Abstract
In fission yeast, the onset of meiosis is triggered by activation of the RNA-binding protein Mei2p. We screened for a high-copy-number suppressor of the ectopic meiosis induced by expression of an active form of Mei2p. Consequently we isolated a truncated form of a novel gene, named mip1, from a fission yeast genomic library. The mip1 gene encoded a protein of 1,313 amino acids which carried a WD-repeat motif in the C-terminal region and was apparently conserved among eukaryotes. Mip1p was cytoplasmic, and two-hybrid and immunoprecipitation analyses demonstrated that Mip1p was bound to Mei2p in vivo. Genetic evidence indicated that wild-type Mip1p was required for the function of Mei2p to induce meiosis and that the truncated form of it (Mip1-15p) dominantly interfered with Mei2p. Mip1p appeared to be involved also in conjugation, associating with Ste11p, which is a key transcription factor for sexual development. Furthermore, Mip1p was essential for cell growth, to which neither Mei2p nor Ste11p is relevant. These results suggest that Mip1p assists functional expression of a number of proteins required for proliferation and sexual development in fission yeast.
Sexual differentiation in the fission yeast Schizosaccharomyces pombe proceeds under nutrient starvation (4, 27, 28). Ste11p, which is a high-mobility-group protein, serves as a key transcription factor in this process. Upon starvation, Ste11p activates a number of genes required for mating and/or meiosis, including the mating-type genes (mat1-P and mat1-M) and mei2 (21). The mei2 gene encodes a pivotal regulator of meiosis (3, 20, 25). Pat1p (Ran1p) kinase (6, 11, 17), which is active during the mitotic cell cycle, phosphorylates Mei2p on two amino acid residues, Ser438 and Thr527, and thereby blocks the function of Mei2p (24). In diploid cells, the gene products of mat1-P and mat1-M cooperate to turn on expression of mei3, which encodes an inhibitor of Pat1p kinase and hence induces derepression of Mei2p (12, 13, 24, 26). Thus, mei2 is activated both transcriptionally and posttranscriptionally in diploid cells under starved conditions. If a mutant Mei2p carrying alanine residues in place of Ser438 and Thr527 (Mei2-SATA) is expressed in proliferating cells, the cells cease growth immediately and enter meiosis, indicating that dephosphorylated Mei2p has the ability to switch the mitotic cell cycle to the meiotic one (24).
Mei2p is an RNA-binding protein with three RNA recognition motifs that is required for both induction of premeiotic DNA synthesis and promotion of the first meiotic division (meiosis I). Cytoplasmic Mei2p performs the former function, and the RNA-binding ability of Mei2p is essential for it. To perform the latter function, Mei2p must move into the nucleus, where it forms a distinct dot structure. A specific RNA species, named meiRNA, has been shown to promote meiosis I as a cofactor that assists nuclear transport of Mei2p (25, 29). However, the molecular function of Mei2p in the cytoplasm as well as that in the nucleus remains unknown.
To identify novel regulators or downstream targets of Mei2p, we designed a genetic screen that exploited the fact that expression of Mei2-SATAp causes ectopic meiotic differentiation. We screened for high-expression suppressors of this phenotype and identified a novel gene, mip1. The mip1 gene encodes a WD-repeat protein which appears to be widely conserved among eukaryotes but whose function is unknown. The original isolate of mip1 was a truncated form and inhibited Mei2p function strongly if overexpressed. However, subsequent genetic and biochemical studies suggested that the product of the wild-type mip1 gene is involved in the activation or functioning of Mei2p, interacting physically with it. Mip1p also appeared to bind to Ste11p, possibly assisting its role in conjugation. In addition, Mip1p plays an essential role in cell growth. We propose that the conserved WD-repeat protein Mip1p is likely to play a general role in the cytoplasm in contributing to functional expression of a certain class of proteins.
MATERIALS AND METHODS
Strains and media.
The S. pombe strains used in this study are listed in Table 1. Yeast media YE, SD, SSA, and MEA were used for routine culture of S. pombe strains (14). Liquid minimal medium MM (14) and its nitrogen-free derivative MM-N were used for growth and starvation experiments. SD, MM, and MM-N used in this study contained only 1% glucose.
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotypea |
|---|---|
| JY362 | h+/h− ade6-M216/ade6-M210 leu1/leu1 |
| JY741 | h− ade6-M216 leu1 ura4-D18 |
| JY765 | h+/h− ade6-M216/ade6-M210 leu1/leu1 ura4-D18/ura4-D18 |
| JZ409 | h− ade6-M216 leu1 pat1-114 |
| JZ725 | h− ade6-704 leu1 ura4-D18 |
| JX383 | h− ade6-M210 leu1::nmt1-mei2-SATA<<ura4+ ura4-D18 |
| JX564 | h+/h− ade6-M216/ade6-M210 leu1/leu1 ura4-D18/ura4-D18 mip1+/mip1::ura4+ |
| JX1017 | h+/h− ade6-M216/ade6-M210 leu1/leu1 ura4+/ura4-D18 |
| JW191 | h+ ade6-704 leu1::nmt1-mei2-SATA<<ura4+ ura4-D18 sup3-5 |
| JW192 | h− ade6-704 leu1::nmt1-mei2-SATA<<ura4+ ura4-D18 adh1-mip1-15-FLAG<<sup3-5 |
| JW193 | h− ade6-704 leu1::nmt1-mei2-SATA<<ura4+ ura4-D18 adh1-mip1-15-FLAG<<sup3-5 mip1::ura4+ |
| JW195 | h90 ade6-704 leu1 adh1-mip1-15-FLAG<<sup3-5 |
| JW196 | h90 ade6-704 leu1 adh1-mip1-15-FLAG<<sup3-5 ura4-D18 mip1::ura4+ |
| JW197 | h− ade6-M216 leu1 ura4-D18 mip1::mip1-310<<ura4+ |
leu1::nmt1-mei2-SATA<<ura4+ indicates that leu1+ is disrupted by the nmt1-mei2-SATA construct and that ura4+ locates in the vicinity of it. adh1-mip1-15-FLAG<<sup3-5 indicates that sup3-5 is in the vicinity of adh1-mip1-15-FLAG. The exact location of this array on the chromosome has not been determined.
Mating and sporulation assay.
Mating and sporulation frequencies were calculated according to equations previously described (8), by counting unmated or unsporulated cells, zygotes, asci, and free spores in a sample.
