Key Points
Lymphoid blast transformation of an MPN with BCR-JAK2 was associated with detection of an IKZF1 deletion and upregulation of IL7R and CRLF2.
Phenotypic shift from cytokine receptor dependence to B-cell receptor–like signaling dependence may represent a mechanism of resistance to ruxolitinib.
Abstract
The basis for acquired resistance to JAK inhibition in patients with JAK2-driven hematologic malignancies is not well understood. We report a patient with a myeloproliferative neoplasm (MPN) with a BCR activator of RhoGEF and GTPase (BCR)–JAK2 fusion with initial hematologic response to ruxolitinib who rapidly developed B-lymphoid blast transformation. We analyzed pre-ruxolitinib and blast transformation samples using genome sequencing, DNA mate-pair sequencing (MPseq), RNA sequencing (RNA-seq), and chromosomal microarray to characterize possible mechanisms of resistance. No resistance mutations in the BCR-JAK2 fusion gene or transcript were identified, and fusion transcript expression levels remained stable. However, at the time of blast transformation, MPseq detected a new IKZF1 copy-number loss, which is predicted to result in loss of normal IKZF1 protein translation. RNA-seq revealed significant upregulation of genes negatively regulated by IKZF1, including IL7R and CRLF2. Disease progression was also characterized by adaptation to an activated B-cell receptor (BCR)–like signaling phenotype, with marked upregulation of genes such as CD79A, CD79B, IGLL1, VPREB1, BLNK, ZAP70, RAG1, and RAG2. In summary, IKZF1 deletion and a switch from cytokine dependence to activated BCR-like signaling phenotype represent putative mechanisms of ruxolitinib resistance in this case, recapitulating preclinical data on resistance to JAK inhibition in CRLF2-rearranged Philadelphia chromosome-like acute lymphoblastic leukemia.
Introduction
Myeloproliferative neoplasms (MPNs) are clonal hematopoietic neoplasms characterized by expansion of 1 or more myeloid lineages.1 Most MPNs are associated with somatic activating fusions or mutations in tyrosine kinase genes, including the BCR-ABL1 gene fusion in chronic myeloid leukemia (CML) and JAK2 V617F in polycythemia vera, primary myelofibrosis, and essential thrombocythemia.
Fusion of JAK2 with various gene partners, including PCM1, ETV6, and BCR has been reported in multiple hematologic malignancies.1-3 BCR-JAK2 fusions have been reported across a spectrum of hematolymphoid malignancies, including MPNs or myelodysplastic/myeloproliferative neoplasms, chronic eosinophilic leukemia, and B-cell acute lymphoblastic leukemia (B-ALL).2,4-10 These neoplasms would be best classified in the World Health Organization category of “Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1 or with PCM1-JAK2.” Ruxolitinib, a JAK1/JAK2 kinase inhibitor, has been shown to induce complete clinical or cytogenetic remissions in neoplasms with JAK2 fusion genes; however, responses tend not to be durable.8,11-13
We report a patient with a CML-like MPN harboring a BCR-JAK2 fusion who initially responded to ruxolitinib, but his disease quickly transformed to lymphoid blast phase. Blast transformation was associated with the detection of an IKZF1 deletion, upregulation of IL7R and CRLF2 RNA expression, and adaptation to an activated B-cell receptor (BCR)-like signaling phenotype, highlighting potential mechanisms of acquired ruxolitinib resistance.
