Abstract

SARS-CoV-2 infects human cells through its surface spike glycoprotein (SgP), which relies on host cell surface heparan sulfate (HS) proteoglycans that facilitate interaction with the ACE2 receptor. Targeting this process could lead to inhibitors of early steps in viral entry. Screening a microarray of 24 HS oligosaccharides against recombinant S1 and receptor-binding domain (RBD) proteins led to identification of only eight sequences as potent antagonists; results that were supported by detailed dual-filter computational studies. Competitive studies using the HS microarray suggested almost equivalent importance of IdoA2S–GlcNS6S and GlcNS3S structures, which were supported by affinity studies. Exhaustive virtual screening on a library of >93 000 sequences led to a novel pharmacophore with at least two 3-O-sulfated GlcN residues that can engineer unique selectivity in recognizing the RBD. This work puts forward the key structural motif in HS that should lead to potent and selective HS or HS-like agents against SARS-CoV-2.
Keywords: Spike protein, glycosaminoglycans, 3-O-sulfation, microarray, molecular docking, pharmacophore modeling
The SARS-CoV-2 pandemic has already taken the lives of millions of individuals across the globe, and although vaccines are key to reducing fear and trepidation, an alternative and effective approach would be to discover small molecule drugs that prevent viral infection and spread.1−5 SARS-CoV-2 is an enveloped, positive-sense RNA virus that possesses considerable sequence similarity to other coronaviruses such as SARS-CoV (∼80%) and MERS-CoV (∼50%).6,7 Unfortunately, prior efforts to develop clinically viable candidates against coronaviruses did not yield highly promising candidates, thereby handicapping quick discovery of small molecule agents against SARS-CoV-2. Further, the role of comorbidities, e.g., diabetes, inflammation, hypertension, and/or cancer, in SARS-CoV-2 etiology, poses a major challenge for the discovery of small molecules that exhibit a pan-etiology solution. Additionally, newly discovered mutations in the virus, which may reduce the efficacy of vaccines,8−11 highlight a critical need for small molecule antagonists of SARS-CoV-2.
SARS-CoV-2 relies on its surface protein, spike glycoprotein (SgP), to recognize receptor angiotensin-converting enzyme 2 (ACE2) and gaining cellular entry into host cells.12−15 In its natural state, SgP exists as a homotrimer of S1 and S2 subunits, of which the former contains the receptor-binding domain (RBD) that mainly interacts with ACE2.13−17 The trimer first undergoes a conformational change that allows one of the S1 subunits to shift from a “closed” to an “open” conformation, thus revealing a more accessible RBD to bind to ACE2.15 Host factors other than ACE2 on the host cell surface are also known to assist SARS-CoV-2 entry including neuropilin.18,19 Heparan sulfate (HS), the sulfated glycosaminoglycan (GAG) chain of HS proteoglycans (HSPGs), is known to contribute to cellular entry of multiple enveloped viruses, including HSV, HIV, CMV, and dengue,20−24 and recent studies by the Esko group have confirmed its key role in SARS-CoV-2 entry as well.25
Over the past year, several groups have studied the interaction of SgP with sulfated GAGs as an approach to develop antagonists of SARS-CoV-2 entry. Natural or chemically modified sulfated polymeric entities such as unfractionated heparin,26−29 fucoidans,27 and enoxaparin28 have been found to bind to SgP monomer and/or trimer with potencies as high as picomolar. In contrast, pixatimod, a lipid-modified sulfated oligosaccharide, was found to bind to RBD, albeit with much lower potency, and inhibited three different isolates of SARS-CoV-2.30 These successes have led the two groups to study an impressive array of 80 HS oligosaccharides of varying lengths and sulfation patterns for binding to SgP proteins.31 They identified a fairly select group of sequences that bind to SgP proteins with affinities in the nanomolar to micromolar range. A major conclusion of these studies was that the common heparin (Hp) sequence, referred to as IdoA2S–GlcNS6S, was preferentially recognized by SgP proteins, e.g., the SgP trimer or the RBD. Our studies on the interaction of SgP with cell surface HSPGs using an in vitro cell-to-cell fusion model have revealed that 3-O-sulfated HSPGs may facilitate viral fusion more than the wild-type HSPGs.32 In fact, overexpression of 3-OST-3B isoform, a sulfotransferase that installs 3-O-sulfate groups in wild-type HS, resulted in cell-to-cell fusion almost equal to that with ACE2.
In this work, we present novel structural features of the HS–SgP system that clarify the observations reported in the literature. We find that structural elements of the common HS sequence as well as the 3-O-sulfate group are both important for binding to the RBD. More importantly, our studies point to the role of multiple 3-O-sulfate groups in high affinity recognition. Overall, our studies lead to the identification of the HS pharmacophore that may lead to HS oligosaccharides, or mimetics thereof, as effective inhibitors of SARS-CoV-2.
Screening of HS Microarray with S1 and RBD Reveals Selective Recognition of Only a Few Sequences
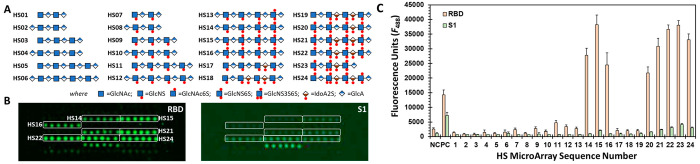
We utilized the HS microarray from Z Biotech containing 24 sequences to rapidly screen SARS-CoV-2 proteins. The sequences were tetra- to nonasaccharide in length and contained as many as nine sulfate groups (Figure 1A). All sequences contained unsulfated glucuronic acid (GlcA) residues, while some had sulfated iduronic acid (IdoA) residues. In addition, two sequences contained one 3-O-sulfated glucosamine (GlcN) residue in the middle of the chain.
Figure 1.
