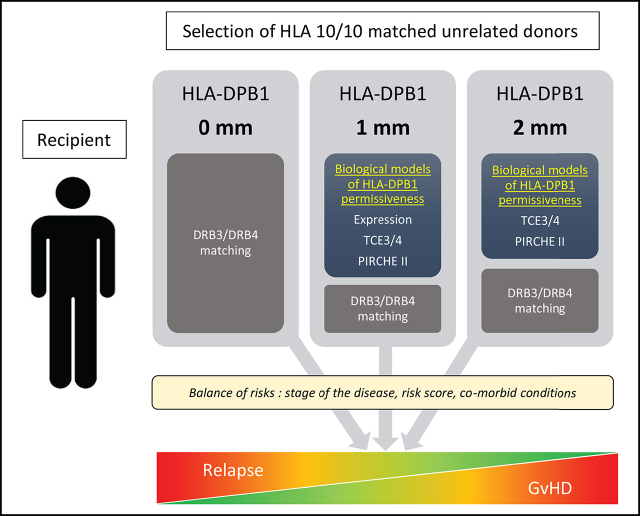
Key Points
Risk of acute GVHD after unrelated HSCT is the highest when single HLA-DPB1 mismatches in the patient have a high cell surface expression.
TCE nonpermissiveness and predicted indirectly recognizable HLA-II epitopes (PIRCHE II) are also predictive of acute GVHD.
Visual Abstract
Abstract
HLA compatibility is a key factor for survival after unrelated hematopoietic stem cell transplantation (HSCT). HLA-A, -B, -C, -DRB1, and -DQB1 are usually matched between donor and recipient. By contrast, HLA-DPB1 mismatches are frequent, although it is feasible to optimize donor selection and DPB1 matching with prospective typing. Because classical DPB1 allele mismatches are often unavoidable, however, several biological models have been developed to predict the optimal DPB1 mismatch combination for less graft-versus-host disease (GVHD) and better overall survival. In 909 recipient/donor pairs, we analyzed the role of 3 biological models: T-cell epitopes (TCEs) based on the immunogenicity of DPB1, cell surface expression of DPB1 molecules based on a single-nucleotide polymorphism located in the 3′ untranslated region, and the Predicted Indirectly ReCognizable HLA Epitopes (PIRCHE) model based on the presentation of allogeneic peptides derived from mismatched HLA, compared with the classical allele mismatch. Matching for both DPB1 alleles remains the best option to prevent acute GVHD. In the situation of one DPB1 allele mismatch, the donor associated with the lowest acute GVHD risks is mismatched for an allele with a low expression profile in the recipient, followed by a permissive TCE3/4 mismatch and/or the absence of PIRCHE II potential against the recipient. In the context of 2 DPB1 mismatches, the same considerations apply for a permissive TCE3/4 mismatch and no PIRCHE II. By combining the biological models, the most favorable DPB1 constellation can be defined. This approach will help optimize donor selection and improve post-HSCT complications and patient prognosis.
Introduction
The significant role of HLA-DPB1 allele mismatches in hematopoietic stem cell transplantation (HSCT) has been well described.1-3 Historically, HLA-DPB1 matching was not considered in the selection of unrelated donors, and mismatches were expected in up to 80% to 85% of otherwise matched unrelated transplant pairs (ie, recipient transplanted with 10/10 matched unrelated donors [MUDs]).2,4 Nowadays, with the introduction of routine HLA-DPB1 typing of patients and upfront typing at donor recruitment, it is feasible to identify HLA 12/12 matched donors for many patients. However, HLA 12/12 matched donors cannot be identified for a substantial number of patients. In this context in which mismatches are often unavoidable, several alternative matching strategies have been sought to define some level of biological permissiveness and to improve clinical outcomes.
The first biological model is based on the immunogenicity of HLA-DPB1 molecules inferred from T-cell epitopes (TCEs) localized in the peptide-binding region. Three (TCE3)5 or four (TCE4)6 functional groups of alleles were defined, respectively, allowing classification of mismatches as permissive or nonpermissive. More attention has been given to TCE3 compared with TCE4,7,8 until recently.9
In a different conceptual model, the risks of acute graft-versus-host disease (aGVHD) in transplants with a single HLA-DPB1 mismatch were associated with a single-nucleotide polymorphism (rs9277534) located in the 3′ untranslated region of the HLA-DPB1 regulatory region shown to significantly influence the quantity of cell surface expression of DPB1 molecules mediating allorecognition (ie, for proof-of-principle that expression is a functional determinant).10 The expression model was not designed for double mismatches, which were considered unacceptable given the high risk of severe grade 3 to 4 aGVHD. The presence of a high expression allele in HLA-DPB1–matched transplantations was also linked to an increased risk of GVHD, probably because of enhanced donor recognition of minor histocompatibility antigens presented by the recipient. Interestingly, a strong correlation between the two rs9277534 variants and TCE grouping has been observed, suggesting that the immunogenicity of HLA-DPB1 molecules could be related, at least to some extent, to their expression levels.11,12
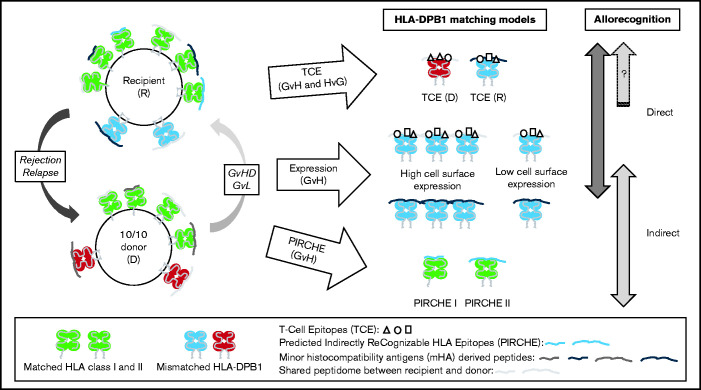
A third model relies on the indirect component of allorecognition and the presentation of allogeneic peptides derived from mismatched HLA molecules to T cells by a shared HLA molecule between the donor and recipient. An in silico approach was developed to predict the number of such peptides labeled PIRCHE (Predicted Indirectly ReCognizable HLA Epitopes).13 The presence of PIRCHE was shown to correlate with clinical outcomes after HSCT.14-16 The biological models and their theoretical and relative contributions to T-cell alloreactivity against HLA-DPB1 incompatibilities are schematically presented in Figure 1. Each biological model considers different aspects of allorecognition (ie, direct and/or indirect), although some information is shared across models. In addition, each model is not applicable to every situation of matching (ie, transplants with 1 or 2 mismatches and vector of incompatibility, as shown in Figure 1).