Case description
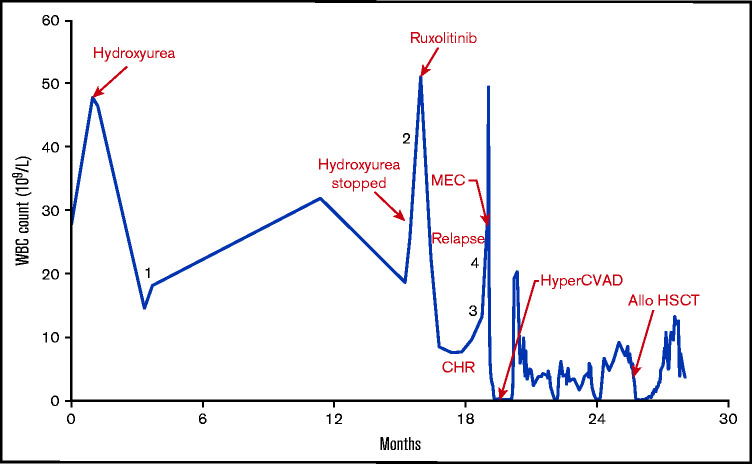
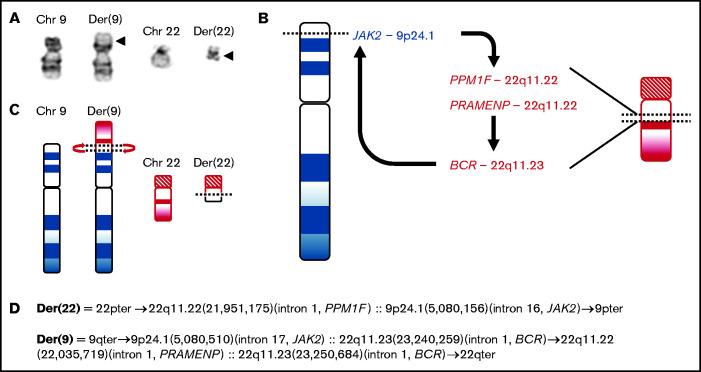
A previously healthy 41-year-old male was found to have leukocytosis and thrombocytopenia on routine complete blood count (white blood cell count [WBC], 27.9 × 109/L, hemoglobin 13.9 g/dL, platelet count, 121 × 109/L) (Figure 1). Approximately 1 month later, his leukocytosis progressed with a left shift without eosinophilia (WBC, 46.6 × 109/L; hemoglobin, 14.4 g/dL; platelet count, 110 × 109/L; granulocytes, 92.7%). Molecular studies were negative for BCR-ABL1 gene fusion, JAK2 V617F, and other JAK2 mutations in exons 12 to 14. Chromosome analysis demonstrated 46,XY,t(9;22)(p24;q11.2)[15]/46,XY[5](Figure 2A), which was further evaluated by metaphase fluorescence in situ hybridization (supplemental Figure 1). A local bone marrow was read as atypical CML, BCR-ABL1-negative. Mast cells were not increased, and lymphoblasts were not present in the marrow. He was started on hydroxyurea and referred to Stanford Hematology for further evaluation.
Figure 1.
WBC count vs time (in months) with major events labeled. The numerals 1 to 4 indicate corresponding time points of correlative analyses. Allo HSCT, allogeneic hematopoietic stem cell transplantation; CHR, complete hematologic remission; hyperCVAD, cytarabine, vincristine, doxorubicin, dexamethasone; MEC, mitoxantrone, etoposide, cytarabine.
Figure 2.
Complex genomic rearrangement resulting in BCR-JAK2. (A) A partial karyotype from a metaphase cell shows an apparently reciprocal translocation between the short (p) arm of chromosome 9 and the long (q) arm of chromosome 22 or a t(9;22)(p24;q11.2). Arrowheads mark the breakpoints on the derivative chromosomes 9 and 22. (B) Genome sequencing defined a more complex rearrangement involving a translocation between 9p24 and 22q11.2 and associated inversion involving 22q11.2. The arrows note the direction of the fusions moving from 5' to 3'. This rearrangement resulted in DNA fusions juxtaposing the intronic regions of BCR-JAK2, JAK2-PPM1F, and PRAMENP-BCR. (C) Idiograms and (D) inferred structure of the derivative chromosomes based on hg38 assembly, NM_014634.4 (PPM1F), NM_004972.3 (JAK2), NM_004327.4 (BCR), and NR_135291.1 (PRAMENP).
After receiving hydroxyurea for 2.5 months, his WBC decreased to 18.2 × 109/L. Genome sequencing of DNA isolated from peripheral blood (time point 1) identified a translocation and inversion event with 3 intronic fusions: BCR-JAK2, JAK2-PPM1F, and PRAMENP-BCR (PRAMENP is a putative pseudogene) (Figure 2B-D; supplemental Table 1). Reverse transcription polymerase chain reaction (RT-PCR) demonstrated an in-frame fusion between exon 1 of BCR and exon 19 of JAK2, which is identical to the fusion transcript previously described14 (supplemental Figure 2; supplemental Table 1). RT-PCR also identified a fusion between exon 16 of JAK2 and exon 2 of PPM1F; however, this results in the introduction of a stop codon at the first complete PPM1F codon. A PRAMENP-BCR fusion transcript was not detected.