SARS-CoV-2 spike glycoprotein (SgP) recognition of a library of HS sequences. (A) Key to the structure of 24 HS sequences printed on the microarray. (B) Images of fluorescence from the bound RBD (left) and S1 (right) proteins (20 μg/mL) detected using Alexa Fluor 488 conjugated to streptavidin. Highlighted rectangles correspond to HS sequences in (A) that exhibit preferential recognition of the two proteins done in six spot replicates (n = 6) for each HS sequence. Two independent experiments were minimally performed for each protein. (C) Plot showing quantitative fluorescence for each HS sequence as numbered in (A). Error bars represent ±1 SE. Negative control (NC) = printing buffer; positive control (PC) = biotinylated mannose. See Supporting Information for detailed experimental conditions.
We utilized four different SARS-CoV-2 proteins including an S1 protein with His- and biotin-tags (S1), an AVI-modified S1 protein with His- and biotin-tags (S1-AVI), an RBD protein with His- and biotin-tags (RBD), and an AVI-modified RBD protein with His- and biotin-tags (RBD-AVI). Whereas SgP exists as a trimer in nature, the S1 and RBD proteins studied here are monomers. In the trimeric form, SgP displays at least one RBD in the “open” form. In contrast, the RBD and S1 proteins used in these studies exist in the “open” form all the time. While the AVI-tagged proteins contain only one site of biotinylation (at the AVI peptide sequence on the C-terminus), the non-AVI proteins are biotinylated at two or more Lys residues. Alexa Fluor 488 labeled streptavidin (S-AF488) was used for detection of each SARS-CoV-2 protein. In a recent publication, Hao et al. have reported the study of the same HS microarray and SARS-CoV-2 proteins except for using a Cy3-based fluorescence.33
Of the 24 sequences, HS14–HS16 and HS20–HS24 were identified as preferentially recognized by RBD (Figure 1B and Figure S1). Essentially the same group of sequences were recognized in the S1 subarray, albeit with much lower fluorescence intensity. Interestingly, neither the S1-AVI nor RBD-AVI subarray produced any signal (not shown). The reason for the lack of signal with the AVI-conjugated proteins is poor sensitivity. Whereas S1-AVI and RBD-AVI can only bind to one fluorophore at the C-terminus, the non-AVI proteins can bind to two or more. Because we sought to minimize nonselective recognition, experimentation at as low protein concentration as possible was highly desirable. Thus, we focused on the non-AVI proteins with higher sensitivity of detection.
A comparative analysis of the relative fluorescence intensities from the HS microarray yields four distinct structure–activity relationships (Figure 1C). (1) Octa- and nonasaccharides (e.g., HS14–HS16, HS20–HS24) are preferred by RBD and S1 over comparable tetra- and hexasaccharides. (2) Comparison of the fluorescence profiles for HS17 and HS18 vs HS20 and HS21, respectively, shows a large preference for the GlcNS6S residue. Likewise, the profiles of HS12 and HS16 also highlight the role of the 6S group on the GlcN residue. In fact, a minimum of two 6S groups on adjacent GlcN residues (e.g., HS13 vs HS14) appears to engineer selectivity for RBD. (3) In contrast, the relative importance of IdoA and GlcA residues is less clear. For example, both HS16 and HS21 display similar RBD recognition preference although the former contains GlcA exclusively, while the latter has two IdoA2S residues (Figure 1C). Nearly similar profiles are observed for other IdoA and GlcA containing sequences, e.g., HS20 and HS22 vs HS14 and HS15. (4) Finally, the presence of a 3S group in GlcNS6S does not appear to further improve recognition for longer sequences, e.g., HS22 vs HS24. Yet the results show that the only hexasaccharide that is recognized well by RBD and S1 is the one with the 3S group, i.e., HS23.
Overall, the HS microarray results indicate that octa/nonasaccharides with multiple GlcNS6S residues are preferentially recognized by RBD. It is important to note that this limited survey of available sequences reveals that uronic acid residues are not differentially recognized. This implies that SARS-CoV-2 RBD may equally recognize both HS and Hp, which primarily contain GlcA and IdoA residues, respectively. With regard to the possible role of a 3S group, its presence in longer sequences perhaps introduces redundancy, but in shorter sequences it engineers binding potency for RBD. In contrast to our results, Hao et al. report a much larger set of S1 and RBD binders including sequences HS01, HS07, HS09–HS13, HS17–HS19, HS14–HS16, and HS20–HS24.33 Yet within this group, HS23 and HS24 were found to be the best binders. Both HS23 and HS24 contain one 3S group. In contrast, Boons and co-workers report that the IdoA2S–GlcNS6S is the preferred binding partner of RBD with not much role for the lone 3S group of GlcNS6S in their HS library.31
Virtual Screening Supports Microarray Results
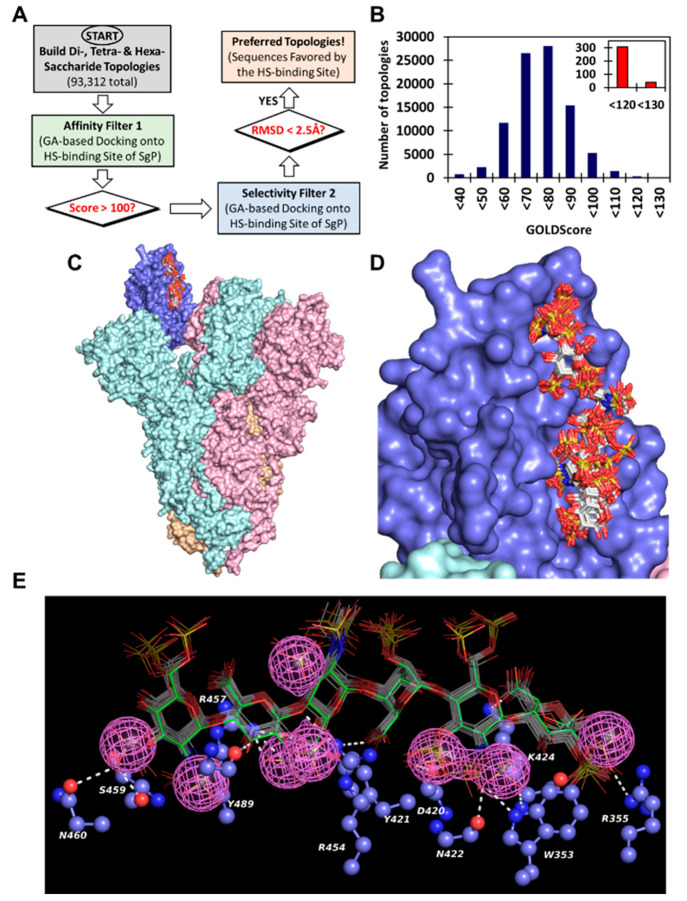
To understand the recognition of HS sequences by SgP, we performed virtual screening using the dual-filter strategy (Figure 2) developed earlier for studying GAG recognition of proteins.34−37 Briefly, the virtual screening strategy utilizes multiple docking runs to evaluate interactions in the form of GOLDScore, which is a surrogate for “in silico affinity”, and root-mean-square deviation (RMSD in Å) between different binding poses, which is a surrogate for “in silico selectivity” of binding. Studies with multiple GAG–protein systems have shown that GOLDScore and RMSD are two reliable, orthogonal parameters for identification of promising HS sequences.35,38−40 Hence, we first generated all topologies of the 24 HS oligosaccharides, arising from two major IdoA puckers (1C4 and 2SO), and docked them into the RBD using our virtual screening algorithm in triplicate (Figure 2A).
