Abstract
The simple sequence repeats (SSRs) of plant chloroplasts show considerable genetic variation and have been widely used in species identification and phylogenetic relationship determination. Whether chloroplast genome SSRs can be used to classify Cyatheaceae species has not yet been studied. Therefore, the chloroplast genomes of eight Cyatheaceae species were sequenced, and their SSR characteristics were compared and statistically analyzed. The results showed that the chloroplast genome structure was highly conserved (genome size: 154,046–166,151 bp), and the gene content (117 genes) and gene order were highly consistent. The distribution characteristics of SSRs (number, relative abundance, relative density, GC content) showed taxon specificity. The primary results were the total numbers of SSRs and mononucleotides: Gymnosphaera (61–67 and 40–47, respectively), Alsophila (121–122 and 95–96), and Sphaeropteris (102–103 and 77–80). Statistical and clustering analyses of SSR characteristics showed that their distribution was consistent with the recent classification of Cyatheaceae, which divided the eight Cyatheaceae species into three genera. This study indicates that the distribution characteristics of Cyatheaceae chloroplast SSRs can provide useful phylogenic information at the genus level.
Keywords: chloroplast SSR, Cyatheaceae, genus, phylogeny
Whether chloroplast genome SSRs can be used to classify Cyatheaceae species has not yet been studied. Therefore, the chloroplast genomes of eight Cyatheaceae species were sequenced by our team, and their SSR characteristics were compared and statistically analyzed. Statistical and clustering analyses of SSR characteristics showed that their distribution was consistent with the recent classification of Cyatheaceae, which divided the eight Cyatheaceae species into three genera.
![]()
1. INTRODUCTION
Simple sequence repeats (SSRs), also known as microsatellites, are short tandem repeat sequences with a motif length of 1–6 bp characterized by high variability and codominant inheritance and have been widely used in species identification, genetic diversity studies, and phylogenetic relationship determination (Chmielewski et al., 2015; Dashnow et al., 2015). SSRs are caused by slipped strand mispairing and subsequent errors during DNA replication, repair, and recombination (Levinson & Gutman, 1987). SSRs are mainly found in intergenic and noncoding regions, with a few present in introns (Li et al., 2004; Liu et al., 2021; Su et al., 2018). Previous studies have shown that the characteristics of genomic SSRs in different taxa (such as their distribution patterns) reflect their phylogenetic relationships (Manee et al., 2020; Srivastava et al., 2019).
The distribution of SSRs in some chloroplast (cp) genomes is nonrandom and dominated by mononucleotides, where A/T bases account for the majority (Ellegren, 2004; George et al., 2015; Ren et al., 2021). Some Polypodiaceae species show similar SSR distribution patterns in their cp genomes (Liu et al., 2021). Recently, Ping et al. (2021) analyzed the distribution pattern of Cupressus SSRs and found that the distribution patterns in Cupressus and Hesperocyparis are highly consistent. In addition, according to the proportions of A/T bases and mononucleotides in Callitropsis funebris, this species is closer to Cupressus. Studies have shown that the number and types of SSRs in cp genomes are conserved within genera, and the types of SSRs differ extensively among genera in Dryopteridaceae (Fan et al., 2021). Furthermore, cpSSRs continue to provide important new clues to explore the phylogeny among lineages.
The Cyatheaceae are an impressive group of ferns because of their arborescent features and a rich number of species, which account for the vast majority of known tree ferns and are mainly distributed in warm and humid tropical and subtropical regions (Korall et al., 2006; Kramer, 1990; PPG I, 2016; Smith et al., 2006). At present, four representative classification systems apply to Cyatheaceae: (a) the Holttum and Edwards system recognizes only one genus (Holttum, 1963); (b) the Tryon system includes six genera (Tryon, 1970); (c) the Lellinger system divides the family into four genera (Lellinger, 1987); and (d) the Pteridophyte Phylogeny Group (PPG I, 2016) system divides Cyatheaceae into three genera. Ching (1978) classified Chinese Cyatheaceae plants into three genera: Sphaeropteris, Gymnosphaera, and Alsophila. On this basis, Xia (1989) treated Alsophila and Gymnosphaera as subgenera and combined them into Alsophila. Recent molecular phylogenetic studies have shown that Cyatheaceae includes four monophyletic groups, namely, Alsophila, Cyathea, Gymnosphaera, and Sphaeropteris (Dong & Zuo, 2018; Janssen & Rakotondrainibe, 2008; Korall et al., 2007; Korall & Pryer, 2014), which provides a framework for us to study the phylogenetic significance of the cpSSRs of Cyatheaceae.
In this study, the cp genomes of eight Cyatheaceae species were sequenced, and the distribution and characteristics of their SSRs were compared. The cp genome sequences of these eight species represent the existing cp genome data for Cyatheaceae, covering most Cyatheaceae genera. Our major objectives were to (a) report the complete cp genomes of A. denticulate and A. metteniana; (b) compare the distribution patterns of the cpSSRs of the eight Cyatheaceae species; and (c) reveal the phylogenetic significance of the SSRs characteristics. Our findings may serve as a foundation for studying the evolutionary cp genomics and phylogeny of Cyatheaceae.
2. MATERIALS AND METHODS
2.1. Sampling
The leaves of Gymnospaera denticulata Baker and Gymnospaera metteniana Hance were collected from Nankunshan in Huizhou and the botanical garden of South China Agricultural University in Guangzhou, respectively. The specimens of Gymnospaera denticulata Baker and Gymnospaera metteniana Hance are stored in the Herbarium of South China Agricultural University (SCAUB; voucher: M Zhu 201910 and M Zhu 201908). The leaves of Gymnospaera podophylla Hook and Gymnospaera gigantea Wall. ex Hook were collected from the South China Botanical Garden of the Chinese Academy of Sciences in Guangzhou (Liu et al., 2018; Wang et al., 2019b). The leaves of Alsophila costularis Baker, Sphaeropteris brunoniana (Hook.) R. M. Tryon, and Sphaeropteris lepifera (Hook.) R. M. Tryon were collected from the Fairy Lake Botanical Garden of the Chinese Academy of Sciences in Shenzhen (Liu et al., 2020; Wang et al., 2019a; Zhu et al., 2020). The leaves of Alsophila spinulosa (Wall. ex Hook.) R. M. Tryon were collected from the Wuhan Botanical Garden of the Chinese Academy of Sciences in Wuhan (Gao et al., 2009). Fresh young leaves from well‐grown plants were collected, wrapped in tin paper, flash‐frozen in liquid nitrogen, and then stored at −80°C before use.
2.2. DNA extraction and sequencing
A plant genomic DNA extraction kit (TIANGEN) was used to extract total DNA from the samples. After the quality of the total DNA samples was confirmed by Shanghai Hanyu Biotechnology Co., Ltd., the samples were subjected to bidirectional sequencing using an Illumina HiSeq 2500, and the raw data obtained were converted into raw reads by CASAVA base‐calling analysis. The clean data obtained after removing the adaptor‐containing, low‐quality sequences were taken for subsequent analysis. Data processing was performed by Trimmomatic v0.32 (Bolger et al., 2014) with the following steps: (a) removal of sequences containing N bases; (b) removal of adaptor sequences in the reads; (c) removal of low‐quality bases (Q value < 20) from the reads in the 3′ to 5′ direction; (d) removal of low‐quality bases (Q value < 20) from the reads in the 5′ to 3′ direction; (e) removal of four bases with an average base quality <20; and (f) removal of the reads and their pairs with a length <50 nt. Velvet v1.2.03 (Zerbino & Birney, 2008) was used to assemble the clean data.
2.3. Characterization of chloroplast genomes
The cp genome of Alsophila spinulosa was used as the reference genome, and Dual Organellar GenoMe Annotator (DOGMA) (Milne et al., 2010) was used to predict the protein‐coding genes, rRNA genes, and tRNA genes in other genomes. Geneious Prime (Kearse et al., 2012) was used for manual correction according to the reference genome. The Shuffle‐Lagan mode in the online software mVISTA (Frazer et al., 2004) was used for genome‐wide comparison. Organellar Genome DRAW (OGDRAW) (Lohse et al., 2007) was used to draw physical cp genome maps, and Sequin software was used for submission of the cp genome of G. denticulata. Microsatellite repeats were predicted using the software MISA (Beier et al., 2017). The threshold repeat number of mononucleotide units was set to 10, the threshold repeat number of dinucleotide units was set to six, the threshold repeat number of trinucleotide units was set to five, and the threshold repeat number of tetra‐, penta‐, and hexanucleotide units was set to three. The minimum distance between two SSRs was set to 0 bp, that is, there was no statistical compound SSR. The distribution characteristics of SSRs of different species in the whole genome and its different regions were compared and analyzed. Among these characteristics, the relative abundance refers to the number of SSRs in the unit sequence length (kb), and the relative density refers to the length of the SSRs (bp) in the unit sequence length (kb).