Cloning and nucleotide sequence determination of mip1.
A heterothallic strain expressing mei2-SATA from the nmt1 promoter (JX383) was transformed with an S. pombe genomic library that carried Sau3AI digests of the chromosomal DNA at the BamHI site of the vector pART1. Transformants were plated on thiamine-free SSA and incubated at 30°C for 4 to 6 days. Colonies formed were isolated, and plasmids recovered from them were transfected into Escherichia coli. The suppression activity of the recovered DNA clones was reexamined by repeating transfection into the host strain. The mip1-15 clone was isolated in this screen. A full-length mip1 clone was isolated by colony hybridization from an S. pombe genomic library that carried on average 5-kb-long BglII fragments at the BamHI site of pBluescript. Nucleotide sequence analysis of each clone was performed by the dideoxy-chain termination method. Subclones for the sequencing were produced by progressive deletion with exonuclease III and S1 nuclease. The open reading frame (ORF) region of mip1 has been sequenced in both directions at least once.
Gene disruption.
One-step gene disruption (19) of mip1 was carried out as follows. A 2.1-kb BglII-EcoRV fragment was replaced by a ura4+ cassette. Cells of a wild-type homothallic diploid strain, JY765 (ura4-D18/ura4-D18), were transformed with a mip1::ura4+ DNA fragment and then spread on SD plates without uracil. Stable Ura+ transformants were selected, and Southern blot analysis confirmed that one of the mip1 alleles was properly disrupted in these Ura+ cells.
Construction of a mip1 temperature-sensitive allele.
To generate a temperature-sensitive allele of mip1, we used the method described by Francesconi et al. (5), with minor modifications. Briefly, a 2.8-kb BglII-XhoI fragment, carrying most of the mip1 ORF including the C terminus and the WD repeat but lacking the N terminus and the promoter region, was cloned into pUC119. The XhoI site was artificially created at the C terminus of the mip1 ORF. The resultant plasmid was used as a template for PCR to introduce replication errors. A pair of oligonucleotide primers for pUC119, M13F (5′-CACGACGTTGTAAAACGAC-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′), were used for PCR amplification. Amplified fragments were digested with SphI and KpnI and cloned between the SphI and KpnI sites of pUC119 carrying the ura4+ cassette. The mutagenized library thus obtained was linearized at the HincII site within the mip1 ORF and transformed into haploid strain JY741. Integration of an entire plasmid at the chromosomal mip1 locus by homologous recombination was expected to result in uracil prototrophy. Ura+ transformants were selected at 25°C on an SD plate. Approximately 3,000 Ura+ transformants were replicated to YE plates and examined for the ability to grow at 36°C. Two strains were found to be temperature sensitive. PCR analysis of the genomic DNA isolated from these Ura+ clones confirmed the integration of the mutagenized plasmid at the proper locus. A plasmid pREP81-mip1 was able to complement the thermosensitive phenotype of these mutant strains at the restrictive temperature. The temperature-sensitive allele of mip1 carried by one of them (JW197) was named mip1-310 and analyzed further.
Construction of FLAG-tagged mip1 clones and adh1-mip1-15-FLAG integrants.
To construct pREP2/42-mip1-FLAG and pREP2/42-mip1-15-FLAG, two synthetic oligonucleotides (5′-TCGACCGACTACAAGGACGACGATGACAAGTGACCC-3′ and 5′-GGGTCACTTGTCATCGTCGTCCTTGTAGTCGG-3′) were annealed and inserted between a synthetic SalI site at the C terminus of the mip1 ORF and the SmaI site on pREP2/42 (10). To obtain a mip1-15 integrant, JZ725 (h− ade6-704 leu1 ura4-D18) was transformed with plasmid pANS1-mip1-15-FLAG. pANS1 contains the adh1 promoter, the sup3-5 marker, and the nmt1 terminator. Transformants that generated white colonies, indicating stable integration of the plasmid, were selected. A resultant h− ade6-704 leu1 ura4-D18 adh1-mip1-15-FLAG<<sup3-5 strain was used in crosses to generate JW192, JW193, JW195, and JW196.
Yeast two-hybrid and three-hybrid assays.
The yeast two-hybrid assay was performed as previously described (1). A DNA fragment encoding the C-terminal region of Mei2p (amino acids [aa] 429 to 733) was cloned into pAS2 to express Mei2p fused with the Gal4 DNA-binding domain (GDB). DNA fragments encoding either full-length Mip1p or its truncated version Mip1-15p were cloned into pACT2 to express Mip1p and Mip1-15p fused with the Gal4 transcriptional activator domain (GAD). The latter plasmids were named pACT2-mip1 and pACT2-mip1-15, respectively. We used a pair of plasmids, pSE1111 (pACT2-SNF4) and pSE1112 (pAS1-SNF1), as a control that exhibits positive interaction in the two-hybrid system (1). Positive results were judged by the appearance of blue color generated by degradation of the substrate by induced β-galactosidase activity.
The yeast three-hybrid assay was performed with the commercially available pBridge vector system (Clontech), with modifications. We replaced the GAD sequence in pBridge with the LexA sequence, producing pLB. The nucleotide sequence for the C-terminal region of either Mei2p or Mei2-SATAp was cloned into pLB and fused to the LexA sequence in frame. This generated pLB-mei2 and pLB-mei2-SATA. The nucleotide sequence for the Pat1p ORF was amplified by PCR and subcloned into pLB-mei2 and pLB-mei2-SATA so that it could be expressed from a promoter. A kinase-negative version of Pat1p ORF, in which the ATP-binding site was disrupted by replacing Lys47 with Arg (12), was constructed by in vitro mutagenesis and cloned similarly. The four versions of pLB-mei2/pat1 plasmids thus constructed were introduced into the tester strain L40 together with pACT2-mip1 to assay the interaction between Mei2p and Mip1p.
Construction of HA-tagged mei2 and ste11 genes.
To construct tagged mei2+, a three-hemagglutinin epitope (3HA) fragment was cloned between a NotI site created at the C terminus of the mei2 ORF and the SacI site on pREP41-mei2, thus generating pREP41-mei2-3HA. pREP41-3HA-ste11 was constructed by inserting a 3HA fragment into the synthetic NdeI site at the initiation codon on pREP41-ste11.
Immunoprecipitation.