Figure 2.
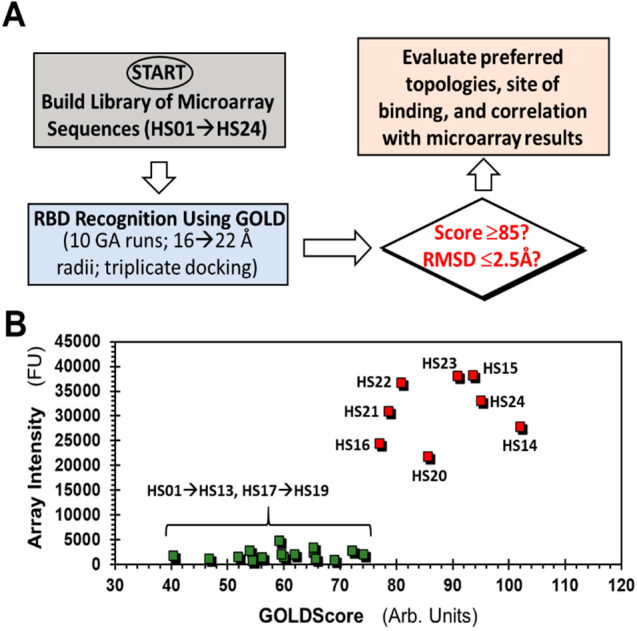
Computational studies for understanding HS recognition of RBD. (A) Virtual screening of the library of HS sequences (HS01–HS24) for binding to the receptor binding domain (RBD) of spike glycoprotein (SgP). GOLD-based docking and scoring were performed in triplicate. Two parameters, GOLDScore and RMSD between the top six poses for each HS sequence, were calculated to evaluate preferential recognition of HS sequences and correlation with microarray data. (B) Correspondence between microarray intensity and GOLDScore for each HS sequence. HS17 → HS24 sequences containing one or more IdoA2S residues were modeled in either all 1C4 or 2SO forms. See Table S1 for details.
Interestingly, only three sequences HS20, HS23, and HS24 displayed high GOLDScores (i.e., >85) and low RMSD values (i.e., ≤2.5 Å) suggesting significant selectivity in RBD recognition (Table S1). In fact, the last two sequences displayed the highest GOLDScores and lowest RMSD values when docked onto RBD, which supports the conclusion that the 3S group on the GlcN residue may confer binding specificity to HS sequences. Several other sequences, e.g., HS14, HS15, and HS21, displayed good GOLDScores (i.e., 75 → 85) but poor RMSD values (i.e., ≥2.5 Å, Table S1), which implied that these sequences are likely to interact well with RBD albeit with poor selectivity. Remaining sequences HS01 → HS13 and HS17 → HS19 bound RBD with much weaker interaction. Alternatively, the group of 24 HS sequences can be segregated into two cohorts of good and poor RBD recognition properties (Figure 2B), which supports the results observed in microarray screening.
HS Microarray-Based Competitive Studies and Biophysical Titrations Reveal Shorter Chains as Antagonists of HS–RBD Interactions
If HS-based antagonists of the HSPG–SgP interaction are to be discovered, it is important to identify the smallest HS sequence that can serve as an effective competitor. Hence, we used fondaparinux (FPX), a 3S-containing pentasaccharide, and HS hexasaccharide (Hexa), a trimer of IdoA2S-GlcNS6S repeating unit, as soluble competitors much smaller than the octa/nonasaccharides preferred by RBD. Whereas the former contained one 3S group, the latter contained three 6S groups, both of which should offer significant competition with HS sequences printed on the microarray.
The competition experiments were performed in a manner similar to that for RBD described above except for the addition of varying concentrations of either FPX or Hexa (0 → 50 μM). Analysis of the FPX and Hexa competition subarrays revealed a clear loss in fluorescence signal for HS14–HS16 and HS20–HS24 sequences (Figure 3 and Figure S2). More specifically, as the concentration of either FPX or Hexa increased, each HS sequence present in the microarray displayed a precipitous drop in bound RBD. Further, the rate of decrease in fluorescence with ligand concentration was fairly consistent and nearly equal for both FPX and Hexa for all RBD preferred HS sequences HS14–HS16 and HS20–HS24 (Figures 3C and S3). This suggests that both FPX and Hexa compete almost equally. Yet a closer review of competitive effects at all ligand concentrations shows that FPX is slightly weaker than Hexa. Alternatively, a single 3S group in a short pentasaccharide sequences is less effective than three 6S groups in a hexasaccharide sequence. However, the shorter chain length of FPX is an attractive feature in the design/discovery of smaller sequences as putative anti-SARS-CoV-2 agents.