2.4. Phylogenetic analysis
The maximum likelihood (ML), Bayesian inference (BI), maximum parsimony (MP), and neighbor‐joining (NJ) methods were used for phylogenetic analysis. MAFFT software (Katoh & Standley, 2013) was used to align the complete cp genome sequences of eight species of Cyatheaceae and one species of Cibotium, Cibotium barometz (Linn.) J. Sm. A phylogenetic tree was constructed using C. barometz (Linn.) J. Sm. as an outgroup. When the ML, MP, and BI trees had been constructed, the whole cp genome was screened in MrModeltest software to obtain the optimal nucleotide substitution model (GTR + I + G) selected based on the Akaike information criterion, and the relevant parameters were estimated. The ML tree was constructed by the software RAxML8.0.20 (Stamatakis, 2014), GTRGAMMAI was selected as the nucleotide substitution model, and the confidence of the branch was completed using the bootstrap analysis in autoMR. The BI tree was constructed by MrBayes v3.2.0 software (Ronquist et al., 2012) and was estimated by running 2,000,000 generations (Nst = 6, rates = invgamma). The MP tree was constructed in PAUP 4.0 software (Swofford, 2002) with the bootstrap value set to 1,000. The NJ tree was constructed in MEGA 7.0 software (Kumar et al., 2016), and the maximum composite likelihood algorithm was selected with the bootstrap value set to 1,000 times. The resulting phylogenetic tree was viewed and edited in Figtree v 1.4.3 software.
2.5. Statistical analysis
When Gymnosphaera is considered an independent taxonomic unit at the genus level, the eight Cyatheaceae species are divided into three genera; that is, G. denticulata, G. podophylla, G. metteniana, and G. gigantea belong to the genus Gymnosphaera; A. spinulosa and A. costularis belong to the genus Alsophila; and S. brunoniana and S. lepifera belong to the genus Sphaeropteris. When Gymnosphaera is classified into the genus Alsophila, Cyatheaceae is divided into two genera. The Kruskal–Wallis H test and Mann–Whitney U test in IBM SPSS v22.0 software (Allen et al., 2014) were used to analyze the significance of differences between taxa when three genera and two genera were assumed, respectively. The statistical results covered the whole cp genome, the SSRs of different unit lengths in the cp genome, and the number, relative abundance, relative density, and GC content of SSRs and SSRs of different unit lengths in the intergenic spacer (IGS), large single‐copy (LSC), intronic, and coding sequence (CDS) regions of the cp genomes of the eight Cyatheaceae species. Photovoltaic (PV) cluster analysis using the ward linkage method in R v3.5.1 (R Core Team, 2013) was performed on the SSRs of each cp genome and its IGS and LSC regions and on the number, relative abundance, relative density, and GC content of mononucleotide SSRs of the cp genomes with the Euclidean distance as the measurement. The number of repetitions was 10,000.
3. RESULTS
3.1. Genome structures and characteristics
The cp genomes of all eight Cyatheaceae species are double‐stranded, closed, circular molecules with a typical tetrad structure (with G. denticulata as an example, as shown in Figure 1). The genome length ranges from 154,046 bp (A. denticulata) to 166,151 bp (A. gigantea). The structure includes a large single‐copy region (LSC) (85,975–92,315 bp), a small single‐copy region (SSC) (23,245–28,874 bp), and an inverted repeat region (IR) (23,245–28,874 bp), where IRA and IRB are two inverted repeat regions. The GC content of each region of the cp genomes of different species varied little, with the total GC content ranging from 40.3% to 41.9% (Table 1). Only LSC, SSC, and one IR were analyzed. The cp genome of each Cyatheaceae species contained 117 genes, which encoded 85 proteins, four rRNAs, and 28 tRNAs. Pseudogenes (ycf66, trnT‐UGU) are also present in these genomes. Among these genes, 13 are located in the IR region. The ndhB gene spans the LSC and IRA regions, and there is a duplicated exon 2 sequence of the ndhB gene is present near the boundary of the IRB. Twelve genes have one intron, and three genes (ycf3, clpP, and rps12) have two introns.
FIGURE 1.
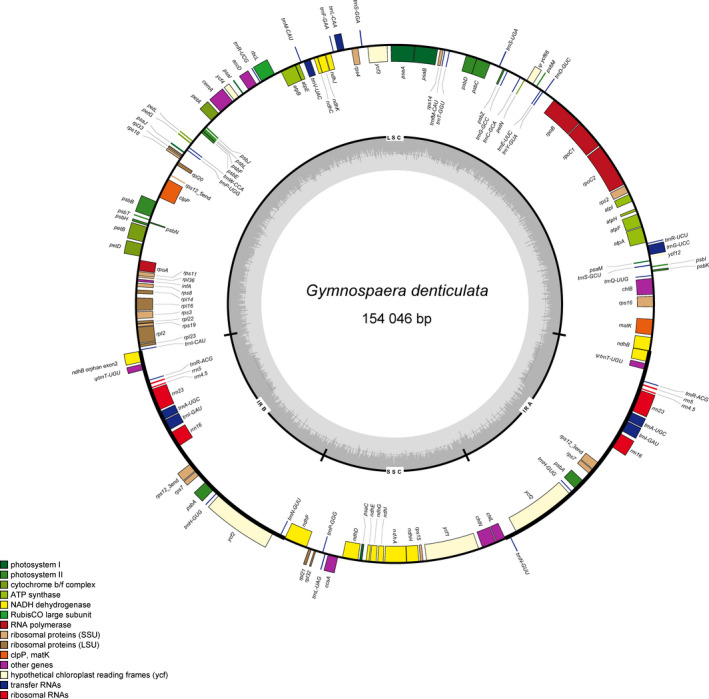
Gene map of the cp genome of Gymnospaera denticulata. Genes located in the outside of the outer circle are transcribed in the counterclockwise direction, whereas those in the inside of the circle are transcribed in the clockwise direction. Color codes represent different functional gene groups. In the middle circle, the GC and AT content variations are indicated by darker and lighter gray, respectively
TABLE 1.
List of the eight Cyatheaceae species, GenBank accession numbers, and structural features of the chloroplast genomes
| Species | GenBank No. | LSC | IR | SSC | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | GC% | Length (%) | Length (bp) | GC% | Length (%) | Length (bp) | GC% | Length (%) | Length (bp) | GC% | ||
| G. denticulata | MT726940 | 85,975 | 40.0 | 55.8 | 23,245 | 40.8 | 15.1 | 21,581 | 38.1 | 14.0 | 154,046 | 40.6 |
| G. podophylla | MG262389 | 86,762 | 40.0 | 52.2 | 28,874 | 46.2 | 17.4 | 21,641 | 38.1 | 13.0 | 166,151 | 41.9 |
| G. gigantea | MH603068 | 92,315 | 41.1 | 57.1 | 23,831 | 43.3 | 14.7 | 21,702 | 38.2 | 13.4 | 161,679 | 41.3 |
| G. metteniana | MN795320 | 92,292 | 41.1 | 57.1 | 23,822 | 43.3 | 14.7 | 21,666 | 38.1 | 13.4 | 161,602 | 41.3 |
| A. costularis | MH684489 | 86,338 | 39.6 | 55.1 | 24,356 | 43.0 | 15.5 | 21,625 | 37.8 | 13.8 | 156,675 | 40.5 |
| A. spinulosa | NC_012818 | 86,308 | 39.6 | 55.1 | 24,365 | 43.0 | 15.6 | 21,623 | 37.9 | 13.8 | 156,661 | 40.4 |
| S. brunoniana | MT543220 | 86,196 | 39.2 | 55.0 | 24,011 | 43.2 | 15.3 | 22,441 | 38.1 | 14.3 | 156,659 | 40.3 |
| S. lepifera | MN623357 | 86,349 | 39.3 | 53.3 | 24,067 | 43.2 | 14.8 | 27,733 | 41.5 | 17.1 | 162,216 | 40.8 |
Abbreviations: IR, inverted repeat region; LSC, Large single copy region; SSC, small single copy region.