Figure 1.
The biological models and their theoretical and relative contributions to T-cell alloreactivity against HLA-DPB1 incompatibilities. Schematic view of HSCT involving a recipient and a 10/10 MUD carrying at least one HLA-DPB1 allele mismatch. Matched HLA class I and II molecules are shown in green; the mismatched HLA-DPB1 molecules in donor and recipient are shown in red and blue, respectively. The donor and recipient differ genetically at genes encoding minor histocompatibility antigens that can be derived into antigenic peptides presented in the peptide-binding groove of HLA molecules (represented by different shades of gray). They can also present some peptides in common (ie, shared peptidome shown in the same gray tone). The 3 biological models for predicting permissive HLA-DPB1 mismatches are illustrated, each one differing in the type of T-cell allorecognition possibly involved (ie, direct and/or indirect) and in the vector of incompatibility (ie, in both directions for TCEs or GvH for expression and PIRCHE). Furthermore, the expression model is limited to situations in which the donor and recipient differ by only one HLA-DPB1 mismatch with an incompatibility in the GvH direction (ie, also including bidirectional mismatches). The expression model was not designed for double mismatches, which were considered unacceptable given the high risk of severe grade 3 to 4 aGVHD. The TCE model is determined by immunogenic variations called TCEs that are located within the peptide-binding region of HLA molecules. These epitopes can be directly recognized by alloreactive T cells. TCE permissiveness and differential immunopeptidome presentation have been proposed to be mediated by HLA-DM peptide editing, thus also potentially involving the indirect pathway of allorecognition in the TCE model. In the expression model, high cell surface expression of the mismatched HLA-DPB1 molecule in the recipient can favor two types of allorecognition, either direct, notably through the TCEs, or indirect, in cases of allopeptides derived from minor histocompatibility antigens and presented in the peptide-binding groove of the mismatched (but also of the matched) HLA-DPB1 molecule. Finally, indirect allorecognition is also expected in cases of allopeptides derived from mismatched HLA-DPB1 molecule(s) that can be presented in the peptide-binding groove of shared HLA class I or II molecules. This type of recognition is described by the PIRCHE model. D, donor; R, recipient.
Based on direct, indirect, or both pathways of antigen recognition, it is unclear if the models can act synergistically or if each model is independent. In the current study, we analyzed clinical outcome in 909 recipient/donor pairs with a focus on the DPB1 matched/mismatched allele(s) stratified according to the classical model compared with the TCE, expression, or PIRCHE models.
Based on the results, we propose an algorithm for the selection of unrelated donors with lowest aGVHD risks that includes every model depending on the DPB1 matched/mismatched allele(s) constellation. Our data could be relevant to further refine the donor search and also to help in the strategy of exploiting the HLA-DPB1 mismatch permissiveness in cellular immunotherapy.17
Materials and methods
Study design, patients, and HLA-DPB1 typing
The role of HLA-DPB1 matching in the Swiss cohort was analyzed retrospectively by considering all 10/10 matched allografts performed from 2008 to 2018. This comprised a total of 909 patients from 4 transplant centers. It mainly consisted of first allografts performed with peripheral blood stem cells as treatment of hematologic malignancies. GVHD prophylaxis was by drugs, mainly cyclosporine with methotrexate or mycophenolate; 71% used serotherapy, mostly antithymocyte globulin added to the drug regimen. Eight percent used in vitro T-cell depletion by Alemtuzumab. No posttransplant cyclophosphamide was used in these patients. Table 1 provides details on patient, donor, and transplant characteristics.
Table 1.
Patient, donor, and transplant characteristics
| Characteristic | Value | Characteristic | Value | Characteristic | Value |
|---|---|---|---|---|---|
| Age of patients | EBMT risk score | Source of stem cells | |||
| <20 y | 123 (14%) | 1-2 | 56 (6%) | Bone marrow | 139 (15%) |
| 20-40 y | 153 (17%) | 3-4 | 560 (62%) | Peripheral blood stem cells | 769 (85%) |
| 40-60 y | 357 (39%) | 5 | 293 (32%) | Cord blood | 1 (0.1%) |
| 60-70 y | 248 (27%) | Comorbid conditions * | Total body irradiation | ||
| >70 y | 28 (3%) | No | 342 (38%) | No | 628 (69%) |
| Year of treatment | Yes | 382 (42%) | Yes | 279 (31%) | |
| 2008 | 50 (5.5%) | Missing | 185 (20%) | NA | 2 (0.2%) |
| 2009 | 58 (6%) | Karnofsky performance scale index | Conditioning | ||
| 2010 | 51 (6%) | 90-100 | 688 (76%) | Myeloablative | 465 (51%) |
| 2011 | 76 (8.5%) | ≤80 | 214 (23%) | Reduced intensity | 443 (49%) |
| 2012 | 80 (9%) | Missing | 7 (1%) | NA | 1 (0.1%) |
| 2013 | 77 (8%) | No. of allograft | Graft manipulation | ||
| 2014 | 93 (10%) | First | 873 (96%) | None | 191 (21%) |
| 2015 | 100 (11%) | Not first | 36 (4%) | Serotherapy/other | 641 (71%) |
| 2016 | 96 (11%) | Sex matching (D/R) | In vitro T-cell depletion | 77 (8%) | |
| 2017 | 115 (13%) | Male/male | 436 (48%) | HLA-DRB3/4/5 matching | |
| 2018 | 113 (12%) | Female/male | 118 (13%) | Matched | 845 (93%) |
| Type of diagnosis | Male/female | 187 (21%) | 1 mismatch DRB3 | 35 (4%) | |
| Acute leukemia | 506 (56%) | Female/female | 168 (18%) | 1 mismatch DRB4 | 28 (3%) |
| MDS/MPN | 181 (20%) | CMV serostatus matching (D/R) | 1 mismatch DRB3 and DRB4 | 1 (0.1%) | |
| Lymphoid malignancy† | 84 (9%) | Negative/negative | 338 (37%) | Transplant center ‡ | |
| NMD | 72 (8%) | Positive/negative | 95 (11%) | 202 | 327 (36%) |
| PCD | 42 (5%) | Negative/positive | 180 (20%) | 208 | 237 (26%) |
| CML | 23 (2%) | Positive/positive | 287 (32%) | 261 | 263 (29%) |
| ST | 1 (0.1%) | Age of donors, y | 334 | 82 (9%) | |
| Status of disease | Median | 31.3 | |||
| Early | 445 (49%) | IQR | 25.2-40.0 | ||
| Intermediate | 279 (31%) | ||||
| Late | 185 (20%) |
CML, chronic myeloid leukemia; CMV, cytomegalovirus; D, donor; IQR, interquartile range; MDS/MPN, myelodysplastic/myeloproliferative syndromes; NA, nonavailable; NMD, all nonmalignant disorders; PCD, plasma cell disorders; R, recipient; ST, solid tumor.