Figure 3.
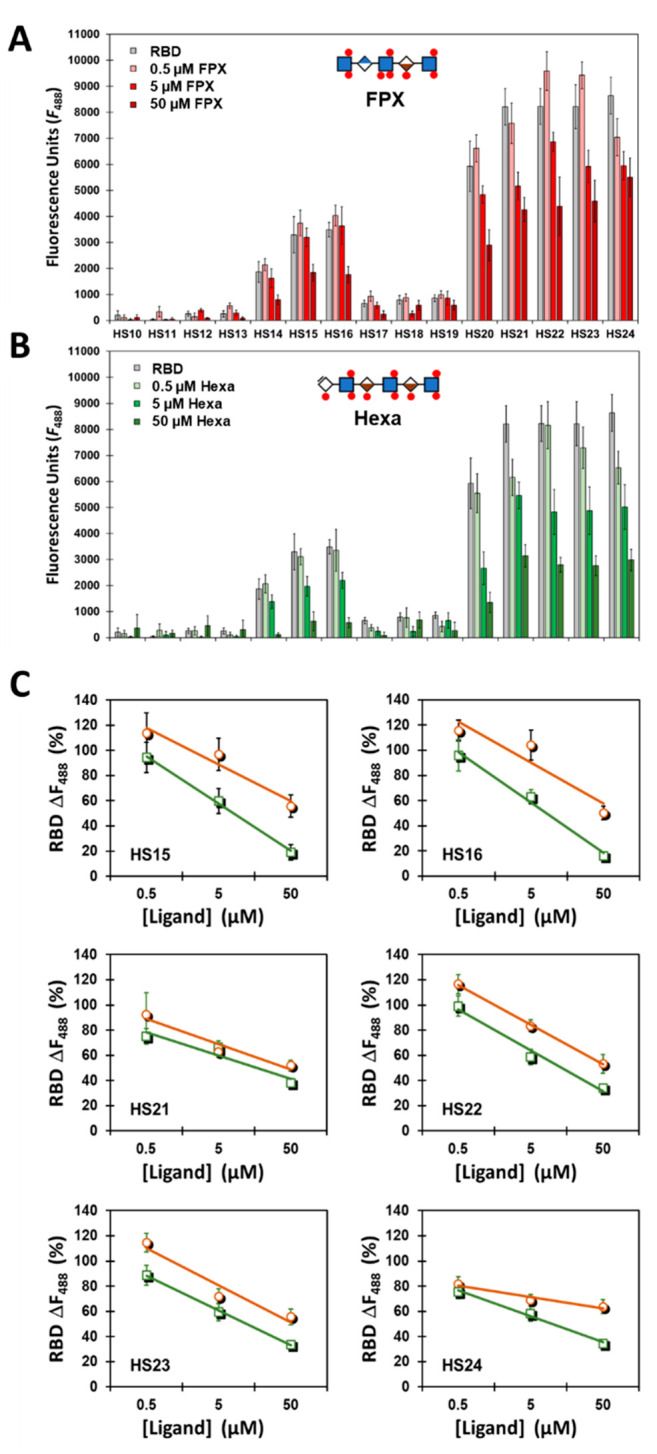
Studies using fondaparinux (FPX, panel A) and HS hexasaccharide (Hexa, panel B) as competitors in RBD recognition of 24 HS sequences on the microarray. The competition experiments were performed at least twice (biological replicates) in the manner similar to that for RBD alone (Figure 1) except for the addition of either 0.5 → 50 μM FPX (green squares/lines) or 0.5 → 50 μM Hexa (brown circles/lines) (n = 6). (A) Plot showing quantitative fluorescence as a function concentration of FPX. The occurrences of slight increases in fluorescence between 0 and 0.5 μM FPX in some cases were determined to be insignificant (p > 0.05). (B) Plot showing quantitative fluorescence as a function of concentration of Hexa. (C) Plots of percent change in fluorescence as a function of increasing ligand concentration (FPX or Hexa) for the six promising sequences (HS15, HS16, HS21, HS22, HS23, and HS24). Error bars represent ±1 SE. See Supporting Information for experimental conditions.
To quantitate the potency of HS sequences, we measured the affinities of HS23, FPX, and Hexa for RBD. By use of fluorescence spectroscopy, the affinities of the three sequences were measured to be 254 ± 26, 320 ± 89, and 130 ± 58 nM, respectively (Figure S4). These affinities are high indicating that RBD–HS interaction can be high affinity if appropriate structures are present in the HS chain. Although the affinities are not exactly identical, they are comparable suggesting that both the common (IdoA2S–GlcNS6S) and GlcNS3S structures are tightly recognized by RBD of the SgP.
Virtual Screening of >93 000 HS Sequences Highlights the Importance of 3S Groups in GlcN Residues
The above results present a strong possibility that smaller HS oligosaccharides should be feasible to a design that can effectively antagonize HS–SgP interaction. The library of natural HS sequences is enormous, and harnessing its diversity to identify possible selective antagonists of this interaction could help in the design of novel anti-SARS-CoV-2 inhibitors. In fact, the GOLDScore and RMSD-based virtual screening algorithm was developed in 2006 to harness the potential of this diversity.37 Recently, we designed a GlcA2S and GlcNS3S containing HS sequence, two rare residues in nature, that selectively activate Hp cofactor II using this technology.39 Likewise, the virtual screening strategy has been used to elucidate the pattern of HS recognition by human neutrophil elastase.35 Thus, we hypothesized that this technology could be used to reveal shorter HS sequence(s) that may bind RBD with higher putative affinity.