3.2. Analysis of the characteristics of SSRs
The number, relative abundance, relative density, and GC content of SSRs in the cp genomes of all eight Cyatheaceae species were systematically compared (Table 2). The number (121–122), relative abundance (0.77–0.78/bp), relative density (9.81–9.82 bp/kb), and GC content (0.18–0.20) of SSRs in the cp genomes of A. spinulosa and A. costularis; the number (102), relative abundance (0.63–0.65/bp), relative density (6.70–8.18 bp/kb), and GC content (0.08–0.10) of SSRs in the cp genomes of S. brunoniana and S. lepifera; and the number (61–67), relative abundance (0.40/bp), relative density (4.11–5.06 bp/kb), and GC content (0.22–0.29) of SSRs of G. denticulata, G. podophylla, G. metteniana, and G. gigantea had similar values, which were not proportional to the sizes of the genomes. When Gymnosphaera was considered as an independent classification unit at the genus level, the eight species of Cyatheaceae were divided into three genera. That is, G. denticulata, G. podophylla, G. metteniana, and G. gigantea belonged to the genus Gymnosphaera, A. spinulosa and A. costularis belonged to the genus Alsophila, and S. brunoniana and S. lepifera belonged to the genus Sphaeropteris, which indicated that in the phylogenetic context of the three genera, the characteristics of SSRs are genus specific at the level of the genome. In Alsophila, the number, relative abundance, and relative density of SSRs were the highest among the eight species, and they were the smallest in Gymnosphaera. The highest GC content was found in Gymnosphaera, and the lowest in Sphaeropteris. The proportions of GC bases in the cp genomes of the eight species of Cyatheaceae were much lower than the proportions of AT. The proportion of SSRs in the IR region was 2–3.3 times the proportion of IR sequences among the whole genome sequence (Figure 2a).
TABLE 2.
Overview of the eight Cyatheaceae chloroplast genomes and characteristics of their SSRs
| G. denticulata | G. podophylla | G. gigantea | G. metteniana | A. costularis | A. spinulosa | S. brunoniana | S. lepifera | |
|---|---|---|---|---|---|---|---|---|
| Sequence analyzed (kb) | 154.05 | 166.15 | 161.68 | 161.60 | 156.68 | 156.66 | 156.66 | 162.22 |
| No. of SSRs | 61 | 67 | 64 | 65 | 121 | 122 | 102 | 103 |
| Relative abundance (No./kb) | 0.40 | 0.40 | 0.40 | 0.40 | 0.77 | 0.78 | 0.65 | 0.63 |
| Total length of SSRs (bp) | 634 | 802 | 818 | 801 | 1539 | 1538 | 1,352 | 1,327 |
| Relative density (bp/kb) | 4.11 | 4.82 | 5.06 | 4.96 | 9.82 | 9.81 | 8.63 | 8.18 |
| GC content | 0.30 | 0.25 | 0.29 | 0.22 | 0.20 | 0.18 | 0.11 | 0.10 |
| Genome content | 0.004 | 0.005 | 0.005 | 0.005 | 0.010 | 0.010 | 0.007 | 0.008 |
FIGURE 2.
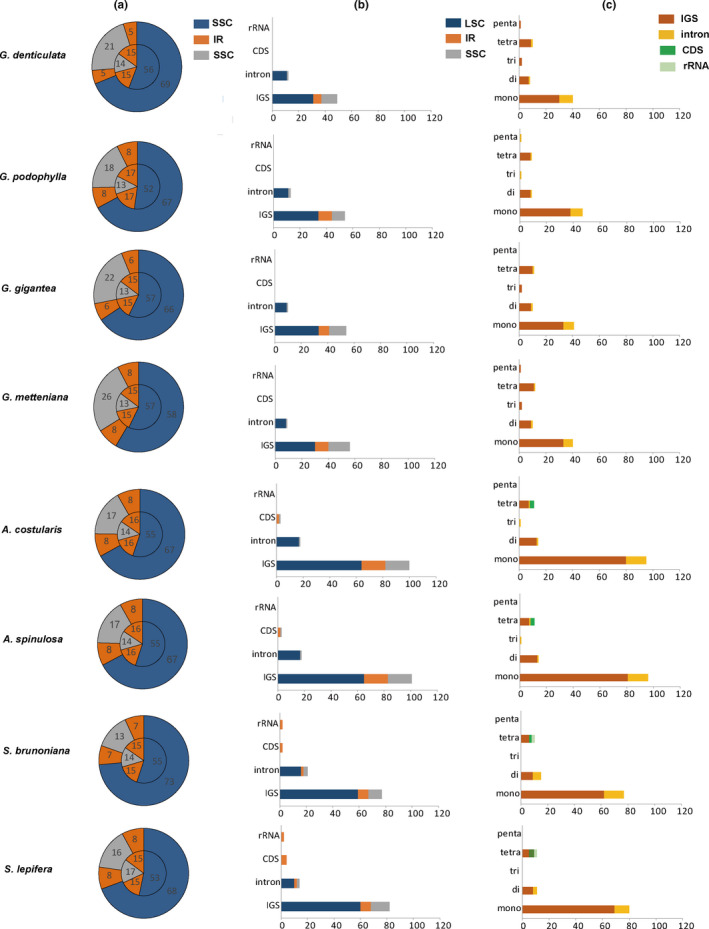
Comparison of microsatellite repeats among the eight Cyatheaceae chloroplast genomes. (a) The inner circle is the distribution ratio of the four regions (LSC, SSC, and IRs) in the genome, and the outer circle is the distribution ratio of SSRs between the four regions. (b) Distribution ratio of SSRs in different regions (LSC, SSC, and IRs) of chloroplast genome. (c) Ratio of mono‐ to pentanucleotide SSRs in different regions (IGS, intron, CDS, and rRNA gene) of the chloroplast genome. Numbers represent the distribution ratio of SSR numbers. LSC: large single copy region; SSC: small single copy region; IR: inverted repeat region; IGS: intergenic spacer region; CDS: coding sequence region
The number, relative abundance, relative density, and GC content of SSRs in the cp genomes of the three Cyatheaceae genera also had similar values in different regions of the genome (LSC, SSC, and IR; IGS, intron, CDS, and rRNA gene regions), indicating that in the phylogenetic context of the three genera, SSR characteristics were genus specific at the level of different regions of the genome (Figure 2; Appendix Tables S9 and S10). For the distributions of SSRs on the genome, the SSRs located in LSC (58.46%–73.52%) were most enriched in each species, followed by SSC (12.65%–21.08%), and the least in IR (9.84%–16.53%). The number of SSRs was highest in LSC and IR in Alsophila (81–82, 20) and lowest in Gymnosphaera (42–45, 8–10). Furthermore, SSRs accounted for 75.5%–86.2%, 13.7%–20.6%, 2.0%–3.9%, and 2.0% of the IGS, intron, CDS, and rRNA gene regions (pseudogenes were treated as IGS regions). Among them, SSRs were detected only in the CDS regions of the cp genomes of Sphaeropteris and Alsophila, and SSRs were detected in the rRNA genes of the cp genomes of Sphaeropteris. The SSRs at IGS regions were most enriched in Alsophila (100–101) and the least in Gymnosphaera (49–56). The number of SSRs located in the intron regions was 18 in Alsophila and 9–13 in Gymnosphaera. These results showed that in the phylogenetic context dividing the eight Cyatheaceae species into three genera, different taxa had different patterns of SSR characteristics in the cp genome and its different regions, namely, the SSR characteristics of the cp genomes of the eight Cyatheaceae species were consistent with their phylogenetic relationship.
