Abstract
Bacterial microcompartments (BMCs) are self-assembling prokaryotic organelles which encapsulate enzymes within a polyhedral protein shell. The shells are comprised of only two structural modules, distinct domains that form pentagonal and hexagonal building blocks, which occupy the vertices and facets, respectively. As all BMC loci encode at least one hexamer- and one pentamer-forming protein, the evolutionary history of BMCs can be interrogated from the perspective of their shells. Here, we discuss how structures of intact shells and detailed phylogenies of their building blocks from a recent phylogenomic survey distinguish families of these domains and reveal clade-specific structural features. These features suggest distinct functional roles that recur across diverse BMCs. For example, it is clear that carboxysomes independently arose twice from metabolosomes, yet the principles of shell the assembly are remarkably conserved.
Keywords: bacterial microcompartment, carboxysome, metabolosome
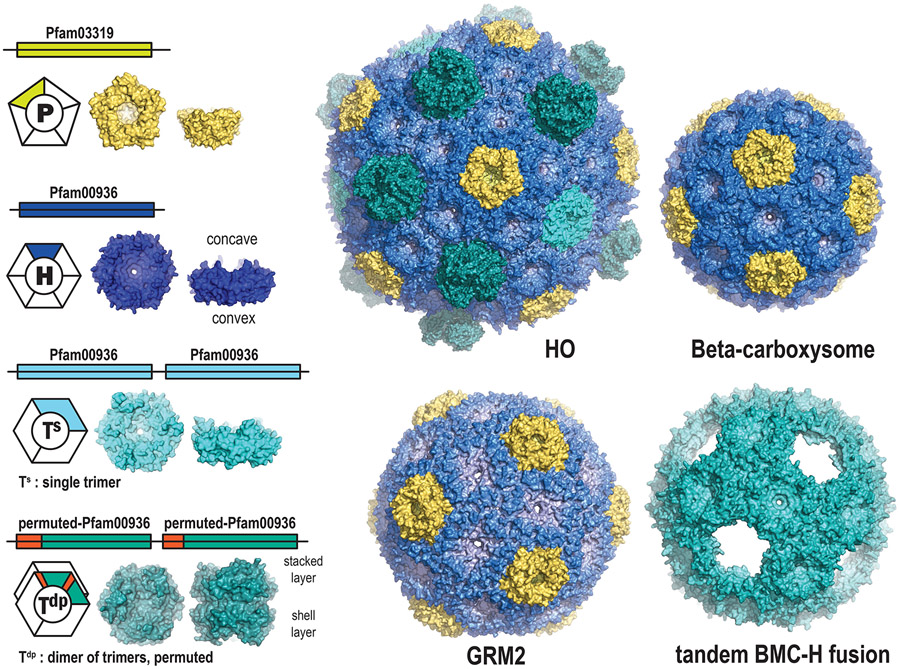
Compartmentalization is a modular organizing principle fundamental to life. The organelles, cells, tissues and organs of eukaryotes are familiar levels of biological subdivision which demonstrate how compartmentalization confers adaptive and complex functionality. It is only relatively recently that it has become evident that bacteria too have intracellular organization. Metabolic diversity underpins microbial diversity and, as in Eukaryotic organisms, compartmentalization in bacterial organelles expands metabolic flexibility by segregating reactions that are incompatible with cytosolic metabolism. These primitive prokaryotic organelles are known as Bacterial Microcompartments (BMCs). However, unlike eukaryotic organelles, the bounding membrane of BMCs is a protein shell. The BMC shell is composed of two basic types of protein – the hexagon-forming Pfam00936 domain and pentagon-forming Pfam03319 domain – that self-assemble into the facets and vertices of polyhedral, often icosahedral shells (Fig. 1). The potential to form BMCs is found across the Bacterial Kingdom where they are involved in both anabolic and catabolic reactions. The homology among shell proteins is readily recognized, which is remarkable in light of the extensive functional diversity of the encapsulated enzymes.
Figure 1. Overview of BMC shell proteins and structures of empty shells.
Shell structures’ PDB codes: HO: model based on 5V74, model beta-carboxysome shell: 6OWG, GRM2 shell: 6QN1, tandem BMC-H fusion shell: 6NER.
