Abstract
Background
Molecular testing for alterations in oncogenic driver genes and targeted therapies have become standard procedures for non‐small cell lung cancer (NSCLC) patients. However, little evidence has shed light on the pattern of co‐existence of driver genes in NSCLC, and whether they may have different tumor features affecting immunotherapy is still unclarified.
Methods
Genomic alterations in 14 lung cancer‐related genes were conducted in 3440 Chinese NSCLC patients using next‐generation sequencing. Meanwhile, tumor mutational burden and immunotherapy dataset from the Memorial sloan kettering cancer center (MSKCC) and lung adenocarcinoma dataset from The Cancer Genome Atlas (TCGA) were utilized for analyzing the impact of the co‐occurring alterations on patients’ survival following immunotherapy.
Results
In this cohort, 90.17% of patients had at least one somatic alteration in the 14 genes, including 51% of co‐occurring alterations. TP53 and epidermal growth factor receptor (EGFR) were the most prevalent genes (54.74% and 53.55%, respectively), followed by KRAS, ERBB2, ALK, PIK3CA, ROS1, RET, MET, BRAF, KIT, FGFR1, PDGFRA, and NRAS. The prevalence of TP53, EGFR, and ERBB2 in our cohort were significantly higher than that from the TCGA database, whereas KRAS, BRAF, and PDGFRA were significantly lower than the latter. Furthermore, the patients who harbored multiple alterations (8.86%, 31/350) in eight driver genes survived longer and have a higher tumor mutation burden compared to the patients with a single alteration. Similar result was found between the patients with co‐occurring alteration of EGFR and other driver genes and the patients with single EGFR alteration. Meanwhile, we found a distinct immune cell infiltration feature between patients with single and multiple driver gene alterations, as well as between patients with only EGFR alteration and co‐occurring groups.
Conclusion
This study identified a unique driver gene feature and found patients harboring co‐occurring alterations of EGFR and other driver genes may benefit from immunotherapy, which may provide more therapeutic selections for EGFR‐mutated NSCLC patients and merit additional investigation.
Keywords: co‐occurring, driver genes, EGFR, immunotherapy, non‐small cell lung cancer, somatic alterations
This study identified a unique driver gene feature and found patients harboring co‐occurring alterations of epidermal growth factor receptor (EGFR) and other driver genes may benefit from immunotherapy, which may provide more therapeutic selections for EGFR‐mutated non‐small cell lung cancer patients and merit additional investigation.

1. INTRODUCTION
Lung cancer is the leading cause of cancer‐related mortality worldwide, causing over 1.7 million deaths annually. 1 Non‐small cell lung cancer (NSCLC) accounts for 85% of lung cancer cases. 2
With the discovery of cancer driver genes, genomic testing has been integrated as a part of the standard diagnostic procedure, and several molecular drugs targeting the driver genes have been applied in the treatment of lung cancer and have shown great effectiveness in increasing the survival of advanced NSCLC. 3 , 4 Epidermal growth factor receptor (EGFR) alterations, including L858R and short insertions/deletions (indels) in exon 19, were identified as the first druggable alterations in NSCLC and proved to be the most robust predictive biomarker for EGFR tyrosine kinase inhibitors (TKIs). 5 Since then, several additional driver gene alterations have been reported, including oncogenic somatic alterations in BRAF, 6 intragenic insertions in ERBB2 (in exon‐20), 7 exon 14 skipping alterations in the MET proto‐oncogene, 8 oncogenic alterations in KRAS, 9 and genes rearrangement of ALK, ROS1, and RET. 10 The National Comprehensive Cancer Network (NCCN) guideline recommends broad molecular profiling, including screening for the presence of activating alterations in EGFR, ALK, ROS1, BRAF, KRAS, MET, ERBB2, and RET to inform the selection of effective targeted therapies for NSCLC patients. Additionally, TP53, PIK3CA, KIT, FGFR1, PDGFRA, or NRAS were previously identified prevalent alterations in patients with NSCLC, and their impacts on target treatment or prognosis have received widespread attention. 11 , 12 , 13 All of the 14 genes mentioned above can be considered lung cancer‐associated genes.
Immunotherapy is considered as a salvage treatment for patients with actionable driver alterations after the progression of related targeted therapies and chemotherapy. 14 However, most clinical trials have shown that immune checkpoint inhibitors (ICIs) have poor activity in patients with driver gene alteration, especially EGFR and ALK. One retrospective study for advanced NSCLC patients with at least one oncogenic driver alteration receiving ICI monotherapy found that the median progression‐free survival (PFS) was only 2.8 months, and the objective response rates by driver alteration were generally low except RET (6%) and ALK (0%). 15 Thus, therapeutic options are restrained in NSCLC patients with driver gene alterations, which is an urgent issue that needs to be addressed.
Recently, studies have found the presence of driver genes' co‐occurring alterations in NSCLC, and its effect on molecularly targeted therapies has attracted focus. 16 Multiple clinical studies have found patients with co‐occurring alterations of TP53 and EGFR alterations had worse prognostic when treated with EGFR‐TKI therapy. 16 Besides, Martín Martorell et al. found that targeted treatment might not be as effective in patients with coexisting of EGFR, KRAS, BRAF alterations, and ALK rearrangement. 17 However, the effect of the co‐existence of driver genes in NSCLC on immunotherapy is still unclarified.
In the present study, genomic alterations of 14 lung cancer‐associated genes were assessed in a cohort of 3440 Chinese NSCLC patients by next‐generation sequencing (NGS). The basic profile of the patient's driver gene alterations was described and compared with corresponding data in The Cancer Genome Atlas (TCGA) to better understand driver gene features in Chinese NSCLC patients. Furthermore, we focused on the patterns of co‐existence of driver genes and their effects on the response to immunotherapy.
2. MATERIALS AND METHODS
2.1. DNA isolation
The formalin fixation and paraffin‐embedding (FFPE) samples and fresh‐frozen tissues were collected and used for gDNA isolation. The specimens selected contained more than 20% tumor cells. The purified gDNA was quantified using the Qubit 3.0 Fluorometer (Life Technologies, Inc.) and StepOnePlus System (Life Technologies, Inc.).