Based on the hematopoietic cell transplantation–specific comorbidity index.
Lymphoid malignancy regroups non-Hodgkin lymphoma, Hodgkin disease, and chronic lymphatic leukemia/prolymphocytic leukemia.
The transplant center code according to European Society for Blood and Marrow Transplantation (EBMT) is listed for the 4 allogeneic centers of Switzerland. All covariables tested in univariate analyses are shown here.
In Switzerland, prospective HLA-DPB1 typing in transplant candidates and selected unrelated donors was introduced at the end of 2016 with the development of high-throughput sequencing. Prospective typing was also performed from 2012 onward for each patient, with several potential 10/10 MUDs identified in the Bone Marrow Donors Worldwide/World Marrow Donor Association database. At the time of the search request, 7.8% of patients in the cohort had only one potential 10/10 MUD identified in the Bone Marrow Donors Worldwide/World Marrow Donor Association database; 28.8% had between 1 and 5 MUDs; and 63.4% had >5 MUDs. For the purpose of the current study, retrospective typing was performed for the donor/recipient pairs not yet fully characterized by using reverse polymerase chain reaction sequence–specific oligonucleotide microbead arrays (One Lambda, Canoga Park, CA) and polymerase chain reaction sequence–specific primers (Genovision, Milan Analytika AG, Rheinfelden, Switzerland). Complementary matching at HLA-DRB3/4/5 was also available and was included as a covariable in the analyses.
This study was approved by the ethical committee of the canton of Geneva and the Geneva University Hospital (CER 06-208 and 08-208R) and was conducted in accordance with the Declaration of Helsinki.
HLA-DPB1 matching models
Several models were considered in this study. The classical approach of counting the number of HLA-DPB1 allele mismatches was first examined, either with or without taking into account the direction of the vector of incompatibility in case of transplants mismatched for one allele. In terms of models inferring biological permissiveness, permissive and nonpermissive TCE mismatches were defined for each pair in the cohort, as previously described for the TCE3 and TCE4 algorithms using Linux/Bash scripts.5,6,18 The nonpermissive mismatches were then split into 2 subgroups (ie, graft-versus-host [GvH] and host-versus-graft [HvG] incompatibilities) or kept together for the analyses. The cell surface expression of HLA-DPB1 alleles in donors and recipients was inferred from the described linkage between exonic variation and the 3′ untranslated region rs9277534-G/A polymorphism.10,19 This allowed the classification of the expression level (ie, respectively high for G-linked alleles and low for A-linked alleles) of all single HLA-DPB1 mismatches with a vector of incompatibility in the GvH direction. In this model, pairs defined by 2 mismatches or by 1 mismatch in the HvG direction only could not be classified and were excluded from the analyses (one-third of the cohort, n = 305). The numbers of PIRCHE derived from the recipient’s mismatched HLA-DPB1 allele(s) and potentially presented in the GvH direction on shared HLA class I (PIRCHE I) or class II (PIRCHE II) molecules between donor and recipient were identified by using the PIRCHE Web tool (www.pirche.com, version 3.1.147) as described elsewhere.13 Of note, only potential binders to HLA-DRB1/3/4/5 molecules were considered in this study because typing for HLA-DQA1 and HLA-DPA1 was not available to predict peptide-binding affinities to HLA-DQ and HLA-DP heterodimers. The distribution of HLA-DPB1 mismatches according to the different biological models is detailed in supplemental Table 1 and is presented for PIRCHE I and II in supplemental Figure 1.
Statistical analysis
The clinical end points considered were overall survival, transplant-related mortality (TRM), grade 2 to 4 aGVHD, grade 3 to 4 aGVHD, chronic GVHD, and relapse/progression (Rel/prog). Initial exploration of the data (ie, single and combined HLA-DPB1 matching models and all covariables listed in Table 1) was done by using univariate analysis (ie, Kaplan-Meier, log-rank test, and cumulative incidence with competing risks). The data were then fitted into Cox multivariable regression models to compare the hazard ratios (HRs) between appropriate HLA-DPB1 matching groups and for each outcome adjusted for relevant covariables. A customary model-building strategy was used in which all covariables somehow associated with outcome and significant in univariate analysis were entered into the model and nonsignificant covariables were eliminated in a backward stepwise model-building procedure. A P value <.05 was considered as significant.
Each of the nine HLA-DPB1 matching models considered in this study was analyzed individually for the different outcomes. The most relevant biological models (ie, with a significant HR retrieved for at least 1 subgroup of patients) were then combined two-by two into 5 models. The subgroups considered at this stage consisted of all possible pairwise combinations from both selected models (eg, TCE3.1 and expression) and were either the same as for the individual analyses (eg, permissive or nonpermissive for TCEs) or pooled subgroups (eg, recipient with a highly expressed mismatch, thus not accounting for the expression level of the mismatch in the donor in contrast to the analyses performed for expression alone). The choice of pooling categories was made firsthand on the basis of the results obtained in the individual matching models (ie, keeping the relevant information for further testing) but also to keep a meaningful number of patients within each subgroup.
Results
HLA-DPB1 matching for predicting HSCT outcomes
Univariate analyses revealed higher risks of aGVHD and lower incidence of relapse in several HLA-DPB1 mismatched groups compared with fully matched allografts. The other clinical end points were not associated with HLA-DPB1 (selected Kaplan-Meier plots are shown for grade 2-4 aGVHD and relapse in supplemental Figures 2-5). Multivariable analyses confirmed this profile (Table 2; supplemental Table 2). Compared with 12/12 transplants (or alternatively to the absence of PIRCHE), the risks of grade 2 to 4 aGVHD were significantly increased: (1) with the presence of one (if bidirectional) or two allele mismatches; (2) with the presence of at least one PIRCHE II; (3) with the presence of a highly expressed mismatched allele in recipient (not the donor); (4) with the presence of nonpermissive TCE3/TCE4 mismatches; and (5) with the presence of permissive TCE3 mismatches, the latest group with a P value of .05. The risks of grade 3 to 4 aGVHD were increased significantly for expression (ie, in pairs with a highly expressed mismatched allele in both the recipient and donor; P = .01), and similar, although not significant, results to grade 2 to 4 aGVHD were observed for most models. Interestingly, HLA-DRB3/4/5 matching was also a risk factor for aGVHD, and the risks were mainly driven by HLA-DRB3 mismatches. However, this observation relies on a very small number of patients (n = 35 and 28 for DRB3 and DRB4 mismatches, respectively, results not shown).
