Abstract
Considerable genetic variation of N-methyl-D-aspartate receptors (NMDARs) has recently become apparent, with many hundreds of de novo variants identified through widely available clinical genetic testing. Individuals with GRIN variants present with neurological conditions such as epilepsy, autism, intellectual disability (ID), movement disorders, schizophrenia and behavioral disorders. Determination of the functional consequence of genetic variation for NMDARs should lead to precision therapeutics. Furthermore, genetic animal models harboring human variants have the potential to reveal mechanisms that are shared among different neurological conditions, providing strategies that may allow treatment of individuals who are refractory to therapy. Preclinical studies in animal models and small open label trials in humans support this idea. However, additional functional data for variants and animal models corresponding to multiple individuals with the same genotype are needed to validate this approach and to lead to thoughtfully designed, randomized, placebo-controlled clinical trials, which could provide data in order to determine safety and efficacy of potential precision therapeutics.
Keywords: GRIN, NMDARs, intellectual disability, epilepsy
Diagnosis and phenotyping
Developmental encephalopathies[1] are disorders associated with delayed childhood development, including impairments in cognition, communication, as well as fine motor and gross motor skills. Impairments in sensory systems such as vision and hearing are also often apparent. When these developmental delays involve multiple modalities, they are often referred to as global delay. When developmental delays persist for more than 5 years, they are almost certain to alter the individual for a prolonged period, thereby compromising their ability to carry out functions needed to live an independent life. When this is the case, the term Intellectual Disability (ID) is used, which is usually categorized as mild, moderate, severe or profound [2]. The alterations in developmental trajectories, including regression or loss of previously acquired skills, can be distinct for each disorder and thus an essential component of phenotyping. Further classification of epilepsy in terms of seizure characteristics and EEG properties is a necessary feature of phenotyping [3, 4]. Individuals harboring GRIN variants also show altered brain structure revealed through imaging, aberrant muscle tone, movement disorders such as ataxia, behavioral disturbances, as well as symptoms that can be understood through autonomic dysfunction. Efforts to harmonize the description of these characteristics has been developed (e.g. Human Phenotype Ontology, HPO, https://hpo.jax.org/app/) and could help synchronize phenotypic descriptions in the peer-reviewed literature[5].
Genotyping
Whole exome sequencing (WES) is widely considered to be the first stage of diagnostic testing when neurodevelopmental disorders are suspected[6], which includes individuals who show concerns for developmental delay, intellectual disability, and/or seizures. While gene panel testing typically utilizes the same technology as next generation sequencing, it still has limitations in terms of analysis compared to WES, such as a reduced capability to detect genomic copy number variants (CNVs, including deletion or duplication). Variation in genomic copy number will usually affect multiple genes, and is detected using chromosomal or SNP microarrays. Genetic diagnosis[7-10] for rare diseases has become an important means to end the cycle of unproductive diagnostic testing, and offers the prospect of catalyzing the development of precision therapies[11-13], even for open label “N=1” trials for the most severely affected individuals [14]. Genetic diagnosis via WES has changed therapeutic treatments, can produce improved outcomes[15, 16], and when applied early, is cost-effective [17]. Although care guidelines for certain well known disorders are continually being refined each year[18-20, 21], diagnosis remains a critical component for rare diseases as it can facilitate natural history studies, provide more accurate prognosis, and help clinicians determine guidelines for the care of individuals with newly discovered genetic disorders. Despite these positive attributes, WES still has several important limitations. These include: causative genes may not be identified for very rare disorders, especially when recessive, some genetic alterations (e.g. repeat expansions, complex rearrangements or very low-grade mosaics) may not be identified due to technical difficulties, and data describing intronic elements capable of controlling translation and RNA splicing are not accessible. For these reasons, many individuals with potential genetic disorders will remain undiagnosed, and this is a driving force arguing for the utility of whole genome sequencing, including for those already subjected to WES.
Classification of Variants
Genetic variation in humans differs for each gene, which necessitates statistical analysis to validate gene constraints as well as the susceptibility of each gene for neurological (or other) diseases. One of the more established scores relevant for this is the Z score for missense variants. In addition, a score for the probability of being loss-of-function intolerant (pLI) and for the observed / expected metric (OE) (https://gnomad.broadinstitute.org/help/constraint) are commonly used. A variant Z score is interpreted as positive if it is greater than 3.09, which suggests that a given transcript is intolerant of variation, and thus constrained. In addition, a pLI score that is greater than 0.9 implies intolerance to null variants, including frameshift, nonsense, and splice variants. Using the upper end of the OE confidence interval (LOEUF, < 0.35) is another way to establish a firm threshold for intolerance to null variants.
Criteria for the classification of sequence variants[22] into five categories has been established by The American College of Medical Genetics and Genomics. These standard terms include 1) pathogenic, 2) likely pathogenic, 3) variants of uncertain significance (VUS), 4) likely benign, and 5) benign. This variant classification considers multiple criteria such as the type of variant, its origin, location, the functional consequences, conservation, allele frequency, predictive computational metrics, and other factors. It is also important to evaluate whether copy number variants (CNV) are present. Indeed, large CNVs affect multiple genes that may contribute to the clinical phenotype in a complex manner, whereas small CNVs could alter the transcription of just a single gene.
Clinical reports describing genetic variation often refer to variants as pathogenic or likely pathogenic, but can also classify a variant as VUS, indicating that it is unclear whether it does or does not participate in clinical aspects of an individual’s disorder. The final determination of whether a variant can alter the function of the encoded protein necessarily requires the evaluation of all avaliable data by a specialist who is familiar with both the clinical disorder as well as the gene. In a subset of cases, determination of the functional properties for the protein encoded by the variant is advisable, even in some cases for null variants that may have dominant negative properties upon the expression of the other allele. The interpretation of genetic variants in a manner that can unequivocally account for an individual’s phenotype remains a goal that awaits a description of the complete spectrum of variation in humans, including across ethnic backgrounds and in health and disease.
GRIN variants
The GRIN gene family contains GRIN1, GRIN2A-D, GRIN3A-B. Of these, variants in GRIN1, GRIN2A, GRIN2B, and GRIN2D genes have been identified in individuals with various neurodevelopmental disorders (Figure 1); the association of GRIN2C[23] and GRIN3[24] with disease remains unclear (see Table 1, Figure 1B). As expected, there are both similarities as well as differences among these genes in terms of the spectrum of genetic variants that appear to contribute to human pathological conditions.