2.2. Target NGS
Hundred nanograms of gDNA were sheared to target 200 bp fragment sizes with a Covaris E210 system (Covaris, Inc.). NGS of tumor gDNA was performed, in which Accel‐NGS 2S DNA Library Kit (Swift Biosciences, Inc.) was used for library preparation and xGen Lockdown Probes Kit (IDT, Inc.) for target enrichment. The custom xGen Lockdown probe was synthesized by IDT, Inc. for the exons and the part of introns of 14 genes of interest (EGFR, ALK, ROS1, TP53, ERBB2, BRAF, KRAS, MET, PIK3CA, NRAS, FGFR1, RET, KIT, and PDGFRA).
The prepared library was quantified by the Qubit 3.0 Fluorometer (Life Technologies, Inc.), and quality and fragment size were measured with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
The samples underwent paired‐end sequencing on an Illumina NextSeq CN500 platform (Illumina, Inc.) with a 150 bp read length. Mean coverage beyond 1300× was achieved for tumor gDNA.
2.3. Data processing
Raw sequencing data were aligned to the reference human genome (UCSC hg19) through Burrows–Wheeler Aligner. 18 After the duplicate removal and local realignment, the Genome Analysis ToolKit (GATK) v3.7 was used for single nucleotide variation (SNV)/indel calling and filtering. 19 Gene fusions were called using Genefuse v0.6.0. 20 The somatic variants were generated for the patient by subtracting the germline variants from the tumor to keep only variants unique to a tumor. The variants were annotated using the ANNOVAR software tool. 21 The somatic alterations were annotated with information from the Catalog of Somatic Alterations in the OncoKB database.
2.4. Data sources
Tumor mutational burden (TMB) and Immunotherapy (MSKCC, Nat Genet 2019) dataset 22 and clinical data were downloaded from cBioPortal (https://www.cbioportal.org), which contains 350 NSCLC samples in total, and all samples with alteration data were selected for alteration and survival analysis. Besides, lung adenocarcinoma (LUAD; TCGA, Firehose Legacy) dataset and mRNA expression data were downloaded from cBioPortal to compare the differences of immune microenvironment between patients with single EGFR alteration and the co‐existing alterations of EGFR and other driver genes.
2.5. Statistical analysis
Statistical analyses were performed using SPSS, GraphPad Prism 7 software, and R language statistical package. The differences between the two groups were assessed using Student's t‐test. The differences were considered significant if p < 0.05. The adjusted odds ratios were calculated. A two‐sided p‐value of <0.05 was considered to be statistically significant if there was no alpha correction. The overall survival (OS) curves were constructed using the Kaplan–Meier method, and the log‐rank test was performed. A p value <0.05 was considered to be statistically significant unless additionally specified.
3. RESULTS
3.1. Samples and clinical data description
A total of 2833 FFPE samples and 607 fresh‐frozen tissues were collected from 3440 patients diagnosed with NSCLC. Adenocarcinoma was the common histological type in this cohort, accounting for 92.7% (3189). Of the total 3440 patients, 1856 were male (53.95%), and 1584 were female (46.05%). The age at diagnosis ranged from 19 to 98 years old, with a median of 62 years (Table 1).
TABLE 1.
Clinical characteristics of 3440 NSCLC patients
| Characteristics | Total, n = 3440 |
|---|---|
| Median age (range) | 62 (19–98) |
| Gender | |
| Male | 1856 |
| Female | 1584 |
| Histology | |
| Adenocarcinoma | 3189 |
| Squamous | 217 |
| Adenosquamous | 23 |
| Large cell | 11 |
| Stage | |
| II–III | 1760 |
| IV | 1680 |
| Smoking history | |
| Yes | 1430 |
| No | 1839 |
| NA | 171 |
Abbreviation: NSCLC, non‐small cell lung cancer; NA: no data.
3.2. Landscape of genomic alterations in 3440 NSCLC patients
Utilizing targeted deep sequencing of all exons and selected introns of 14 lung cancer‐related genes in 3440 NSCLC tissue samples, we found that 90.17% (3102 out of 3440) of patients had at least one somatic alteration. Among the 14 genes, 39.16% (1347/3440) of NSCLC patients were found to have a single alteration. 51.02% (1755/3440) harbored multiple alterations: 36.02% (1239/3440) had double alterations, 12.21% (420/3440) had triple alterations, and 2.79% (96/3440) had more than three alterations (Table S1).
In this study, the most prevalent genes were TP53 (54.74%), EGFR (53.55%), and KRAS (13.40%) (Figure 1A), followed by ERBB2 (9.51%), ALK (7.82%), PIK3CA (6.34%), ROS1 (5.78%), RET (4.01%), MET (3.92%), BRAF (3.14%), KIT (3.05%), FGFR1 (1.98%), PDGFRA (1.86%), and NRAS (0.55%). Among the 14 genes in our cohort, except for PIK3CA (6.3% vs. 12.0%) and FGFR1 (1.98% vs. no data), the prevalence of the other 12 genes was similar to the results reported in a previous Chinese NSCLC population. 11 The variant classification spectrum showed that missense alteration type was the most common, followed by frameshift deletion and nonsense alteration (Figure 1B). Comparing the prevalence of 14 genes in the LUAD patients from the TCGA database identified significant differences in TP53 (54.74% vs. 46.09%), EGFR (53.55% vs. 14.35%), KRAS (13.40% vs. 32.61%), ERBB2 (9.51% vs. 2.61%), BRAF (3.14% vs. 9.57%), and PDGFRA (1.86% vs. 6.09%) in our cohort (Figure 1C). Furthermore, the prevalence of 14 genes in both LUAD and lung squamous cell carcinoma (LUSC) patients was calculated and compared (Figure S1). We found that the alteration of EGFR, KRAS, and ALK occurred more often in patients with LUAD than LUSC (p < 0.05), however, the frequencies of TP53, PIK3CA, and FGFR1 were significantly lower than the latter (p < 0.01), which was similar to the previous report. 23
FIGURE 1.