BMCs were first discovered as polyhedral bodies in cyanobacteria [1] and later named carboxysomes for their role in fixing CO2 [2]. Now, comprehensive phylogenomic surveys of BMC loci reveal a widespread taxonomic distribution, across 45 different bacterial phyla [3,4]. Moreover, classification of BMC loci by clustering their domain profiles distinguishes at least 65 different types or subtypes of catabolic BMC loci (metabolosomes), including a large number of unknown function [3,4]. In contrast, only two types of carboxysomes are found, differentiated by the type of carbonic anhydrase and RuBisCO encapsulated. Crystal structures of many individual shell proteins from carboxysomes and metabolosomes [5] have shown that the cyclic symmetry axis of the hexagonal assembly is typically circumscribed by conserved residues that dictate the size and charge of the pore opening. These pores serve as the conduits for substrates and products of the encapsulated reactions.
In the last decade the study of BMCs has been tremendously advanced by the discovery that empty shells could be formed in the absence of cargo [6-11]. These tend to be smaller than their native (cargo replete) counterparts. Structural studies of empty shells from several functionally distinct BMCs [9,12,13] show that the BMC-H hexamers make up the bulk of the shell (Fig. 1). Interactions of the hexamers with each other and with the pentamers are highly conserved across types, with the sidedness/orientation of the faces of the shell proteins conserved [9,12,13]. In one structure, the BMC-T proteins were also visualized and were found to have slightly different angles of interaction with BMC-H proteins [12,14], indicating that they fulfill a special structural and/or functional role.
Collectively these structures constitute an architectural map of the universal features of BMC shell assembly. Nevertheless, it remains challenging to predict the assembly properties for many of the newly-identified BMC types, which exhibit a large combinatorial diversity of shell protein types encoded by their loci, including many paralogs and type-specific variants.
In the most recent phylogenomic BMC locus survey [4], in which phylogenies were constructed for approximately 9500 BMC-H, 4300 BMC-T, and 4900 BMC-P protein sequences, unexpected relationships among shell proteins from functionally distinct BMC types were found, indicating that a classification system based on sequence features could be devised. By assigning descriptively-named RGB hexcodes to identifiable tree clades, using similar colors for closely-related clades, a function-agnostic rankless classification system was developed for discussing the evolutionary history of BMC shell proteins, complementary to and independent of prior assumptions about function. Here we review these data and situate our findings in the context of conserved architectural principles and the evolution and functional diversity of BMCs.