To test this expectation, we generated a combinatorial library of all possible sequences that are di- to hexasaccharide long. The HS sequences included all common residues such as IdoA, IdoA2S, GlcA, GlcNS, GlcNAc, GlcNS6S, and GlcNAc6S. In addition, residues rarely found in nature including GlcA2S, GlcNS3S, GlcNAc3S, GlcNS3S6S, and GlcNAc3S6S were also included. We also studied Amine(NRE)–Acid and Acid(NRE)–Amine sequences, which contained either GlcN or IdoA/GlcA residues, respectively, at the nonreducing end (NRE). Finally, the IdoA and IdoA2S residues were considered explicitly in either 1C4 or 2SO forms. In all, the combinatorial library contained 95 976 topologies made up of 72 di-, 2592 tetra-, and 93 312 hexasaccharide topologies. This comprehensive study was undertaken to ensure that the vast topological virtual space of HS hexasaccharides is fully studied.
Each of these topologies were docked onto RBD domain of the SgP trimer using the GOLD-based enrichment strategy (Figure 4A). The cryo-EM structure of SgP trimer contains one RBD domain in an open conformation (Figure S5). Analysis of the results following application of the “in silico affinity” filter indicated that none of the di- or tetrasaccharide sequences displayed reasonably high GOLDScores (i.e., >85). This implied a weak interaction of small oligosaccharides with the RBD of the trimer. In contrast, 345 hexasaccharides presented GOLDScores of >120, which reflects high affinity for the protein (Figure 4B).37,39,40 In the second filter study, the 345 high affinity sequences were docked multiple times and assessed for consistency of binding (i.e., RMSD between the docked poses) to identify 45 sequences with RMSD ≤ 2.5 Å (Figure 4C). Except for one, each of these sequences contained at least one 3S group. More importantly, 71% of these, i.e., 32 sequences, contained two or more 3S groups per chain (Table S2). In fact, 31 of the 32 sequences belonged to the Amine(NRE)–Acid group, suggesting a remarkably high level of selectivity for these multi-3S containing hexasaccharides.
Figure 4.
Computational screening of a library of di-, tetra-, and hexasaccharide sequences of HS (total 95 976 topologies) against the RBD of SgP for identification of origin of selectivity at the atomistic level. (A) Flowchart of the dual-filter algorithm used in computational screening, which included GOLDScore as the first filter and RMSD (consistency of binding) as second filter. (B) Results following application of the first filter in the form of a histogram of the number of HS hexasaccharide topologies for every 10-unit change in GOLDScore. Inset shows promising high affinity topologies. (C) High-affinity, high-specificity sequences (shown as sticks) bound to the “open” RBD (blue rendering) of trimeric SgP (pink, cyan, and orange rendering). (D) Zoomed version of the 45 high selectivity HS hexasaccharides (sticks in white color by atom) binding to the RBD (blue rendering). See Table S2 for details on the structure of these sequences. (E) Pharmacophoric representation of the highly selective sequences (colored sticks) shown with interacting residues (ball and stick representation) of the RBD. The pink mesh engulfing sulfate groups is the pharmacophore. Hydrogen bonds are shown as white dotted lines.
Analysis of the binding poses of the best hexasaccharides indicates highly consistent patterns of recognition (Figure 4D). Such consistency typically arises if a pharmacophore is present within these structurally distinct sequences. In fact, clustering of these HS sequences reveals a pharmacophore, which is predicted to originate from 2S, 3S, and 6S groups of GlcN and 2S groups of IdoA/GlcA residues (Figure 4E). Detailed analyses of the binding poses of these sequences reveal the ionic/hydrogen-bonding forces governing their interactions with RBD residues including Trp353, Arg355, Tyr421, Gln422, Lys424, Arg454, Arg457, Gln460, Tyr489, etc., which engage almost all the residues of the hexasaccharide, thus explaining the reason for their high “in silico affinity” and “in silico selectivity”. As an example, the predicted atomistic interactions for the highest scoring hexasaccharide are shown in Figure S6. Unfortunately, this HS sequence and other sequences containing multiple 3S groups are not commercially available, making it difficult to quickly validate the prediction. However, the pharmacophore generated from this comprehensive study will aid the development of a mimetic that potently and selectively antagonizes the HS–SgP system.
Considering the recent rise in the number of infections arising from the δ variant of SARS-CoV-2, we sought to evaluate its HS recognition properties. The RBD of the δ variant contains two mutations including L452R and T478K. L452R and T478K are located outside the predicted binding site of FPX and Hexa (Figure S7), which suggests that their interaction is unlikely to be impacted. In fact, CVLS studies of FPX, HS23, and Hexa indicated minimal effect (Figure S7C). Likewise, spectrofluorimetric studies showed minimal impact on the affinity of binding for FPX and Hexa. Yet T478K is about 23 Å away from the predicted site of FPX, which in principle could be engaged by full-length HS. Thus, the absence of impact with the δ variant supports the predicted interaction of FPX and Hexa; it does not eliminate the possibility of better interaction with polymeric HS in the proteoglycan form, which is the binding partner of SgP on cell surfaces.
Overall, this work presents a rather interesting observation. The RBD domain of SARS-CoV-2 SgP tends to preferentially recognize HS hexasaccharides with more than one 3-O-sulfated (3S) GlcN residue. This is the first detailed insight into how RBD recognizes the entire library of HS sequences.
A key conclusion from this exhaustive screening is that RBD recognition by a distinct group of HS sequences is highly selective. These sequences show a common pattern of sulfate group distribution with predicted binding to both ionic and nonionic (polar) residues of the RBD in a highly consistent manner. Polar residues, especially Gln, in a highly electropositive site have been implicated in engineering selectivity in HS–protein systems.41 Thus, it is highly significant that the HS binding site of the RBD contains two Gln residues (Figure 4E), which recognize the 3S groups.
