In the original article, there was a mistake in Table 1 as published. The references indicated in row B are wrong. The corrected Table 1 appears in the attached below.
Table 1.
(A–F) The advantages and disadvantages of different integration strategies.
| Integration strategies | Advantages | Disadvantages | References | ||
|---|---|---|---|---|---|
| A | Endogenous locus |
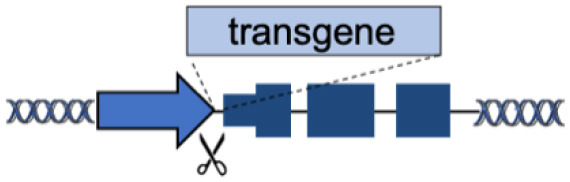
|
Physiological transgene expression Corrects multiple mutations |
Gene-specific strategy Limited to gene body mutations |
Urnov et al., 2005; Lombardo et al., 2007; Li et al., 2011; Genovese et al., 2014; Voit et al., 2014; Dever et al., 2016; Hubbard et al., 2016; Schiroli et al., 2017; Sweeney et al., 2017; Kuo et al., 2018; Wang et al., 2019; Rai et al., 2020; Wang L. et al., 2020 |
| B | Superactive promoters (ALB, HBA) |
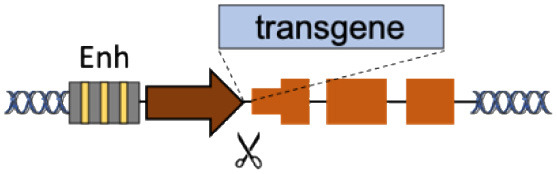
|
Accommodates different transgenes Supraphysiological expression Few integrations required |
Partial gene disruption Limited to non-cell autonomous disorders Extensive validation required |
Barzel et al., 2015; Sharma et al., 2015; Davidoff and Nathwani, 2016; Laoharawee et al., 2018; Chen et al., 2019; Conway et al., 2019; De Caneva et al., 2019; Ou et al., 2019, 2020; Zhang et al., 2019; Wang Q. et al., 2020 |
| C | Tolerant to integration (AAVS1, CCR5, Rosa26) |
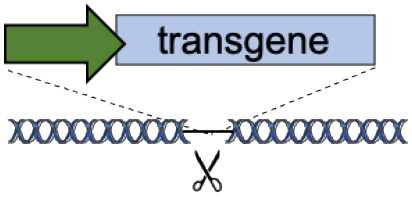
|
Accommodates different transgenes | Artificial promoters required Variable expression |
De Ravin et al., 2016; Diez et al., 2017; Stephens et al., 2018, 2019; Gomez-Ospina et al., 2019; Scharenberg et al., 2020 |
| D | Chromatin domains (NAD) |
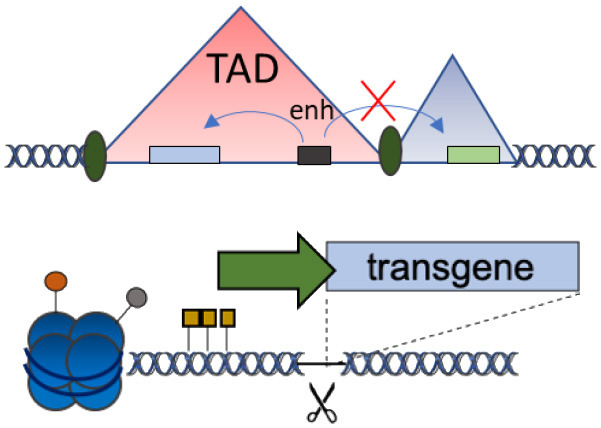
|
Fine gene regulation Far from oncogenic genes |
No proof-of-principle in clinically relevant models | Schenkwein et al., 2020 |
| E | Disease-modifier genes (CCR5, HBA) |
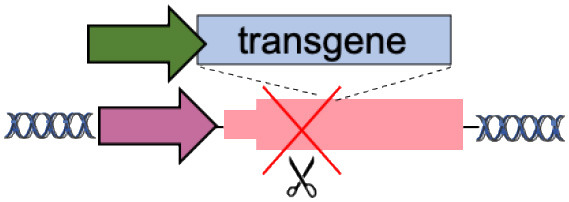
|
Improve therapeutic effect Lower therapeutic threshold |
Extensive validation required Limited to well-known diseases |
Voit et al., 2013; Wiebking et al., 2018 |
| F | Specificity Exchange (TCR, BCR) |
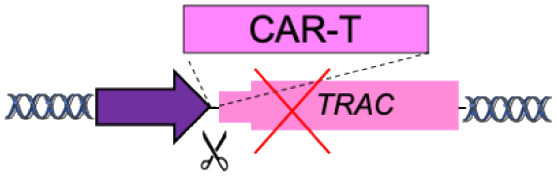
|
Improved CAR expression and potency | Off-targets Translocations risk (for multiple edits) |
Eyquem et al., 2017; MacLeod et al., 2017; Greiner et al., 2019; Hartweger et al., 2019; Moffett et al., 2019; Voss et al., 2019 |
Scissors: nuclease; Solid arrows: promoters; Enh, enhancers; TAD, topologically associating; d, domain; Solid ovals: histone modifications; Solid squares: DNA modifications.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
- Barzel A., Paulk N. K., Shi Y., Huang Y., Chu K., Zhang F., et al. (2015). Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364. 10.1038/nature13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shi M., Gilam A., Zheng Q., Zhang Y., Afrikanova I., et al. (2019). Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human factor. Sci. Rep. 9:16838. 10.1038/s41598-019-53198-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway A., Mendel M., Kim K., McGovern K., Boyko A., Zhang L., et al. (2019). Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. 27, 866–877. 10.1016/j.ymthe.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff A. M., Nathwani A. C. (2016). Genetic targeting of the albumin locus to treat Hemophilia. N. Engl. J. Med. 374, 1288–1290. 10.1056/NEJMcibr1600347 [DOI] [PubMed] [Google Scholar]