Table 2.
Multivariable analyses for aGVHD grade 2 to 4 and Rel/prog and association with each HLA-DPB1 matching model
| HLA-DPB1 matching | Categories | aGVHD grade 2 to 4 (N = 860, events = 297)* | Rel/prog (N = 837, events = 302)*† | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Events/n | HR | 95% CI | P | Events/n | HR | 95% CI | P | ||||
| Classical matching.1 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| 1 mismatch | 145/398 | 1.40 | 1.05 | 1.87 | .02 | 145/393 | 0.81 | 0.62 | 1.05 | .11 | |
| 2 mismatch | 85/212 | 1.52 | 1.10 | 2.10 | .01 | 61/207 | 0.62 | 0.45 | 0.86 | .004 | |
| Classical matching.2 | matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| 1 mismatch bidirectional | 101/254 | 1.60 | 1.17 | 2.19 | .003 | 96/248 | 0.84 | 0.63 | 1.11 | .22 | |
| 1 mismatch GvH | 23/79 | 1.01 | 0.63 | 1.63 | .96 | 23/76 | 0.66 | 0.42 | 1.05 | .08 | |
| 1 mismatch HvG | 21/65 | 1.20 | 0.73 | 1.95 | .48 | 26/69 | 0.88 | 0.57 | 1.36 | .56 | |
| 2 mismatch | 85/212 | 1.52 | 1.10 | 2.10 | .01 | 61/207 | 0.62 | 0.45 | 0.86 | .004 | |
| TCE3.1 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive | 115/288 | 1.54 | 1.13 | 2.09 | .006 | 91/284 | 0.68 | 0.51 | 0.91 | .01 | |
| Permissive | 115/322 | 1.36 | 1.00 | 1.84 | .05 | 115/316 | 0.80 | 0.61 | 1.05 | .10 | |
| TCE3.2 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive GvH | 71/165 | 1.76 | 1.25 | 2.46 | .001 | 50/163 | 0.67 | 0.48 | 0.95 | .02 | |
| Nonpermissive HvG | 44/123 | 1.28 | 0.87 | 1.88 | .21 | 41/121 | 0.70 | 0.48 | 1.01 | .06 | |
| Permissive | 115/322 | 1.36 | 1.00 | 1.84 | .05 | 115/316 | 0.80 | 0.61 | 1.05 | .10 | |
| TCE4.1 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive | 156/403 | 1.48 | 1.11 | 1.97 | .008 | 127/403 | 0.66 | 0.51 | 0.86 | .002 | |
| Permissive | 74/207 | 1.37 | 0.99 | 1.91 | .06 | 79/197 | 0.92 | 0.68 | 1.24 | .58 | |
| TCE4.2 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive GvH | 87/228 | 1.45 | 1.05 | 1.99 | .02 | 69/230 | 0.63 | 0.46 | 0.86 | .003 | |
| Nonpermissive HvG | 69/175 | 1.52 | 1.08 | 2.13 | .02 | 58/173 | 0.71 | 0.51 | 0.98 | .04 | |
| Permissive | 74/207 | 1.37 | 0.99 | 1.91 | .06 | 79/197 | 0.92 | 0.68 | 1.24 | .58 | |
| Expression | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| R-high, D-high | 24/47 | 2.84 | 1.76 | 4.58 | <.001 | 13/44 | 0.74 | 0.41 | 1.33 | .31 | |
| R-high, D-low | 47/121 | 1.56 | 1.07 | 2.27 | .02 | 31/116 | 0.61 | 0.41 | 0.92 | .02 | |
| R-low, D-high | 13/57 | 0.81 | 0.45 | 1.48 | .50 | 33/59 | 1.11 | 0.75 | 1.66 | .60 | |
| R-low, D-low | 40/106 | 1.38 | 0.93 | 2.05 | .11 | 39/102 | 0.76 | 0.52 | 1.10 | .15 | |
| PIRCHE I | 0 | 137/438 | 1.00 | — | 161/424 | 1.00 | — | ||||
| 1-3 | 116/297 | 1.26 | 0.98 | 1.62 | .07 | 99/284 | 0.83 | 0.64 | 1.06 | .13 | |
| >3 | 44/125 | 1.17 | 0.83 | 1.66 | .37 | 42/129 | 0.84 | 0.60 | 1.18 | .32 | |
| PIRCHE II | 0 | 110/389 | 1.00 | — | 151/378 | 1.00 | — | ||||
| 1-10 | 97/250 | 1.51 | 1.15 | 1.99 | .003 | 83/236 | 0.76 | 0.58 | 0.99 | .04 | |
| >10 | 90/221 | 1.46 | 1.09 | 1.95 | .01 | 68/223 | 0.73 | 0.55 | 0.97 | .03 | |
.05 > P ≥ .01 are shown in bold and italic; P < 0.01 are shown in bold, italic, and underlined. Significant covariables retained for aGVHD: HLA-DRB3/4/5 matching, graft manipulation, and transplant center.
Significant covariables retained for relapse/progression (Rel/prog): European Society for Blood and Marrow Transplantation risk score, graft manipulation, and transplant center. CI, confidence interval; D, donor; Events, number of events in the risk category for the specified outcome; n, number of patients in the risk category for the specified outcome; R, recipient.
The number of patients/events for the regressions with expression is N = 579/191 and 558/212 for aGVHD ≥2 and Rel/prog, respectively.
Patients with nonmalignant disorder are excluded from analyses on relapse.
Mirroring closely the increased risk of aGVHD, a lower incidence of relapse/progression was observed for the same groups compared with 12/12 allografts, except for one allele mismatches, R-high/D-high mismatched pairs, and TCE3 permissive mismatches, which were not statistically different (Table 2).
Less convincing results were observed for the other outcomes, as defined by P values very close to .05. These results are presented in supplemental Table 2 and are not discussed further here.