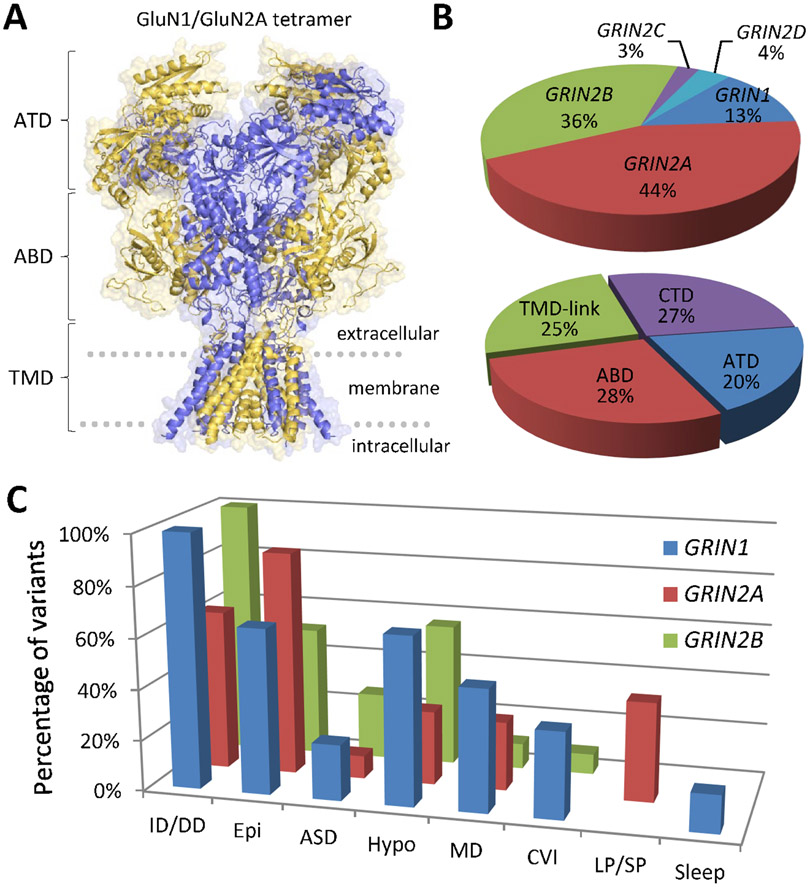
Figure 1. GRIN variants are associated with various neurologic disorders.
A), Architecture and domain organization for the NMDAR. B), Summary of NMDAR variants in different GRIN subunits and domains. C), Summary of GRIN variant-associated phenotypes. ATD: amino terminal domain, ABD: agonist binding domain, TMD-link: transmembrane domains (M1-4) and linker regions, CTD: intracellular carboxy-terminal domain. ASD: Autism Spectrum Disorder, CVI: cortical visual impairment, Epi: epilepsy/seizures, Hypo: hypotonia, ID/DD: intellectual disability/developmental delay; LP/SP: language/speech problems; MD: movement disorders; Sleep: sleep problems. Many individuals showed multiple phenotypes, which are only a snapshot of the current literature, which is disproportionally weighted by different diagnostic approaches and ascertainment.
Table 1.
NMDAR subunit null mouse models and prevalence of null mutations in affected individuals.
| Protein Subunit Name |
Heterozygous Null Phenotype | Homozygous Null Phenotype | Individuals with Large- Scale Disruptions (LSD) or Premature Stops |
|---|---|---|---|
| Severe Phenotypes in Mice | |||
| GluN1 | Viable and fertile[61, 90]; accelerated cortical neuron development[90] | Lethal – No suckling response and poor lung development[61]; altered brainstem nuclei development[60] | 4 total (4/91; 4.4%): 4 preSTOP |
| GluN2B | Viable and fertile[62] | Lethal – No suckling response with altered brainsteam nuclei development[62] | 36 total (36/258; 14%): 14 LSD and 22 preSTOP |
| Viable, but with deficits | |||
| GluN2A | Viable and fertile[63]; minute and transient changes in brain microstructure assessed via diffusion tensor imaging[74] | Viable and fertile [63]; diminished spatial learning[63] with largely transient epileptiform activity[74, 75]; alterations in pup vocalizations[74] | 53 total (53/311; 17%): 29 LSD and 24 preSTOP |
| GluN2C | Viable and fertile[65]; alterations in sensorimotor gating [91] | Viable and fertile[65]; some alterations in working memory[70] and sensorimotor gating [91] | 1 total (1/19; 5.3%): 1 preSTOP |
| GluN2D | Viable and fertile[92] | Viable and fertile; hypolocomotion and increased anxiety-like behaviors[72, 73, 92] | Yet to be reported |
| GluN3A | Viable and fertile[93] | Viable and fertile[93]; impaired locomotor activity[94]; changes to spatial and aversion learning[94, 95] | 2 total (2/13; 15%): 2 preSTOP |
| GluN3B | Viable and fertile[96] | Viable and fertile[96]; impaired motor learning and coordination[96] | Yet to be reported |
Heterozygous and homozygous null (knockout) mouse models have been generated for each of the seven GRIN genes. Mice containing homozygous null mutations for GluN1 and GluN2B are postnatal lethal, while heterozygous null offspring survive normally. In general, homozygous null mice, regardless of which gene has been disrupted, display some sort of aberrant phenotype that could be extrapolated to patients. It should be noted, however, only one large-scale chromosomal disruption has been reported to be homozygous in human, an inherited GRIN2A deletion affecting both alleles, with all others reported being heterozygous. Given the overall lack of characterization of heterozygous null GRIN mouse models, it is difficult to determine whether they can truly mimic features of the human condition. Large-scale disruptions (LSD) refers to chromosomal deletions, duplications, inversions, insertions, or translocations; premature stops (preSTOP) refers to a nonsense mutation resulting in a premature stop codon in the mRNA.
GRIN genes code for different NMDAR GluN subunits. Functional NMDARs (Figure 1A) require 2 subunits coded by GRIN1 and 2 subunits coded by any GRIN2 or GRIN3. This specificity is regulated in anatomical, developmental, and neuronal subtype specific manners. NMDARs are assembled from subunits within the endoplasmic reticulum; the spectrum of features that regulate assembly, such as the relative availability of any given subunit, are not fully understood. NMDARs are essentail to multiple key roles including neuronal migration, synaptic connectivity, neuronal pruning and survival, and synaptic plasticity (reviewed in this Special Edition and see [25]). Thus, it is not surprising that variation of NMDARs plays a role in human disease.
GRIN1 is a phylogenetically conserved gene. Through statistical representations in the neurotypical population, GRIN1 appears intolerant of both missense (Z = 6.22) and null variation (pLI = 0.98; LOEUF = 0.31), assessed via gnomAD (v2.1.1, https://gnomad.broadinstitute.org/). Pathogenic heterozygous missense variants have arisen de novo, and are clustered near one another in a limited number of domains with important functions, such as the agonist binding domain (made up of two different portions of the polypeptide chain), pore-forming transmembrane domains, and the linker regions that connect the agonist binding domain to the channel pore and control the opening of the channel following agonist binding[26, 27] (Figure 1). The amino-terminal domain (also know as N-terminal domain, or NTD) that resides distal to the membrane and the C-terminal domain (CTD), which resides intracellularly, are more tolerant to variation, and thus appear less likely to contribute to GRIN1-related developmental disorder. However, this notion may change with further study of the actions and binding partners for these two regions as we still have an incomplete understanding of their roles.
Despite the high constraint metrics, null GRIN1 variants have not yet been associated with neurological disease. Several individuals who possess heterozygous GRIN1 null variants are reported to not show any distinct clinically relevant phenotypes[26]. One family with a homozygous GRIN1 null variant that was associated with fatal epileptic encephalopathy has been reported[26], suggesting that null variants can contribute to GRIN1-related disorders but only when affecting both alleles. This is currently the only report of an autosomal recessive form of any GRIN-related disorder. Despite the description of two families with rare homozygous missense variants [26, 28], autosomal recessive GRIN1-related developmental disorders due to missense variation have not yet been unequivocally confirmed.