- De Caneva A., Porro F., Bortolussi G., Sola R., Lisjak M., Barzel A., et al. (2019). Coupling AAV-mediated promoterless gene targeting to SaCas9 nuclease to efficiently correct liver metabolic diseases. JCI Insight. 5:128863. 10.1172/jci.insight.128863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ravin S. S., Reik A., Liu P. Q., Li L., Wu X., Su L., et al. (2016). Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 34, 424–429. 10.1038/nbt.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever D. P., Bak R. O., Reinisch A., Camarena J., Washington G., Nicolas C. E., et al. (2016). CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389. 10.1038/nature20134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez B., Genovese P., Roman-Rodriguez F. J., Alvarez L., Schiroli G., Ugalde L., et al. (2017). Therapeutic gene editing in CD34(+) hematopoietic progenitors from Fanconi anemia patients. EMBO Mol. Med. 9, 1574–1588. 10.15252/emmm.201707540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S. J., Hamieh M., Cunanan K. M., et al. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. 10.1038/nature21405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese P., Schiroli G., Escobar G., Tomaso T. D., Firrito C., Calabria A., et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240. 10.1038/nature13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N., Scharenberg S. G., Mostrel N., Bak R. O., Mantri S., Quadros R. M., et al. (2019). Human genome-edited hematopoietic stem cells phenotypically correct Mucopolysaccharidosis type. Nat. Commun. 10:4045. 10.1038/s41467-019-11962-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner V., Bou Puerto R., Liu S., Herbel C., Carmona E. M., Goldberg M. S. (2019). CRISPR-mediated editing of the B cell receptor in primary human B cells. iScience 12, 369–378. 10.1016/j.isci.2019.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweger H., McGuire A. T., Horning M., Taylor J. J., Dosenovic P., Yost D., et al. (2019). HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med. 216, 1301–1310. 10.1084/jem.20190287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard N., Hagin D., Sommer K., Song Y., Khan I., Clough C., et al. (2016). Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood 127, 2513–2522. 10.1182/blood-2015-11-683235 [DOI] [PubMed] [Google Scholar]
- Kuo C. Y., Long J. D., Campo-Fernandez B., de Oliveira S., Cooper A. R., Romero Z., et al. (2018). Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. 23, 2606–2616. 10.1016/j.celrep.2018.04.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoharawee K., DeKelver R. C., Podetz-Pedersen K. M., Rohde M., Sproul S., Nguyen H. O., et al. (2018). Dose-dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol. Ther. 26, 1127–1136. 10.1016/j.ymthe.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Haurigot V., Doyon Y., Li T., Wong S. Y., Bhagwat A. S., et al. (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221. 10.1038/nature10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Genovese P., Beausejour C. M., Colleoni S., Lee Y. L., Kim K. A., et al. (2007). Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306. 10.1038/nbt1353 [DOI] [PubMed] [Google Scholar]
- MacLeod D. T., Antony J., Martin A. J., Moser R. J., Hekele A., Wetzel K. J., et al. (2017). Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther. 25, 949–961. 10.1016/j.ymthe.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett H. F., Harms C. K., Fitzpatrick K. S., Tooley M. R., Boonyaratanakornkit J., Taylor J. J. (2019). B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4:aax0644. 10.1126/sciimmunol.aax0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L., DeKelver R. C., Rohde M., Tom S., Radeke R., St Martin S. J., et al. (2019). ZFN-mediated in vivo genome editing corrects murine hurler syndrome. Mol. Ther. 27, 178–187. 10.1016/j.ymthe.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L., Przybilla M. J., Ahlat O., Kim S., Overn P., Jarnes J., et al. (2020). A highly efficacious PS gene editing system corrects metabolic and neurological complications of Mucopolysaccharidosis type I. Mol. Ther. 28, 1442–1454. 10.1016/j.ymthe.2020.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Romito M., Rivers E., Turchiano G., Blattner G., Vetharoy W., et al. (2020). Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott - Aldrich Syndrome. Nat. Commun. 