Hydroxyurea was discontinued because of disease progression after 14 months (progressive leukocytosis and myeloid immaturity without eosinophilia). Similar to his initial presentation, neither palpable lymphadenopathy or splenomegaly were detected. He provided informed consent for an institutional review board–approved single-patient compassionate use protocol with ruxolitinib, initiated at a dose of 15 mg twice per day in 28-day cycles. Peripheral blood specimens were collected under protocols approved and overseen by the Stanford University Administrative Panel for the Protection of Human Subjects and the Stanford Cancer Institute Scientific Review Committee. The participant was counseled and gave consent for genomic sequencing studies. Specimens were collected before the start of treatment with ruxolitinib (time point 2). At this time, peripheral blood demonstrated left-shift neutrophilia and leukocytosis with myeloid predominance without an increase in blasts. Bone marrow aspirate and core biopsy demonstrated hypercellular marrow without an increase in blasts (supplemental Figure 3A-D).
After cycle 2 of ruxolitinib, he achieved WBC normalization with minimal myeloid immaturity. After cycle 3, peripheral blood showed return of myeloid immaturity with increased blasts (time points 3 and 4, collected 1 week apart). A bone marrow biopsy showed lymphoid blast transformation. The blasts were positive for CD10 and CD79a and negative for myeloperoxidase by immunohistochemistry (supplemental Figure 3E-I). Flow cytometric analysis reiterated these findings, with the CD34+ blast population expressing CD79a and terminal deoxynucleotidyltransferase. During the course of ruxolitinib therapy, no cytogenetic response was observed, and no additional karyotype abnormalities were noted at the time of lymphoid blast disease. We performed analysis of the genome, DNA mate-pair sequencing (MPseq), RNA sequencing (RNA-seq), and chromosomal microarray (CMA) data from the pre-ruxolitinib (time point 2) and blast transformation specimens (time points 3 and 4) to identify acquired genetic changes associated with disease progression. The BCR-JAK2 fusion was persistent throughout the treatment course up to this point, detected at time points 1 to 4 (Figure 1).
The patient underwent induction chemotherapy with mitoxantrone, etoposide, and cytarabine followed by hyperfractionated cytarabine, vincristine, doxorubicin, and dexamethasone for residual disease. He achieved a complete hematologic and cytogenetic remission but suffered a cytogenetic relapse before proceeding to a matched unrelated donor allogeneic hematopoietic stem cell transplantation with busulfan-cyclophosphamide conditioning. His transplant course was complicated by infections and acute graft-versus-host disease of the skin, but he achieved a complete hematologic and cytogenetic remission. As of the writing of this article, he is still alive and without evidence of disease 7 years after hematopoietic stem cell transplantation.
Methods
To characterize potential molecular causes of acquired resistance, we performed genome sequencing, MPseq, RNA-seq, and CMA on the lymphoid blast transformation and pre-ruxolitinib samples as described in the supplemental Materials.
Results and discussion
MPseq demonstrated a subclonal 90-kb IKZF1 deletion encompassing exons 2 to 7 (NM_006060.6; chr7:50 306 990-50 395 984; hg38) in the blast transformation sample (time point 3), with no evidence of the IKZF1 deletion in the pre-ruxolitinib sample (time point 2) (data not shown). Because exon 2 contains the ATG translational start site, these deletions are predicted to result in loss of normal IKZF1 protein translation from these deletion alleles. We cannot distinguish whether the IKZF1 deletion was acquired near the time of lymphoid blast transformation or represents the outgrowth of a minor preexisting clone. No other deletions were detected in other genes commonly altered in B-ALL with IKZF1 deletion, including CDKN2A, CDKN2B, EBF1, and PAX5 before or after blast transformation. Genome sequencing did not identify any new candidate coding variants in the blast transformation sample when compared with the pre-ruxolitinib sample.
RNA-seq data were compared in the blast transformation specimen (time point 3) relative to the pre-ruxolitinib sample (time point 2). Overall, JAK2 expression decreased by approximately twofold in the blast transformation specimen. RT-PCR followed by Sanger sequencing and RNA-seq failed to detect any newly acquired or potential resistance mutations in the BCR-JAK2 fusion transcript, and the level of fusion transcript expression remained consistent between samples based on normalized junction-spanning read counts. Given the finding of an IKZF1 deletion in the blast transformation specimen by MPseq, we also examined IKZF1 splicing and expression. We did not detect evidence of aberrant IKZF1 splicing. IKZF1 expression demonstrated a minor but statistically significant decrease (1.2-fold; adjusted P = 5.7 × 10−8), and significant upregulation was observed for genes negatively regulated by IKZF1,15,16 including IL7R (8.5-fold increase) and CRLF2 (3.4-fold increase) (supplemental Table 2). Pre-BCR signaling genes, including CD79A, CD79B, IGLL1, VPREB1, BLNK, ZAP70, RAG1, and RAG2 exhibited significant upregulation from 4.1-fold to 11.0-fold (supplemental Table 3) in a pattern similar to that reported in Philadelphia chromosome-like ALL (Ph-like ALL).17 Upregulation of PAX5 (4.5-fold) and CD19 (4.7-fold) was also observed, further supporting the diagnosis of a lymphoid blast phenotype.