Another conclusion from this exhaustive screening is that a large majority of HS sequences bind to the RBD with rather weak affinity (low GOLDScores) and nonselectively (high RMSD). Alternatively, most HS sequences that are small (di- → hexasaccharides) would interact rather moderately. This does not imply that these structures are not important. In fact, longer HS chains, which display much higher affinity to SgP29 and contain non-3S structures, are involved in the recruitment of virus to host cell surface, as shown recently.25
This work shows for the first time that if the focus is on smaller oligosaccharides, then 3S containing structures are especially important. In fact, a 3S-containing pentasaccharide FPX competes well with longer HS sequences (hexa → nona) in competitive studies (Figure 3). This implies that smaller, structurally rich oligosaccharides could be developed as potent inhibitors of HS–SgP interaction. In this connection, the structure of the pharmacophore is highly valuable.
Unfortunately, oligosaccharides with multiple 3S groups are difficult to obtain. Both synthetic and chemoenzymatic methods of obtaining homogeneous 3S-containing sequences are challenging to implement and difficult to scale up.42−45 An alternative is to utilize functional mimetics of HS, such as non-saccharide GAG mimetics (NSGMs), which have been developed as disruptors of endogenous GAG–receptor systems.46−49 Thus, translating the pharmacophore (Figure 4E) discovered in this work to a synthetic NSGM will also be a major goal of the future.
Various other challenges regarding HS oligosaccharides will have to be overcome including synthesis and off-target effects. This includes elucidating the interactome of the sequences identified in this study, which is difficult because the GAG interactome technology is still being developed.50 In the interim, screening against proteins known to bind GAGs51 against 31 hexasaccharides of Table S2 would be advisible.
Acknowledgments
This work was supported in part by NIH Grants HL107152, CA241951, and HL151333 to U.R.D. We also thank the computing resources made available through Grant S10RR027411 from the National Center for Research Resources to Virginia Commonwealth University.
Glossary
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- FPX
fondaparinux
- GAG
glycosaminoglycan
- GlcA
glucuronic acid
- GlcN
glucosamine
- Hexa
heparan sulfate hexasaccharide
- Hp
heparin
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
- IdoA
iduronic acid
- NSGM
non-saccharide glycosaminoglycan mimetic
- RBD
receptor-binding domain
- RMSD
root-mean-square deviation
- S-AF488
Alexa Fluor 488 labeled streptavidin
- SgP
spike glycoprotein
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.1c00343.
Figures S1–S7, which present microarray, computational, and biophysical results, and Tables S1 and S2, which present scores and RMSD results for HS sequences binding to RBD and SgP (PDF)
Author Contributions
§ J.E.C. and N.V.S. contributed equally. J.E.C. did the microarray experimentation and analysis and manuscript first draft and revisions. N.V.S. did the computational design, analysis, and writeup. C.P.O. did the biophysical studies. U.R.D. did the hypothesis generation, experimental design and analysis, supervision, manuscript revision and finalization, and funding acquisition.
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Sivaraman H.; Er S. Y.; Choong Y. K.; Gavor E.; Sivaraman J. Structural basis of SARS-CoV-2-and SARS-CoV-receptor binding and small-molecule blockers as potential therapeutics. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 465–493. 10.1146/annurev-pharmtox-061220-093932. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Brindisi M.; Shahabi D.; Chapman M. E.; Mesecar A. D. Drug development and medicinal chemistry efforts toward SARS-Coronavirus and Covid-19 therapeutics. ChemMedChem 2020, 15, 907–932. 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos I. d. A.; Grosche V. R.; Bergamini F. R. G.; Sabino-Silva R.; Jardim A. C. G. Antivirals against coronaviruses: candidate drugs for SARS-CoV-2 treatment?. Front. Microbiol. 2020, 11, 1818. 10.3389/fmicb.2020.01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Horani R. A.; Kar S. Potential anti-SARS-CoV-2 therapeutics that target the post-entry stages of the viral life cycle: A comprehensive review. Viruses 2020, 12, 1092. 10.3390/v12101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V.; Beer J. C.; Sankaranarayanan N. V.; Swanson-mungerson M.; Desai U. R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discovery Today 2020, 25, 1535–1544. 10.1016/j.drudis.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L.; Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol., Immunol. Inf. 2021, 54, 159–163. 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J.; Wang D.; Xu W.; Holmes E. C.; Gao G. F.; Wu G.; Chen W.; Shi W.; Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Wang M.; Yu J.; Cerutti G.; Nair M. S.; Huang Y.; Kwong P. D.; Shapiro L.; Ho D. D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. BioRxiv 2021, 10.1101/2021.03.01.433466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.; Garcia-Knight M. A.; Khalid M. M.; Servellita V.; Wang C.; Kate Morris M.; Sotomayor-González A.; Reyes K. R.; Gliwa A. S.; Reddy N. P.; Sanchez C.; Martin S.; Federman S.; Cheng J.; Balcerek J.; Taylor J.; Streithorst A.; Miller S.; Renuka Kumar G.; Sreekumar B.; Chen P.-Y.; Schulze-Gahmen U.; Taha T. Y.; Hayashi J.; McMahon S.; Lidsky P. V.; Xiao Y.; Hemarajata P.; Green N. M.; Espinosa A.; Kath C.; Haw M.; Bell J.; Hanson C.; Wadford D. A.; Anaya C.; Ferguson D.; Frankino P. A.; Shivram H.; Wyman S. K.; Andino R.; Chiu C. Y. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-1 2 variant in california carrying a L452R spike protein mutation 2. MedRxiv 2021, 10.1101/2021.03.07.21252647. [DOI] [Google Scholar]