3.3. Analysis of the types and characteristics of SSRs of different nucleotide numbers
The proportions of mono‐, di‐, tri‐, tetra‐, and pentanucleotide SSRs in each species were 62.5%–78.0%, 10.6%–15.6%, 0%–3.3%, 9.0%–18.5%, and 0%–1.5%, respectively. In the distribution of species, the number of mononucleotide had obvious genus specificity: Gymnosphaera (40–47), Sphaeropteris (77–80), and Alsophila (95–96). No hexanucleotide SSRs were detected. Among mononucleotide repeats, more A/T motifs were observed, and the dinucleotide repeats were dominated by AT/TA motifs. There were more tetranucleotide SSRs than tri‐, and pentanucleotide SSRs. Trinucleotide SSRs do not exist in Sphaeropteris, and pentanucleotide SSRs do not exist in Sphaeropteris, Alsophila, and G. gigantea. The mono‐, di‐, tri‐, tetra‐, and pentanucleotide SSRs of the cp genomes of the three genera were present in similar numbers, relative abundance, relative density, and GC content at the level of the genome and in the specific regions of the genome (LSC, SSC, and IRs; IGS, intron, CDS, and rRNA gene regions), which was especially true for mononucleotide and dinucleotide SSRs (Table 3; Appendix Tables S9 and S10).
TABLE 3.
The number, relative abundance, relative density, and GC content of mono‐ to pentanucleotide SSRs in the eight chloroplast genomes of Cyatheaceae
| Repeat type | Characteristics of SSR | G. denticulata | G. podophylla | G. gigantea | G. metteniana | A. costularis | A. spinulosa | S. brunoniana | S. lepifera |
|---|---|---|---|---|---|---|---|---|---|
| Mononucleotide | No. of SSRs | 40 | 47 | 41 | 40 | 95 | 96 | 77 | 80 |
| Abundance (No./kb) | 0.26 | 0.28 | 0.25 | 0.25 | 0.61 | 0.61 | 0.49 | 0.49 | |
| Density (bp/kb) | 2.93 | 3.16 | 3.13 | 2.88 | 7.67 | 7.60 | 6.41 | 6.23 | |
| GC | 0.36 | 0.35 | 0.40 | 0.3 | 0.22 | 0.19 | 0.08 | 0.09 | |
| Dinucleotide | No. of SSRs | 8 | 9 | 10 | 10 | 14 | 14 | 15 | 11 |
| Abundance (No./kb) | 0.05 | 0.05 | 0.06 | 0.06 | 0.09 | 0.09 | 0.10 | 0.07 | |
| Density (bp/kb) | 0.69 | 0.83 | 0.94 | 0.92 | 1.35 | 1.35 | 1.40 | 1.04 | |
| GC | 0.06 | 0 | 0.08 | 0.05 | 0 | 0 | 0.23 | 0.12 | |
| Trinucleotide | No. of SSRs | 2 | 1 | 2 | 2 | 1 | 1 | 0 | 0 |
| Abundance (No./kb) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0 | 0 | |
| Density (bp/kb) | 0.16 | 0.07 | 0.15 | 0.15 | 0.08 | 0.08 | 0 | 0 | |
| GC | 0 | 0 | 0 | 0 | 0.33 | 0.33 | 0 | 0 | |
| Tetranucleotide | No. of SSRs | 10 | 9 | 11 | 12 | 11 | 11 | 10 | 12 |
| Abundance (No./kb) | 0.06 | 0.05 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.07 | |
| Density (bp/kb) | 0.80 | 0.67 | 0.84 | 0.92 | 0.87 | 0.87 | 0.82 | 0.91 | |
| GC | 0.18 | 0.14 | 0.18 | 0.16 | 0.29 | 0.29 | 0.3 | 0.21 | |
| Pentanucleotide | No. of SSRs | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Abundance (No./kb) | 0.01 | 0.01 | 0 | 0.01 | 0 | 0 | 0 | 0 | |
| Density (bp/kb) | 0.10 | 0.09 | 0 | 0.09 | 0 | 0 | 0 | 0 | |
| GC | 0 | 0.4 | 0 | 0.4 | 0 | 0 | 0 | 0 |
The number, relative abundance, relative density, and GC content of SSRs of different unit lengths in the cp genome and its different regions had genus specificity in the phylogenetic context of dividing the eight Cyatheaceae species into three genera. In addition, the number, relative abundance, and relative density of SSRs of different base types in the cp genomes of the three genera of plants also had genus specificity, which was especially true for mono‐ and dinucleotide SSRs (Figure 3; Appendix Table S9). Alsophila had the highest A/T and C/G motif content (77, 18–19), Gymnosphaera had the least A/T motif content (26–32), and Sphaeropteris had the least C/G motif content (6).
FIGURE 3.
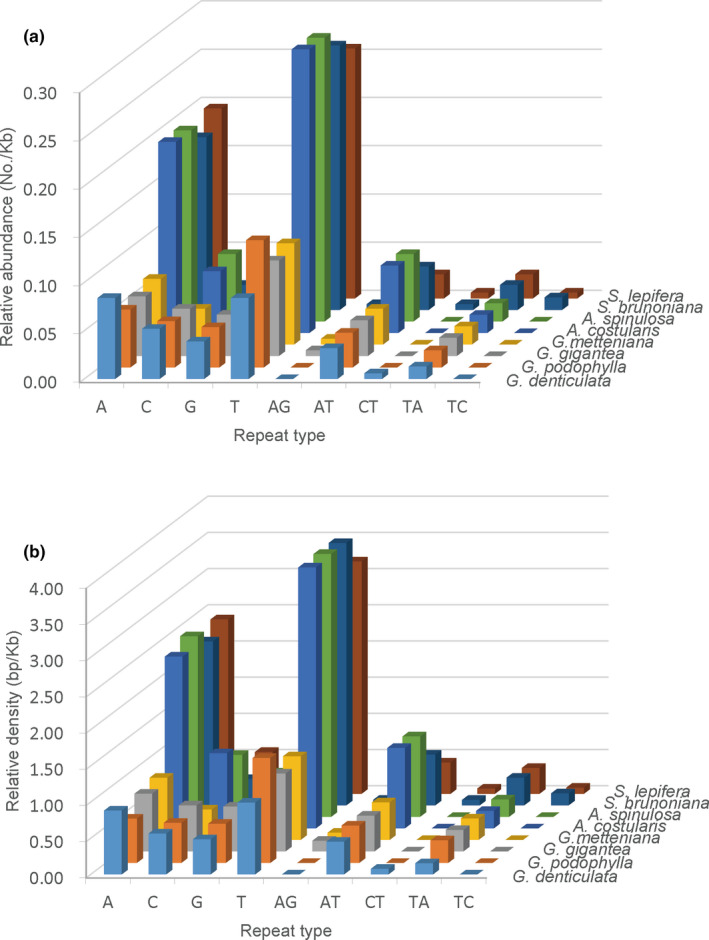
Relative abundance and relative density of mono‐ and dinucleotide SSRs in the eight chloroplast genomes of Cyatheaceae
3.4. Phylogenetic analysis
The cp genomes of the eight species of Cyatheaceae were compared globally, and the phylogenetic trees were constructed with four methods (ML, BI, MP, and NJ) using C. barometz (Linn.) J. Sm. as an outgroup, as shown in Figure 4. The topologies of the four trees were consistent, except that the support rate of the branches of the G. denticulata and G. gigantea was lower (the bootstrap values with the ML, MP, and NJ methods were 55%, 59.2%, and 99%, respectively, and the posterior probability of BI was 0.935). The support rate of the other branches was higher (the bootstrap values with the ML, MP, and NJ methods were all 100%, and the posterior probability of BI was 1.00). Closely related S. brunoniana and S. lepifera were clustered into one branch, which was located at the base of the phylogenetic tree, indicating that this group diverged earlier in this family. G. denticulata, G. podophylla, G. gigantea, and G. metteniana were clustered into one branch, which was located inside the branch of S. brunoniana and S. lepifera and was a sister group of the branch formed by A. spinulosa and A. costularis.
FIGURE 4.