BMC-P Proteins
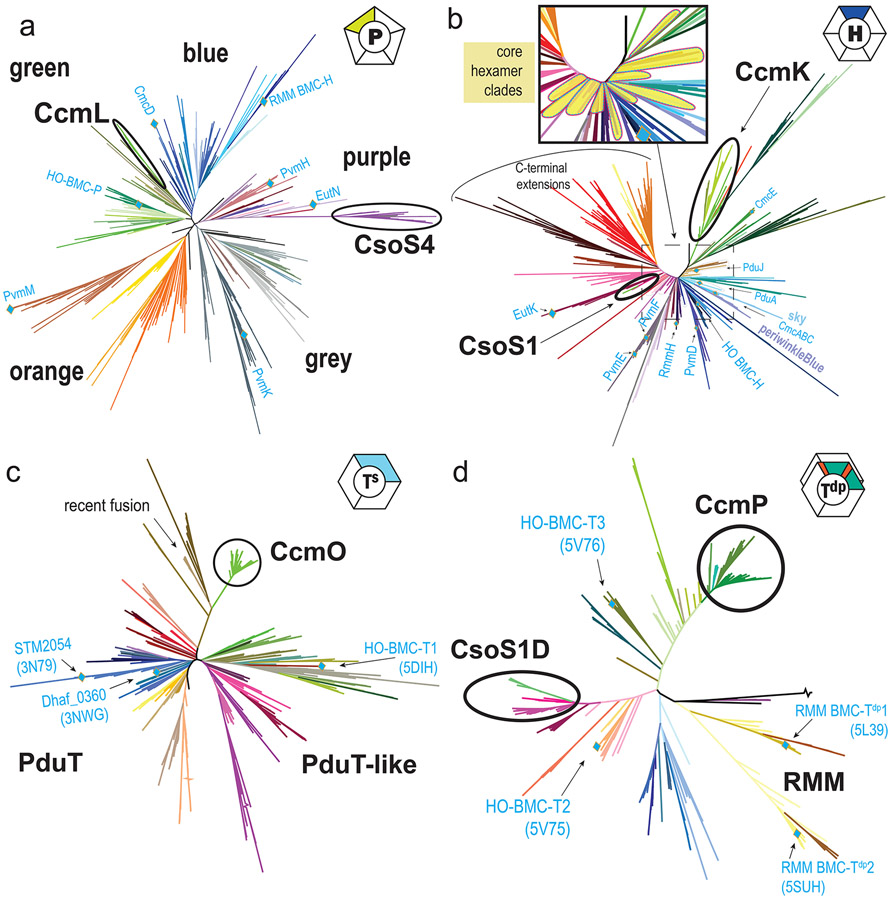
Pentamers are presumed to be confined to vertices of fully-assembled shells, and have been shown to be essential for the function of the shell as a diffusive barrier [15]. They generally exhibit strong sequence similarity, including several universally-conserved amino acid positions which correspond to the hexamer-pentamer interface (Fig. 4ab). They resolve into four or five distinct phylogenetic groups: blue/green-type, purple-type, orange-type, and grey-type (Fig. 2a). The majority of BMC loci encode a single BMC-P from the blue/green or purple-type clades, representing a “standard” pentamer. Interestingly, many loci (particularly SPUs, FRAGs, ACI, HO, and PVM, see Fig. 2,3 legends for type nomenclature) encode BMC-P “triplets” containing a PvmM-like orange-type and a PvmK-like grey-type together with a standard pentamer [4]. Given that BMC shells are assumed to require only 12 pentamers, and that the pore formed at the cyclic symmetry axis is small (~4 Å in diameter), they are assumed not to play a significant role in metabolite conductance. However, a recent study of the protein stoichiometry of beta-carboxysomes showed varying occupation of the vertices by CcmL, depending on environmental conditions [16]. Shells without BMC-P proteins at the vertices, referred to as “wiffle ball” shells, have also been obtained in synthetic model systems (Fig. 1) [17,18]. The diversity of roles for BMC-P proteins, and their dynamics, warrants further study.
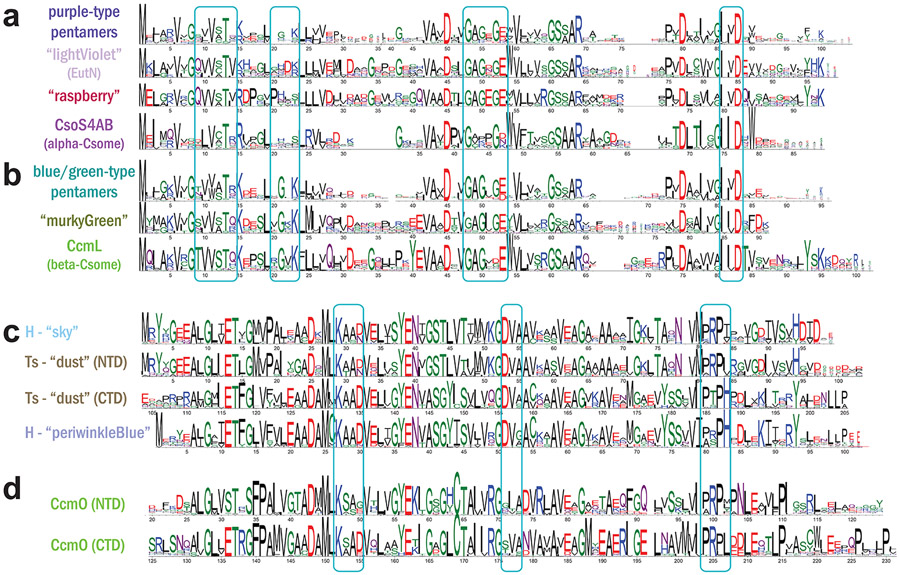
Figure 4. Clade-specific sequence features of BMC shell proteins.
Sequence conservation logos are shown for sequences of selected BMC-P (a-b) and BMC-H and BMC-T clades (c-d). Boxes indicate regions of residues involved in lateral inter-protein interactions based on assembled shell structures. Logos were generated using WebLogo software upon multiple sequence alignment with MAFFT.
Figure 2. Unrooted phylogenetic trees of BMC shell proteins.
Trees were generated with RAxML for non-redundant sets of shell protein sequences. (a) BMC-P (b) BMC-H (c), BMC-Ts proteins, which are simple trimers that do not dimerize and (d) BMC-Tdp proteins, this family dimerizes across the concave face. Position of characterized proteins are labeled with their gene or model system name. Protein structure PDB codes are indicated in parentheses. PDU: propanediol utilization BMC; EUT: ethanolamine utilization BMC; HO: Haliangium ochraceum BMC; RMM: Rhodococcus and Mycobacterium microcompartment (functional type nomenclature as used in [3,4]).