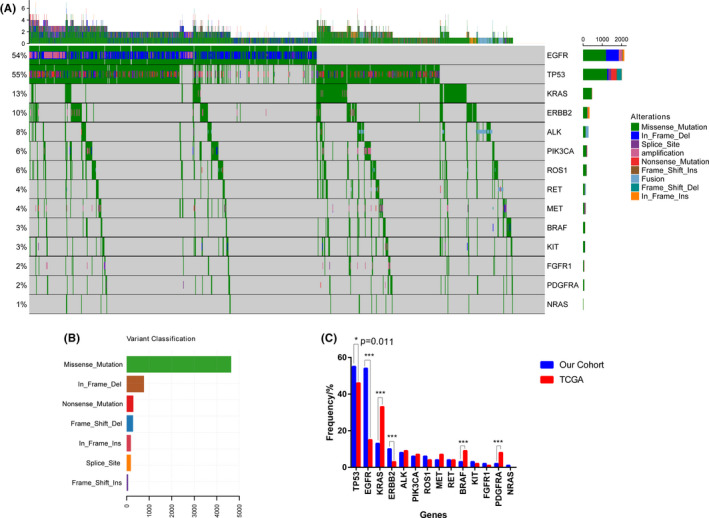
Landscape of somatic alterations in NSCLC involved in this study. (A). The landscape of alteration in 3440 NSCLC patients (B). Variant classification of all alterations (C). Comparison of the alteration frequencies of the 14 cancer‐related genes between our cohort and the TCGA cohort. Two‐sided Fisher's tests were conducted to compare the different frequencies between two cohorts. *** p ≤ 0.001, *p ≤ 0.05. NSCLC, non‐small cell lung cancer; TCGA, The Cancer Genome Atlas
3.3. Alteration analysis of 14 genes
In this study, 53.55% of NSCLC patients in our cohort had EGFR alternations, most of which had been well developed as actionable variants, such as L858R, exon 19 del, exon 20‐ins, L861Q, G719X, S768I, and T790 M (Figure 2A). Besides, 112 rare EGFR alterations were found in the cohort (Table S2). As profiled in Figure 2B, alterations of EGFR are distributed relatively throughout the whole protein. Multiple EGFR alterations were found in 18.72% of patients (644/3440). Of the 644 patients, 78.73%, 15.37%, and 5.12% of them had double alterations, triple alterations, and quadruple alterations, respectively. KRAS alteration was detected in 13.40% of patients (461/3440), and most of the alterations were located in exon 2 (11.66%, 401/3440); the remaining ones were detected in exon 3 (1.16%, 40 out of 3440) and exon 4 (0.55%, 19 out of 3440). The most prevalent alterations included G12C (4.62%), G12D (2.38%), and G12V (2.56%) (Figure 2A,C). ERBB2 alterations were detected in 327 patients (9.51%), distributed throughout the whole protein, of which a quarter (26.00%, 85/327) located in exon 20. The ERBB2‐positive cases featured samples with more nonsynonymous SNV (6.40%, 220/3440), nonframeshift insertion (2.33%, 80/3440), and amplification (0.90%, 31/3440) (Figure 2A,D). A total of 135 (3.92%, 135/3440) MET alterations were detected, of which 46 (1.34%, 46/3440) located in exon 14, 25 (0.73%, 25/3440) exon 21, 31 (0.9%, 31/3440) other location, and 31 (0.9%, 31/3440) amplifications (Figure 2A,E). We identified 3.14% of patients (108/3440) harbor BRAF alteration, and 0.93% (32/3440) were V600E. Most of the remaining were located in exon 15 (0.99%, 34/108) and exon 11 (0.73%, 25/108) (Figure 2A,F). TP53 was the most frequently mutated gene, detected in 54.74% of patients (1883/3440), which contains more nonsense alteration. The sites of TP53 alteration were mostly located in exon 5–8 (46.74%, 1608/3440) (Figure 2A,G). PIK3CA alteration was detected in 6.34% of patients (218/3440), of which 1.60% were located in exon 9 (55/3440) and exon 20 (1.83%, 63 /3440). There were 25 (0.73%, 25/3440) cases of E545K, 35 (1.02%, 35/3440) cases of H1047R/L/Q, and 22 cases (0.64%, 22/3440) of E542K (Figure 2A,H).
FIGURE 2.

Somatic alteration frequencies of 14 cancer‐related genes (A). The main alteration sites and frequency of 14 genes. The lollipop plot shows the genomic distribution of EGFR (B), KRAS (C), ERBB2 (D), MET (E), BRAF (F), TP53 (G) PIK3CA (H), and ALK (I). The gray bar represents the entire protein with the different amino acid positions. The length of the gray lines indicates the number of alterations detected at the specified position, and the colored circles on the gray bar represent the corresponding alteration types. The colored boxes are different functional domains. Amp, amplification; Fus, Fusion; Tol, Total alteration frequency; EGFR, epidermal growth factor receptor
Other genomic alterations were as follows: ALK (7.82%, 269/3440), ROS1 (5.78%, 199/3440), RET (4.01%, 138/3440), KIT (3.05%, 105/3440), FGFR1 (1.98%, 68/3440), PDGFRA (1.86%, 64/3440), and NRAS (0.55%, 19/3440) (Figure 2A,I; Figure S2).
3.4. ALK, ROS1, and RET fusions in NSCLC
In the cohort, 147 patients (4.27%) had ALK rearrangements, of which 97.28% (143/147) were EML4‐ALK, and 4 other ALK fusions (2 CLIP1‐ALK, 1 HIP1‐ALK, and 1 KIF5B‐ALK). The frequency of EML4‐ALK subtypes is shown in Figure 3 as the most common subtypes of EML4‐ALK were E6:A20 (variant 3; 43.42%) and E13:A20 (variant 1; 31.58%), whereas E20:A20 (variant 2) accounted for 11.18% (Figure 3).
FIGURE 3.
