. 10.1371/journal.pone.0030760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, B. J. , Mclean, A. H. C. , Hrček, J. , Gerardo, N. M. , & Godfray, H. C. J. (2017). Establishment and maintenance of aphid endosymbionts after horizontal transfer is dependent on host genotype. Biology Letters, 13, 20170016. 10.1098/rsbl.2017.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabatel, A. , Febvay, G. , Gaget, K. , Duport, G. , Baa‐Puyoulet, P. , Sapountzis, P. , Bendridi, N. , Rey, M. , Rahbé, Y. , Charles, H. , Calevro, F. , & Colella, S. (2013). Tyrosine pathway regulation is host‐mediated in the pea aphid symbiosis during late embryonic and early larval development. BMC Genomics, 14, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J. A. , & Moran, N. A. (2006). Costs and benefits of symbiont infection in aphids: Variation among symbionts and across temperatures. Proceedings of the Royal Society B: Biological Sciences, 273, 603–610. 10.1098/rspb.2005.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J. A. , Weldon, S. , Smith, A. H. , Kim, K. L. , Hu, Y. , Łukasik, P. , Doll, S. , Anastopoulos, I. , Novin, M. , & Oliver, K. M. (2013). Uncovering symbiont‐driven genetic diversity across North American pea aphids. Molecular Ecology, 22, 2045–2059. 10.1111/mec.12211 [DOI] [PubMed] [Google Scholar]
- Singh, K. S. , Troczka, B. J. , Duarte, A. , Balabanidou, V. , Trissi, N. , Carabajal Paladino, L. Z. , Nguyen, P. , Zimmer, C. T. , Papapostolou, K. M. , Randall, E. , Lueke, B. , Marec, F. , Mazzoni, E. , Williamson, M. S. , Hayward, A. , Nauen, R. , Vontas, J. , & Bass, C. (2020). The genetic architecture of a host shift: An adaptive walk protected an aphid and its endosymbiont from plant chemical defenses. Science Advances, 6(19), eaba1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaljac, M. , Kirfel, P. , Grotmann, J. , & Vilcinskas, A. (2018). Fitness costs of infection with Serratia symbiotica are associated with greater susceptibility to insecticides in the pea aphid Acyrthosiphon pisum . Pest Management Science, 74, 1829–1836. [DOI] [PubMed] [Google Scholar]
- Smith, T. E. , & Moran, N. A. (2020). Coordination of host and symbiont gene expression reveals a metabolic tug‐of‐war between aphids and Buchnera. Proceedings of the National Academy of Sciences, 117, 2113–2121. 10.1073/pnas.1916748117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Q. , Oliver, K. M. , Xie, W. , Wu, Q. J. , Wang, S. L. , & Zhang, Y. J. (2015). The whitefly‐associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Functional Ecology, 29, 1007–1018. [Google Scholar]
- Teixeira, L. , Ferreira, A. , & Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster . PLoS Biology, 6, e2. 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, P.‐P. , Chang, C.‐Y. , Miao, N.‐H. , Li, M.‐Y. , & Liu, X.‐D. (2019). Infections with Arsenophonus facultative endosymbionts alter performance of Aphids (Aphis gossypii) on an Amino‐Acid‐Deficient Diet. Applied and Environmental Microbiology, 85(23), e01407‐19. 10.1128/aem.01407-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Horikawa, M. , Tsunoda, T. , Maoka, T. , Matsumoto, S. , Simon, J. C. , & Fukatsu, T. (2010). Symbiotic bacterium modifies aphid body color. Science, 330, 1102–1104. 10.1126/science.1195463 [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Shibao, H. , Matsumoto, T. , & Fukatsu, T. (2002). Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum . Molecular Ecology, 11, 2123–2135. 10.1046/j.1365-294X.2002.01606.x [DOI] [PubMed] [Google Scholar]
- Van Den Bosch, T. J. M. , & Welte, C. U. (2017). Detoxifying symbionts in agriculturally important pest insects. Microbial Biotechnology, 10, 531–540. 10.1111/1751-7915.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ham, R. C. H. J. , Kamerbeek, J. , Palacios, C. , Rausell, C. , Abascal, F. , Bastolla, U. , Fernández, J. M. , Jiménez, L. , Postigo, M. , Silva, F. J. , Tamames, J. , Viguera, E. , Latorre, A. , Valencia, A. , Morán, F. , & Moya, A. (2003). Reductive genome evolution in Buchnera aphidicola . Proceedings of the National Academy of Sciences of the United States of America, 100, 581–586. 10.1073/pnas.0235981100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger, C. , & Gouskov, A. (2011). Only helpful when required: A longevity cost of harbouring defensive symbionts. Journal of Evolutionary Biology, 24, 1611–1617. 10.1111/j.1420-9101.2011.02292.x [DOI] [PubMed] [Google Scholar]