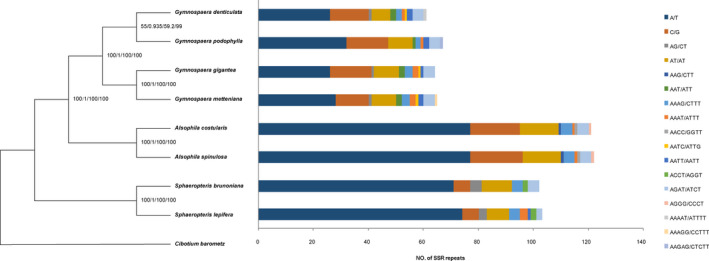
Ml, BI, MP, and NJ phylogenetic trees based on eight complete chloroplast genome sequences in Cyatheaceae and the distribution of different types of SSR motifs. The outgroup is Cibotium barometz
3.5. Statistical analysis of the results
The Kruskal–Wallis H test and Mann–Whitney U test values are shown in Appendix Table S11. In this study, only SSRs in the IGS and LSC regions of the cp genome, as well as mononucleotide SSRs in the whole cp genomes, were considered. Significant differences in the number, relative abundance, relative density, and GC content of SSRs were observed when the eight Cyatheaceae species were divided into three genera (Kruskal–Wallis H, p < .05). When Gymnosphaera was included in the genus Alsophila, two genera were defined, and only the difference in the GC content was significant (Table 4). The number of SSRs in other regions of the cp genome and the number of SSRs of other unit lengths were small, so they are not discussed in this study. The clustering results for the number, relative abundance, relative density, and GC content of the SSRs in the cp genomes and their IGS regions and the mononucleotide SSRs of the whole cp genomes of the eight Cyatheaceae species (Figure 5) showed that the eight species were divided into two groups. That is, S. brunoniana, S. lepifera, A. spinulosa, and A. costularis were in one group, and G. denticulata, G. podophylla, G. metteniana, and G. gigantea were in the other group.
TABLE 4.
Significance test of the number, relative abundance, relative density, and GC content of SSRs in the whole cp genome, IGS, and LSC and mononucleotide SSRs in the whole cp genomes of the eight Cyatheaceae species
| Classification treatment | Genus | Species | SSR Characteristics | P | Literature cited | |||
|---|---|---|---|---|---|---|---|---|
| Genome | IGS | LSC | Mononucleotide | |||||
| Three genera | Gymnosphaera |
G. denticulata G. podophylla |
No. of SSRs | 0.050* | 0.048* | 0.048* | 0.050* | Smith et al. (2006); Korall et al. (2007); Janssen and Rakotondrainibe (2008); Korall and Pryer (2014); Ching (1978); Dong and Zuo (2018) |
|
G. gigantea G. metteniana |
Relative abundance (No./kb) | 0.033* | 0.050* | 0.050* | 0.050* | |||
| Alsophila |
A. costularis A. spinulosa |
Relative density (bp/kb) | 0.050* | 0.050* | 0.050* | 0.050* | ||
| Sphaeropteris |
S. brunoniana S. lepifera |
GC content | 0.050* | 0.050* | 0.050* | 0.050* | ||
| Two genera | Alsophila |
G. G. denticulata G. podophylla |
No. of SSRs | 0.502 | 0.502 | 0.502 | 0.502 | PPG I (2016); Xia (1989); Zhang and Nishida (2013) |
|
G. gigantea G. metteniana |
Relative abundance (No./kb) | 0.478 | 0.505 | 0.505 | 0.505 | |||
|
A. costularis A. spinulosa |
Relative density (bp/kb) | 0.505 | 0.505 | 0.505 | 0.505 | |||
| Sphaeropteris |
S. brunoniana S. lepifera |
GC content | 0.046* | 0.046* | 0.046* | 0.046* | ||
When Cyatheaceae plants are treated as three genera, the Kruskal–Wallis H test is used; when treated as two genera, the Mann–Whitney U test is used; *p ≤ .05.
Abbreviations: IGS, intergenic spacer region; LSC, large single copy region.
FIGURE 5.
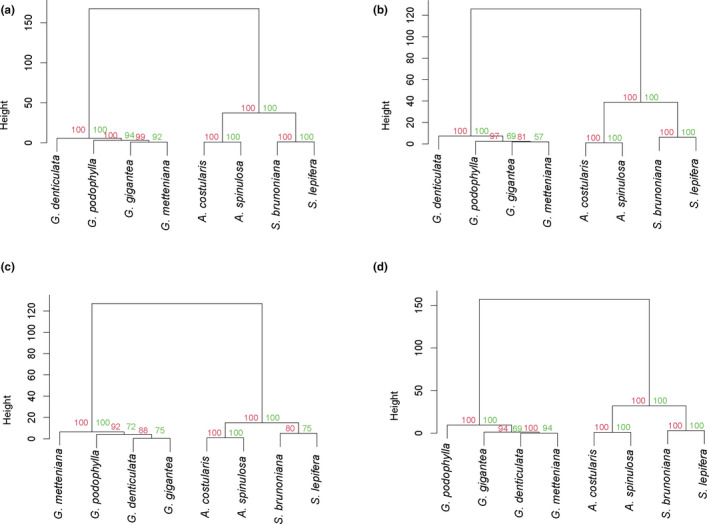
Clustering analysis of eight Cyatheaceae species based on the number, relative abundance, relative density, and GC content of SSRs across the whole chloroplast genome (a), IGS (b), LSC (c), and mononucleotide (d) SSRs in the chloroplast genomes
4. DISCUSSION
4.1. Characteristics of the cpSSRs of the eight Cyatheaceae species
The cp plastomes of the eight Cyatheaceae species are very conservative and are similar in structure and gene content (117 genes). The types and order of genes are the same. The lower distribution of SSRs (Figure 2a) in the IR region may be related to the higher mismatch repair rate and lower mutation rate in the IR region (Ellegren, 2004; Li et al., 2016). The lower GC content (Tables 2 and 3) of SSRs may be associated with the tendency of GC‐rich regions toward AT mutations (Kuang et al., 2011; Ren et al., 2007). SSRs are mainly located in intergenic and noncoding regions, with a few present in exons (Li et al., 2004; Su et al., 2018), and the results of this study are consistent with this information. This phenomenon is related to negative selection against frameshift mutations in coding regions (Metzgar et al., 2000). CpSSRs are characterized by high variability and codominant inheritance (Chmielewski et al., 2015; George et al., 2015); therefore, their sequences can be used to effectively classify low taxonomic and closely related groups and variant plant subspecies.
In this study, we used MISA to scan the recently assembled Cyatheaceae cp genomes for microsatellites of 1–6 bp. The cp genomes of eight Cyatheaceae species were similarly analyzed using the same bioinformatics tool and search parameters to compare our results. The number, relative abundance, relative density, and GC content of the cpSSRs of A. spinulosa and A. costularis; the number, relative abundance, relative density, and GC content of the cpSSRs of S. brunoniana and S. lepifera; and the number, relative abundance, relative density, and GC content of the cpSSRs of G. denticulata, G. podophylla, G. metteniana, and G. gigantea had similar values. Based on these findings, the eight Cyatheaceae species were divided into three groups, which is consistent with the recent studies (Ching, 1978; Dong & Zuo, 2018; Janssen & Rakotondrainibe, 2008; Korall et al., 2007; Korall & Pryer, 2014; Smith et al., 2006), in which eight Cyatheaceae species were divided into three genera, indicating that the characteristics of the cpSSRs showed genus specificity at the genome level in the phylogenetic context of the three genera (Figure 2, Tables 2 and 3, and Appendix Tables S9–S11).
The number, relative abundance, and relative density of the cpSSRs of Alsophila were lower than those of Gymnosphaera, while those of Sphaeropteris fell between the two. The numbers of SSRs of Alsophila, Gymnosphaera, and Sphaeropteris distributed in the LSC region were 81–82 (66.94%–67.21%), 38–45 (58.46%–68.85%), and 70–75 (67.96%–73.52%), respectively, which was the most in each species, followed by the SSC region, and the least in the IR region (Appendix Table S10). The numbers of SSRs of Alsophila, Gymnosphaera, and Sphaeropteris distributed in the IGS region were 99–100 (81.81%–81.97%), 49–56 (80.32%–86.15%), and 77–83 (75.49%–80.58%), respectively, which was the most in each species, followed by the intron region. SSRs were detected in the CDS region of Alsophila and Sphaeropteris, and SSRs were detected only in the rRNA gene of Sphaeropteris (Appendix Table S10). The cpSSRs of each species were mainly distributed in the IGS region and LSC regions, which is consistent with that reported in Adrinandra (Nguyen et al., 2021), Blumea (Abdullah et al., 2021), Mikania (Su et al., 2018), Prunus (Huang et al., 2021), Cupressaceae (Ping et al., 2021), Poaceae (Wei et al., 2021), Polypodiaceae (Liu et al., 2021), and ferns (Fan et al., 2021). Alsophila had the highest relative abundance and density (0.77–0.78/kb, 9.81–9.82 bp/kb, respectively) of the SSRs in the cp genome, and the lowest relative abundance and density (0.40/kb, 4.11–5.06 bp/kb) were found in Gymnosphaera. The number, relative abundances, and density of SSRs showed great similarity across eukaryotic genomes among taxonomic groups (Manee et al., 2020; Qi et al., 2015; Srivastava et al., 2019).