Combined biological models of permissiveness
The most relevant biological models analyzed in the previous section (ie, PIRCHE II, expression, TCE3.1 and TCE4.1) were combined two-by-two to investigate potential additive or synergistic effects on the primary outcomes. Again, highly significant HRs were observed for grade 2 to 4 aGVHD and relapse but not for the other outcomes (Table 3; supplemental Table 3). Foremost, the presence of a highly expressed mismatched allele in the recipient was associated with an increased risk of aGVHD compared with 12/12 allografts, whereas no difference was observed for lower expressed HLA-DPB1 mismatches. This observation was consistent across all subgroups; that is, combined with permissive or nonpermissive TCE3/TCE4 mismatches or with the absence or presence of PIRCHE II. In other words, considering TCE or PIRCHE II was not informative for stratifying the risks of aGVHD within groups with different levels of expression. By contrast, the results for TCEs and PIRCHE II were less straightforward. The presence of PIRCHE II was associated with a 67% or 73% risk increase of aGVHD compared with 12/12 transplants when combined with nonpermissive TCE4 or TCE3 mismatches, respectively, and a slightly less increase in combination with TCE3 permissive mismatches (40%; P = .04) but not with TCE4 (P = .1). Moreover, the absence of PIRCHE II in the context of either permissive or nonpermissive TCE3/TCE4 mismatches was not significantly different from 12/12 transplants (pairwise log-rank tests, data not shown; supplemental Figure 4). Taken together, our results suggest that PIRCHE II could add information to the TCE model, at least in case of nonpermissiveness. The results for grade 3 to 4 aGVHD were very similar to those for grade 2 to 4 aGVHD, but significant differences were not retrieved because these events were much rarer in our cohort (supplemental Table 3).
Table 3.
Multivariable analyses for acute GVHD grade 2 to 4 and Rel/prog and association with combined biological models
| HLA-DPB1 matching | Categories | aGVHD grade 2 to 4 (N = 860, events = 297)* | Rel/prog (N = 837, events = 302)*† | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Events/n | HR | 95% CI | P | Events/n | HR | 95% CI | P | ||||
| TCE3.1 and PIRCHE II | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive, no PIRCHE II | 13/51 | 0.85 | 0.47 | 1.53 | .58 | 20/49 | 0.87 | 0.53 | 1.42 | .57 | |
| Nonpermissive, PIRCHE II | 102/237 | 1.73 | 1.27 | 2.37 | <.001 | 71/235 | 0.65 | 0.47 | 0.88 | .005 | |
| Permissive, no PIRCHE II | 31/91 | 1.28 | 0.84 | 1.97 | .25 | 36/95 | 0.88 | 0.59 | 1.29 | .50 | |
| Permissive, PIRCHE II | 84/231 | 1.40 | 1.01 | 1.93 | .04 | 79/221 | 0.77 | 0.57 | 1.03 | .08 | |
| TCE4.1 and PIRCHE II | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| Nonpermissive, no PIRCHE II | 25/91 | 0.93 | 0.59 | 1.48 | .77 | 33/92 | 0.75 | 0.50 | 1.12 | .15 | |
| Nonpermissive, PIRCHE II | 131/312 | 1.67 | 1.24 | 2.25 | .001 | 94/311 | 0.64 | 0.48 | 0.85 | .002 | |
| Permissive, no PIRCHE II | 19/51 | 1.48 | 0.89 | 2.48 | .13 | 23/52 | 1.15 | 0.72 | 1.82 | .56 | |
| Permissive, PIRCHE II | 55/156 | 1.35 | 0.94 | 1.93 | .10 | 56/145 | 0.85 | 0.61 | 1.19 | .34 | |
| Expression and PIRCHE II | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| R-high, no PIRCHE II‡ | 9/13 | NI | NI | NI | NI | 4/11 | NI | NI | NI | NI | |
| R-high, PIRCHE II | 62/155 | 1.72 | 1.21 | 2.44 | .002 | 40/149 | 0.61 | 0.42 | 0.88 | .008 | |
| R-low, no PIRCHE II | 10/42 | 0.86 | 0.44 | 1.67 | .66 | 16/42 | 0.72 | 0.42 | 1.23 | .23 | |
| R-low, PIRCHE II | 43/121 | 1.30 | 0.88 | 1.92 | .19 | 56/119 | 0.95 | 0.68 | 1.32 | .75 | |
| Expression and TCE3.1 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| R-high, nonpermissive | 39/89 | 1.87 | 1.26 | 2.80 | .002 | 26/86 | 0.73 | 0.47 | 1.13 | .15 | |
| R-high, permissive | 32/79 | 1.79 | 1.17 | 2.73 | .008 | 18/74 | 0.55 | 0.33 | 0.92 | .02 | |
| R-low, nonpermissive | 11/36 | 1.10 | 0.58 | 2.10 | .77 | 19/35 | 1.00 | 0.61 | 1.65 | 1.00 | |
| R-low, permissive | 42/127 | 1.21 | 0.82 | 1.79 | .34 | 53/126 | 0.85 | 0.61 | 1.20 | .36 | |
| Expression and TCE4.1 | Matched | 67/250 | 1.00 | — | 96/237 | 1.00 | — | ||||
| R-high, nonpermissive | 43/98 | 1.88 | 1.28 | 2.77 | <.001 | 29/95 | 0.71 | 0.47 | 1.09 | .12 | |
| R-high, permissive | 28/70 | 1.76 | 1.13 | 2.75 | .01 | 15/65 | 0.54 | 0.31 | 0.93 | .03 | |
| R-low, nonpermissive | 27/89 | 1.11 | 0.71 | 1.74 | .65 | 35/91 | 0.71 | 0.48 | 1.05 | .08 | |
| R-low, permissive | 26/74 | 1.28 | 0.81 | 2.02 | .29 | 37/70 | 1.16 | 0.79 | 1.70 | .45 | |
.05 > P ≥ .01 are shown in bold and italic; P < .01 are shown in bold, italic, and underlined. Significant covariables retained for relapse/progression (Rel/prog): European Society for Blood and Marrow Transplantation risk score, graft manipulation, and transplant center. Significant covariables retained for aGVHD: HLA-DRB3/4/5 matching, graft manipulation, and transplant center. CI, confidence interval; D, donor; Events, number of events in the risk category for the specified outcome; n, number of patients in the risk category for the specified outcome; R, recipient.
The number of patients/events for the regressions with expression is N = 579/191 and 558/212 for aGVHD ≥2 and Rel/prog, respectively.
Patients with nonmalignant disorder are excluded from analyses on relapse.
Not interpretable (NI) because of the very small number of patients in this group.