IKZF1 encodes for the protein Ikaros, a transcription factor integral in normal lymphopoiesis.18 Acquired deletions of IKZF1 have been associated with lymphoid blast transformation in CML and at least 1 case of a BCR-JAK2 MPN.5,18,19 In BCR-ABL1-positive ALL, IKZF1 deletions have been hypothesized to contribute to tyrosine kinase inhibitor resistance by altering cell adhesion pathways, which results in leukemic cells being relocated to the bone marrow.18 In Ph-like ALL, which is frequently associated with CRLF2 rearrangement and concomitant activating JAK2 mutations,20,21 preclinical models demonstrate short-term response to ruxolitinib before leukemic cells overcome the effects of JAK1/JAK2 inhibition.17 This resistance to JAK1/JAK2-inhibition in CRLF2 rearranged Ph-like ALL is mediated by a genomic and phenotypic shift from cytokine receptor dependence to BCR-like signaling dependence.17
In summary, we report a rare case of a BCR-JAK2 fusion MPN with initial near complete hematologic remission with ruxolitinib followed by rapid lymphoid blast transformation, a pattern suggestive of acquired resistance that has been observed in other JAK2-rearranged neoplasms.5,8,13 The molecular basis for resistance to ruxolitinib in neoplasms with JAK2 fusion genes has not been characterized, but IKZF1 deletion and a switch from a cytokine dependence to an activated BCR-like signaling phenotype are strong candidates for contributors to the development of ruxolitinib resistance in this case. Although combinatorial approaches to treatment with agents such as ruxolitinib, dasatinib, idelalisib, and dexamethasone overcome the BCR-like signaling pattern of resistance in preclinical models,17 this requires clinical evaluation in patients with myeloid or lymphoid neoplasms with JAK2 fusion genes and Ph-like ALL.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgment
J.G. thanks the Charles and Ann Johnson Foundation for supporting research on MPNs.
Authorship
Contribution: J.G. treated the patients; J.A.C., J.D.M., and J.G. designed the study; D.A.A., C.D.B., A.M.C., M.D.E., and R.S.O. performed histopathologic, cytogenetic, and fluorescence in situ hybridization analyses; Y.H., K.M.R., L.B.B., A.Z.F., L.F., H.M.K., S.B.M., K.E.P., B.A.P., and J.D.M. performed the genome sequencing, DNA MPseq, RNA-seq, and CMA experiments and analyzed the data; J.A.C., J.D.M., and J.G. wrote the initial manuscript; and all authors approved the final version except for A.M.C. who died before the manuscript was drafted.
Conflict-of-interest disclosure: J.A.C. received the AACR-AstraZeneca clinical immune-oncology research fellowship grant. D.A.A. has served on advisory boards and consulted for AbbVie, Amgen, Jazz Pharmaceuticals, Monsanto, and Roche. M.D.E. has served on advisory boards for Acceleron Pharma. S.B.M. is on the scientific advisory board for MyOme. J.D.M. has served on advisory boards and consulted for AbbVie, Bristol Myers Squibb, Illumina, and PierianDx and serves on the Board of Directors for the Association for Molecular Pathology. J.G. has served on advisory boards and received honoraria from Incyte (the manufacturer of ruxolitinib)and has received funding from Incyte for conducting clinical trials using ruxolitinib in MPNs. The remaining authors declare no competing financial interests.