For Mip1p-Mei2p immunoprecipitation experiments, we transformed JY741 doubly with pREP41-mei2-3HA and either pREP42-mip1-FLAG, pREP42-mip1-15-FLAG, or the vector pREP42. For Mip1p-Ste11p immunoprecipitation experiments, we transformed JY741 doubly with pREP41-3HA-ste11 and either pREP42-mip1-FLAG, pREP42-mip1-15-FLAG, or the vector pREP42. Cells were harvested and washed once with STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3 [pH 8.0]) and kept at −70°C. HBIP buffer (25 mM morpholinepropane sulfonic acid [MOPS], 5 mM EGTA, 15 mM MgCl2, 50 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 1 mM dithiothreitol, 0.1 mM sodium vanadate, 0.8% NP-40, 10% glycerol, 150 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 40 μg of aprotinin per ml, 20 μg of leupeptin per ml, 2 protease inhibitor cocktail tablets [Complete Mini; Boehringer Mannheim] per ml) was then added to the cells. They were disrupted by glass beads and centrifuged at 15,000 rpm for 5 min at −5°C. Immunoprecipitation was performed by incubating the supernatant on ice for 40 min with mouse anti-FLAG monoclonal antibody M2 (Sigma) and 25 μl of 50% (vol/vol) protein G-Sepharose (4FF; Pharmacia Biotech).
Fluorescence microscopy of Mei2p-green fluorescent protein (GFP).
Diploid cells transformed doubly with pGFT81-mei2 (24) and either pREP2-mip1-FLAG or pREP2-mip1-15-FLAG were cultured in MM at 30°C to mid-log phase and then shifted to MM-N and incubated further for 4 h at 29°C. Cells were fixed with methanol at −20°C overnight. After washed once with PEMS buffer, they were stained with Hoechst 33342 to visualize nuclei and observed with a chilled charge-coupled device camera (Hamamatsu) attached to an Axiophot fluorescence microscope (Carl Zeiss).
Immunofluorescence microscopy.
Cells were fixed with 3% paraformaldehyde at 30°C for 40 min. For double staining of Mei2p-GFP and Mip1-15p-FLAG, rabbit anti-GFP polyclonal antibodies (Clontech) and mouse anti-FLAG monoclonal antibody M2 (Sigma) were used. They were further detected by BODIPY-FL-conjugated anti-rabbit immunoglobulin G and Cy3-conjugated anti-mouse immunoglobulin G, respectively.
Nucleotide sequence accession number.
The sequence data for mip1 are available from EMBL/GenBank/DDBJ under accession no. AB032552.
RESULTS
Screening for high-copy-number suppressors of the activated mei2 allele.
Haploid strain JX383 carried an integrated mei2-SATA allele driven by the nmt1 promoter. In the absence of thiamine, the nmt1 promoter was derepressed and mei2-SATA was expressed. JX383 then underwent ectopic meiotic development and failed to form colonies on a thiamine-depleted plate (24). To isolate novel factors relevant to mei2 function, we screened for high-copy-number suppressors of JX383 which could recover growth of the strain on a thiamine-free SSA plate. We obtained five efficient high-copy-number suppressors, but three of them were found to suppress the activity of the nmt1 promoter and were discarded. One of the remaining two was rcd1, which encodes a regulator of transcription of ste11 (18). The functional relationship between rcd1 and mei2 is under investigation. Here we report the analysis of the last clone, mip1.
By sequencing the 5-kb-long insert of the clone, we found a large ORF which apparently lacked the initiation codon. Then we isolated the 5′-terminal region of the gene by colony hybridization and determined the complete ORF sequence. We named this gene mip1 (Mei2p-interacting protein). It turned out to correspond to SPAC57A7.11 defined by the genome sequence project. The mip1 gene encoded 1,313 amino acid residues, and the original suppressor clone lacked the N-terminal 173 residues. We named this truncated mip1 allele mip1-15.
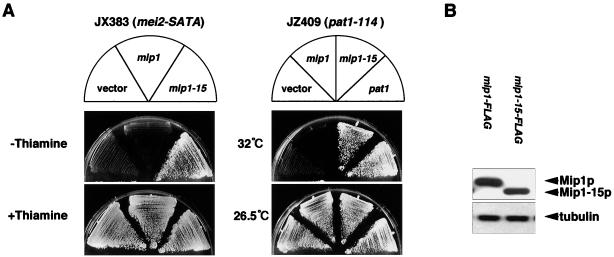
To confirm that mip1-15 was responsible for the suppression, we subcloned mip1-15 ORF to pREP41, which carried the moderate nmt1 promoter. This plasmid efficiently suppressed mei2-SATA. Surprisingly, however, the full-length mip1 ORF connected to the same promoter could not suppress mei2-SATA (Fig. 1A). This difference was not due to the stability of the protein products (separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) (Fig. 1B), indicating that the property of Mip1p in relation to mei2-SATA was changed by the deletion of the N-terminal residues. Overexpression of mip1-15, but not mip1, also suppressed the pat1-driven meiosis (Fig. 1A). The pat1ts mutant enters meiosis at the restrictive temperature (2, 7) because inactivation of Pat1p kinase leads to loss of inhibitory phosphorylation of Mei2p (24). Thus, these results suggest that Mip1-15p may interfere with the function of Mei2p itself or its downstream factor.
FIG. 1.
Suppression of ectopic meiosis of the nmt1-mei2-SATA and pat1-114 strains by overexpression of mip1-15. (A) JX383 (h− nmt1-mei2-SATA) was transformed with either pREP41-mip1, pREP41-mip1-15, or the vector pREP41. JZ409 (h− pat1-114) was also transformed with either pREP41-mip1, pREP41-mip1-15, pREP41-pat1, or the vector pREP41. Each transformant of JX383 was grown on a plate containing thiamine, restreaked on an SSA plates with or without 2 μM thiamine, and incubated at 32°C. Each transformant of JZ409 was grown at 26.5°C, spread on an SSA plate, and incubated at either 32 or 26.5°C. (B) To examine protein stability, the wild-type strain JY741 was transformed with either pREP41-mip1-FLAG or pREP41-mip1-15-FLAG, and total extracts were prepared from the transformants grown in thiamine-free MM for 16 h. Proteins extracted from 106 cells were separated by SDS-PAGE and subjected to Western blot analysis using an anti-FLAG antibody (upper panel) or the antitubulin antibody Tat1 (lower panel).
mip1 encodes a novel protein carrying a WD-40 repeat.