The number, relative abundance, relative density, and GC content of SSRs of different unit length in Cyatheaceae cp genomes were also genus specific (Table 3; Appendix Tables S9–S11), which was especially true for mononucleotide and dinucleotide SSRs (Figure 3), possibly because of the lower content of SSRs of other unit lengths. The number of mononucleotide repeats of Alsophila, Gymnosphaera, and Sphaeropteris was the largest at 77–80 (75.49%–77.67%), 40–47 (61.54%–70.15%), and 95–96 (78.51%–78.69%), and the proportions of the A/T repeat motif were 80.21%–81.05%, 63.41%–70%, and 92.21%–92.50%, respectively. Dinucleotide repeats of Alsophila, Gymnosphaera, and Sphaeropteris showed two types of repeat motifs (AG/CT, AT/AT), with numbers of 9–14, 8–14, and 11–15 and AT/AT accounting for 100%, 87.50%–100%, and 72.73%–73.33% of the motifs, respectively (Appendix Tables S9–S11). Mononucleotide SSRs which exist in a large numbers in cp genomes (George et al., 2015; Liang et al., 2019) are the most abundant (Table 3), and A/T motifs are the most common (Figure 3; Appendix Table S9). This finding is similar to previously reported patterns of land plants (George et al., 2015; Huang et al., 2021; Ren et al., 2021; Vieira et al., 2014; Wei et al., 2021). However, in Polypodiaceae plastomes, most repeats were C/G mononucleotides. This increase in GC content may be related to the adaptation mechanism of Polypodiaceae to the environment (Gao et al., 2018; Liu et al., 2021). These results indicate that SSRs in cp genomes can reflect genetic variation between different taxa.
The distribution of different repeat types (from mononucleotide to hexanucleotide) of motifs in coding and noncoding regions, introns, and intergenic regions displayed a high degree of genus specificity (Appendix Tables S10 and S11), which can be partially explained by the interaction of mutation mechanisms and differential selection (Toth et al., 2000). The most common mutation mechanism affecting SSRs is slipped replication. Other mechanisms, such as unequal crossing over, nucleotide substitution, and duplication events, are also responsible for SSR variation (Hancock, 1999; Schlotterer & Tautz, 1992). The SSRs of different groups of genomes have specific distribution patterns, which are related to their common ancestors. Evolutionary trends have been linked to the inclusion of SSRs, which may have been preserved because of their ability to adapt to novel regulatory mechanisms (Srivastava et al., 2019). Analysis of the characteristics of SSRs provides useful clues for the phylogenetic study of Cyatheaceae and facilitates an understanding of the evolution of SSRs in plant genomes.
4.2. Phylogenetic significance of SSR characteristics of the cp genomes of the eight Cyatheaceae species
Dong and Zuo (2018) pointed out that Gymnosphaera and Alsophila were significantly different in morphological traits such as petiole color, the presence or absence of degenerated pinnae at the base, and the presence or absence of indusium and sporogenesis, and advocated for the restoration of the hierarchical status of the genus Gymnosphaera to reflect the divergence mechanisms of this group of plants in terms of molecular phylogeny, morphology, and sporogenesis. Based on five chloroplast DNA regions (rbcL, rbcL‐accD, rbcL‐atpB, trnG‐trnR, and trnL‐trnF), the results of the phylogenetic analysis of Cyatheaceae conducted by Dong and Zuo (2018) support the independence of Gymnosphaera. Since the stipe of Sphaeropteris is stramineous or purple, not black, and the cells of the scale are all essentially similar to the edge of the scale, which is often toothed or even ciliate, Sphaeropteris can be clearly distinguished from other genera in the family Cyatheaceae. Molecular phylogenetic studies further confirm this point (Dong & Zuo, 2018; Janssen & Rakotondrainibe, 2008; Korall & Pryer, 2014; Wang et al., 2003).
In the recent studies, eight Cyatheaceae species were divided into three genera (Ching, 1978; Dong & Zuo, 2018; Janssen & Rakotondrainibe, 2008; Korall et al., 2007; Korall & Pryer, 2014; Smith et al., 2006) or two genera (PPG I, 2016; Xia, 1989; Zhang & Nishida, 2013), in which case SSRs were compared by the Kruskal–Wallis H test or the Mann–Whitney U test, respectively, and the results showed that the statistical and clustering results showed that their distribution was consistent with the recent classification of Cyatheaceae which divided the eight Cyatheaceae species into three genera, which was confirmed by the PV clustering analysis of SSRs. In this study, phylogenetic trees were constructed using the cp genomes of eight Cyatheaceae species, which showed that G. denticulata, G. podophylla, G. gigantea, G. metteniana, A. costularis, and A. spinulosa formed a monophyletic clade. In general, phylogenetic relationships from sequence construction are not sufficient to indicate whether two taxa are distinct. The phylogenetic relationships from SSR characteristics cluster analysis can distinguish the two groups. The different clustering results may be related to the use of different clustering methods and different data. In this phylogenetic context of the three genera, the SSR characteristics have genus specificity, which may reflect a universal law among Cyatheaceae genera.
Recent studies have shown minimal differences in the distribution patterns and numbers of SSRs among cpSSRs of the related species (Fan et al., 2021; Liu et al., 2021; Ping et al., 2021; Wei et al., 2021). Comparative analysis of cpSSRs from broad plant groups may be useful for a better understanding of the diversity and evolutionary trends of cp genomes (George et al., 2015). This study indicates that the distribution characteristics of cpSSRs of Cyatheaceae can provide useful phylogeny information at the genus level. Relatively few studies have explored phylogenetic relationships in ferns by analyzing cpSSRs. Since the software programs that identify SSRs are limited by their efficiency and parameter settings and may also be affected by the quality of the SSR dataset generated, their accuracy requires improvement (Ellegren, 2004; Lim et al., 2013). Given the limitations of the current plant genome sequences, we did not analyze the large‐scale SSR characteristics of the cp genomes of Cyatheaceae, nor did we obtain the plant materials of Cyathea that are generally distributed in South America. However, this study is based on the existing chloroplast genomes of Cyatheaceae and the eight species encompass most Cyatheaceae genera. Our results demonstrate that the distribution characteristics of the cpSSRs of the existing Cyatheaceae are genus specific. Our aim was not to solve the phylogenetic problem of Cyatheaceae but to identify traits that can be used for phylogenetics. This study provides a new basis for the classification of Cyatheaceae at the species and genus levels, thus advancing the phylogenetic study of Cyatheaceae. In the future, more genomic and transcriptomic data are needed to validate these results.
5. CONCLUSION
The cp genomes of the eight Cyatheaceae species have the same gene types in the same order and similar structure and gene content. The distribution characteristics of the cpSSRs of the eight species are consistent with the recent classification of Cyatheaceae, which divides the eight Cyatheaceae species into three genera, indicating that in the phylogenetic context of the three genera, the distribution characteristics of SSRs in their cp genomes are genus specific, which may be a general rule among Cyatheaceae. Analyzing the characteristics of SSRs provides clues and new ideas for research on the phylogeny of Cyatheaceae.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTION
Ming Zhu: Conceptualization (equal); Data curation (lead); Formal analysis (lead); Writing‐original draft (lead). Peipei Feng: Data curation (equal); Writing‐original draft (equal). Jingyao Ping: Data curation (equal); Writing‐original draft (equal). Jinye Li: Data curation (supporting); Writing‐original draft (supporting). YingJuan Su: Conceptualization (lead); Writing‐original draft (equal). Ting Wang: Conceptualization (lead); Writing‐original draft (equal).