In contrast to GRIN1, through similar statistical representations in the neurotypical population (gnomAD v2.1.1, https://gnomad.broadinstitute.org/), GRIN2A is intolerant of null variation (pLI = 1.00; LOEUF = 0.19), but not necessarily intolerant for missense variants (Z = 2.83). Presumed pathogenic heterozygous missense variants are typically de novo and are localized in a similar fashion as for GRIN1 and other GRIN genes to the regions encoding the agonist binding domain, the pore-forming transmembrane domain, and the short linkers that connect the agonist binding domain and the channel pore (Figure 1). Similar to GRIN1, the NTD as well as the intracellular CTD show more tolerance to missense variation [29].
In addition, GRIN2A null variants are associated with a disease spectrum that can be clinically mild, with affected individuals in some cases able to reproduce (Table 1). Thus, GRIN2A null variants are unique among pathogenic variants in the GRIN gene family, as they are occassionally found to be inherited.
Similar to GRIN1, the GRIN2B gene through similar statistical representations in the neurotypical population (gnomAD v2.1.1, https://gnomad.broadinstitute.org/), is intolerant for missense (Z = 5.42) and null variation (pLI = 1.00; LOEUF = 0.06). Pathogenic heterozygous missense variants are typically de novo and show a similar pattern of localization to critical domains in the subunit encoded by GRIN2B (Figure 1). Variants are highly concentrated in the agonist binding sites, in addition to the transmembrane and linker regions, with minimal pathogenic variants in the NTD or the CTD [30-32]. GRIN2B null variants are associated with GRIN2B-related neurodevelopmental disorder, and are usually found to arise de novo [30, 31].
Like GRIN1 and GRIN2B, the GRIN2D gene through similar statistical representations in the neurotypical population (gnomAD v2.1.1, https://gnomad.broadinstitute.org/), is intolerant for missense (Z = 4.85) and null variation (pLI = 1.00; LOEUF = 0.17). Pathogenic heterozygous missense variants are typically de novo[33, 34]. GRIN2D-related disorders are the least frequently observed among GRIN disorders, and thus it is premature to draw conclusions about potential clustering of pathogenic missense variants in any region of the protein encoded by GRIN2D (Figure 1B and Table 1). Null variants in GRIN2D gene (other than large scale deletions) have not yet been reported.
Clinical characteristics
Patients with GRIN1-related neurodevelopmental disorder show multiple deficits, including ID, epilepsy, hypotonia, and for some individuals, movement disorders. All affected individuals evaluated to date show variable levels of ID:, including 5% mild, 7% moderate, 71% severe, or 17% profound [31].
Sixty-five percent of individuals presented with epilepsy (Figure-1C, Table 2). The onset ranges from birth to 11 years of age, and two thirds demonstrated resistance to conventional antiseizure treatment. Seizure types include generalized seizures (68 %; with multiple semiologies), focal seizures (20 %), and epileptic spasms (13 %). Additional clinical characteristics (Figure-1C, Table 2) include hypotonia (66 %), movement disorders (48 %), cortical visual impairment (CVI, 34 %), as well as oculogyric crises (11 %). Some individuals show features of autism spectrum disorders, or exhibit other behavior problems such as stereotypic movement disorder (32 %), sleep problem (15 %), and self-harm behavior (7 %) [31]. A subset of individuals showed an unusual type of cortical malformation that consisted of extensive bilateral polymicrogyria together with lateral ventriculomegaly, enlarged extra-axial spaces, reduced thickness of the corpus callosum, basal ganglia dysplasia, and decreased white matter volume [35].
Table-2.
Summary of phenotypes associated with GRIN variants
| GRIN1 | GRIN2A | GRIN2B | |
|---|---|---|---|
| Intellectual disability | 100% | 63% | 100% |
| Epilepsy/seizures | 65% | 88% | 57% |
| Muscular hypotonia | 66% | 29% | 56% |
| Movement disorders | 48% | 27% | 56% |
| Autism spectrum disorder | 22% | 9% | 26% |
| Cortical visual impairment | 34% | – | 8% |
| Language/speech problems | – | 39% | – |
| Schizophrenia | – | 3% | – |
| Sleep problems | 15% | – | – |
Many patients showed multiple phenotypes, which are only snapshot of the current literature, which is disproportionally weighted by different diagnostic and ascertainment procedures.
Neurodevelopmental disorders in GRIN2A individuals are associated predominantly with epilepsy and ID. However, as many as 37 % of the individuals demonstrate normal intelligence and 63 % have ID (46% mild, and 22%with moderate, 11% severe, 21% profound) [29]. Brain imaging is usually normal and only a minority (14 %) reveal nonspecific changes[29]. Epilepsy is present in almost all GRIN2A individuals (Figure-1C, Table 2) with onset from birth to 8 years of age. Interestingly, seizures may resolve between 8 and 20 years of age. Fifty-seven percent present with focal seizures; 40 % showed a centrotemporal focus similar to Rolandic epilepsy. EEG demonstrated in 34 % continuous spikes and waves during slow wave sleep (CSWS). Additional challenges including hypotonia (29 %), movement disorders (27 %), autism spectrum disorders (9 %), and/or psychiatric disorders, such as schizophrenia (3 %) [29] (Table 2). A unique feature associated within the GRIN-associated disorders is the breadth of language/speech problems observed in GRIN2A-related developmental disorders, which include dysarthria, dysphasia, speech dyspraxia, speech regression with residual impairments in more than a third (Table 2) and 19 % had aphasia [29, 36].
Individuals affected with GRIN2B-associated disorders exhibit ID, hypotonia, epilepsy, and movement disorders. All affected (so far) have DD preceding certain degrees of ID (Figure-1C, Table 2). A wide range of ID is observed, which includes 6% mild, 21% moderate, and 73% severe-to-profound (http://grin-portal.broadinstitute.org/#tab-1201-3).
Some form of epilepsy is present in half of the affected individuals and shows an onset between birth to 9 years of age. Seizures are medically refractory for half. The spectrum of seizure characteristics is similar to that observed for GRIN1 variants, which include 35% epileptic spasms, 48% focal seizures, and 58% generalized seizures [31].
Additional clinical characteristics are perhaps less frequent and/or are somewhat milder then GRIN1-associated neurodevelopmental disorders. However, the spectrum of clinical characteristics is similar to GRIN1 with hypotonia (56%) and spasticity (23%), autism spectrum disorder (26%), movement disorders (10%), and cortical visual impairment (8%) [31] (Table 2). Cortical malformations with polymicrogyria and basal ganglia dysplasia in GRIN1 is mirrored in a subset of individuals with GRIN2B disorders [30].
As described above, variation in GRIN2D appears far less frequent than that in GRIN1, GRIN2A, or GRIN2B [37, 38]. One population-based study reported no truncated GRIN2D variants, suggesting a crucial role in early development and survival [39]. However, a different conclusion was reached by other investigators[24], who raised the idea that intronic variations (i.e. missense) might be related to the risk for schizophrenia[23]. GRIN2D missense variants have been observed in individuals with severe, drug-resistant epileptic encephalopathy with an early onset [33, 34, 38, 40, 41]. Functional analysis of variants introduced into GRIN2D cDNA have shown gain of function characteristics[33, 38], possibly with a compensatory reduced expression[34]. Only 28 GRIN2D variants are currently documented in the literature[25].