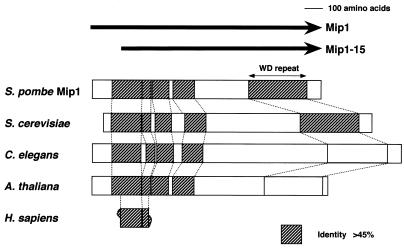
Using the BLAST homology search algorithm, we found that Mip1p was highly similar to the products of putative genes assigned in Saccharomyces cerevisiae, Caenorhabditis elegans, and Arabidopsis thaliana by the respective genome projects. These proteins all carried a WD repeat (15) in the C terminus and another highly conserved domain in the N terminus (Fig. 2). A short nucleotide sequence found in the human cancer genome anatomy project also appeared to encode the N-terminal region of these proteins (Fig. 2). Thus, this protein family is likely to be widespread among eukaryotes. The Mip1p homologues of other organisms have been identified only in the sequencing project, and their functions remain unknown.
FIG. 2.
Schematic illustration of the structure of S. pombe Mip1 protein (SPAC57A7.11) and its S. cerevisiae (SCYHR186C), C. elegans (C10C5.6), and A. thaliana (T16O11.22) homologues. A fragmentary sequence derived from the human cancer genome anatomy project is also shown (ng92c11.s1). The regions highly conserved (>45% identity) between Mip1p and the homologues are hatched.
mip1 is essential for cell proliferation.
Using the ura4+ cassette, we disrupted one copy of the mip1 gene in diploid strain JY765. The resultant mip1+/mip1::ura4+ diploid JX564 was subjected to sporulation and tetrad analysis. Each ascus generated only two viable spores, both of which were Ura−. These results strongly suggest that disruption of mip1 is lethal. Microscopic observation revealed that ∼40% of mip1Δ spores germinated and underwent one to three rounds of cell division before halting growth. The length of the arrested cells was smaller than that of the wild type (data not shown; see below).
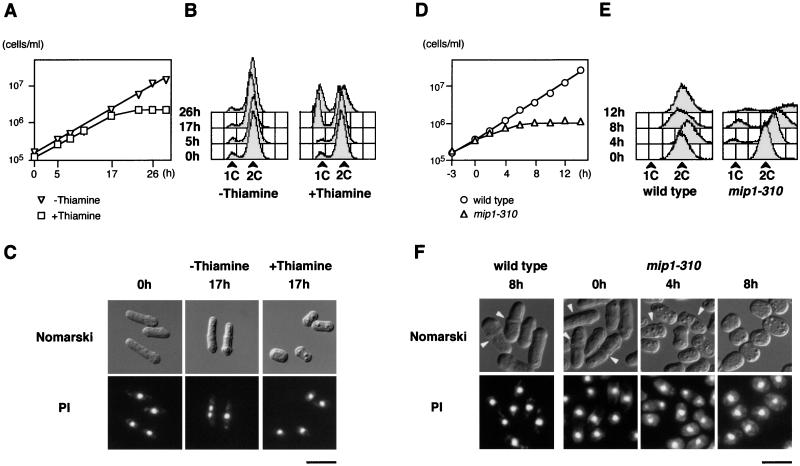
We constructed haploid mip1Δ strains carrying either pREP81-mip1 or pREP81-mip1-15. Both strains could grow in the absence of thiamine, although cells with pREP81-mip1-15 grew more slowly (doubling time of 6.0 h in MM) than cells with pREP81-mip1 (3.4 h). To investigate the terminal phenotype of mip1Δ, we shifted them to thiamine-containing medium to shut off the nmt1 promoter. The pREP81-mip1 cells, however, continued proliferation, indicating that the repressed level of mip1 expression from this plasmid was still enough to sustain cell growth. In contrast, pREP81-mip1-15 cells could not grow on the thiamine-containing medium. Upon addition of thiamine to the pREP81-mip1-15 liquid culture, most cells started to arrest growth after approximately three rounds of cell division (Fig. 3A). The arrested cells were short, and flow cytometry indicated that nearly 40% of the cells arrested with 1C DNA content (Fig. 3B and C).
FIG. 3.
mip1+ is essential for cell growth. (A) Growth of mip1Δ cells transformed with pREP81-mip1-15. Cells were grown in MM to a concentration of about 105/ml; cell number was measured chronologically after the addition of 2 μM thiamine to the culture (□). Results for a control culture with no thiamine addition are also shown (▿). (B) Flow cytometry of the cells at 0, 5, 17, and 26 h after the addition of thiamine. (C) Nomarski micrographs of mip1Δ cells carrying the mip1-15 plasmid cultured with or without thiamine for the indicated time. Fluorescence micrographs of the same cell population stained with the DNA-binding dye propidium iodide (PI) are also shown. Bar, 10 μm. (D) Growth of mip1-310 cells. Cells were grown in YE medium to a concentration of about 105/ml at 26.5°C. They were shifted up to 36°C (0 h), and cell number was measured chronologically (▵). Results for a control wild-type strain are also shown (○). (E) Flow cytometry of the cells at 0, 4, 8, and 12 h after the shift. (F) Morphology of mip1-310 and control wild-type cells cultured at 36°C for the indicated time. Arrowheads indicate dividing cells. Fluorescence micrographs of the same cells stained with propidium iodide are also shown. Bar, 10 μm.
To further explore the mitotic role of mip1, we aimed to construct a temperature-sensitive mip1 allele and succeeded in isolating mip1-310 as such (see Materials and Methods). At 26.5°C, mip1-310 cells were indistinguishable from wild-type cells in either growth rate or morphology. Upon a shift to 36°C, however, mip1-310 cells began to grow at a lower rate and continued cell division with reduced cell size for some while (Fig. 3D and F at 4 h). They finally terminated cell division, exhibiting a very short cell length (Fig. 3D and F at 8 h). The viability of these cells was less than 10% at 4 h after the shift. The mip1-310 mutant could proliferate only below 34°C.
During the shutoff or thermal inactivation of Mip1p, a significant number of cells with 1C DNA content appeared (Fig. 3B and E). This may be explained by supposing that cell division with reduced cell size indirectly led to the accumulation of cell population with 1C DNA content, as was originally noted in the wee1 cell cycle mutant (16). Unlike the wee1 mutant, however, the mip1 mutant was deficient in cell growth and inviable. Thus, Mip1p is likely to be required for facilitating cell cycle control in addition to supporting cell growth.
mip1 plays a positive role in meiosis.