Regarding relapse/progression, the risks were decreased compared with 12/12 transplants in the following 3 groups: nonpermissive TCE3/TCE4 mismatches combined with the presence of PIRCHE II, highly expressed mismatched allele in recipient and presence of PIRCHE II, or when combined with permissive TCE3/TCE4 mismatches.
Discussion
This retrospective study is in line with the growing amount of evidence pointing to HLA-DPB1 matching and its related biological models of permissiveness as important parameters to consider for optimizing donor selection to improve prognosis. Among the several primary clinical end points examined here, aGVHD and relapse were shown to be significantly influenced by HLA-DPB1 matching. A balance between deleterious and protective effects (ie, higher risk of aGVHD vs lower incidence of relapse) has been proposed to explain why DPB1 allele mismatches have usually not been associated with a significant difference in survival,3,9 with few exceptions.4,20
Allowing for a more fine-tuned approach than just counting the number of mismatches, the biological models examined here were also informative regarding the risks of aGVHD and relapse, except for PIRCHE I. Interestingly, we found no significant differences for the other outcomes (overall survival, TRM, and chronic GVHD). In agreement with our results, a high cell surface expression of one mismatched allele in recipient has previously been associated with increased risks of aGVHD9,10,21,22 and sometimes with a decreased incidence of relapse.10 Furthermore, our results are consistent with previous findings that the level of expression of the patient’s mismatched HLA-DPB1 allele correlates with outcome more so than the expression level of the donor’s mismatched allele and follows a biological GvH recognition.22 Differences between fully matched transplants, permissive and nonpermissive TCE mismatches were previously described, with some heterogeneity across studies, for survival and TRM,4-8 aGVHD,5,7,8,22,23 and relapse.8,23,24 Some studies also did not observe any significant differences for TCE3 or TCE4 for these outcomes.20,25 Our results are thus consistent regarding aGVHD and relapse; we do not retrieve a signal for the other clinical end points.
For the most part, our results on relapse accompanied the ones observed for aGVHD, although with minor differences regarding groups that were associated with a significant P value. We thus focus on aGVHD to discuss in more detail the specific contributions of each biological model. The concordance between expression and TCEs (ie, at classifying high-risk vs low-risk mismatches) in our cohort was 68%, a percentage similar to those already reported.12,22 We detected more PIRCHE I and II with TCE nonpermissive GvH mismatches than with permissive and nonpermissive HvG mismatches, similar to a previous report by Thus et al.15 In addition, the number of PIRCHE I and II was the largest in recipients with a highly expressed mismatched allele, especially when the donor-mismatched allele had a low expression (supplemental Figure 1). It is thus possible that PIRCHE acts as a partial surrogate for TCEs and expression regarding clinical outcomes. Our results suggest that the presence of at least one PIRCHE II was sufficient to affect significantly the risks of GVHD and that this was not driven by the number of potential binders (Tables 2 and 3); this theory remains to be formally investigated. The study of Thus et al15 reported a significant correlation between the presence of PIRCHE I and PIRCHE II and the increased incidence of aGVHD. However, this was observed in a much smaller group of patients (n = 88). Of interest, we observed a trend toward increased incidence of aGVHD with the presence of PIRCHE I early posttransplant, but the differences were not significant globally and at later stages (results not shown). The importance of PIRCHE I therefore warrants confirmation by independent studies and needs to be contrasted with the influence of PIRCHE II.
Recent comparative analyses using expression and TCEs,9,12,21,22 on the one hand, or TCEs and PIRCHE,15 on the other hand, have proposed that their combination provides more information than either model alone and helps to better stratify the risks. This is because each model emphasizes only part of the complex mechanisms of T-cell alloreactivity against incompatible HLA-DPB1 molecules. Indeed, the biological models are analyzing distinct components of the alloreactive response but with some overlap (Figure 1). For instance, recent evidence suggests that TCE permissive mismatches present less divergent immunopeptidomes than nonpermissive mismatches, with a role for HLA-DM mediating peptide editing.26 Components of both direct and indirect allorecognition are thus probably involved in the TCE model, similarly to the expression model, whereas PIRCHE is strictly restricted to the indirect pathway of recognition. Moreover, the different models do not cover the same breadth of information; for example, expression and PIRCHE focus specifically on incompatibilities in the GvH direction, and expression does not account for more than one HLA-DPB1 mismatch. Actually, their relative contributions were not straightforward to interpret when combined two-by-two in multivariable regressions. Each model seemed to play a significant role but more on an individual basis rather than by acting in concert. Previous studies reported similar complex relationships,21,22 whereas others have suggested that the biological models could be prioritized according to their performance for different clinical outcomes (eg, expression with aGVHD or TCE4 with survival, respectively).9 A recent analysis observed increased risks for GVHD/relapse-free survival, nonrelapse mortality, and aGVHD and reduced risks for relapse in grafts with two DPB1 mismatches combined to TCE3 nonpermissiveness in GvH direction.23 An additive effect of expression combined with a TCE3 nonpermissive allotype was associated with aGVHD and relapse. In contrast to our data, the risks were assessed by combining both 10/10 and 9/10 grafts. A synthetic look at aGVHD in Table 3 found that expression adds new information beyond the information provided by PIRCHE II or by the TCE status, whereas the reverse situation is not true (ie, the TCE status or presence of PIRCHE II does not add new information once the level of expression is determined).
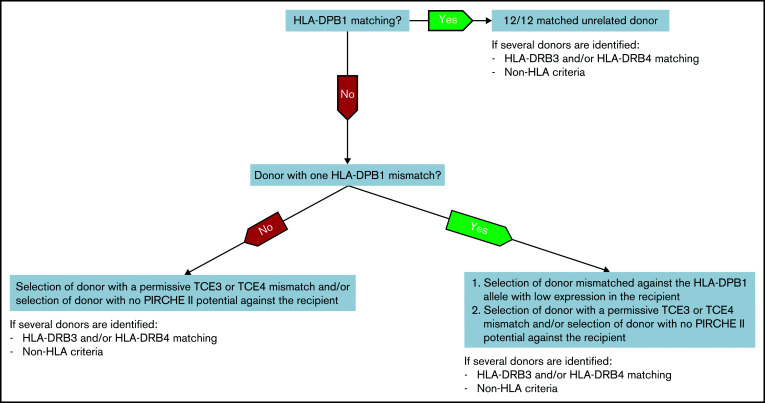
Also, once the TCE status is defined, the presence of PIRCHE II adds new information, whereas TCEs provide additional information only in the presence of PIRCHE II. This led us to propose a tentative algorithm for selecting unrelated donors with lowest aGVHD risks in Switzerland, as presented in Figure 2. Although the decision tree is mainly devised for patients with a low risk of relapse, adapting the selection to a donor carrying either two DPB1 mismatches, a mismatch against a highly expressed allele in the recipient, a nonpermissive TCE mismatch, or having a high PIRCHE II potential could also be beneficial in situations in which the risk of relapse/progression in the patient is preponderant over the risk of acute GVHD and should be minimized. For instance, permissive mismatches have been proposed to be associated with a limited alloreactivity sufficient to elicit GVL, thus maintaining treatment efficacy, without the deleterious effects of clinically uncontrollable GVHD.17 However, we do not see any effect on overall survival of the different models, but reduction of the risks of aGVHD would be associated with less immunosuppression, better immune reconstitution, lower risk of concomitant infections, and other complications.27-30 Thus, a flexible and individualized approach should always be considered along these general guidelines. Feasibility of TCE permissive matching for selecting prospectively among unrelated donors who were equally matched has previously been shown,31 and several donor algorithms (eg, HapLogic,32 OptiMatch [https://search.wmda.info/login]) already include information about TCE permissiveness in their match grade. Considerations about incorporating the other models in these algorithms should arise given our results and other recent studies addressing this issue.9,22
Figure 2.