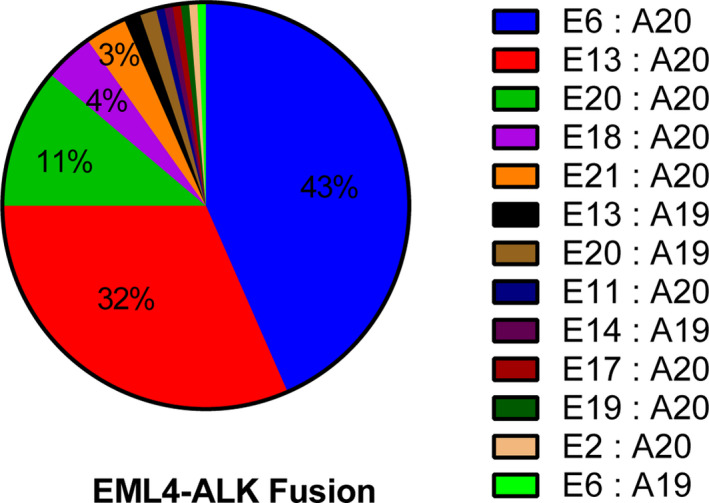
The frequency and distribution of EML4‐ALK fusion subtypes identified in the NSCLC cohort. NSCLC, non‐small cell lung cancer
We also find 1.51% (52/3440) of patients have RET rearrangement (36 KIF5B‐RET, 12 CCDC6‐RET, 2 ERC1‐RET, and 2 NCOA4‐RET) and 0.76% (26/3440) of patients harbor ROS1 rearrangement (14 CD74‐ROS1, 5 EZR‐ROS1, 2 LRIG3‐ROS1, 2 SLC34A2‐ROS1, 2 SDC4‐ROS1, and 1 ERC1‐ROS1) in this cohort (Figure 2A).
3.5. Patients’ characteristics and somatic alterations
We evaluated the association between alteration in 14 genes and gender and found that the alteration rate of EGFR (male vs. female: 41.47% vs. 67.91%, p < 0.0001) and ALK (male vs. female: 6.59% vs. 9.30%, p = 0.003) were higher in female than in male patients in our cohort. Whereas, significantly higher prevalence of the TP53 (63.12% vs. 44.94%, p < 0.0001), KRAS (17.17% vs. 8.92%, p < 0.0001), KIT (3.62% vs. 2.28%, p = 0.027), and FGFR1 (2.43% vs. 1.46%, p = 0.049) was found in male patients. There was no significant difference between male and female NSCLC patients for the alterations rates of other genes (ERBB2, PIK3CA, ROS1, RET, MET, BRAF, PDGFRA, and NRAS). Similar to EGFR alteration, the rearrangements of ALK (male vs. female: 3.40% vs. 5.32%, p = 0.007) and ROS1 (male vs. female: 0.43% vs. 1.14%, p = 0.028) were enriched in females patients. Besides, we found the median age in the ALK (median age 57, range 31–84) and ROS1 (median age 58.5, range 30–76) rearrangements‐positive cohort was lower than the whole cohort (median age 62, range 19–98), which demonstrates that younger patients were more likely to harbor ALK and ROS1 rearrangements.
3.6. Co‐occurring alterations of driver gene in NSCLC
The frequencies of co‐occurring alterations in 14 cancer‐related genes were identified as 51.02% (1755/3440) in our cohort (Table S1). The more common genes co‐occurring with EGFR were TP53 (28.26%), ERBB2 (3.66%), PIK3CA (3.14%), ROS1 (2.38%), and KRAS (28.26%). Besides, 7.15% of patients carried co‐occurring alterations of KRAS and TP53. Mutually exclusive or co‐occurring set of 14 genes were detected using the somatic interactions function of the maftools package, which performs pair‐wise Fisher's Exact test to detect such significant pair of genes. As a result, six pairs of significantly co‐altered genes were found in the study, including the co‐occurring in KRAS and RET/KIT/ALK; NRAS and ALK; MET and RET; PDGFRA and FGFR1. It is worth noting that EGFR alterations were mutually exclusive with the other 12 gene alterations except for PIK3CA (Figure 4). Besides, TP53 was mutually exclusive with MET/KRAS.
FIGURE 4.
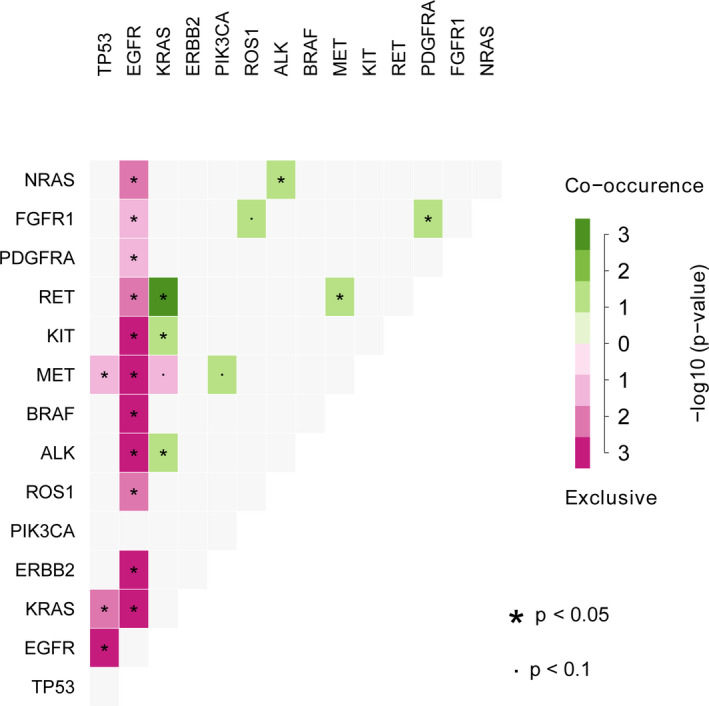
Co‐occurrence or exclusivity of 14 genes alterations events in the NSCLC (n = 3440). The green box represents the pair genes are significantly co‐altered, whereas the purple box indicates the two genes are significantly mutually exclusive. The depth of the color reflects the size of the p value that the darker the color, the smaller the p value. NSCLC, non‐small cell lung cancer
Eight driver genes, EGFR, ROS1, MET, RET, ALK, ERBB2, KRAS, and BRAF, are recommended by the NCCN guideline to inform the selection of effective targeted therapies for NSCLC patients. For the eight driver genes, approximately 80.87% (2782/3440) of Chinese NSCLC patients harbored at least one alteration, and single and multiple alterations (co‐occurring alterations) were accounted for 77.89% (2167/2782) and 22.11% (615/2782), respectively (Table 2). To find the profile (frequency) of Western populations, we calculated that in the corresponding TCGA (n = 230) and the MSKCC (n = 350) datasets. As shown in Table 2, fewer patients of Western populations (our cohort vs. TCGA: 80.87% vs. 65.65%, p < 0.0001; our cohort vs. MSKCC: 80.87% vs. 62.57%, p < 0.0001) carried alteration in eight driver genes compared with our cohort. Among the eight driver genes, EGFR and KRAS were the more common alterations, and the co‐occurring alterations, including EGFR or KRAS, have attracted wide attention. Thus, patients from the three cohorts were divided into eight groups according to the type and number of the altered gene they carried, including EGFR/KRAS_S (Patients with single EGFR/KRAS alteration), EGFR/KRAS_M (Patients with co‐occurring alterations of EGFR/KRAS and other seven driver genes), ALL/Others_S [Patients with single alteration in eight driver genes/others six driver genes (ROS1, MET, RET, ALK, ERBB2, BRAF)], and ALL/Others_M (Patients with multiple alterations in eight driver genes/other six driver genes) (Table 2).