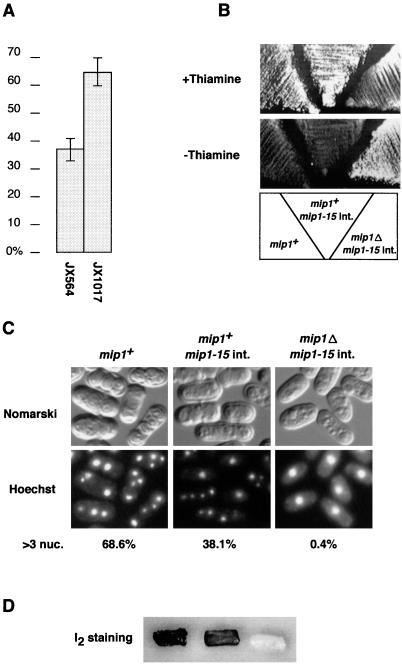
To see whether mip1 has some role relevant to meiosis, we examined sporulation of an h+/h− mip1Δ/mip1+ diploid strain JX564. This strain proliferated normally but sporulated at only 37% efficiency, while the efficiency of the wild-type JX1017 was 65% (Fig. 4A). Most of the unsporulated JX564 cells were arrested with a single nucleus, suggesting that the entry to meiosis was impaired in them. Despite their clear difference in meiotic proficiency, JX564 and JX1017 proliferated at nearly the same rate. This suggests that Mip1p plays a positive role in promoting the entry to meiosis but that its amount is not much in excess of the level needed to meet this end in wild-type diploid cells.
FIG. 4.
Mip1p is required for the entry to meiosis. (A) Lowered sporulation efficiency in diploid cells heterozygous for mip1. The wild-type strain JX1017 and the mip1+/mip1Δ strain JX564 were streaked on sporulation medium MEA and incubated at 30°C for 3 days. The sporulation frequency of each strain was scored under the microscope. The error bar for each data represents the standard deviation of three independent measurements. (B to D) Requirement of mip1+ for haploid meiosis induced by mei2-SATA. JW192 (nmt1-mei2-SATA adh1-mip1-15) and the isogenic mip1Δ strain JW193 were streaked on an SSA plate with or without thiamine and incubated at 32°C for 3 days (+Thiamine) or 5 days (−Thiamine) (B). The same strains were cultured in liquid MM at 30°C for 48 h. Cells were fixed with ethanol, stained with Hoechst 33342, and photographed (C). They were also spread in patches on an SSA plate, incubated at 30°C for 4 days, and stained with I2 vapor (D). Nuc., nuclei.
To further demonstrate the participation of Mip1p in meiosis, we compared meiotic development induced by mei2-SATA in mip1+ and mip1Δ backgrounds. Two haploid strains were constructed for this purpose. One was a mip1Δ strain in which both nmt1-mei2-SATA and adh1-mip1-15 were integrated (JW192), and the other was an isogenic mip1+ strain (JW193). The adh1-mip1-15 construct was integrated in order to support mitotic growth of the mip1Δ strain. They were shifted to thiamine-depleted medium to express mei2-SATA. Strikingly, even when mei2-SATA was maximally expressed, mip1Δ cells did not initiate meiosis and continued proliferation, whereas mip1+ entered meiosis efficiently (Fig. 4B and C). These results support the idea that the wild-type mip1 allele performs a positive function in mei2-driven meiosis. The mip1-15 allele is apparently defective in this function.
Mip1p interacts physically with Mei2p.
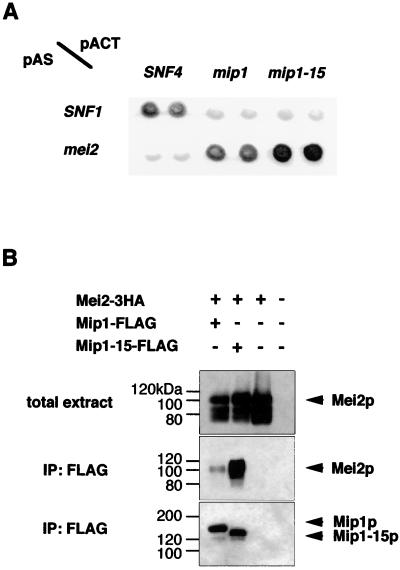
A number of genes are involved in the genetic cascade controlling meiotic entry in fission yeast. Among them, mei2 is apparently the most downstream factor whose activation is sufficient for the induction of meiosis. As shown above, Mip1p is required for the mei2-driven meiosis, whereas its truncated version Mip1-15p interferes with it. These results suggested the possibility that Mip1p might directly interact with Mei2p. To examine this, we performed a yeast two-hybrid assay (1). The C-terminal region of Mei2p (aa 429 to 733), which could perform the essential function of Mei2p (24), was fused to GDB. Full-length Mip1p and its truncated version Mip1-15p were fused to GAD and tested for interaction with the Mei2p bait by assaying β-galactosidase activity. Both versions of Mip1p were found to interact with Mei2p specifically. Apparently Mip1-15p interacted with Mei2p more strongly than full-length Mip1p (Fig. 5A).
FIG. 5.
Mip1p and Mei2p form a complex in vivo. (A) Yeast two-hybrid analysis. The interaction between Mip1p and Mei2p and that between Mip1-15p and Mei2p are positive, as judged by the blue coloring of the host cells. The intensity of the blue color is reproduced in black and white here. The pair SNF1 and SNF4 represents a standard positive control. (B) Coimmunoprecipitation of Mip1p and Mei2p. Crude extracts were prepared from vegetative cells expressing tagged proteins as indicated and subjected to immunoprecipitation (IP) with an anti-FLAG antibody. Each precipitate was separated by SDS-PAGE and examined for the presence of Mei2 protein by Western blotting using an anti-HA antibody. The immunoblotting patterns of total crude extracts are also shown.
To confirm the association of Mip1p and Mei2p in vivo, we expressed FLAG-tagged Mip1p and 3HA-tagged Mei2p in wild-type cells. Mip1p was immunoprecipitated using an anti-FLAG antibody, and coprecipitation of Mei2p was investigated. As shown in Fig. 5B, Mei2p was coimmunoprecipitated efficiently with Mip1-15p and less efficiently with full-length Mip1p. Therefore, we conclude that Mip1p interacts with Mei2p in vivo and that Mip1-15p has acquired a strengthened affinity to Mei2p.