Supporting information
AppendixTable S1‐S11
ACKNOWLEDGMENTS
We would like to thank the Wuhan Botanical Garden, South China Botanical Garden, and Fairy Lake Botanical Garden of the Chinese Academy of Sciences for providing samples of Cyatheaceae plants.
Zhu, M. , Feng, P. , Ping, J. , Li, J. , Su, Y. , & Wang, T. (2021). Phylogenetic significance of the characteristics of simple sequence repeats at the genus level based on the complete chloroplast genome sequences of Cyatheaceae. Ecology and Evolution, 11, 14327–14340. 10.1002/ece3.8151
Funding information
This work was funded by the National Natural Science Foundation of China (31670200, 31770587, 31872670, and 32071781).
Contributor Information
YingJuan Su, Email: suyj@mail.sysu.edu.cn.
Ting Wang, Email: tingwang@scau.edu.cn.
DATA AVAILABILITY STATEMENT
The chloroplast genomes of Gymnosphaera denticulata Baker and Gymnosphaera metteniana Hance were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and under the accession numbers of MT726940 and MN795320, respectively.
REFERENCES
- Abdullah, Mehmood, F. , Rahim, A. , Heidari, P. , Ahmed, I. , & Poczai, P. (2021). Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecology and Evolution, 11(12), 7810–7826. 10.1002/ece3.7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, P. , Bennett, K. , & Heritage, B. (2014). SPSS version 22: A practical guide. Cengage Learning. [Google Scholar]
- Beier, S. , Thiel, T. , Muench, T. , Scholz, U. , & Mascher, M. (2017). MISA‐web: A web server for microsatellite prediction. Bioinformatics, 33(16), 2583–2585. 10.1093/bioinformatics/btx198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching, R. C. (1978). The Chinese fern families and genera: Systematic arrangement and historical origin. Acta Phytotaxonomica Sinica, 16(4), 16–37. [Google Scholar]
- Chmielewski, M. , Meyza, K. , Chybicki, I. J. , Dzialuk, A. , Litkowiec, M. , & Burczyk, J. (2015). Chloroplast microsatellites as a tool for phylogeographic studies: The case of white oaks in Poland. Iforest‐Biogeosciences and Forestry, 8(6), 765–771. 10.3832/ifor1597-008 [DOI] [Google Scholar]
- Dashnow, H. , Tan, S. , Das, D. , Easteal, S. , & Oshlack, A. (2015). Genotyping microsatellites in next‐generation sequencing data. BMC Bioinformatics, 16(Suppl 2), A5. 10.1186/1471-2105-16-s2-a5 [DOI] [Google Scholar]
- Dong, S. Y. , & Zuo, Z. Y. (2018). On the recognition of gymnosphaera as a distinct genus in Cyatheaceae. Annals of the Missouri Botanical Garden, 103(1), 1–23. 10.3417/2017049 [DOI] [Google Scholar]
- Ellegren, H. (2004). Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics, 5(6), 435–445. 10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- Fan, R. , Ma, W. , Liu, S. , & Huang, Q. (2021). Integrated analysis of three newly sequenced fern chloroplast genomes: Genome structure and comparative analysis. Ecology and Evolution, 11(9), 4550–4563. 10.1002/ece3.7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K. A. , Pachter, L. , Poliakov, A. , Rubin, E. M. , & Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Research, 32, W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L. , Yi, X. , Yang, Y. X. , Su, Y. J. , & Wang, T. (2009). Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evolutionary Biology, 9(1), 1–14. 10.1186/1471-2148-9-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, R. , Wang, W. , Huang, Q. , Fan, R. , Wang, X. , Feng, P. , Zhao, G. , Bian, S. , Ren, H. , & Chang, Y. (2018). Complete chloroplast genome sequence of Dryopteris fragrans (L.) Schott and the repeat structures against the thermal environment. Scientific Reports, 8(1), 1–11. 10.1038/s41598-018-35061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, B. , Bhatt, B. S. , Awasthi, M. , George, B. , & Singh, A. K. (2015). Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Current Genetics, 61(4), 665–677. 10.1007/s00294-015-0495-9 [DOI] [PubMed] [Google Scholar]
- Hancock, J. M. (1999). Microsatellites and other simple sequences: Genomic contextand mutational mechanisms. In Goldstein D., & Schlötterer C. (Eds.), Microsatellites: Evolution and applications (pp. 1–9). Oxford University Press. [Google Scholar]
- Holttum, R. E. (1963). Cyatheaceae. In Steenis V. C., & Holttum R. E. (Eds.), Flora Malesiana, Ser.2 (Vol. 1, pp. 65–176). Wolters‐Noordhoff Publishing. [Google Scholar]
- Huang, X. , Tan, W. , Li, F. , Liao, R. , Guo, Z. , Shi, T. , & Gao, Z. (2021). The chloroplast genome of Prunus zhengheensis: Genome comparative and phylogenetic relationships analysis. Gene, 793, 145751. 10.1016/j.gene.2021.145751 [DOI] [PubMed] [Google Scholar]
- Janssen, T. , & Rakotondrainibe, F. (2008). A revision of the indusiate scaly tree ferns (Cyatheaceae, Cyathea subgen. Alsophila sect. Alsophila) in Madagascar, the Comoros and the Seychelles. Adansonia, 30(2), 221–374. [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. , Cooper, A. , Markowitz, S. , Duran, C. , Thierer, T. , Ashton, B. , Meintjes, P. , & Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korall, P. , Conant, D. S. , Metzgar, J. S. , Schneider, H. , & Pryer, K. M. (2007). A molecular phylogeny of scaly tree ferns (Cyatheaceae). American Journal of Botany, 94(5), 873–886. 10.3732/ajb.94.5.873 [DOI] [PubMed] [Google Scholar]
- Korall, P. , & Pryer, K. M. (2014). Global biogeography of scaly tree ferns (Cyatheaceae): Evidence for Gondwanan vicariance and limited transoceanic dispersal. Journal of Biogeography, 41(2), 402–413. 10.1111/jbi.12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korall, P. , Pryer, K. A. , Metzgar, J. S. , Schneider, H. , & Conant, D. S. (2006). Tree ferns: Monophyletic groups and their relationships as revealed by four protein‐coding plastid loci. Molecular Phylogenetics and Evolution, 39(3), 830–845. 10.1016/j.ympev.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Kramer, K. U. (1990). Cyatheaceae. In Kubitzki K. & Green P. S. (Eds.) The families and genera of vascular plants, Vol. 1: Pteridophytes and gymnosperms (pp. 69–74). SpringerVerlag. [Google Scholar]
- Kuang, D. Y. , Wu, H. , Wang, Y. L. , Gao, L. M. , Zhang, S. Z. , & Lu, L. (2011). Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome, 54(8), 663–673. 10.1139/g11-026 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellinger, D. B. (1987). The disposition of Trichopteris (Cyatheaceae). American Fern Journal, 77(3), 90–94. 10.2307/1547497 [DOI] [Google Scholar]
- Levinson, G. , & Gutman, G. A. (1987). Slipped‐strand mispairing: A major mechanism for DNA sequence evolution. Molecular Biology and Evolution, 4(3), 203–221. 10.1093/oxfordjournals.molbev.a040442 [DOI] [PubMed] [Google Scholar]
- Li, F. W. , Kuo, L. Y. , Pryer, K. M. , & Rothfels, C. J. (2016). Genes translocated into the plastid inverted repeat show decelerated substitution rates and elevated GC content. Genome Biology and Evolution, 8(8), 2452–2458. 10.1093/gbe/evw167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C. , Korol, A. B. , Fahima, T. , & Nevo, E. (2004). Microsatellites within genes: Structure, function, and evolution. Molecular Biology and Evolution, 21(6), 991–1007. 10.1093/molbev/msh073 [DOI] [PubMed] [Google Scholar]
- Liang, C. , Wang, L. , Lei, J. , Duan, B. , Ma, W. , Xiao, S. , Qi, H. , Wang, Z. , Liu, Y. , Shen, X. , Guo, S. , Hu, H. , Xu, J. , & Chen, S. (2019). A comparative analysis of the chloroplast genomes of four salvia medicinal plants. Engineering, 5(5), 907–915. 10.1016/j.eng.2019.01.017 [DOI] [Google Scholar]
- Lim, K. G. , Kwoh, C. K. , Hsu, L. Y. , & Wirawan, A. (2013). Review of tandem repeat search tools: A systematic approach to evaluating algorithmic performance. Briefings in Bioinformatics, 14(1), 67–81. 10.1093/bib/bbs023 [DOI] [PubMed] [Google Scholar]