Decision tree for optimizing donor selection for HSCT candidates based on the results of this study and developed to reduce aGVHD risks in patients with a low risk of relapse. Classical HLA-DPB1 matching is the first parameter considered if a 10/10 donor can be identified. In case a 12/12 matched donor is not identified, the feasibility of selecting a donor with one or two biological permissive mismatches is explored. In cases when one or several donors carrying only one HLA-DPB1 allele mismatch are available, a low expression mismatched allele should be preferred in the recipient, followed by the feasibility to select a permissive TCE3 or TCE4 mismatch and/or to avoid PIRCHE II. The prioritization of expression over the 2 other biological models is based on the combined analyses in Table 3 showing that expression adds new information beyond TCEs and PIRCHE II. Of course, TCEs and PIRCHE II in addition to expression or as an alternative strategy are also informative. When allele matching cannot be achieved (ie, for selecting among unrelated donors with two HLA-DPB1 mismatches), the feasibility to select a permissive TCE3 or TCE4 mismatch and/or to avoid PIRCHE II applies. According to our data, TCEs and PIRCHE II should be considered equally during donor selection. The role of HLA-DRB3/4 matching and additional non-HLA factors should also be considered.
Our data also sustain a role of HLA-DRB3/4 matching for predicting the risks of aGVHD as an independent factor and as a covariable in the univariate and multivariable analyses, respectively.
Our study has some limitations despite the large size of the cohort. For instance, a few groups were relatively small for the analyses that combined the biological models two-by-two. We performed power and sample size calculations (not shown), and we used 2 different groupings for the TCE model with distinct sample sizes, which make us confident that our results are robust. For the same reason, we could not perform analyses combining the 3 models together. Also, grade 3 to 4 aGVHD is of main clinical importance due to its potential for severe sequelae, including death. However, this concerns rather rare events and because of this, we could not retrieve significant signals in our data, although the results resembled the significant observations made on grade 2 to 4 aGVHD. In addition, the tool for PIRCHE II is based on peptides derived from the mismatched DPB1 alleles presented on DRB1, DRB3, DRB4, and DRB5 and not on DQB1 or DPB1 because DRA is monomorphic and the polymorphic DQA1 and DPA1 genes were not typed to allow the software to make binding predictions for DQ and DP heterodimers. A transplant center effect was detected with several clinical outcomes. Although the centers share common practices (eg, donor selection process, HLA compatibility), clinical protocols can differ among them. The multivariable analyses were adjusted for this effect.
Although several studies have already compared the TCEs and the expression model9,12,21-23 or PIRCHE and TCEs,15 the current study, to the best of our knowledge, is the first to include the 3 models. In summary, HLA-DPB1 matching for both alleles (ie,12/12) remains the best option to prevent aGVHD. In the context of one HLA-DPB1 allele match, the donor with the lowest aGVHD risk would be mismatched to an allele with low cell surface expression in the recipient, followed by a permissive TCE3 or TCE4 mismatch and/or by the absence of PIRCHE II potential against the recipient. In the context of 2 DPB1 allele mismatches, the donor with the lowest aGVHD risk would have a permissive TCE3 or TCE4 mismatch and/or no PIRCHE II potential against the recipient. Because the 3 models are significant and exhibit complex relationships, this should be confirmed by independent studies, at least for the PIRCHE model, which has not been extensively tested in large cohorts. Donor selection includes immunogenetic and other factors, and the more we know, the more we will be able to personalize the choice for the best outcome. It would also be interesting to see if those findings will still be observed with the growing use of cyclophosphamide posttransplantation as GVHD prophylaxis. Finally, T cells from a donor with a DPB1 mismatch could be an interesting tool in the future for cellular immunotherapy after HSCT.17,33 The initial choice of such a donor could be part of a global strategy, including prevention of complications and potential for post-HSCT therapy to intervene in case of relapse.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors are grateful to the technicians of the National Reference Laboratory for Histocompatibility (LNRH) for their most efficient support for HLA typing.
This study was supported by the Swiss National Science Foundation (grant #310030_173237/1), IRGHET (International Research Group on Unrelated Hematopoietic Stem Cell Transplantation), and the Dr. Henri Dubois-Ferrière Dinu Lippatti foundation.
Authorship
Contribution: J.V., S.F.-L., and S.B. designed the study; J.R.P., H.B., and S.B. performed statistical analysis; and S.B. and J.V. drafted the manuscript; and all authors assembled the data, critically reviewed and edited the manuscript, and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean Villard, Transplantation Immunology Unit and National Reference Laboratory for Histocompatibility, Geneva University Hospitals, Gabrielle-Perret-Gentil 4, 1211 Geneva 4, Switzerland; e-mail: jean.villard@hcuge.ch.