Correspondence: Jason Gotlib, Stanford Cancer Institute, 875 Blake Wilbur Dr, Room 2324, Stanford, CA 94305-6555; e-mail: jason.gotlib@stanford.edu; and Jason D. Merker, UNC Lineberger Comprehensive Cancer Center, 125 Mason Farm Rd, Marsico Hall 5108, CB #7525, Chapel Hill, CA 27599-7525; e-mail: jason_merker@med.unc.edu.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al, eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Vol 2. 4th ed. Lyon, France: International Agency for Research on Cancer; 2017. [Google Scholar]
- 2.Bain BJ, Ahmad S.. Should myeloid and lymphoid neoplasms with PCM1-JAK2 and other rearrangements of JAK2 be recognized as specific entities? Br J Haematol. 2014;166(6):809-817. [DOI] [PubMed] [Google Scholar]
- 3.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 4.Chamseddine AN, Etancelin P, Penther D, et al. Transformation of an unclassified myeloproliferative neoplasm with a rare BCR-JAK2 fusion transcript resulting from the translocation (9;22)(p24;q11). Case Rep Hematol. 2015;2015:252537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duployez N, Nibourel O, Ducourneau B, et al. Acquisition of genomic events leading to lymphoblastic transformation in a rare case of myeloproliferative neoplasm with BCR-JAK2 fusion transcript. Eur J Haematol. 2016;97(4):399-402. [DOI] [PubMed] [Google Scholar]
- 6.He R, Greipp PT, Rangan A, et al. BCR-JAK2 fusion in a myeloproliferative neoplasm with associated eosinophilia. Cancer Genet. 2016;209(5):223-228. [DOI] [PubMed] [Google Scholar]
- 7.Kantarcioglu B, Kaygusuz-Atagunduz I, Uzay A, Toptas T, Tuglular TF, Bayik M.. Myelodysplastic syndrome with t(9;22)(p24;q11.2), a BCR-JAK2 fusion: case report and review of the literature. Int J Hematol. 2015;102(3):383-387. [DOI] [PubMed] [Google Scholar]
- 8.Schwaab J, Knut M, Haferlach C, et al. Limited duration of complete remission on ruxolitinib in myeloid neoplasms with PCM1-JAK2 and BCR-JAK2 fusion genes. Ann Hematol. 2015;94(2):233-238. [DOI] [PubMed] [Google Scholar]
- 9.Tang G, Sydney Sir Philip JK, Weinberg O, et al. Hematopoietic neoplasms with 9p24/JAK2 rearrangement: a multicenter study. Mod Pathol. 2019; 32(4):490-498. [DOI] [PubMed] [Google Scholar]
- 10.Snider JS, Znoyko I, Lindsey KG, et al. Integrated genomic analysis using chromosomal microarray, fluorescence in situ hybridization and mate pair analyses: Characterization of a cryptic t(9;22)(p24.1;q11.2)/BCR-JAK2 in myeloid/lymphoid neoplasm with eosinophilia. Cancer Genet. 2020;246-247:44-47. [DOI] [PubMed] [Google Scholar]
- 11.Lierman E, Selleslag D, Smits S, Billiet J, Vandenberghe P.. Ruxolitinib inhibits transforming JAK2 fusion proteins in vitro and induces complete cytogenetic remission in t(8;9)(p22;p24)/PCM1-JAK2-positive chronic eosinophilic leukemia. Blood. 2012;120(7):1529-1531. [DOI] [PubMed] [Google Scholar]
- 12.Rumi E, Milosevic JD, Casetti I, et al. Efficacy of ruxolitinib in chronic eosinophilic leukemia associated with a PCM1-JAK2 fusion gene. J Clin Oncol. 2013;31(17):e269-e271. [DOI] [PubMed] [Google Scholar]
- 13.Schwaab J, Naumann N, Luebke J, et al. Response to tyrosine kinase inhibitors in myeloid neoplasms associated with PCM1-JAK2, BCR-JAK2 and ETV6-ABL1 fusion genes. Am J Hematol. 2020;95(7):824-833. [DOI] [PubMed] [Google Scholar]
- 14.Griesinger F, Hennig H, Hillmer F, et al. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes Chromosomes Cancer. 2005;44(3):329-333. [DOI] [PubMed] [Google Scholar]
- 15.Ge Z, Gu Y, Zhao G, et al. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget. 2016;7(31):49722-49732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge Z, Gu Y, Xiao L, et al. Co-existence of IL7R high and SH2B3 low expression distinguishes a novel high-risk acute lymphoblastic leukemia with Ikaros dysfunction. Oncotarget. 2016;7(29):46014-46027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurtz C, Wertheim GB, Loftus JP, et al. Oncogene-independent BCR-like signaling adaptation confers drug resistance in Ph-like ALL. J Clin Invest. 2020;130(7):3637-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marke R, van Leeuwen FN, Scheijen B.. The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2018;103(4): 565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191): 110-114. [DOI] [PubMed] [Google Scholar]
- 20.Herold T, Schneider S, Metzeler KH, et al. Adults with Philadelphia chromosome-like acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102(1):130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.