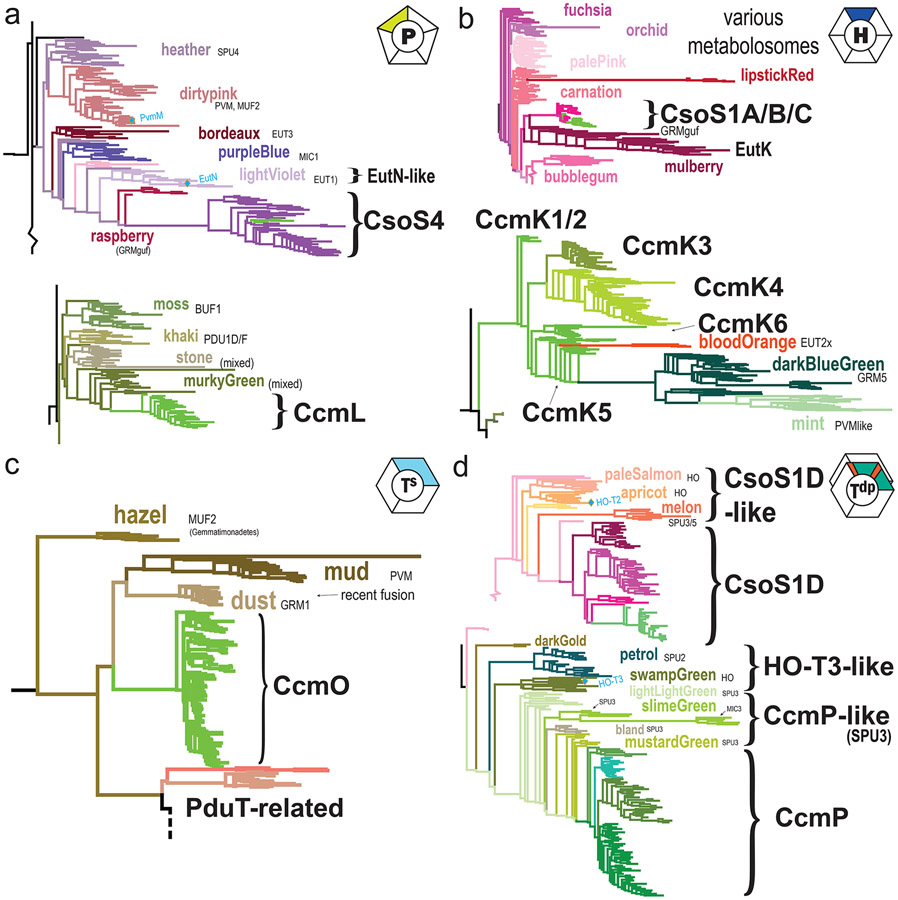
Figure 3. Relationships of carboxysome shell proteins to nearest metabolosome relatives.
Subtrees are shown for carboxysome and related clades of (a) BMC-P, (b) BMC-H, (c) BMC-Ts and (d) BMC-Tdp phylogenies from Figure 2. Clades with a predominant BMC type are indicated; BMC type abbreviations: SPU: sugar phosphate BMC, PVM: Planctomycete and Verrumicrobia BMC; MUF: Metabolosome of unknown function, MIC: Metabolosome with incomplete core, BUF: BMC of unknown function, GRM: Glycyl radical BMC (functional type nomenclature as used in [3,4]).
Uniquely among all BMC loci, alpha-carboxysomes encode two very similar BMC-P proteins, CsoS4A/CsoS4B [19], from a single purple-type clade that is strongly divergent from other BMC-P clades (Fig. 2a,4a). With the alpha-cyanobacterial CsoS4 sequences virtually indistinguishable from their orthologs in chemoautotrophs, the whole clade maps closest to other Proteobacteria-specific metabolosome purple-type clades, nearest to raspberry GRMguf-type pentamers from Deltaproteobacteria as well as the well-studied EutN/lightViolet clade from Gammaproteobacteria (Fig. 3a), supporting a Proteobacterial origin for the alpha-carboxysome.
In contrast, beta-carboxysome loci encode a single pentamer, CcmL, from a completely different section of the tree (Fig. 2a). CcmL is most closely related to green-type metabolosome pentamers from Firmicutes (Fig. 3a,4b). The mixed nature of the nearest clade (murkyGreen), representing a variety of BMC types (PDU1E, PDU1C, EUT2A, and GRM3A), suggests that beta-carboxysomes co-opted a metabolosome pentamer. Clade-specific features may possibly represent unique interaction surfaces for BMC-type-specific binding partners, or particular cytoplasmic features such as cytoskeletal elements.