GRIN3A, which encodes a glycine binding subunit that can coassemble with GluN1 to form a glycine sensitive receptor in neurons[42], is expressed throughout the CNS[25]. Various studies suggest a role in multiple behaviors[43]. Genetic variations in GRIN3A genes have been associated with bipolar disorder [44], however, no further evidence has suggested GRIN3A is involved in neurological disorders. GRIN3B-encoding the GluN3B subunit is expressed in brainstem and spinal cord [45]. One study of the function of GRIN3B gene in motor neuron diseases reported a SNP that was caused a null allele was present in about 10% of the general population [46]. Additional research evaluating truncating variants in individuals with neurodevelopmental disorders identified GRIN3A and GRIN3B truncation variants in the control cohorts[24, 39]. These observations cannot determine whether or not GRIN3 plays a role in human disease.
Functional assessment of GRIN variants
The determination of how a GRIN variant might alter protein function usually requries electrophysiological and biochemical assays. The goal of these functional assays is to determine whether a variant results in a loss-of-function (LoF), a gain-of-function (GoF), does not influence receptor function, or produces some complex mixture of effects on protein function. NMDARs have many different functional properties, so testing typically needs to be a comprehensive evaluation of agonist potency, receptor function, endogenous modulation, and protein expression and trafficking. Further, these assessments are limited to what we currently know about NMDAR function. It is important to take this comprehensive approach because, a priori, it is impossible to know which function(s) a given variant will impact. A case in point is the recently decribed GRIN1_p.P532H variant, which resides in the glycine binding domain but exerts its effects on glutamate binding to the GluN2 subunit[47]. Functional testing often occurs on several levels. The first tier of experimentation typically assesses how a variant alters function of the NMDAR as expressed in a a non-neuronal heterologous expression system, for example, Xenopus oocytes or cultured fibroblasts (e.g. HEK293 cells) that can be manipulated to express the recombinant subunits of interest, thereby allowing the study of a purified population of receptor by various assays. The second tier of experimentation can include experiments to assess how a variant might alter NMDAR function in a cultured neuron. This can be followed by study of the variant introduced into a whole animal either by knockin methods or viral-mediated gene transfer. The secondary tier of study involving native neurons are meant to extrapolate the phenotype and neuronal characteristics that are expected to be observed in affected individuals. Whether or not a given variant is ultimately proven to contribute to disease characteristics may involve studies that go beyond the expertise of a neurologist or clinical geneticist tasked with making a determination of likely pathogenticity.
Functional analysis is a necessary step that provides a wealth of opportunities to advance understanding of the condition. It allows stratification of individuals for future approaches to precision therapies, as individuals with variants that produce similar changes in receptor function are more likely to yield similar results in clinical trials and clinical practice than variants that simply happen to be close to each other on the polypeptide chain. Precision therapeutics requires objective evidence of benefit through well-designed clinical trials (safety and efficacy), and stratification of similar individuals increases the likelihood that clinical trials will be able to reach meaningful conclusions.
Testing in animals often does recapitulate key features observed in affected individuals. However, in other situations, the variant might perturb neurological systems that are altered in a way that does not fully reproduce the observed clinical features. For example, a missense variant from one individual with epilepsy might, when knocked into mice, elevate or reduce seizure threshold. Failure of complete reproduction of the clinical features does not invalidate in vivo models showing a clear and measureable deficit[48]. Moreover, such deficits can be studied for altered neurodevelopment, rescue pharmacology, or for the refinement of genetic strategies. That is, these models can be useful tools to gain insight into various paths to mitigate deficits in affected individuals.
The number of known genetic variants in NMDARs vastly outnumbers those for which we have some functional information. However, an appreciation of the utility of comprehensive functional characterization in terms of diagnoses, stratification, and future potential treatments is driving increased effort toward functional characterization. Among the more than 700 GRIN variants, published functional evaluation exists for for less than half [25] (see http://functionalvariants.emory.edu/ and http://grin-portal.broadinstitute.org/). Much of this work has progressed at the level of tier 1 in heterologous expression systems, and several parameters can be measured in vitro in recombinant receptors. However, clearly this initial approach has the limitations that heterologous expression does not enable detection of functional changes that require the unique developmental and anatomical context and features of the neuronal environment explored in tier 2 studies. Variants in the intracellular CTD[49, 50] do not typically alter functional parameters in heterologous systems. However, the CTD is known to interact with a large number of scaffolding proteins and intracellular signaling systems in neurons, and thus neuronal assays are needed to potentially see the ramifications of variation in the CTD. In this regard, cells or tissues from transgenic animals are needed to evaluate how CTD variants alter NMDAR function. Accessible functional parameters that can be assessed include glutamate and co-agonists glycine potency, voltage-dependence and potency for channel block by extracellular Mg2+, sensitivity to endogenous extracellular modulators such as Zn2+, single channel open probability, receptor deactivation time course in order to predict the synaptic response time course, desensitization, synaptic plasticity, and receptor trafficking, including both receptor subcellular localization and cell surface expression [29, 34, 37, 51-59].
Missense variants can in principle alter any properties of the NMDAR. For this reason, it is important to study as many functional attributes of the receptor as possible, since one variant can (and often does) alter multiple parameters. For example, a variant can produce a change in one parameter that increases current flow, and a unrelated change in another parameter that reduces current flow. While human variation is heterozygous for missense variants of NMDAR subunits, in tier 1 testing missense variants are typically tested in a homozygous state and co-expressed with a wild-type co-subunit. For example, wild-type GluN1 will be co-expressed with a GluN2B variant. For clarity, this demonstrates the potential for the variant to alter NMDAR function.
To recap the prior descriptions, human NMDAR subunit variations include missense (which could be GoF, LoF or unknown), nonsense (resulting in a truncated subunit) or deletion of the entire subunit. These are heterozygous alterations, meaning there is another unaffected allele coding for a normal subunit. It remains unclear if heterozygous deletion of an entire NMDAR subunit will behave simply as a heterozygous missense LoF variants, the latter of which may have a dominant negative effect by assembling with other GluN subunits in the endoplasmic reticulum. In support of this dominant negative role of LoF missense variants, as noted previously and in Table 1, heterozygous loss of GRIN1 does not show any distinct clinically relevant phenotypes[26]. Further, nonsense variation that results in a truncated NMDAR subunit may have variable effects, including as dominant negative, that may depend on the length of truncation. This could occur if variants that reduced function or truncated the receptor depleted the pool of partner GluN subunits, and making instead receptor complexes that do not reach the cell surface. The determination of functional consequences in this situation may require knock-in animals that contain the variant in one allele, and thus mimic more closely the human variation, allowing assessment of trafficking as well as circuit development and normal behaviors. This construct validity is the necessary first step towards face validity of effective precision therapy applied to an animal model, to ensure that ultimately precision therapy (see “Preclinical pharmacological studies“) can be translated into safe and efficacious clinical trials and ultimately applications in affected individuals.