Mip1p is located in the cytoplasm.
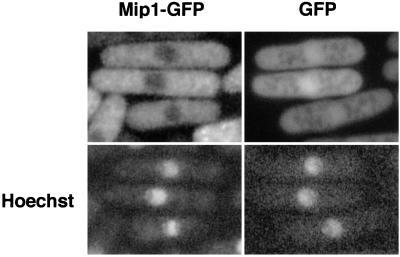
To examine subcellular localization of Mip1p, we fused GFP to the C terminus of Mip1p. The Mip1p-GFP fusion protein was expressed from the nmt1 promoter on pREP41. This fusion protein was functional because it could complement mip1Δ. Fluorescence microscopy revealed that Mip1p-GFP was distributed uniformly in the cytoplasm but was scarce in the nucleus (Fig. 6). This localization of Mip1p was similar to that of Mei2p in the premeiotic stage, suggesting that Mip1p is likely to interact with Mei2p at this stage (24, 29).
FIG. 6.
Subcellular localization of Mip1p tagged with GFP. Wild-type strain JY362 transformed with either pREP41-mip1-GFP or pREP41-GFP was grown in liquid MM and examined for GFP fluorescence without fixation. A fluorescence micrograph of the same cells stained with the DNA-binding dye Hoechst 33342 is also shown for each transformant.
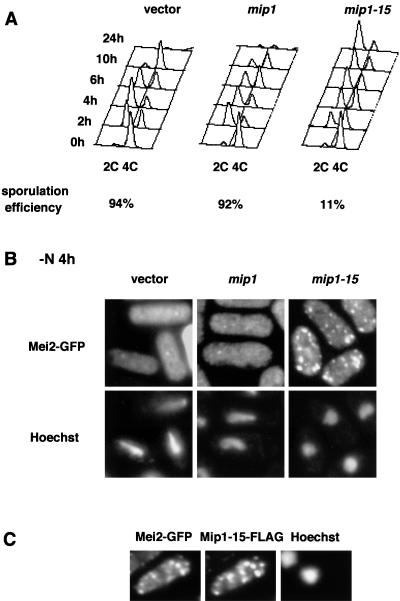
In contrast to wild-type Mip1p, overexpression of Mip1-15p counteracted Mei2p and blocked the initiation of meiosis (Fig. 1). To gain a deeper insight into this observation, we compared the effect of overexpression of the two versions of Mip1p on Mei2p localization. We coexpressed Mei2p-GFP together with either Mip1p-FLAG or Mip1-15p-FLAG in diploid strain JY765. Each tagged protein was indistinguishable from its untagged prototype in function. The diploid transformants were shifted to the nitrogen-depleted medium and monitored for progression of meiosis and localization of Mei2p-GFP (Fig. 7). Control cells carrying only the vector exhibited horse-tail nuclei during meiotic prophase, as previously reported (18a). Mei2p formed a dot in the nucleus at this phase, being located also in the cytoplasm homogeneously. Cells overproducing Mip1p-FLAG behaved like the control cells (Fig. 7A and B). In contrast, most cells overproducing Mip1-15p-FLAG failed to enter meiosis and arrested at G1 (Fig. 7A). Mei2p-GFP was mislocated in the latter cells, forming patches mostly in the cytoplasm (Fig. 7B). These patches were apparently colocated with Mip1-15p-FLAG (Fig. 7C), consistent with the observed coimmunoprecipitation of the two proteins. The inhibition of meiotic entry by Mip1-15p overproduction was alleviated by elevated expression of Mei2p (data not shown). Thus, we conclude that Mip1-15p associates Mei2p tightly and induces abnormal aggregation of the proteins, thereby blocking Mei2p function.
FIG. 7.
Aggregation of Mei2p induced by Mip1-15p overproduction. (A) Flow cytometry of the wild-type diploid strain JY765 carrying pGFT81-mei2 and either pREP2-mip1-FLAG, pREP2-mip1-15-FLAG, or the vector pREP2. Each strain was grown to 4 × 106 cells/ml in MM and shifted to MM-N to induce meiosis. Cells were harvested at intervals after the shift and subjected to flow cytometry. Sporulation efficiency of each strain was determined 24 h after the shift. (B) The same strains as in panel A were fixed by methanol at 4 h after the shift and observed for fluorescence of Mei2p-GFP. Fluorescence micrographs of cells stained with the DNA-binding dye Hoechst 33342 are also shown. (C) Cells producing Mei2p-GFP and Mip1-15p-FLAG were fixed by formaldehyde at 4 h after the shift. They were doubly immunostained with anti-GFP antibodies and a monoclonal anti-FLAG antibody and subjected to fluorescence microscopy.
Mip1p interacts with Ste11p, which is required for conjugation.
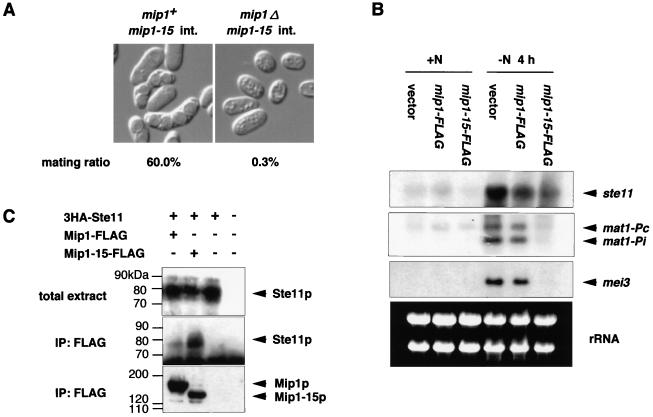
We observed that overexpression of mip1-15 reduced efficiency of conjugation in wild-type haploid cells (data not shown). Furthermore, haploid mip1Δ cells carrying the integrated adh1-mip1-15 allele hardly underwent mating, while mip1+ cells carrying the same allele could mate considerably (Fig. 8A). These results suggested that, as was true with meiosis, Mip1p might have a positive role in conjugation that is inhibited by Mip1-15p. Overexpression of Mip1-15p could inhibit conjugation in mei2Δ cells, suggesting that this effect is not mediated by Mei2p (data not shown). It has been known that ste11 and some of its target genes are essential to initiate conjugation in S. pombe (21). Therefore, we examined transcription of ste11 itself and its target genes mat1-Pc and mat1-Pi in the diploid Mip1-15p overproducer. Expression of ste11 was not much affected in this strain, but expression of mat1-Pc and mat1-Pi was impaired (Fig. 8B). Consistently, expression of mei3, which depends on the function of mat1-Pi (22, 26), was not detectable in this strain (Fig. 8B). The conjugation defect caused by Mip1-15p overproduction was alleviated by elevated expression of Ste11p (data not shown). These results suggested that the function of Ste11p was impaired by Mip1-15p. We examined whether Mip1-15p and Ste11p could be coimmunoprecipitated. As shown in Fig. 8C, Ste11p was precipitated efficiently with Mip1-15p and less efficiently with Mip1p. These results support the conclusion that Mip1p plays a positive role in conjugation, possibly by activating or assisting Ste11p directly.