TABLE 2.
Comparison of co‐occurring alterations in Chinese, TCGA, and MSKCC cohorts
| Groups |
Chinese (n = 3440) |
TCGA (n = 230) |
p value Chinese versus TCGA |
MSKCC (n = 350) |
p value Chinese versus MSKCC |
|---|---|---|---|---|---|
| Positive | 80.87% (2782/3440) | 65.65% (151/230) | <0.0001 | 62.57% (219/350) | <0.0001 |
| Negative | 19.13% (658/3440) | 34.35% (79/230) | 37.43% (131/350) | ||
| ALL_S | 77.89% (2167/2782) | 76.82% (116/151) | 0.763 | 85.84% (188/219) | 0.005 |
| ALL_M | 22.11% (615/2782) | 23.18% (35/151) | 14.16% (31/219) | ||
| EGFR_S | 50.86% (1415/2782) | 17.88% (27/151) | 0.839 | 16.89% (37/219) | 0.199 |
| EGFR_M | 15.35% (427/2782) | 4.64% (7/151) | 2.74% (6/219) | ||
| KRAS_S | 9.49% (264/2782) | 33.77% (51/151) | 0.054 | 46.58% (102/219) | <0.0001 |
| KRAS_M | 7.08% (197/2782) | 14.57% (22/151) | 10.05% (22/219) | ||
| Others_S | 17.54% (488/2782) | 25.17% (38/151) | 0.227 | 22.37% (49/219) | 0.641 |
| Others_M | 2.19%(61/2782) | 5.30% (8/151) | 1.83% (4/219) |
Positive, At least one alteration in eight driver genes (EGFR, KRAS, ROS1, MET, RET, ALK, ERBB2, and BRAF); Negative, Non alteration in eight driver genes; ALL/Others_S, Single alteration in eight driver genes/others six driver genes (ROS1, MET, RET, ALK, ERBB2, and BRAF); ALL/Others_M, Multiple alterations in eight driver genes/other six driver genes; EGFR/KRAS_S, Single EGFR/KRAS alteration; EGFR_M, Co‐occurring alterations of EGFR and other seven driver genes; KRAS_M, Co‐occurring alterations of KRAS and other seven driver genes.
Abbreviations: EGFR, epidermal growth factor receptor; TCGA, The Cancer Genome Atlas; MSKCC, Memorial Sloan Kettering Cancer Center.
3.7. Patients with multiple alterations have a longer survival time and higher TMB score
To study the effects of the co‐existing driver alterations on the survival of immunotherapy, we compared the difference in survival between patients with single alteration (ALL_S group) and patients with multiple alterations (ALL_M group) in the MSKCC cohort and found an interesting result that the latter has a significantly longer survival time (median survival: 12 months vs. unreach, p = 0.026) (Figure 5A). Similarly, group EGFR_M survived significantly longer than group EGFR_S (median survival: unreach vs. 11 months; p = 0.038) (Figure 5B), group KRAS_M survived longer than group KRAS_S (median survival: 14 vs. 12 months; p = 0.330) (Figure 5C), and group Others_M survived longer than Others_S (median survival: unreach vs. 14 months; p = 0.248) (Figure 5D). In summary, among immunotherapy patients, those with multiple alterations in the eight driver genes have a longer survival time. Meanwhile, we revealed that the patients with multiple alterations in eight driver genes had higher TMB levels (Figure 5E; Table S3).
FIGURE 5.

Multiple alterations are associated with longer survival and higher TMB score in patients after immunotherapy (A). Patients with multiple alterations in eight driver genes (ALL_M Group) (EGFR, ROS1, MET, RET, ALK, ERBB2, KRAS, and BRAF) have better overall survival (p = 0.026) (B). Patients with co‐occurring alterations of EGFR and the other seven driver genes (EGFR_M Group) have better overall survival (p = 0.038) (C). Patients with co‐occurring alterations of KRAS and other seven driver genes (KRAS_M group) have better overall survival (p = 0.330) (D). Patients with multiple alterations in the other six driver genes (Others_M Group) (ROS1, MET, RET, ALK, ERBB2, and BRAF) have better overall survival (p = 0.248) (E). Patients with multiple alterations (ALL_M, EGFR_M, KRAS_M, and Others_M Group) have higher TMB levels. *** p ≤ 0.001; EGFR, epidermal growth factor receptor; TMB, tumor mutational burden
All 350 NSCLC patients in the MSKCC dataset received PD‐1/PD‐L1‐targeted monotherapy (atezolizumab, durvalumab, nivolumab, or pembrolizumab) or combination immunotherapy (Combo; 6%, 21/350). 22 Interestingly, the patients who received the combination therapy had better survival than those who were treated with PD‐1/PD‐L1‐targeted monotherapy (median survival: 46 vs. 11 months; p = 0.009) (Figure S3). However, Fisher's exact test results proved that the difference in survival between patients with single alteration (All_S/EGFR_S) and those with multiple alterations (All_M/EGFR_M) was independent of the drug class of patients received (all p > 0.05, Table S4).