In vivo models of GRIN variation
In an effort to move a step closer to human physiology, the generation of mouse models containing human-specific variants allows for the detailed exploration of GRIN variants. More specifically, genetically-modified mice are essential for the elucidation of how these variants impact the brain on a developmental, circuit, cellular, and molecular level, as well as provide investigators with a mammalian platform for testing therapeutic approaches. Although homozygous null (knockout) mutations for each of the seven GRIN genes have been generated, information gleaned from the study of these mice should be viewed cautiously, as the majority with GRIN variants only have one impacted allele As discussed above, gene deletions can affect individuals differently than LoF missense and nonsense variants, which suggests that LoF missense and nonsense variants have the capacity to act as dominant negatives. Thus, heterozygous null (knockout) or variant knock-in mouse models- whether nonsense, missense, or deletion – are highly relevant to the human condition and should thus be given precedent when making comparisons to affected individuals.
The first targeted mutation in the GRIN gene family was the homozygous null mutation in the Grin1 gene, which resulted in perinatal lethality[60, 61]. Grin1 null mice die due to respiratory failure as well as failure to suckle, highlighting the omnipresent roles of NMDARs in all facets of early brain function, including those in the brainstem. Like homozygous Grin1 null mice, homozygous Grin2b null mice also suffer perinatal lethality[62]. Although Grin2b knockout mice do breathe, they do not suckle and thus do not survive the neonatal period. The homozygous null mutations of Grin2a[63, 64], Grin2c [65], and Grin2d [66] appear less severe than those homozygous null Grin1 and Grin2b mice, with each being fertile and viable, albiet with abberations in a variety of behavoiral and cognitive tasks[38, 67-73]. However, these characterizations largely preceded an appreciation of the scope of GRIN disorders, and additional study of null mutations, especially in heterozygous animals, could provide meaningful insights into the impact of LoF GRIN variants. In this vein, subsequent studies of homozygous and heterozygous Grin2a null mice revealed that these mice displayed epileptiform discharges, although these changes in circuit excitability appear to be developmentally transient [74, 75].
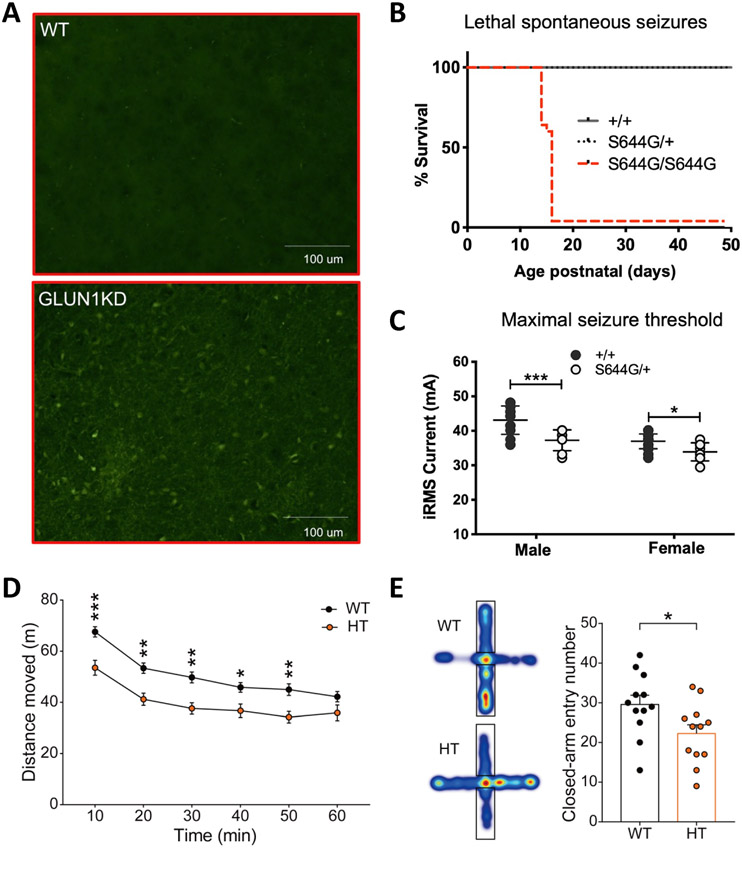
To date, multiple mouse models carry missense mutations or human variants in Grin1, Grin2a, and Grin2b genes, with additional models of human GRIN variants actively being developed. The first missense variants of Grin1 (p.N616Q and p.N616R, which reside within the ion channel) revealed two important concepts for GRIN disorders [76]. First, heterozygous mice with a Grin1 loss-of-function missense variant are capable of generating a more severe phenotype than heterozygous null mice (however see [77]). While heterozygous Grin1 null mice have no clear phenotype, heterozygous p.N616Q mice have diminished maternal behaviours and heterozygotes for p.N616R are unable to suckle, with both mouse models dying in the early neonatal periods. The only similar mapped GRIN1 human variant in the pore-forming region, p.N616K, is likely GoF (http://grin-portal.broadinstitute.org/), but further details are unpublished. It should be noted, however, that data needed to make strong conclusions regarding LoF variants and heterozygous null mice are still lacking, as the location of this variant along the different domains of the receptor likely plays a decisive role in determining its severity (i.e. variants in the pore-forming region are predicted to be more severe than those in the NTD)[29]. Second, missense variants for Grin1 can demonstrate a broad range of phenotype characteristics that depends on the the amino acid exchange encoded by the missense variant. Additional studies of homozygous missense Grin1 mutations (that do not currently map to human variants) were either lethal[78, 79] or showed altered behaviors [78, 80, 81]. In studies of Grin1 haploinsufficiency(Figure-2A) [77], behavioral abnormalities were rescued in adult mice by Cre recombinase gene editing [82].
Fig. 2. Mouse models of human GRIN variants.
A), Fluoro-Jade C staining showed that neurons of Grin1-KD mice in the striatum surrounding the anterior commissure are degenerating in the adult brain. B), Survival curve showing the rate and onset between postnatal days 15 and 17 of Grin2ap. S644G genotype-dependent in F2 hybrid male and female mice. C), Heterozygous (het) Grin2a-p.S644G adult mice have lower seizure threshold in minimal seizure end points. D), Hypoactivity in Grin2b+/C456Y mice (P68–78) in the open-field test. E), Anxiolytic-like behavior in Grin2b+/C456Y mice (P70–124) in the elevated plus-maze, as shown by entries into in closed arms. Modified with permission from (Intson et al., 2019) (A), (Amador et al., 2020) (B,C), and (Shin et al., 2020) (D,E).