11:4034. 10.1038/s41467-020-17626-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharenberg S. G., Poletto E., Lucot K. L., Colella P., Sheikali A., Montine T. J., et al. (2020). Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing. Nat. Commun. 11:3327. 10.1038/s41467-020-17148-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkwein D., Afzal S., Nousiainen A., Schmidt M., Yla-Herttuala S. (2020). Efficient nuclease-directed integration of lentivirus vectors into the human ribosomal DNA locus. Mol. Ther. 28, 1858–1875. 10.1016/j.ymthe.2020.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., et al. (2017). Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 9:aan0820. 10.1126/scitranslmed.aan0820 [DOI] [PubMed] [Google Scholar]
- Sharma R., Anguela X. M., Doyon Y., Wechsler T., DeKelver R. C., Sproul S., et al. (2015). In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126, 1777–1784. 10.1182/blood-2014-12-615492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C. J., Kashentseva E., Everett W., Kaliberova L., Curiel D. T. (2018). Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 25, 139–156. 10.1038/s41434-018-0003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C. J., Lauron E. J., Kashentseva E., Lu Z. H., Yokoyama W. M., Curiel D. T. (2019). Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J. Contr. Release. 298, 128–141. 10.1016/j.jconrel.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney C. L., Zou J., Choi U., Merling R. K., Liu A., Bodansky A., et al. (2017). Targeted repair of CYBB in X-CGD iPSCs requires retention of intronic sequences for expression and functional correction. Mol. Ther. 25, 321–330. 10.1016/j.ymthe.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F. D., Miller J. C., Lee Y. L., Beausejour C. M., Rock J. M., Augustus S., et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. 10.1038/nature03556 [DOI] [PubMed] [Google Scholar]
- Voit R. A., Hendel A., Pruett-Miller S. M., Porteus M. H. (2014). Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucl. Acids Res. 42, 1365–1378. 10.1093/nar/gkt947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R. A., McMahon M. A., Sawyer S. L., Porteus M. H. (2013). Generation of an HIV resistant T-cell line by targeted “stacking” of restriction factors. Mol. Ther. 21, 786–795. 10.1038/mt.2012.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss J. E., Gonzalez-Martin A., Andrabi R., Fuller R. P., Murrell B., McCoy L. E., et al. (2019). Reprogramming the antigen specificity of B cells using genome-editing technologies. Elife 8:42995. 10.7554/eLife.42995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang Y., Breton C., Bell P., Li M., Zhang J., et al. (2020). A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci. Adv. 6:eaax5701. 10.1126/sciadv.aax5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang Y., Breton C. A., White J., Zhang J., Che Y., et al. (2019). CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX-knockout mice. Blood 133, 2745–2752. 10.1182/blood.2019000790 [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhong X., Li Q., Su J., Liu Y., Mo L., et al. (2020). CRISPR-Cas9-mediated in vivo gene integration at the albumin locus recovers hemostasis in neonatal and adult hemophilia B mice. Mol. Ther. Methods Clin. Dev. 18, 520–531. 10.1016/j.omtm.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebking V., Patterson J. O., Martin R., Chanda M. K., Lee C. M., Srifa W., et al. (2018). Metabolic engineering generates a transgene-free safety switch for cell therapy. Nat. Biotechnol 2020:6. 10.1038/s41587-020-0580-6 [DOI] [PubMed] [Google Scholar]
- Zhang J. P., Cheng X. X., Zhao M., Li G. H., Xu J., Zhang F., et al. (2019). Curing hemophilia A by NHEJ-mediated ectopic F8 insertion in the mouse. Genome Biol. 20:276. 10.1186/s13059-019-1907-9 [DOI] [PMC free article] [PubMed] [Google Scholar]