3.8. The differences of immune microenvironment between patients with single and multiple alterations
To find why patients with multiple alterations (the ALL_M and EGFR_M group) had better survival outcomes compared to the patients with single EGFR alteration (the ALL_S and the EGFR_S group) after immunotherapy, we investigated the fractions of tumor‐infiltrated immune cells (TIICs) between these groups in the TCGA cohort. The expression signature matrix of the 22 infiltrated immune cell types was analyzed based on CIBERSORT software. M2 macrophages accounted for a large proportion of NSCLC immune cell infiltration both in the four groups (Figure 6A,B). The fractions of five TIICs varied significantly among ALL_S and ALL_M groups. Three TIICs (T cell CD8+, activated memory T cell CD4+, and activated natural killer [NK] cell) were in a higher proportion in the ALL_M group than those in the ALL_S group (p < 0.05), whereas resting memory CD4+ T cells and activated mast cells were in a higher proportion in the ALL_S group (p < 0.05). Similarly, resting memory T cell CD4+, regulatory T cell (Tregs), activated myeloid dendritic cell, and activated mast cells were more common in the EGFR_S group compared with the EGFR_M group (p < 0.05), and the EGFR_M group generally contained a higher fraction of resting mast cell than the EGFR_S group (p < 0.05). The results showed the heterogeneity of immune cell infiltration between patients with single and multiple alterations.
FIGURE 6.

Tumor‐infiltrated immune cells in NSCLC patients with co‐occurring alterations from the TCGA cohort. (A) Patients with single driver gene alteration (ALL_S) versus co‐occurring alterations in eight driver genes (ALL_M). (B) The patients with only EGFR (EGFR_S) versus co‐occurring alterations of EGFR and other seven driver genes (EGFR_M). p < 0.05 for all eligible samples. ** p ≤ 0.01, * p ≤ 0.05. EGFR, epidermal growth factor receptor; NSCLC, non‐small cell lung cancer; TCGA, The Cancer Genome Atlas
4. DISCUSSION
This study performed 14 cancer‐related gene alternation analyses in a lager Chinese NSCLC cohort (n = 3440), and identified 90.17% (3102/3440) of patients with at least one alteration, including TP53 (54.74%), EGFR (53.55%), KRAS (13.40%), ERBB2 (9.51%), ALK (7.82%), PIK3CA (6.34%), ROS1 (5.78%), RET (4.01%), MET (3.92%), BRAF (3.14%), KIT (3.05%), FGFR1 (1.98%), PDGFRA (1.86%), and NRAS (0.55%).
Previous studies have found that the alterations of driver genes are related to ethnicity. For example, in KRAS‐positive NSCLC, the patients in Western countries (about 25%) are much more than in Asia (10%–15%). 9 , 24 , 25 , 26 We also found this prevalence, that Chinese patients with NSCLC had a much higher frequency of EGFR, ERBB2, and TP53 alterations but a significantly lower frequency of KRAS, BRAF, and PDGFRA alterations than the Western patient population. The alterations of KRAS, KIT, FGFR1, and TP53 were significantly higher in males, while EGFR alterations and ALK rearrangement are more common in females. The genomic alterations profiling of Chinese NSCLC patients in this study was consistent with previous studies. 11 , 27
Non‐small cell lung cancer is the most commonly diagnosed cancer and the leading cause of cancer death. Fortunately, driver gene screening is widely use to guide molecular targeted therapy, which has shown great effectiveness in improved the prognosis. The patients of NSCLC with EGFR alteration may benefit from treatment using EGFR TKIs. In this study, 53.55% of NSCLC patients harbored EGFR alterations, and 43.11% of patients with EGFR‐L858R and exon 19 del alterations, which was consistent with another report. 11 Less common alterations, such as L861Q, S768I, and G719X, accounted for approximately 7% of patients. Although these alterations are not sensitive to the EGFR‐TKI as same as L858R and exon 19 del, they had been proved to have a benefit from afatinib therapy. 28
KRAS alterations are associated with a poor NSCLC prognosis. 13.40% of patients harbored KRAS alterations in this cohort, consistent with previous reports. 29 PIK3CA plays a pivotal role in cell metabolism and proliferation and whose alterations are commonly found in a variety of cancers. 5.4% of patients harbored PIK3CA alterations in this cohort, and most of that is located in the helical binding domain (exon 9, E545K, or E542K) or the catalytic subunit (exon 20, H1047R, or H1047L), which are considered oncogenic and targetable. 30 , 31 , 32 , 33 BRAF alteration frequency is 3.14% in this cohort, 0.9% (22 out of 3440) harbored V600E alterations, which were significantly associated with shorter disease‐free and OS rates. 34 , 35 TP53 gene was initially found to be essential for the DNA‐damage checkpoint, encodes a tumor suppressor protein (p53 protein) containing transcriptional activation, DNA binding, and oligomerization domains. 36 , 37 Most mutant p53 proteins have lost their DNA‐binding activity, leading to the loss of their growth inhibition and apoptotic properties. 38 In this cohort, TP53 (54.74%) was the most frequently altered gene and mainly on the DNA‐binding domain. Studies on primary East Asian patient populations have detected the EML4‐ALK fusion gene in 3%–7% of NSCLCs, 39 , 40 , 41 , 42 most commonly in adenocarcinomas and females. Similar to the previous studies, the incidence of ALK rearrangement was 4.27% in this cohort. Due to different breakpoints on EML4, several subtypes of the EML4‐ALK alteration have been described. 42 , 43 , 44 The most common subtypes were E6:A20 (variant 3), E13:A20 (variant 1), and E20:A20 (variant 2), accounting for 43.42%, 31.58%, and 11.18% of all EML4‐ALK cases in our cohort, respectively. EML4‐ALK fusion serves as a therapeutic target for ALK TKIs and has shown promising results when treating NSCLC patients carrying ALK rearrangement. 45 However, studies have suggested differential clinical responses to ALK inhibitors among different subtypes of EML4‐ALK. EML4‐ALK variant 3 may be a major source of ALK inhibitor resistance in the clinic. The stratification of patients with advanced ALK rearrangement‐positive NSCLC by the variant‐specific genotype should help to predict clinical responses to ALK inhibitors. 11
There is mounting evidence that the presence of co‐occurring alterations in patients with NSCLC, analyzed the 3440 NSCLC Chinese patient cohort, we also identified 51.02% of NSCLC patients with co‐occurring alterations in 14 genes. Recently, some reports demonstrated that the presence of co‐occurring alterations presented challenges for NSCLC targeted therapy. For example, among EGFR‐altered NSCLC patients, TP53 alterations reduce responsiveness to EGFR‐TKIs and worsen prognosis, 46 , 47 KRAS alteration was significantly associated with an absence of response to EGFR‐TKI, 48 and PIK3CA alteration was associated with shorter OS in some studies but do not appear to impact response rates and PFS with first‐line or second‐line EGFR‐TKI therapy. Therefore, the EGFR alteration test alone may not be sufficient to determine a patient's sensitivity to TKI therapy. Among EGFR‐altered patients, the co‐occurring frequencies of TP53, KRAS, and PIK3CA were 28.26%, 2.15%, and 3.14%, respectively, and they may not benefit equally from EGFR‐TKI compared with patients with only EGFR alteration.