Recently, patient-specific knock-in mouse models have been generated for variants in GRIN2A (p.S644G, GoF[58]; p.N615K, GoF[83]) and GRIN2B (p.C456Y, mixed[59]). In both Grin2a GoF variants, homozygosity was either preadolescent lethal, dying around P15, (Figure-2B) [58] or associated with a worsening phenotype [83]. Notably, heterozygous GoF variants displayed a range of phenotypes, with either reduced seizure thresholds (Figure-2C) and hippocampal thinning [58] to changes in circuit output, such as a reduction in EEG power in the γ-range [83]. The study of Grin2b p.C456Y highlighted a limitation of our current classification system, as this variant presented with mixed (gain and loss) functional changes to receptor function when studied in vitro[30, 51, 55]. However, the totality of these changes resulted in a decrease in Grin2b expression, coupled with reduced hippocampal long-term depression (Figure-2D,E). With the mixed functional effects observed in vitro, there were no a priori ways to predict how this variant might impact neurodevelopment and total brain function in a mouse, calling to attention the importance of patient-derived animal model testing. Moreover, this study suggested that treatment with cycloserine early, (but not late), in development could improve behavioral abnormalities. These data seem to be in contrast to those described in the knockdown Grin1 mouse model where behavorial deficits could largely be restored by gene correcting therapy in adulthood (Figure-3A) [77]. These differences could be due to the different roles that GluN1 and GluN2B play in brain function or differences in temporal expression patterns. These results also reveal a disparity in chemical versus genetic therapeutic strategy. Regardless, the sum of our data on patient-derived mouse models suggest that each human variant might require its own unique model to identify the optimal therapeutic approches. In order to expedite this process, future in vivo model development and characterization should build a map of examplar human GRIN variants with gain, loss, and mixed function variants spanning each domain of a particular subunit. This approach might allow future prediction of phenotypic consequences, best clinical trial stratification, and best therapeutic options for each affected individual based on where their variant lies along the different modular domains of the receptor.
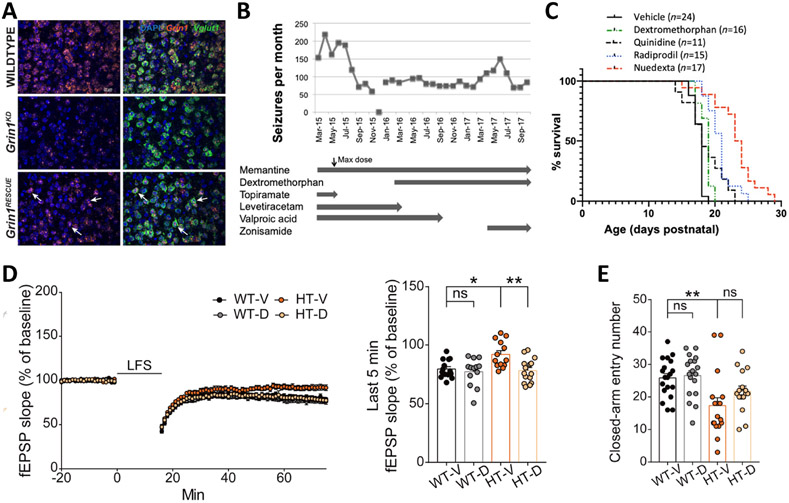
Figure 3. Therapeutic strategies for the treatment of GRIN variants.
A), Grin1 mRNA expression in Vglut+ cells in the adult mouse somatosensory cortex (1.53 mm lateral from midline). Grin1 (orange) and Vglut1 (green) mRNA was visualized in mouse sagittal sections (20 μm) with fluorescent in situ hybridization in WT, Grin1KD, and Grin1RESCUE mice. B), Impact of drug therapy on clinical seizures for the individual with GRIN2A-p.S644G variant. C), Pharmacological rescue of lethal seizures of p.S644G/p.S644G homozygotes, showing the respective survival after daily injections of dextromethorphan, quinidine, radiprodil, and Nuedexta®. D), Early chronic oral D-cycloserine (DCS) treatment (40 mg/kg) normalizes LFS-LTD at SC-CA1 synapses in juvenile Grin2b+/C456Y mice (P17–20). E), Early chronic oral DCS treatment (40 mg/kg) improves anxiolytic-like behavior in adult Grin2b+/C456Y mice (P63–73). Modified with permission from[77] (A), [58](B,C), and [59](D,E).
Preclinical pharmacological studies
Given that a GRIN variant can often be defined as gain or loss of function, how best can pharmacological modulation be identified that can alleviate symptoms? Pharmacological modulation of clinical symptoms secondary to modulation of NMDAR properties could be beneficial if, and only if, current symptoms are actively being mediated by the GoF or LoF for the NMDAR. However, this is likely an overly simplistic viewpoint, and it seems almost certain that compensatory changes and/or maladaptive plasticity due to the variant will have altered some aspect of overall brain development (e.g., findings noted with altered brain imaging). Such compensation may produce unintended consequences of pharmacological treatments when they alter NMDAR function. In this situation, precision medicine based on correcting the NMDAR variants function in vitro may not be helpful to affected individuals (although classifying individuals based on variant function and location along the affected subunit would likely bring more empirical evidence to improve existing treatments). This is the main reason why increased emphasis on rodent models of GRIN variants is needed.
Using in vitro heterologous systems, NMDAR channel blockers and negative allosteric modulators have shown to retain high potency and efficacy at some GoF variants, and this in vitro data is a powerful tool to inform pharmacological agent testing in mouse models [25, 30, 33, 51, 53, 58, 84-87]. Differential sensitivity of each GoF GRIN variant [53, 84, 85, 87]. emphasizes the need to determine the sensitivity of each GRIN variant to potential pharmacological treatments. However, compounds with the highest potency or those that are the most selective in vitro seldom prove to be the best for use in animals. For example, as mentioned previously, mice with a homozygous variant in Grin2a (p.S644G, GoF) die before the third week of life. Chronic treatment with either dextromethorphan (channel blocker), Nuedexta (dextromethorphan + quinidine), or memantine (channel blocker) significantly delayed the onset of lethal seizures (Figure-3C) [58]. Surprisingly, the most efficacious treatment was Nuedexta, even though the IC50 for dextromethorphan on homozygous GluN2A-p.S644G recombinant receptors is 22 μM, which is very similar to that of memantine at 30 μM[58]. Although, the difference in efficacy is likely due to quinidine’s effect at prolonging dextromethorphan’s half-life, this study demonstrates a pressing need for drug validation in rodent models of GRIN variants in order to better understand the functional specificity of NMDAR channel blockers and negative allosteric modulators.
The treatment and pharmacological outlook for the use of NMDAR positive allosteric modulators may be more straightforward. Relying on endogenous chemical backbones, such as neurosteroids (i.e. endogenous neurosteroid 24(S)-hydroxycholesterol), FDA-approved aminoglycosides or co-agonists at the glycine-binding site (i.e. D-serine, L-serine, and D-cycloserine) can help to mitigate common reasons why pharmacological agents fail in vivo (i.e. brain penetration and bioavailability), while still being highly efficacious on a set of loss-of-function GRIN1, GRIN2A and GRIN2B variants[47, 51, 52, 56, 88, 89]. Early treatment (before adulthood) with D-cycloserine on young transgenic mice harboring a patient-derived loss-of-function GRIN2B-p.C456Y variant mitigated NMDAR-dependent synaptic long-term depression and corrected aberrant anxiety behavior (Figure-3D,E) [59]. These preclinical studies provide foundational evidence for clinically useful pharmacological agents for the treatment of GRIN variants as stand-alone options or synergist options in addition to genetic manipulation.
Human studies
Multiple pathogenic variants in GRIN genes have been described as GoF or LoF for the NMDAR, which provides clinicians with opportunities to potentially mitigate dysfunction through the use of either NMDAR-specific blockers/inhibitors or enhancers/potentiators. An initial proof of principle was established by treating a male individual with early-onset epileptic encephalopathy and drug-resistant seizures due to a pathogenic de novo GOF missense variant in GRIN2A (p.L812M) with the FDA-approved NMDAR channel blocker memantine. This treatment resulted in a significant reduction in seizure frequency[84, 87]. Additional individuals with GRIN variants have been treated with memantine, but only several of them have been reported in the peer-reviewed literature.