BMC-H Proteins
The BMC-H hexamer comprises the main building block of the shell. They have long been assumed to form only homohexamers, but recent evidence suggests that heterohexamer assemblies also can be formed [20,21]. Most BMC shells are composed from multiple BMC-H paralogs; these genes may be encoded distal from the main organelle locus in satellite loci [3]. These are presumed to be differentially regulated from the main locus, and may provide additional flexibility in the permeability properties of the shell.
The comprehensive phylogenomic survey [4] revealed a core set of tightly-conserved clades, centered around the blue cluster, which appear to contain the fundamental, and presumably numerically dominant, building blocks of the metabolosome shell (Fig. 2b, inset). This is consistent with the most abundant BMC-H proteins from biochemical studies of intact metaboloomes such as PduA/J [22,23] and EutM [24]. Other clades, which typically constitute additional paralogs, seem to be more specialized and show greater sequence divergence or sequence extensions (Fig. 2b). Alpha-carboxysome hexamers are all derived from a single evolutionary type, despite many alpha-carboxysomes utilizing up to three (CsoS1A,B,C) paralogs that are nearly identical within a given locus [25](Fig. 3b). They constitute a very simple shell consisting almost exclusively of CsoS1 paralogs [26]. This compact clade again exhibits a long internal stem which separates CsoS1s from their closest metabolosome homologs (mulberry, EutK-like), implying a large sequence divergence and strong conservation upon the establishment of alpha-carboxysome hexamers.
Beta-carboxysomes are also composed of multiple BMC-H paralogs (CcmKs), clustering at the periphery of the hexamer tree, however very far from the CsoS1 clade (Fig. 2b). CcmK1 and CcmK2 paralogs map basally within the CcmK-like subtree and cannot be resolved phylogenetically due to extremely strong sequence constraint (Fig. 3b), in accordance with their essentiality [27-29] and stoichiometric prevalence in intact, natively-purified carboxysomes [16]. CcmK3 and CcmK4, which are encoded together in a satellite locus, appear as sister clades diverging together from the middle of the CcmK subtree (Fig. 3b), were recently recent demonstrated to form heterohexamers capable of dimerizing across their concave faces to form dodecamer in a pH-dependent manner [20], potentially serving as a metabolite channel subject to regulation. With additional genome sequence data now available for ecophysiologically diverse cyanobacteria, additional CcmK paralogs have been discovered [30]. For example, the newly identified CcmK5, from taxonomically-diverse genomes that are distinctive in lacking CcmK3 and CcmK4, structurally resembles CcmK1/2 and CcmK4; while CcmK6, found predominantly in heterocyst-forming cyanobacteria, resembles CcmK3, with which it often co-occurs. These observations suggest the potential for additional plasticity in shell permeability properties. While CcmK paralogs likely arose by a series of gene duplications after the establishment of beta-carboxysomes, three groups of CcmK5-like hexamers are found in heterotrophs (Fig. 3b), including two large sister clades from GRM5 and PVMlike metabolosomes as well as a smaller clade of EUT2x-type metabolosomes (a EUT2 related BMC that lacks the genes for ethanolamine lyase) from the Firmicute genus Sporosarcina, suggesting horizontal gene transfer from beta-cyanobacteria. In contrast to the ocean-dwelling alpha-cyanobacteria, beta-cyanobacteria live in diverse habitats subject to environmental dynamics (e.g., nutrient availability, desiccation, symbioses); suggesting that the multiplicity of CcmK paralogs arose as an adaptation for altering carboxysome permeability in response to fluctuating environments [20] experienced by ecophysiologically diverse cyanobacteria.
BMC-T proteins
BMC-T proteins are a tandem fusion of two copies of the Pfam00936 domain (Fig. 1), having originated from gene duplication and/or fusion events. Crystal structures BMC-T trimers reveal pseudohexameric assemblies that are similar in size and shape as BMC-H hexamers. The three-fold instead of six-fold symmetry allows for twice as many distinct conserved residues converging at the pores, and trimers also have two distinct edges that can interact with (different) surrounding shell proteins in a facet. Each domain in a BMC-T protein can diverge separately and at differing rates, resulting in extensive diversification. One clade from GRM1A/B loci, “Ts_dust” (Fig. 3c) appears to be a recent fusion of two adjacently-encoded BMC-H paralogs (Fig. 4c) [31]. This provides inspiration for the design of shell building blocks that are poised for adaptive evolution for new functions in bioengineering, such as the synthetic BMC-T protein that self-assembles into 20 nm wiffle ball shells even without a BMC-P [18] (Fig. 1). In general, BMC-T proteins to have specialized functions such binding an Fe-S cluster at the pore, as in PduT [32,33](Fig. 2c). Some extreme variants such as PduB, EutL and EtuB exhibit circular sequence permutations, which reorders the secondary structure elements within each domain, and have off-axis pores that possibly regulate opening of the central pore [34-37]. BMC-Ts proteins are widespread, found in 52 of the 68 presently-identified BMC types, although they are conspicuously absent from several BMC types such as GRM2, RMM, PVMlike, EUT2x, BUF1B, MIC1/MIC2, and alpha-carboxysomes [4].