- Wang, L. , Zhang, S. , Luo, J.‐Y. , Wang, C.‐Y. , Lv, L.‐M. , Zhu, X.‐Z. , Li, C.‐H. , & Cui, J.‐J. (2016). Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS One, 11, e0146345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, T. L. , Fukatsu, T. , & Ishikawa, H. (2003). Transmission of symbiotic bacteria Buchnera to parthenogenetic embryos in the aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Arthropod Structure & Development, 32, 241–245. 10.1016/S1467-8039(03)00036-7 [DOI] [PubMed] [Google Scholar]
- Xia, J. (1997). Biological control of cotton aphid (Aphis gossypii Glover) in cotton (inter)cropping systems in China: a simulation study. Wageningen University, Wageningen University dissertation. [Google Scholar]
- Xia, J. Y. , Van Der Werf, W. , & Rabbinge, R. (1999). Influence of temperature on bionomics of cotton aphid, Aphis gossypii, on cotton. Entomologia Experimentalis Et Applicata, 90, 25–35. [Google Scholar]
- Xu, S. F. , Jiang, L. Y. , Qiao, G. X. , & Chen, J. (2020). The bacterial flora associated with the polyphagous Aphid Aphis gossypii Glover (Hemiptera: Aphididae) is strongly affected by host plants. Microbial Ecology, 79, 971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Leonard, S. P. , Li, Y. , & Moran, N. A. (2019). Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proceedings of the National Academy of Sciences of the United States of America, 116, 24712–24718. 10.1073/pnas.1915307116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. X. , & Zhong, T. S. (1990). Experimental studies on some aphid life‐cycle patterns and the hybridization of two sibling species. In Campbell R. K., & Eikenbarys R. D. (Eds.), Aphid‐plant genotype interactions (pp. 37–50). Elsevier. [Google Scholar]
- Zhang, J. , Pan, Y. , Zheng, C. , Gao, X. , Wei, X. , Xi, J. , Peng, T. , & Shang, Q. (2016). Rapid evolution of symbiotic bacteria populations in spirotetramat‐resistant Aphis gossypii glover revealed by pyrosequencing. Comparative Biochemistry and Physiology Part D: Genomics & Proteomics, 20, 151–158. 10.1016/j.cbd.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Luo, J. Y. , Jiang, W. L. , Wu, L. K. , Zhang, L. J. , Ji, J. C. , Wang, L. , Ma, Y. , & Cui, J. J. (2019). Response of the bacterial community of Propylea japonica (Thunberg) to Cry2Ab protein. Environmental Pollution, 254, 113063. 10.1016/j.envpol.2019.113063 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Luo, J. Y. , Wang, L. , Wang, C. Y. , Lu, L. M. , Zhang, L. J. , Zhu, X. Z. , & Cui, J. J. (2018). The biotypes and host shifts of cotton‐melon aphids Aphis gossypii in northern China. Journal of Integrative Agriculture, 17, 2066–2073. 10.1016/S2095-3119(17)61817-3 [DOI] [Google Scholar]
- Zhang, S. , Luo, J. Y. , Wang, L. , Zhang, L. J. , Zhu, X. Z. , Jiang, W. L. , & Cui, J. J. (2019). Bacterial communities in natural versus pesticide‐treated Aphis gossypii populations in North China. Microbiologyopen, 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. C. , Cao, W. J. , Zhong, L. R. , Godfray, H. C. J. , & Liu, X. D. (2016). Host plant determines the population size of an obligate symbiont (Buchnera aphidicola) in Aphids. Applied and Environmental Microbiology, 82, 2336–2346. 10.1128/AEM.04131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhang, S. , Luo, J. Y. , Wang, C. Y. , Lv, L. M. , & Cui, J. J. (2016). Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Scientific Reports, 6, 22958. 10.1038/srep22958 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Appendix S3
Appendix S4
Appendix S5
Data Availability Statement
All raw sequences were deposited in the NCBI Sequence Read Archive under accession number SRA Accession no. PRJNA725955.