References
- 1.Fernández-Viña MA, Klein JP, Haagenson M, et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 2013;121(22):4603-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersdorf EW, Gooley T, Malkki M, et al. The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br J Haematol. 2001;112(4):988-994. [DOI] [PubMed] [Google Scholar]
- 3.Shaw BE, Gooley TA, Malkki M, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110(13):4560-4566. [DOI] [PubMed] [Google Scholar]
- 4.Ludajic K, Balavarca Y, Bickeböller H, et al. Impact of HLA-DPB1 allelic and single amino acid mismatches on HSCT. Br J Haematol. 2008;142(3):436-443. [DOI] [PubMed] [Google Scholar]
- 5.Zino E, Frumento G, Marktel S, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103(4):1417-1424. [DOI] [PubMed] [Google Scholar]
- 6.Crocchiolo R, Zino E, Vago L, et al. Italian Bone Marrow Donor Registry. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114(7):1437-1444. [DOI] [PubMed] [Google Scholar]
- 7.Fleischhauer K, Shaw BE, Gooley T, et al. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pidala J, Lee SJ, Ahn KW, et al. Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood. 2014;124(16):2596-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorentino F, Sacchi N, Oldani E, et al. Comparative evaluation of biological human leukocyte antigen DPB1 mismatch models for survival and graft-versus-host disease prediction after unrelated donor hematopoietic cell transplantation. Haematologica. 2020;105(4):e186-e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersdorf EW, Malkki M, O’Huigin C, et al. High HLA-DP expression and graft-versus-host disease. N Engl J Med. 2015;373(7):599-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischhauer K. Immunogenetics of HLA-DP—a new view of permissible mismatches. N Engl J Med. 2015;373(7):669-672. [DOI] [PubMed] [Google Scholar]
- 12.Meurer T, Arrieta-Bolaños E, Metzing M, et al. Dissecting genetic control of HLA-DPB1 expression and its relation to structural mismatch models in hematopoietic stem cell transplantation. Front Immunol. 2018;9:2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geneugelijk K, Thus KA, Spierings E.. Predicting alloreactivity in transplantation. J Immunol Res. 2014;2014:159479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geneugelijk K, Thus KA, van Deutekom HWM, et al. Exploratory study of predicted indirectly recognizable HLA epitopes in mismatched hematopoietic cell transplantations. Front Immunol. 2019;10:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thus KA, Ruizendaal MT, de Hoop TA, et al. Refinement of the definition of permissible HLA-DPB1 mismatches with predicted indirectly recognizable HLA-DPB1 epitopes. Biol Blood Marrow Transplant. 2014;20(11):1705-1710. [DOI] [PubMed] [Google Scholar]
- 16.Thus KA, Te Boome L, Kuball J, Spierings E.. Indirectly recognized HLA-C mismatches and their potential role in transplant outcome. Front Immunol. 2014;5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischhauer K, Shaw BE.. HLA-DP in unrelated hematopoietic cell transplantation revisited: challenges and opportunities. Blood. 2017;130(9):1089-1096. [DOI] [PubMed] [Google Scholar]
- 18.Crivello P, Zito L, Sizzano F, et al. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2015;21(2):233-241. [DOI] [PubMed] [Google Scholar]
- 19.Schöne B, Bergmann S, Lang K, et al. Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: linkage analysis of over 32 000 samples [published correction appears in Hum Immunol. 2020;81(10-11):661]. Hum Immunol. 2018;79(1):20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettens F, Passweg J, Schanz U, et al. Impact of HLA-DPB1 haplotypes on outcome of 10/10 matched unrelated hematopoietic stem cell donor transplants depends on MHC-linked microsatellite polymorphisms. Biol Blood Marrow Transplant. 2012;18(4):608-616. [DOI] [PubMed] [Google Scholar]
- 21.Morishima S, Shiina T, Suzuki S, et al. Japan Marrow Donor Program. Evolutionary basis of HLA-DPB1 alleles affects acute GVHD in unrelated donor stem cell transplantation. Blood. 2018;131(7):808-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersdorf EW, Bengtsson M, De Santis D, et al. International Histocompatibility Working Group in Hematopoietic-Cell Transplantation. Role of HLA-DP expression in graft-versus-host disease after unrelated donor transplantation. J Clin Oncol. 2020;38(24):2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mytilineos D, Tsamadou C, Neuchel C, et al. The human leukocyte antigen-DPB1 degree of compatibility is determined by its expression level and mismatch permissiveness: a German multicenter analysis. Front Immunol. 2021;11:614976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagne K, Loiseau P, Dubois V, et al. Is there any impact of HLA-DPB1 disparity in 10/10 HLA-matched unrelated hematopoietic SCT? Results of a French multicentric retrospective study. Bone Marrow Transplant. 2015;50(2):232-236. [DOI] [PubMed] [Google Scholar]
- 25.Loiseau P, Busson M, Balere ML, et al. HLA association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13(8):965-974. [DOI] [PubMed] [Google Scholar]
- 26.Meurer T, Crivello P, Metzing MF, et al. Permissive HLA-DPB1 mismatches in HCT depend on immunopeptidome divergence and editing by HLA-DM. Blood. 2021;137(7):923-928. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhry MS, Velardi E, Malard F, van den Brink MR.. Immune reconstitution after allogeneic hematopoietic stem cell transplantation: time to T up the thymus. J Immunol. 2017;198(1):40-46. [DOI] [PubMed] [Google Scholar]
- 28.Hakki M, Riddell SR, Storek J, et al. Immune reconstitution to cytomegalovirus after allogeneic hematopoietic stem cell transplantation: impact of host factors, drug therapy, and subclinical reactivation. Blood. 2003;102(8):3060-3067. [DOI] [PubMed] [Google Scholar]
- 29.Miller HK, Braun TM, Stillwell T, et al. Infectious risk after allogeneic hematopoietic cell transplantation complicated by acute graft-versus-host disease. Biol Blood Marrow Transplant. 2017;23(3):522-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan TL, Pritchett JC, Leifer C, et al. HHV-6B infection, T-cell reconstitution, and graft-vs-host disease after hematopoietic stem cell transplantation. Bone Marrow Transplant. 2018;53(12):1508-1517. [DOI] [PubMed] [Google Scholar]
- 31.Tram K, Stritesky G, Wadsworth K, Ng J, Anasetti C, Dehn J.. Identification of DPB1 permissive unrelated donors is highly likely. Biol Blood Marrow Transplant. 2017;23(1):81-86. [DOI] [PubMed] [Google Scholar]
- 32.Dehn J, Setterholm M, Buck K, et al. HapLogic: a predictive human leukocyte antigen-matching algorithm to enhance rapid identification of the optimal unrelated hematopoietic stem cell sources for transplantation. Biol Blood Marrow Transplant. 2016;22(11):2038-2046. [DOI] [PubMed] [Google Scholar]
- 33.Chabannon C, Kuball J, Bondanza A, et al. Hematopoietic stem cell transplantation in its 60s: a platform for cellular therapies. Sci Transl Med. 2018;10(436):eaap9630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.