There are a limited numbers of publications if solely considering cases with a demonstrably pathogenic GRIN variant with a confirmed GoF consequence for the NMDAR. For GRIN1, recently a GOF missense variant that showed enhanced potency for a set of FDA-approved NMDAR channel blockers was described (GRIN1-p.M641I), which creates a situation where clinical treatments would preferentially block variant but not wild type receptors [87]. The affected individual responded favorably to memantine with a significant reduction in seizure frequency and severity of spasms, with evidence of efficacy when seizures/spasms worsened during accidental discontinuation of memantine [87]. For GRIN2A, only one other individual (p.S644G) has been reported in the literature to have a significant reduction in seizure frequency after adding memantine and later dextromethorphan, similar to the GRIN2A variant p.L812M [58]. For GRIN2B, four affected individuals have been reported [30]. All of these individuals showed subtle and subjective improvements in awareness, behavior or sleep. However, none had quantifiable improvements in these modalities by objective assessment. For GRIN2D, two affected individuals have been reported that showed mild to moderate improvement in seizure frequency following the addition of memantine to their treatment regimen. The older one subsequently developed refractory status epilepticus, which showed dramatic electroclinical improvement during treatment with ketamine and magnesium [33]. Only one affected person with LOF GRIN variant has been reported to experience subjective behavioral improvements as well as improved sleep and motor development during administration of L-serine [89]. Across these studies, a safety assessment cannot be provided due to the small number of individuals studied; it may be dangerous to generalize further. These early but limited studies suggest that precision medicine for GRIN-related disorders may be possible, but there remains a pressing need to assess these treatments in well-designed, double-blinded, placebo-controlled clinical trials that systematically and quantitatively assess multiple parameters of safety and efficacy.
Highlights.
We review genetic variation of NMDA receptors associated with neurological disease
Genetic variation of NMDA receptors can alter their function
Initial studies suggest links between functional alterations and treatment strategies
Additional studies, including animal models, are needed to validate this approach
Acknowledgements
CURE, GRIN2B Foundation, Simons Foundation, and Ponzio Family Chair in Neurology Research/Children’s Hospital Colorado Foundation.
None reported
CIHR MOP119298
NINDS NS111619 (SFT) and NS113530 (CRC), NICHD R01HD082373 (HY), GRIN2B Foundation (HY), Simons Foundation (SFT)
CURE, GRIN2B Foundation and Simons Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Clinical trials sponsored by Rett Syndrome Research Trust, Neuren, Acadia, Ovid, and Marinus. He has consulted for RettSyndrome.org, AveXis, Ovid, Takeda, and Marinus. Additional unrelated funding support from NIH
-
JL: Member of SAB for CureGRIN Foundation, GRIN2B Foundation.IK: none
- None reported
- SFT: Member of SAB for Sage Therapeutics, Eumentis Therapeutics, CureGRIN Foundation, GRIN2B Foundation. Co-founder of NeurOp Inc, and Agrithera Inc, PI on grants to Emory from Janssen and Biogen, and co-inventor on Emory-owned IP. HY: PI on grants to Emory from Sage Therapeutics and co-inventor on Emory-owned IP.
- None reported.
References
- 1.Paciorkowski AR, Seltzer LE, and Neul JL, Developmental Encephalopathies, in Swaiman's Pediatric Neurology, Swaiman KF, et al. , Editors. 2018, Mosby: Philadelphia. p. 242–248. [Google Scholar]
- 2.Institute of Medicine (U.S.). Committee to Evaluate the Supplemental Security Income Disability Program for Children with Mental Disorders, et al. , Mental disorders and disabilities among low-income children. 2015, National Academies Press: Washington, D.C. p. 169–178. [PubMed] [Google Scholar]
- 3.Scheffer IE, et al. , ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia, 2017. 58(4): p. 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McTague A, et al. , The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol, 2016. 15(3): p. 304–16. [DOI] [PubMed] [Google Scholar]
- 5.Lewis-Smith D, et al. , Modeling seizures in the Human Phenotype Ontology according to contemporary ILAE concepts makes big phenotypic data tractable. Epilepsia, 2021. 62(6): p. 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava S, et al. , Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med, 2019. 21(11): p. 2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bick D, et al. , Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J Med Genet, 2019. 56(12): p. 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith HS, et al. , Clinical Application of Genome and Exome Sequencing as a Diagnostic Tool for Pediatric Patients: a Scoping Review of the Literature. Genet Med, 2019. 21(1): p. 3–16. [DOI] [PubMed] [Google Scholar]
- 9.Wise AL, et al. , Genomic medicine for undiagnosed diseases. Lancet, 2019. 394(10197): p. 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Happ HC and Carvill GL, A 2020 View on the Genetics of Developmental and Epileptic Encephalopathies. Epilepsy Curr, 2020. 20(2): p. 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truty R, et al. , Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open, 2019. 4(3): p. 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symonds JD, Zuberi SM, and Johnson MR, Advances in epilepsy gene discovery and implications for epilepsy diagnosis and treatment. Curr Opin Neurol, 2017. 30(2): p. 193–199. [DOI] [PubMed] [Google Scholar]
- 13.Tidball AM, et al. , Variant-specific changes in persistent or resurgent sodium current in SCN8A-related epilepsy patient-derived neurons. Brain, 2020. 143(10): p. 3025–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, et al. , Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med, 2019. 381(17): p. 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuperberg M, et al. , Utility of Whole Exome Sequencing for Genetic Diagnosis of Previously Undiagnosed Pediatric Neurology Patients. J Child Neurol, 2016. 31(14): p. 1534–1539. [DOI] [PubMed] [Google Scholar]
- 16.Berg AT, Loddenkemper T, and Baca CB, Diagnostic delays in children with early onset epilepsy: impact, reasons, and opportunities to improve care. Epilepsia, 2014. 55(1): p. 123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell KB, et al. , A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia, 2018. 59(6): p. 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akunne HC, et al. , An Electrophilic Affinity Ligand Based on (+)-Mk801 Distinguishes PCP Site 1 from PCP Site 2. Neurochem Res, 1994. 19: p. 385–389. [DOI] [PubMed] [Google Scholar]
- 19.Fu C, et al. , Multisystem comorbidities in classic Rett syndrome: a scoping review. BMJ Paediatr Open, 2020. 4(1): p. e000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagli AI, Mathews J, and Williams CA, Angelman Syndrome, in GeneReviews((R)), Adam MP, et al. , Editors. 1993: Seattle (WA). [Google Scholar]
- 21.Wirrell EC, et al. , Optimizing the Diagnosis and Management of Dravet Syndrome: Recommendations From a North American Consensus Panel. Pediatr Neurol, 2017. 68: p. 18–34 e3. [DOI] [PubMed] [Google Scholar]
- 22.Richards S, et al. , Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 2015. 17(5): p. 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, et al. , Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl Psychiatry, 2018. 8(1): p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos-Gomez A, et al. , Disease-associated GRIN protein truncating variants trigger NMDA receptor loss-of-function. Hum Mol Genet, 2021. 29(24): p. 3859–3871. [DOI] [PubMed] [Google Scholar]