All beta-carboxysome loci encode an enigmatic BMC-Ts, CcmO, which is relatively poorly characterized, even lacking experimental demonstration that it forms trimers/pseudohexamerss. CcmO maps at the periphery of the tree in a tight cluster, indicative of evolutionary constraint, with a long internal stem representing significant sequence divergence, most closely related to three other outlier clades from metabolosomes (Fig. 3c), potentially a result of longbranch attaction. In a homology model, CcmO has a standard CcmK-like (KIGS) motif that would surround the pore in a trimer/pseudohexamer, and typical edge-interaction interfaces in both domains (Fig. 4d). CcmO however is essential for formation of functional carboxsyomes [27,38] so it clearly fulfills an important structural function in beta-carboxysomes. There is a second class of trimers that obligately dimerize across their concave surfaces (BMC-Tdp Proteins, Fig. 1) The pores in these trimer are relatively large (~14 Å) and are gated by absolutely conserved surrounding residues [39-42]. For the beta-carboxysomal protein CcmP there is structural evidence of metabolites bound in a pocket on the inside cavity that was modeled as 3-PGA [39] or ADP [41] and its presence was correlated with pore opening [41]. With more functionally diverse BMC loci are identified, genes encoding BMC-Tdp proteins are found to be widespread, occurring in 21 of the 68 locus types, including HO, RMM, EUT3, and SPU2/3/6, as well as both the alpha- and beta-carboxysomes (Fig. 2d).
Four families of BMC-Tdp can be distinguished phylogenetically (Fig. 2d), with the alpha-carboxysome type, CsoS1D, again very distant from the beta-carboxysome type, CcmP. Unlike the BMC-H and BMC-P sequences of alpha-carboxysomes, the CsoS1D orthologs from cyanobacteria are readily distinguished from their chemoautotroph counterparts, which comprise three distinct clades (Fig. 3d). While each contains a variety of taxa, only Gammaproteobacteria can be found in all three, suggesting this taxonomic class may have been associated with the origins of the alpha-carboxysome. This supports the idea that BMC-Tdp proteins contribute a more specialized function compared to standard hexamers – perhaps in regulation and/or metabolite transport across the shell, or in the context of their different redox dynamics ( chemoautotrophs do not generate O2). Nevertheless, the entire CsoS1D clade again diverges strongly from metabolosome orthologs, with the long-stemmed melon clade as their closest relatives, encoded mostly by SPU-type loci [3,4] from Proteobacteria.
In contrast, the CcmP clade from beta-carboxysomes is adjacent to several clades from SPU3-type metabolosomes, almost indistinguishably, with the closest derived from Chloroflexi (Fig. 3d). The association of SPU metabolosomes with both carboxysome clades is interesting because the class of substrates that they catabolize, sugar phosphate, is also the immediate product of carboxysomes, 3-phosphoglycerate; thus perhaps these BMC-Tdp proteins conduct this kind of metabolite. The very close connection with these CcmP-like tandem domains is unclear, however, especially as the taxonomy and BMC-type of the nearest metabolosome ancestor is completely different from that of CcmL, CcmK, and CcmO. Because CcmP is always encoded in a satellite locus, it is likely that the protein was horizontally acquired, although such a transfer would have had to predate the establishment of the modern cyanobacteria, as it is rarely missing from any genomes and is found even in the early-branching genera such as Gloeobacter. Nevertheless, again the BMC-Tdp sequences from beta-carboxysomes are more closely affiliated with their metabolosome ancestors and map to a very different region of the tree than the alpha-cyanobacterial BMC-Tdp homologues.