FIG. 8.
mip1 is required for the mating process. (A) h90 mip1-15 integrant (int.) strains with or without the authentic mip1+ gene (JW195 or JW196) were streaked on sporulation medium SSA and incubated at 30°C for 4 days. The mating frequency of each strain was scored under the microscope. (B) Northern blot analysis of mip1-15-overexpressing cells. Total RNA was extracted from the same strains as analyzed in Fig. 7, either growing exponentially (0 h) or starved of nitrogen for 4 h. RNA (10 μg) was loaded in each lane after denaturation by formamide, electrophoresed, transferred to a nylon membrane, and hybridized with the indicated probes. rRNA in each sample, stained with ethidium bromide, are shown in the lower panel to verify nearly equal loading of RNA in each lane. (C) Coimmunoprecipitation of Mip1p and Ste11p. Crude extracts were prepared from cells expressing tagged proteins as indicated and growing mitotically. They were subjected to immunoprecipitation (IP) with an anti-FLAG antibody. The precipitates were subjected to SDS-PAGE and examined for the presence of Ste11 protein by Western blotting using an anti-HA antibody. Immunoblotting patterns of the crude extracts are also shown.
We investigated the behavior of the temperature-sensitive mip1-310 mutant in mating and meiosis. At every tested temperature up to 33°C, which was close to the limit for mitotic proliferation of the mutant, mip1-310 cells performed mating and meiosis almost as efficiently as wild-type cells. This analysis hence turned out to be uninformative.
Mip1p interacts only with unphosphorylated Mei2p.
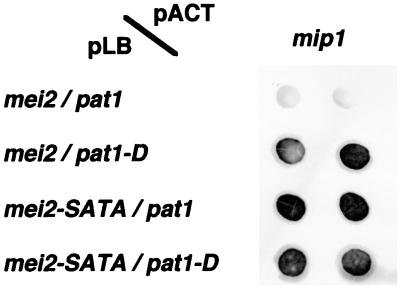
As shown above, Mip1p interacted with two different proteins, Mei2p and Ste11p. It is noteworthy that these proteins are both phosphorylated by Pat1p kinase (9, 24), implying that Mip1p may recognize the phosphorylation sites in these proteins. Therefore, we examined whether interaction of Mei2p with Mip1p could be affected by the phosphorylation by Pat1p. To do so, we used a yeast three-hybrid assay. As demonstrated in Fig. 9, Mei2p lost the ability to interact with Mip1p when Pat1p kinase was expressed in the host cell. Expression of kinase-negative Pat1p (Pat1-Dp) had no effect. Unphosphorylatable Mei2-SATAp interacted with Mip1p efficiently even in the presence of Pat1p kinase. These results strongly suggest that Mip1p interacts with Mei2p when it is not phosphorylated by Pat1p.
FIG. 9.
Interaction of Mip1p with unphosphorylated Mei2p, examined by yeast three-hybrid analysis. Either LexA-Mei2p or LexA-Mei2-SATAp was expressed together with either active Pat1p or kinase-negative Pat1-Dp from a single plasmid. GAD-Mip1p was expressed from another plasmid. For each combination, two independent transformants were assayed for β-galactosidase activity. The intensity of the blue color is reproduced in black and white here.
Consistent with the above idea, a shortened form of the Mei2p C-terminal region (aa 532 to 750) which lacked the two phosphorylation sites (Ser438 and Thr527) failed to interact with Mip1p in two-hybrid assay (data not shown), whereas the Mei2p C-terminal region carrying aa 429 to 733 could do so (Fig. 5A). This observation may suggest further that these phosphorylation sites, in their unphosphorylated form, serve as the binding sites for Mip1p.
DISCUSSION
We have identified a novel WD-repeat protein, Mip1p, which interacts with the meiotic regulator Mei2p in fission yeast. In the screening for high-copy-number suppressors of ectopic meiosis induced by active Mei2p, we isolated a truncated form of mip1 missing the N-terminal region of the ORF. This mip1-15 allele inhibited meiotic differentiation if overexpressed. However, we concluded that wild-type Mip1p plays a positive role in meiotic differentiation for the following reasons: (i) unlike mip1-15, overexpression of wild-type mip1 does not interfere with meiosis; (ii) reduction of the mip1 gene dosage by half in a diploid cell lowered the frequency of meiotic entry; and (iii) the negative effect of mip1-15 on mei2-driven meiosis can be alleviated by overproduction of Mei2-SATAp in the presence of wild-type mip1 but not in its absence.
Two-hybrid and immunoprecipitation experiments demonstrated that Mip1p interacts physically with Mei2p in vivo. Thus, Mip1p may bind and assist Mei2p in the process of its activation or function, thereby facilitating meiotic entry. The N-terminal truncation in Mip1-15p likely converted this protein into a dominant negative form inhibiting Mei2p function. This model is consistent with the observation that Mip1-15p but not Mip1p causes abnormal aggregation of Mei2p in the cell. In addition, Mip1-15p was bound to Mei2p more tightly than Mip1p, and the meiotic defect caused by Mip1-15p was alleviated by elevated expression of Mei2p from a plasmid.