- Liu, S. S. , Ping, J. Y. , Wang, Z. , Wang, T. , & Su, Y. J. (2018). Complete chloroplast genome of the tree fern Alsophila podophylla (Cyatheaceae). Mitochondrial DNA Part B‐Resources, 3(1), 48–49. 10.1080/23802359.2017.1419095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. S. , Wang, Z. , Su, Y. J. , & Wang, T. (2021). Comparative genomic analysis of Polypodiaceae chloroplasts reveals fine structural features and dynamic insertion sequences. BMC Plant Biology, 21(1), 1–15. 10.1186/s12870-020-02800-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. S. , Wang, Z. , Wang, H. , Su, Y. J. , & Wang, T. (2020). Patterns and rates of plastid rps12 gene evolution inferred in a phylogenetic context using plastomic data of ferns. Scientific Reports, 10(1), 1–12. 10.1038/s41598-020-66219-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse, M. , Drechsel, O. , & Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high‐quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics, 52(5–6), 267–274. 10.1007/s00294-007-0161-y [DOI] [PubMed] [Google Scholar]
- Manee, M. M. , Algarni, A. T. , Alharbi, S. N. , Al‐Shomrani, B. M. , Ibrahim, M. A. , Binghadir, S. A. , & Al‐Fageeh, M. B. (2020). Genome‐wide characterization and analysis of microsatellite sequences in camelid species. Mammal Research, 65(2), 359–373. 10.1007/s13364-019-00458-x [DOI] [Google Scholar]
- Metzgar, D. , Bytof, J. , & Wills, C. (2000). Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Research, 10(1), 72–80. [PMC free article] [PubMed] [Google Scholar]
- Milne, I. , Bayer, M. , Cardle, L. , Shaw, P. , Stephen, G. , Wright, F. , & Marshall, D. (2010). Tablet‐next generation sequence assembly visualization. Bioinformatics, 26(3), 401–402. 10.1093/bioinformatics/btp666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H. Q. , Nguyen, T. N. L. , Doan, T. N. , Nguyen, T. T. N. , Phạm, M. H. , Le, T. L. , Sy, D. T. , Chu, H. H. , & Chu, H. M. (2021). Complete chloroplast genome of novel Adrinandra megaphylla Hu species: Molecular structure, comparative and phylogenetic analysis. Scientific Reports, 11(1), 11731. 10.1038/s41598-021-91071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping, J. Y. , Feng, P. P. , Li, J. Y. , Zhang, R. J. , Su, Y. J. , & Wang, T. (2021). Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis funebris . Ecology and Evolution, 11(9), 4786–4802. 10.1002/ece3.7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PPG I (2016). A community‐derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution, 54, 563–603. 10.1111/jse.12229 [DOI] [Google Scholar]
- Qi, W. H. , Jiang, X. M. , Du, L. M. , Xiao, G. S. , Hu, T. Z. , Yue, B. S. , & Quan, Q. M. (2015). Genome‐wide survey and analysis of microsatellite sequences in bovid species. PLoS One, 10(7), 10.1371/journal.pone.0133667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing (p. 2013). R Foundation for Statistical Computing. [Google Scholar]
- Ren, F. , Wang, L. , Li, Y. , Zhuo, W. , Xu, Z. , Guo, H. , Liu, Y. , Gao, R. , & Song, J. (2021). Highly variable chloroplast genome from two endangered Papaveraceae lithophytes Corydalis tomentella and Corydalis saxicola . Ecology and Evolution, 11(9), 4158–4171. 10.1002/ece3.7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, L. , Gao, G. , Zhao, D. , Ding, M. , Luo, J. , & Deng, H. (2007). Developmental stage related patterns of codon usage and genomic GC content: Searching for evolutionary fingerprints with models of stem cell differentiation. Genome Biology, 8(3), R35. 10.1186/gb-2007-8-3-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D. L. , Darling, A. , Höhna, S. , Larget, B. , Liu, L. , Suchard, M. A. , & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer, C. , & Tautz, D. (1992). Slippage synthesis of simple sequence DNA. Nucleic Acids Research, 20(2), 211–215. 10.1093/nar/20.2.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. R. , Pryer, K. M. , Schuettpelz, E. , Korall, P. , Schneider, H. , & Wolf, P. G. (2006). A classification for extant ferns. Taxon, 55(3), 705–731. 10.2307/25065646 [DOI] [Google Scholar]
- Srivastava, S. , Avvaru, A. K. , Sowpati, D. T. , & Mishra, R. K. (2019). Patterns of microsatellite distribution across eukaryotic genomes. BMC Genomics, 20(1), 1–14. 10.1186/s12864-019-5516-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30(9), 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Huang, L. , Wang, Z. , & Wang, T. (2018). Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata: Structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Molecular Phylogenetics and Evolution, 126, 181–195. 10.1016/j.ympev.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Swofford, D. (2002). PAUP4: Phylogenetic analysis using parsimony (and other methods), version 4.0. Sinauer. [Google Scholar]
- Toth, G. , Gaspari, Z. , & Jurka, J. (2000). Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Research, 10(7), 967–981. 10.1101/gr.10.7.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon, R. M. (1970). The classification of Cyatheaceae. Contributions from the Gray Herbarium, 200, 1–53. [Google Scholar]
- Vieira, L. N. , Faoro, H. , Rogalski, M. , Fraga, H. P. F. , Cardoso, R. L. A. , de Souza, E. M. , de Oliveira Pedrosa, F. , Nodari, R. O. , & Guerra, M. P. (2014). The complete chloroplast genome sequence of Podocarpus lambertii: Genome structure, evolutionary aspects, gene content and SSR detection. PLoS One, 9(3), e90618. 10.1371/journal.pone.0090618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , He, Z. , Wang, Z. , Sun, X. , & Su, Y. (2019). The first complete chloroplast genome of Alsophila costularis (Cyatheaceae), a least concerned relict tree fern. Mitochondrial DNA Part B‐Resources, 4(1), 1897–1898. 10.1080/23802359.2019.1614888 [DOI] [Google Scholar]
- Wang, T. , Hong, Y. , Wang, Z. , & Su, Y. (2019). Characterization of the complete chloroplast genome of Alsophila gigantea (Cyatheaceae), an ornamental and CITES giant tree fern. Mitochondrial DNA Part B‐Resources, 4(1), 967–968. 10.1080/23802359.2019.1580162 [DOI] [Google Scholar]
- Wang, T. , Su, Y. J. , Zheng, B. , Li, X. Y. , Chen, G. P. , & Zeng, Q. I. (2003). Phylogenetic analysis of the chloroplast trnL intron and trnL‐trnF intergenic spacer sequences of the Cyatheaceae plants from China. Journal of Tropical and Subtropical Botany, 11(2), 137–142. [Google Scholar]
- Wei, X. , Li, X. , Chen, T. , Chen, Z. , Jin, Y. , Malik, K. , & Li, C. (2021). Complete chloroplast genomes of Achnatherum inebrians and comparative analyses with related species from Poaceae. FEBS Open Bio, 11(6), 1704–1718. 10.1002/2211-5463.13170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Q. (1989). The classification of the cyatheaceae in china. Acta Phytotaxonomica Sinica, 27, 1–16. [Google Scholar]
- Zerbino, D. R. , & Birney, E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 18(5), 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. C. , & Nishida, H. (2013). Cyatheaceae, Flora of China (Vol. 2–3, pp. 134–138). Science Press; Missouri Botanical Garden Press. [Google Scholar]
- Zhu, M. , Zhao, G. , Ping, J. , Liang, Y. , Feng, P. , Su, Y. , & Wang, T. (2020). Complete chloroplast genome of Sphaeropteris brunoniana (Cyatheaceae). Mitochondrial DNA. Part B Resources, 5(3), 2938–2939. 10.1080/23802359.2020.1787893 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AppendixTable S1‐S11
Data Availability Statement
The chloroplast genomes of Gymnosphaera denticulata Baker and Gymnosphaera metteniana Hance were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and under the accession numbers of MT726940 and MN795320, respectively.