Conclusions
The structure and selective permeability of BMC shells constitutes the interface between the encapsulated reactions and the surrounding metabolism and is intrinsic to BMC function. Bioinformatic analysis suggests that the BMC shell is an ancient innovation and the modularity of the components used in its construction enabled it to evolve for diverse functions, and sometimes more than once for the same function. Multiple lines of evidence suggest that alpha- and beta-type carboxysomes arose independently. Likewise, the shell protein building blocks of most metabolosomes are drawn from multiple lineages that arose from BMC-H, BMC-T and BMC-P evolution, indicating frequent exchange of the modular components. Bioinformatic analyses combined with structural characterization of model shell systems are pointing to some universal principles of shell architectures, such as the role of at least one identifiable paralog from the central Blue BMC-H clade as the standard hexamer, that assembles with other hexamers and trimers oriented with the concave faces outward. The modular assembly from a catalog of functionally distinct but structurally interchangeable building blocks that assemble into a predictable orientation is fundamental to the native functions of shells and their engineering into new contexts.
HIGHLIGHTS:
Phylogenomic analysis of shell proteins uncovers supra-functional relationships
Alpha-carboxysome shell proteins arose independently from beta-carboxysome shells
Beta-carboxysome shell proteins retain similarities with metabolosome shells
Alpha-carboxysome shells likely diverged from Proteobacterial metabolosomes
Interactions among building blocks in intact shells are conserved across all types
Acknowledgements
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-01 and by the U.S. Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Drews G, Niklowitz W: Beiträge zur Cytologie der Blaualgen. II. Zentroplasma und granuläre Einschlüsse von Phormidium uncinatum. Archiv für Mikrobiologie 1956, 24:147–162. [PubMed] [Google Scholar]
- 2.Shively JM, Ball F, Brown DH, Saunders RE: Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 1973, 182:584–586. [DOI] [PubMed] [Google Scholar]
- 3.Axen SD, Erbilgin O, Kerfeld CA: A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 2014, 10:e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutter M, Melnicki MR, Schulz F, Woyke T, Kerfeld CA: A Catalog of the Diversity and Ubiquity of Metabolic Organelles in Bacteria. bioRxiv 2021:2021.2001.2025.427685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa JM, Bair K, Holton T, Bobik TA, Yeates TO: MCPdb: The Bacterial Microcompartment Database. bioRxiv 2021:2021.2001.2009.426059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC: Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One 2008, 3:e3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA: Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. Journal of Molecular Biology 2014, 426:2217–2228. [DOI] [PubMed] [Google Scholar]
- 8.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ: Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 2010, 38:305–315. [DOI] [PubMed] [Google Scholar]
- 9. Kalnins G, Cesle EE, Jansons J, Liepins J, Filimonenko A, Tars K: Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nat Commun 2020, 11:388. The authors present a Cryo-EM structure of a GRM2 metabolosome shell and investigate the structures formed by co-expression of different combinations of shell proteins and cargo enzymes.
- 10.Cai F, Bernstein SL, Wilson SC, Kerfeld CA: Production and Characterization of Synthetic Carboxysome Shells with Incorporated Luminal Proteins. Plant Physiol 2016, 170:1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer MJ, Juodeikis R, Brown IR, Frank S, Palmer DJ, Deery E, Beal DM, Xue WF, Warren MJ: Effect of bio-engineering on size, shape, composition and rigidity of bacterial microcompartments. Sci Rep 2016, 6:36899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutter M, Greber B, Aussignargues C, Kerfeld CA: Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 2017, 356:1293–1297. The crystal structure described here represents the first atomic resolution structure of a complete BMC shell produced from co-expressed BMC-H, BMC-P, BMC-Ts and BMC-Tdp subunits. It reveals all the native interactions between those and shows the concave outwards facing orientation of the BMC-H and BMC-Ts subunits.
- 13. Sutter M, Laughlin TG, Sloan NB, Serwas D, Davies KM, Kerfeld CA: Structure of a Synthetic beta-Carboxysome Shell. Plant Physiol 2019, 181:1050–1058. This manuscript describes the Cryo-EM structure of a beta-carboxysome shell obtained from recombinant expression and reveals that orientation of and interfaces among the shell proteins are conserved between carboxysome and metabolosome shells.
- 14.Greber BJ, Sutter M, Kerfeld CA: The Plasticity of Molecular Interactions Governs Bacterial Microcompartment Shell Assembly. Structure 2019, 27:749–763 e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S: The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 2009, 4:e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun YQ, Wollman AJM, Huang F, Leake MC, Liu LN: Single-Organelle Quantification Reveals Stoichiometric and Structural Variability of Carboxysomes Dependent on the Environment. Plant Cell 2019, 31:1648–1664. This study analyzes stoichiometry of individual components of the beta-carboxysome and finds an unexpected correlation of BMC-P abundance with environmental conditions.