We presently have no firm picture about the molecular function of Mip1p, but the following discussion may be noteworthy. Although interaction of wild-type Mip1p with Mei2p was demonstrated, this interaction appeared to be relatively weak, suggesting that Mip1p may interact with Mei2p transiently rather than permanently. Results of the three-hybrid analysis indicate that Mip1p interacts with unphosphorylated Mei2p but not with Mei2p phosphorylated by Pat1p. This may mean that Mip1p interacts with Mei2p when the protein is produced de novo in the cytoplasm, prior to its phosphorylation by Pat1p. If this is the case, Mip1p may assist protein folding of nascent Mei2p and/or a subsequent crucial step for Mei2p activation. The period of their interaction will be very short, and once Mei2p is phosphorylated by Pat1p, Mip1p may dissociate from it. This scenario is consistent with the idea that Mip1p is a kind of molecular chaperon, acting to assist protein folding and subsequent processing required for the proper function of the target protein.
In addition to Mei2p, a crucial transcription factor for sexual development, Ste11p, is associated with Mip1p in vivo. Coimmunoprecipitation demonstrated that Ste11p interacts weakly with Mip1p but more strongly with Mip1-15p. Mip1-15p, but not Mip1p, inhibited conjugation if overexpressed. Furthermore, cells expressing mip1-15 did not conjugate in the absence of wild-type mip1 allele but became proficient in mating if mip1+ was present, indicating that mip1+ plays a positive role in conjugation. Thus, the relationship of Mip1p with Ste11p appears very similar to the relationship of Mip1p with Mei2p, suggesting that Mip1p participates in conjugation through the interaction with Ste11p. It is interesting that Mip1p can recognize and regulate two totally different proteins, Mei2p and Ste11p, the former an RNA-binding protein and the latter a high-mobility-group transcription factor. A common feature of these two proteins is their possession of conserved phosphorylation motifs by Pat1p kinase, which are relevant to their nuclear localization (9, 24, 29). We have demonstrated that the interaction of Mip1p with Mei2p depends on the phosphorylation state of the motifs. Thus, these motifs themselves may be the sites of interaction with Mip1p.
As discussed above, Mip1p plays a crucial role for sexual development by interacting with Mei2p and Ste11p, which are key factors for meiosis and conjugation. However, they are not the only targets of Mip1p in the cell. Gene disruption of mip1 indicated that the function of mip1 is essential for cell proliferation. Neither Mei2p nor Ste11p is the target molecule interacting with Mip1p during mitotic proliferation, because these proteins are dispensable for asexual growth (21, 23). Therefore, there must be another target(s) of Mip1p required for mitotic cell growth. A notable feature of the truncated mip1-15 allele is that it dominantly inhibits meiosis and conjugation if overexpressed but does not interfere with, and even to some extent supports, mitotic growth. It is hence presumable that Mip1p functions in somewhat different modes in mitotic growth and in sexual development.
It is particularly intriguing that Mip1p is evolutionarily conserved among eukaryotes. Several Mip1p homologues have been identified in other organisms by genome sequencing projects, but none of them have been analyzed for function. The present work with fission yeast is the first example of a functional analysis of this family. Given that Mip1p is required for cell growth as well as sexual development and therefore is likely to interact with several proteins in fission yeast, its molecular function is probably ubiquitous and fundamental. We propose that fission yeast Mip1p plays a critical role in the cytoplasm in assisting a certain class of proteins to express their function. Although more extensive analysis is required to elucidate the precise molecular function of Mip1p, this report provides an important basis for further studies of this conserved WD-repeat protein family.
ACKNOWLEDGMENTS
We thank K. Gull for providing the Tat1 antitubulin antibody, S. J. Elledge for providing the yeast two-hybrid system, and M. Sato for construction of some plasmids.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (A) and for Specially Promoted Research from the Ministry of Education, Science, Sports and Culture of Japan and by the Mitsubishi Foundation. S.S.-Y. was a recipient of a JSPS Fellowship for Japanese Junior Scientist.
REFERENCES
- 1.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 2.Beach D, Rodgers L, Gould J. RAN1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 3.Bresch C, Muller G, Egel R. Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- 4.Egel R. Mating-type genes, meiosis, and sporulation. In: Nasim A, et al., editors. Molecular biology of the fission yeast. San Diego, Calif: Academic Press; 1989. pp. 31–73. [Google Scholar]
- 5.Francesconi S, Park H, Wang T S-F. Fission yeast with DNA polymerase δ temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 7.Iino Y, Yamamoto M. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1985;82:2447–2451. doi: 10.1073/pnas.82.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunitomo H, Sugimoto A, Wilkinson C R M, Yamamoto M. Schizosaccharomyces pombe pac2+ controls the onset of sexual development via a pathway independent of the cAMP cascade. Curr Genet. 1995;28:32–38. doi: 10.1007/BF00311879. [DOI] [PubMed] [Google Scholar]
- 9.Li P, McLeod M. Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell. 1996;87:869–880. doi: 10.1016/s0092-8674(00)81994-7. [DOI] [PubMed] [Google Scholar]
- 10.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 11.McLeod M, Beach D. Homology between the ran1+ gene of fission yeast and protein kinase. EMBO J. 1986;5:3665–3671. doi: 10.1002/j.1460-2075.1986.tb04697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLeod M, Beach D. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature. 1988;332:509–514. doi: 10.1038/332509a0. [DOI] [PubMed] [Google Scholar]
- 13.McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 15.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 16.Nurse P. Genetic control of cell size at cell division in fission yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 17.Nurse P. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497–502. [Google Scholar]
- 18.Okazaki N, Okazaki K, Watanabe Y, Kato-Hayashi M, Yamamoto M, Okayama H. Novel factor highly conserved amang eukaryotes controls sexual development in fission yeast. Mol Cell Biol. 1998;18:887–895. doi: 10.1128/mcb.18.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Robinow C F. The number of chromosomes in Schizosaccharomyces pombe: light microscopy of stained preparations. Genetics. 1977;87:491–497. doi: 10.1093/genetics/87.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothstein R. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 20.Shimoda C, Uehira M, Kishida M, Fujioka H, Iino Y, Watanabe Y, Yamamoto M. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1987;169:93–96. doi: 10.1128/jb.169.1.93-96.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 22.van Heeckeren W J, Dorris D R, Struhl K. The mating-type proteins of fission yeast induce meiosis by directly activating mei3 transcription. Mol Cell Biol. 1998;18:7317–7326. doi: 10.1128/mcb.18.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 26.Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci. 1996;21:18–22. [PubMed] [Google Scholar]
- 28.Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J R, Broach J B, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- 29.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]