- Wang P.; Liu L.; Iketani S.; Luo Y.; Guo Y.; Wang M.; Yu J.; Zhang B.; Kwong P. D.; Graham B. S.; Mascola J. R.; Chang J. Y.; Yin M. T.; Sobieszczyk M.; Kyratsous C. A.; Shapiro L.; Sheng Z.; Nair M. S.; Huang Y.; Ho D. D. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. BioRxiv 2021, 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Weisblum Y.; Schmidt F.; Zhang F.; DaSilva J.; Poston D.; Lorenzi J. C. C.; Muecksch F.; Rutkowska M.; Hoffmann H. H.; Michailidis E.; Gaebler C.; Agudelo M.; Cho A.; Wang Z.; Gazumyan A.; Cipolla M.; Luchsinger L.; Hillyer C. D.; Caskey M.; Robbiani D. F.; Rice C. M.; Nussenzweig M. C.; Hatziioannou T.; Bieniasz P. D. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Shang J.; Graham R.; Baric R. S.; Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C. L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M.; Sarma P.; Shekhar N.; Prakash A.; Avti P.; Bhattacharyya A.; Kaur H.; Kumar S.; Bansal S.; Sharma A. R.; Medhi B. Update on the target structures of SARS-CoV-2: A systematic review. Indian J. Pharmacol. 2020, 52, 142–149. 10.4103/ijp.IJP_338_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L.; Ojha R.; Pedro L. D.; Djannatian M.; Franz J.; Kuivanen S.; van der Meer F.; Kallio K.; Kaya T.; Anastasina M.; Smura T.; Levanov L.; Szirovicza L.; Tobi A.; Kallio-Kokko H.; Österlund P.; Joensuu M.; Meunier F. A.; Butcher S. J.; Winkler M. S.; Mollenhauer B.; Helenius A.; Gokce O.; Teesalu T.; Hepojoki J.; Vapalahti O.; Stadelmann C.; Balistreri G.; Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J. L.; Simonetti B.; Klein K.; Chen K. E.; Williamson M. K.; Antón-Plágaro C.; Shoemark D. K.; Simón-Gracia L.; Bauer M.; Hollandi R.; Greber U. F.; Horvath P.; Sessions R. B.; Helenius A.; Hiscox J. A.; Teesalu T.; Matthews D. A.; Davidson A. D.; Collins B. M.; Cullen P. J.; Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Thorp S. C. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 2002, 22, 1–25. 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- Song B. H.; Lee G. C.; Moon M. S.; Cho Y. H.; Lee C. H. Human cytomegalovirus binding to heparan sulfate proteoglycans on the cell surface and/or entry stimulates the expression of human leukocyte antigen class I. J. Gen. Virol. 2001, 82, 2405–2413. 10.1099/0022-1317-82-10-2405. [DOI] [PubMed] [Google Scholar]
- Tyagi M.; Rusnati M.; Presta M.; Giacca M. Internalization of HIV-1 Tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- Hilgard P.; Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology 2000, 32, 1069–1077. 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- Shieh M. T.; WuDunn D.; Montgomery R. I.; Esko J. D.; Spear P. G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 1992, 116, 1273–1281. 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. M.; Sandoval D. R.; Spliid C. B.; Pihl J.; Perrett H. R.; Painter C. D.; Narayanan A.; Majowicz S. A.; Kwong E. M.; McVicar R. N.; Thacker B. E.; Glass C. A.; Yang Z.; Torres J. L.; Golden G. J.; Bartels P. L.; Porell R. N.; Garretson A. F.; Laubach L.; Feldman J.; Yin X.; Pu Y.; Hauser B. M.; Caradonna T. M.; Kellman B. P.; Martino C.; Gordts P. L. S. M.; Chanda S. K.; Schmidt A. G.; Godula K.; Leibel S. L.; Jose J.; Corbett K. D.; Ward A. B.; Carlin A. F.; Esko J. D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 2020, 183, 1043–1057. 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Chen C. Z.; Swaroop M.; Xu M.; Wang L.; Lee J.; Wang A. Q.; Pradhan M.; Hagen N.; Chen L.; Shen M.; Luo Z.; Xu X.; Xu Y.; Huang W.; Zheng W.; Ye Y. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discovery 2020, 6, 80. 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P. S.; Oh H.; Kwon S. J.; Jin W.; Zhang F.; Fraser K.; Hong J. J.; Linhardt R. J.; Dordick J. S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discovery 2020, 6, 50. 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R.; Sharp J. S.; Zhang F.; Pomin V. H.; Ashpole N. M.; Mitra D.; Jin W.; Liu H.; Sharma P.; Linhardt R. J. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. BioRxiv 2020, 10.1101/2020.06.08.140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y.; Jin W.; Sood A.; Montgomery D. W.; Grant O. C.; Fuster M. M.; Fu L.; Dordick J. S.; Woods R. J.; Zhang F.; Linhardt R. J. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res. 2020, 181, 104873. 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond S.; Mycroft-West C.; Gandhi N.; Tree J.; Buttigieg K.; Coombes N.; Elmore M.; Nyström K.; Said J.; Setoh Y. X.; Amarilla A.; Modhiran N.; Sng J.; Chhabra M.; Watterson D.; Young P.; Khromykh A.; Lima M.; A.Yates E.; Karlsson R.; Chen Y.-H.; Zhang Y.; Hammond E.; Dredge K.; Carroll M.; Trybala E.; Bergström T.; Ferro V.; Skidmore M.; Turnbull J. Synthetic heparan sulfate mimetic pixatimod (PG545) potently inhibits SARS-CoV-2 By disrupting the spike-ACE2 interaction. BioRxiv 2020, 10.1101/2020.06.24.169334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Chopra P.; Li X.; Bouwman K.; Tompkins S. M.; Wolfert M.; de Vries R.; Boons G.-J. Heparan sulfate proteoglycans as attachment factor for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1009–1018. 10.1021/acscentsci.1c00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V.; Tandon R.; Sankaranarayanan N. V.; Beer J. C.; Kohlmeir E. K.; Swanson-Mungerson M.; Desai U. R. Preferential recognition and antagonism of SARS-CoV-2 spike glycoprotein binding to 3-O-sulfated heparan sulfate. BioRxiv 2020, 10.1101/2020.10.08.331751. [DOI] [Google Scholar]