Many studies have shown that patients with EGFR alterations are unable to benefit from immunotherapy and that may be associated with the development of hyper progressive disease and lead to increased toxic effects. 49 , 50 Furthermore, previous studies have indicated that EGFR‐TKI might not be as effective in NSCLC patients with co‐occurring alterations of EGFR and other driver genes. 16 , 17 Thus, effective treatment is urgently needed for these NSCLC patients. Intriguingly, we found that NSCLC patients with co‐occurring alterations of EGFR and other driver genes have higher TMB levels and longer OS than patients with a single EGFR alteration after immunotherapy, and similar results were found between patients with multiple driver gene alterations and single alteration in eight driver genes. The results demonstrate that the coexistence of other gene alterations affects the effectiveness of immunotherapy, the underlying molecular mechanism of which needs further study. Meanwhile, we discovered that the fractions of TIICs varied among the EGFR_M and EGFR_S groups as well as between the ALL_M group and the ALL_S group. Patients harboring coexisting alterations of EGFR and other driver genes have lower fractions of resting memory CD4 T cell, regulatory T cell (Tregs), activated myeloid dendritic cell, and activated mast cell, and have higher fractions of resting mast cell. Previous studies found the differences in immune cell composition in NSCLC are associated with survival. For example, the higher fraction of resting mast cells is associated with longer survival time, but a higher fraction of active dendritic cells or activated tumor Tregs is correlated with a poor prognosis. 51 , 52 Cho et al. analyzed the immune cell composition in peripheral blood mononuclear cells from nine NSCLC patients pre‐ and post‐treatment with immunotherapy and found that NK cells were enriched in the immunotherapy responder group and with higher overall activity compared with that of non‐responders. 53 In summary, the patients carried co‐occurring alterations of EGFR and other driver genes with longer survival and higher TMB score and had features of immune cell infiltration associated with better prognosis. Taken together, the patients with co‐occurring alterations of EGFR and other driver genes may benefit from immunotherapy, which may be associated with the immune microenvironment, and clinical research with a larger sample size is required to verify this result.
In conclusion, we performed NGS on a cohort of 3440 NSCLC patients to present a clear feature of driver gene alterations in Chines NSCLC patients. Besides, we identified that the co‐occurring of driver genes are associated with longer survival on immunotherapy. Importantly, patients harboring co‐occurring alterations of EGFR and other driver genes may benefit from immunotherapy, which may provide more therapeutic selections for EGFR‐mutated NSCLC patients and merit additional investigation.
ETHICS STATEMENT
This study was approved by the ethics committee of Fifth Medical Center of Chinese PLA General Hospital and conducted under the principles of the Declaration of Helsinki and the Good Clinical Practice guidelines. All enrolled patients provided written informed consent.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Tables S1‐4
ACKNOWLEDGMENTS
The authors sincerely thank Lifehealthcare Clinical Laboratories, Hangzhou, China for providing help in genetic sequencing and results interpretation.
Sun S, Du W, Sun Q, et al. Driver gene alterations profiling of Chinese non‐small cell lung cancer and the effects of co‐occurring alterations on immunotherapy. Cancer Med. 2021;10:7360–7372. 10.1002/cam4.4178
Shengjie Sun and Wenjuan Du contributed equally to this work and share first authorship.