- 17.Hagen A, Sutter M, Sloan N, Kerfeld CA: Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nature Communications 2018, 9:2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutter M, McGuire S, Ferlez B, Kerfeld CA: Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies. ACS Synth Biol 2019, 8:668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao YY, Jiang YL, Chen Y, Zhou CZ, Li Q: Crystal structure of pentameric shell protein CsoS4B of Halothiobacillus neapolitanus alpha-carboxysome. Biochem Biophys Res Commun 2019, 515:510–515. [DOI] [PubMed] [Google Scholar]
- 20. Sommer M, Sutter M, Gupta S, Kirst H, Turmo A, Lechno-Yossef S, Burton RL, Saechao C, Sloan NB, Cheng X, et al. : Heterohexamers Formed by CcmK3 and CcmK4 Increase the Complexity of Beta Carboxysome Shells. Plant Physiology 2019, 179:156. This work demonstrates for the first time evidence for heterohexamer formation that was long missed because most shell protein studies had been focused on single purified proteins.
- 21.Garcia-Alles LF, Root K, Maveyraud L, Aubry N, Lesniewska E, Mourey L, Zenobi R, Truan G: Occurrence and stability of hetero-hexamer associations formed by beta-carboxysome CcmK shell components. PLoS One 2019, 14:e0223877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havemann GD, Bobik TA: Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 2003, 185:5086–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Simpson DM, Wenner N, Brownridge P, Harman VM, Hinton JCD, Beynon RJ, Liu LN: Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nat Commun 2020, 11:1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slininger Lee MF, Jakobson CM, Tullman-Ercek D: Evidence for Improved Encapsulated Pathway Behavior in a Bacterial Microcompartment through Shell Protein Engineering. ACS Synthetic Biology 2017. This study exemplifies the modularity of the BMC shell by showing that the standard (blue clade) BMC-H from a PDU BMC can be exchanged with the EUT paralog structurally, but concomitantly alters shell permeability.
- 25.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM: Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol 2001, 67:5351–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S: Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J Bacteriol 2012, 194:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA: Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 2013, 155:1131–1140. [DOI] [PubMed] [Google Scholar]
- 28.Price GD, Howitt SM, Harrison K, Badger MR: Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol 1993, 175:2871–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faulkner M, Szabo I, Weetman SL, Sicard F, Huber RG, Bond PJ, Rosta E, Liu LN: Molecular simulations unravel the molecular principles that mediate selective permeability of carboxysome shell protein. Scientific Reports 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sommer M, Cai F, Melnicki M, Kerfeld CA: beta-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J Exp Bot 2017, 68:3841–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferlez B, Sutter M, Kerfeld CA: Glycyl Radical Enzyme-Associated Microcompartments: Redox-Replete Bacterial Organelles. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SE, Warren MJ: Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One 2010, 5:e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang A, Warren MJ, Pickersgill RW: Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallographica Section D-Biological Crystallography 2011, 67:91–96. [DOI] [PubMed] [Google Scholar]
- 34.Sagermann M, Ohtaki A, Nikolakakis K: Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl. Acad. Sci. U.S.A 2009, 106:8883–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang A, Liang M, Prentice MB, Pickersgill RW: Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 2012, 68:1642–1652. [DOI] [PubMed] [Google Scholar]
- 36.Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW: Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. The Biochemical journal 2009, 423:199–207. [DOI] [PubMed] [Google Scholar]
- 37.Thompson MC, Cascio D, Leibly DJ, Yeates TO: An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Sci 2015, 24:956–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marco E, Martinez I, Ronen-Tarazi M, Orus MI, Kaplan A: Inactivation of ccmO in Synechococcus sp. Strain PCC 7942 Results in a Mutant Requiring High Levels of CO(2). Appl Environ Microbiol 1994, 60:1018–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA: The Structure of CcmP, a Tandem Bacterial Microcompartment Domain Protein from the beta-Carboxysome, Forms a Subcompartment Within a Microcompartment. Journal of Biological Chemistry 2013, 288:16055–16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA: Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. Journal of Molecular Biology 2009, 392:319–333. [DOI] [PubMed] [Google Scholar]
- 41.Larsson AM, Hasse D, Valegard K, Andersson I: Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. J Exp Bot 2017, 68:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallette E, Kimber MS: A Complete Structural Inventory of the Mycobacterial Microcompartment Shell Proteins Constrains Models of Global Architecture and Transport. J Biol Chem 2017, 292:1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]