- Hao W.; Ma B.; Li Z.; Wang X.; Gao X.; Li Y.; Qin B.; Shang S.; Cui S.; Tan Z. Binding of the SARS-CoV-2 spike protein to glycans. Sci. Bull. 2021, 66, 1205–1214. 10.1016/j.scib.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan N. V.; Nagarajan B.; Desai U. R. So you think computational approaches to understanding glycosaminoglycan–protein interactions are too dry and too rigid? Think again!. Curr. Opin. Struct. Biol. 2018, 50, 91–100. 10.1016/j.sbi.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummarapurugu A. B.; Afosah D. K.; Sankaranarayanan N. V.; Gangji R. N.; Zheng S.; Kennedy T.; Rubin B. K.; Voynow J. A.; Desai U. R. Molecular principles for heparin oligosaccharide-based inhibition of neutrophil elastase in cystic fibrosis. J. Biol. Chem. 2018, 293, 12480–12490. 10.1074/jbc.RA118.002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan B.; Sankaranarayanan N. V.; Patel B. B.; Desai U. R. A molecular dynamics-based algorithm for evaluating the glycosaminoglycan mimicking potential of synthetic, homogenous, sulfated small molecules. PLoS One 2017, 12, e0171619. 10.1371/journal.pone.0171619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman A.; Mosier P. D.; Desai U. R. Finding a needle in a haystack: Development of a combinatorial virtual screening approach for identifying high specificity heparin/heparan sulfate sequences. J. Med. Chem. 2006, 49, 3553–3562. 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan N. V.; Bi Y.; Kuberan B.; Desai U. R. Combinatorial virtual library screening analysis of antithrombin binding oligosaccharide motif generation by heparan sulfate 3-O-sulfotransferase 1. Comput. Struct. Biotechnol. J. 2020, 18, 933–941. 10.1016/j.csbj.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan N. V.; Strebel T. R.; Boothello R. S.; Sheerin K.; Raghuraman A.; Sallas F.; Mosier P. D.; Watermeyer N. D.; Oscarson S.; Desai U. R. A hexasaccharide containing rare 2-O-sulfate-glucuronic acid residues selectively activates heparin cofactor II. Angew. Chem., Int. Ed. 2017, 56, 2312–2317. 10.1002/anie.201609541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan N. V.; Desai U. R. Toward a robust computational screening strategy for identifying glycosaminoglycan sequences that display high specificity for target proteins. Glycobiology 2014, 24, 1323–1333. 10.1093/glycob/cwu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.; Desai U. R. A simple method for discovering druggable, specific glycosaminoglycan-protein systems. Elucidation of key Principles from heparin/heparan sulfate-binding proteins. PLoS One 2015, 10, e0141127 10.1371/journal.pone.0141127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende M.; Bednarek C.; Wawryszyn M.; Sauter P.; Biskup M. B.; Schepers U.; Bräse S. Chemical synthesis of glycosaminoglycans. Chem. Rev. 2016, 116, 8193–8255. 10.1021/acs.chemrev.6b00010. [DOI] [PubMed] [Google Scholar]
- Lu W.; Zong C.; Chopra P.; Pepi L. E.; Xu Y.; Amster I. J.; Liu J.; Boons G. J. Controlled chemoenzymatic synthesis of heparan sulfate oligosaccharides. Angew. Chem., Int. Ed. 2018, 57, 5340–5344. 10.1002/anie.201800387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Pagadala V.; Jester H. M.; Lim A. M.; Pham T. Q.; Goulas A. M. P.; Liu J.; Linhardt R. J. Chemoenzymatic synthesis of heparan sulfate and heparin oligosaccharides and NMR analysis: Paving the way to a diverse library for glycobiologists. Chem. Sci. 2017, 8, 7932–7940. 10.1039/C7SC03541A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Linhardt R. J. Chemoenzymatic synthesis of heparan sulfate and heparin. Nat. Prod. Rep. 2014, 31, 1676–1685. 10.1039/C4NP00076E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai U. R. The promise of sulfated synthetic small moelcules are modulators of glycosaminoglycan function. Future Med. Chem. 2013, 5, 1363–1366. 10.4155/fmc.13.117. [DOI] [PubMed] [Google Scholar]
- Boothello R. S.; Patel N. J.; Sharon C.; Abdelfadiel E. I.; Morla S.; Brophy D. F.; Lippman H. R.; Desai U. R.; Patel B. B. A Unique nonsaccharide mimetic of heparin hexasaccharide inhibits colon cancer Stem cells via P38 MAP kinase activation. Mol. Cancer Ther. 2019, 18, 51–61. 10.1158/1535-7163.MCT-18-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morla S. Glycosaminoglycans and glycosaminoglycan mimetics in cancer and inflammation. Int. J. Mol. Sci. 2019, 20, 1963. 10.3390/ijms20081963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afosah D. K.; Al-Horani R. A. Sulfated non-saccharide glycosaminoglycan mimetics as novel drug discovery platform for various pathologies. Curr. Med. Chem. 2020, 27, 3412–3447. 10.2174/0929867325666181120101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffrin A. M.; Hsieh-Wilson L. C. Photoaffinity probes for the identification of sequence-specific glycosaminoglycan-binding proteins. J. Am. Chem. Soc. 2020, 142, 13672–13676. 10.1021/jacs.0c06046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet S. D.; Clerc O.; Ricard-Blum S. Glycosaminglycan-protein interactions: the first draft of the glycosaminoglycan interactome. J. Histochem. Cytochem. 2021, 69, 93–104. 10.1369/0022155420946403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.