DATA AVAILABILITY STATEMENT
The datasets analyzed for this study can be found in the cBioPortal [https://www.cbioportal.org]. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non‐small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non‐small‐cell lung cancer. Lancet. 2016;388:1012‐1024. [DOI] [PubMed] [Google Scholar]
- 4. Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non‐small‐cell lung cancer annual update 2014. J Thorac Oncol. 2015;10:S1‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497‐1500. [DOI] [PubMed] [Google Scholar]
- 6. Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046‐2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910‐4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850‐859. [DOI] [PubMed] [Google Scholar]
- 9. Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early‐stage resected non‐small‐cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173‐2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378‐381. [DOI] [PubMed] [Google Scholar]
- 11. Wen S, Dai L, Wang L, et al. Genomic signature of driver genes identified by target next‐generation sequencing in Chinese non‐small cell lung cancer. Oncologist. 2019;24:e1070‐e1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill A, Gupta R, Zhao D, Vankina R, Amanam I, Salgia R. Targeted therapies in non‐small‐cell lung cancer. Cancer Treat Res. 2019;178:3‐43. [DOI] [PubMed] [Google Scholar]
- 14. Berghoff AS, Bellosillo B, Caux C, et al. Immune checkpoint inhibitor treatment in patients with oncogene‐ addicted non‐small cell lung cancer (NSCLC): summary of a multidisciplinary round‐table discussion. ESMO Open. 2019;4:e000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30:1321‐1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skoulidis F, Heymach JV. Co‐occurring genomic alterations in non‐small‐cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495‐509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martín Martorell P, Huerta M, Compañ Quilis A, et al. Coexistence of EGFR, KRAS, BRAF, and PIK3CA mutations and ALK rearrangement in a comprehensive cohort of 326 consecutive Spanish nonsquamous NSCLC patients. Clin Lung Cancer. 2017;18:e395‐e402. [DOI] [PubMed] [Google Scholar]
- 18. Li H, Durbin R. Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics. 2010;26:589‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next‐generation DNA sequencing data. Nat Genet. 2011;43:491‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S, Liu M, Huang T, Liao W, Xu M, Gu J. GeneFuse: detection and visualization of target gene fusions from DNA sequencing data. Int J Biol Sci. 2018;14:843‐848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samstein RM, Lee C‐H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gou LY, Wu YL. Prevalence of driver mutations in non‐small‐cell lung cancers in the People's Republic of China. Lung Cancer (Auckl). 2014;5:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small‐cell lung cancer: meta‐analyses by ethnicity and histology (mutMap). Ann Oncol. 2013;24:2371‐2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol. 2013;8:52‐61. [DOI] [PubMed] [Google Scholar]
- 26. Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking‐related KRAS‐mutant cancers. Clin Cancer Res. 2012;18:6169‐6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang H, Zhang J, Zhang L, et al. Comprehensive analysis of genomic alterations detected by next‐generation sequencing‐based tissue and circulating tumor DNA assays in Chinese patients with non‐small cell lung cancer. Oncol Lett. 2019;18:4762‐4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J‐H, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non‐small‐cell lung cancer harbouring uncommon EGFR mutations: a combined post‐hoc analysis of LUX‐Lung 2, LUX‐Lung 3, and LUX‐Lung 6. Lancet Oncol. 2015;16:830‐838. [DOI] [PubMed] [Google Scholar]
- 29. Li S, Li L, Zhu Y, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110:2812‐2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheffler M, Bos M, Gardizi M, et al. PIK3CA mutations in non‐small cell lung cancer (NSCLC): genetic heterogeneity, prognostic impact and incidence of prior malignancies. Oncotarget. 2015;6:1315‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351‐1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3‐kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802‐807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oxnard GR, Binder A, Jänne PA. New targetable oncogenes in non‐small‐cell lung cancer. J Clin Oncol. 2013;31:1097‐1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non‐small‐cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574‐3579. [DOI] [PubMed] [Google Scholar]
- 35. Warth A, Penzel R, Lindenmaier H, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J. 2014;43:872‐883. [DOI] [PubMed] [Google Scholar]
- 36. Yin Y, Stephen CW, Luciani MG, Fåhraeus R. p53 Stability and activity is regulated by Mdm2‐mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462‐467. [DOI] [PubMed] [Google Scholar]
- 37. Polse KA. S. Hickson‐Curran, et al. Patient attitudes and behavior regarding hygiene and replacement of soft contact lenses and storage cases [Contact Lens Anterior Eye (2011), doi: 10.1016/j.clae.2010.12.005]. Cont Lens Anterior Eye. 2012;35:92‐83; author reply 4‐5. [DOI] [PubMed] [Google Scholar]
- 38. Soussi T, Béroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233‐240. [DOI] [PubMed] [Google Scholar]
- 39. Suh JH, Johnson A, Albacker L, et al. Comprehensive genomic profiling facilitates implementation of the National Comprehensive Cancer Network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism‐driven clinical trials. Oncologist. 2016;21:684‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paik JH, Choi C‐M, Kim H, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK‐rearranged adenocarcinoma. Lung Cancer. 2012;76:403‐409. [DOI] [PubMed] [Google Scholar]
- 41. Horn L, Pao W. EML4‐ALK: honing in on a new target in non‐small‐cell lung cancer. J Clin Oncol. 2009;27:4232‐4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non‐small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011;6:466‐472. [DOI] [PubMed] [Google Scholar]
- 43. Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4‐ALK transforming gene in non‐small cell lung cancer. Cancer Res. 2008;68:4971‐4976. [DOI] [PubMed] [Google Scholar]
- 44. Li T, Maus MKH, Desai SJ, et al. Large‐scale screening and molecular characterization of EML4‐ALK fusion variants in archival non‐small‐cell lung cancer tumor specimens using quantitative reverse transcription polymerase chain reaction assays. J Thorac Oncol. 2014;9:18‐25. [DOI] [PubMed] [Google Scholar]
- 45. Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non‐small‐cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12:1004‐1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Canale M, Petracci E, Delmonte A, et al. Impact of TP53 mutations on outcome in EGFR‐mutated patients treated with first‐line tyrosine kinase inhibitors. Clin Cancer Res. 2017;23:2195‐2202. [DOI] [PubMed] [Google Scholar]
- 47. Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co‐mutation in patients with EGFR‐mutated non‐small cell lung cancer (NSCLC). Lung Cancer. 2017;111:23‐29. [DOI] [PubMed] [Google Scholar]
- 48. Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soo RA, Lim SM, Syn NL, et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non‐small cell lung cancer: Current controversies and future directions. Lung Cancer. 2018;115:12‐20. [DOI] [PubMed] [Google Scholar]
- 50. Li X, Lian Z, Wang S, Xing L, Yu J. Interactions between EGFR and PD‐1/PD‐L1 pathway: implications for treatment of NSCLC. Cancer Lett. 2018;418:1‐9. [DOI] [PubMed] [Google Scholar]
- 51. Tamminga M, Hiltermann TJN, Schuuring E, Timens W, Fehrmann RS, Groen HJ. Immune microenvironment composition in non‐small cell lung cancer and its association with survival. Clin Transl Immunol. 2020;9:e1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non‐small‐cell lung cancer by single‐cell sequencing. Nat Med. 2018;24:978‐985. [DOI] [PubMed] [Google Scholar]
- 53. Cho Y‐H, Choi MG, Kim DH, et al. Natural killer cells as a potential biomarker for predicting immunotherapy efficacy in patients with non‐small cell lung cancer. Target Oncol. 2020;15:241‐247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Tables S1‐4
Data Availability Statement
The datasets analyzed for this study can be found in the cBioPortal [https://www.cbioportal.org]. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