- 25.Hansen KB, et al. , Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacological Reviews, 2021. 73: p. 1–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemke JR, et al. , Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology, 2016. 86(23): p. 2171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platzer K and Lemke JR, GRIN1-Related Neurodevelopmental Disorder, in GeneReviews((R)), Adam MP, et al. , Editors. 1993: Seattle (WA). [PubMed] [Google Scholar]
- 28.Rossi M, et al. , Novel homozygous missense variant of GRIN1 in two sibs with intellectual disability and autistic features without epilepsy. Eur J Hum Genet, 2017. 25(3): p. 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strehlow V, et al. , GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain, 2019. 142(1): p. 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platzer K, et al. , GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet, 2017. 54(7): p. 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platzer K and Lemke JR, GRIN2B-Related Neurodevelopmental Disorder, in GeneReviews((R)), Adam MP, et al. , Editors. 1993: Seattle (WA). [PubMed] [Google Scholar]
- 32.Hu C, et al. , Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci, 2016. 132(2): p. 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, et al. , GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet, 2016. 99(4): p. 802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.XiangWei W, et al. , Heterogeneous clinical and functional features of GRIN2D-related developmental and epileptic encephalopathy. Brain, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fry AE, et al. , De novo mutations in GRIN1 cause extensive bilateral polymicrogyria. Brain, 2018. 141(3): p. 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner SJ, et al. , GRIN2A: an aptly named gene for speech dysfunction. Neurology, 2015. 84(6): p. 586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.XiangWei W, Jiang Y, and Yuan H, De Novo Mutations and Rare Variants Occurring in NMDA Receptors. Curr Opin Physiol, 2018. 2: p. 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camp CR and Yuan H, GRIN2D/GluN2D NMDA receptor: Unique features and its contribution to pediatric developmental and epileptic encephalopathy. Eur J Paediatr Neurol, 2020. 24: p. 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarabeux J, et al. , Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry, 2011. 1: p. e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuchida N, et al. , GRIN2D variants in three cases of developmental and epileptic encephalopathy. Clin Genet, 2018. 94(6): p. 538–547. [DOI] [PubMed] [Google Scholar]
- 41.Jiao J, et al. , Identification of a novel GRIN2D variant in a neonate with intractable epileptic encephalopathy-a case report. BMC Pediatr, 2021. 21(1): p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grand T, et al. , Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun, 2018. 9(1): p. 4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawley O, Conde-Dusman MJ, and Perez-Otano I, GluN3A NMDA receptor subunits: more enigmatic than ever? J Physiol, 2021. [DOI] [PubMed] [Google Scholar]
- 44.Gaynor SC, et al. , A targeted sequencing study of glutamatergic candidate genes in suicide attempters with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet, 2016. 171(8): p. 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishi M, et al. , Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci, 2001. 21(23): p. RC185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niemann S, et al. , Motoneuron-specific NR3B gene: no association with ALS and evidence for a common null allele. Neurology, 2008. 70(9): p. 666–76. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, et al. , A de novo GRIN1 variant associated with myoclonus and developmental delay: from molecular mechanism to rescue pharmacology. Frontiers in Genetics, 2021. doi: 10.3389/fgene.2021.694312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fallah MS and Eubanks JH, Seizures in Mouse Models of Rare Neurodevelopmental Disorders. Neuroscience, 2020. 445: p. 50–68. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, et al. , A Rare Variant Identified Within the GluN2B C-Terminus in a Patient with Autism Affects NMDA Receptor Surface Expression and Spine Density. J Neurosci, 2017. 37(15): p. 4093–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mota Vieira M, et al. , An Epilepsy-Associated GRIN2A Rare Variant Disrupts CaMKIIalpha Phosphorylation of GluN2A and NMDA Receptor Trafficking. Cell Rep, 2020. 32(9): p. 108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanger SA, et al. , Mechanistic Insight into NMDA Receptor Dysregulation by Rare Variants in the GluN2A and GluN2B Agonist Binding Domains. Am J Hum Genet, 2016. 99(6): p. 1261–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Addis L, et al. , Epilepsy-associated GRIN2A mutations reduce NMDA receptor trafficking and agonist potency - molecular profiling and functional rescue. Sci Rep, 2017. 7(1): p. 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogden KK, et al. , Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet, 2017. 13(1): p. e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amin JB, et al. , Divergent roles of a peripheral transmembrane segment in AMPA and NMDA receptors. J Gen Physiol, 2017. 149(6): p. 661–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fedele L, et al. , Disease-associated missense mutations in GluN2B subunit alter NMDA receptor ligand binding and ion channel properties. Nat Commun, 2018. 9(1): p. 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vyklicky V, et al. , Surface Expression, Function, and Pharmacology of Disease-Associated Mutations in the Membrane Domain of the Human GluN2B Subunit. Front Mol Neurosci, 2018. 11: p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, et al. , De novo GRIN variants in NMDA receptor M2 channel pore-forming loop are associated with neurological diseases. Hum Mutat, 2019. 40(12): p. 2393–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amador A, et al. , Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain, 2020. 143(7): p. 2039–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin W, et al. , Early correction of synaptic long-term depression improves abnormal anxiety-like behavior in adult GluN2B-C456Y-mutant mice. PLoS Biol, 2020. 18(4): p. e3000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, et al. , Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell, 1994. 76(3): p. 427–37. [DOI] [PubMed] [Google Scholar]
- 61.Forrest D, et al. , Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron, 1994. 13: p. 325–338. [DOI] [PubMed] [Google Scholar]
- 62.Kutsuwada T, et al. , Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor î2 subunit mutant mice. Neuron, 1996. 16: p. 333–344. [DOI] [PubMed] [Google Scholar]
- 63.Sakimura K, et al. , Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor 2A subunit. Nature, 1995. 373: p. 151–155. [DOI] [PubMed] [Google Scholar]
- 64.Kadotani H, et al. , Motor discoordination results from combined gene disruption of the NMDA receptor NR2A and NR2C subunits, but not from single disruption of the NR2A or NR2C subunit. J Neurosci, 1996. 16(24): p. 7859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebralidze AK, et al. , Modification of NMDA receptor channels and synaptic transmission by targeted disruption of the NR2C gene. J Neurosci, 1996. 16: p. 5014–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ikeda K, et al. , Reduced spontaneous activity of mice defective in the epsilon 4 subunit of the NMDA receptor channel. Brain Res Mol Brain Res, 1995. 33(1): p. 61–71. [DOI] [PubMed] [Google Scholar]
- 67.Boyce-Rustay JM and Holmes A, Genetic inactivation of the NMDA receptor NR2A subunit has anxiolytic- and antidepressant-like effects in mice. Neuropsychopharmacology, 2006. 31(11): p. 2405–14. [DOI] [PubMed] [Google Scholar]
- 68.Brigman JL, et al. , Impaired discrimination learning in mice lacking the NMDA receptor NR2A subunit. Learn Mem, 2008. 15(2): p. 50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bannerman DM, et al. , NMDA receptor subunit NR2A is required for rapidly acquired spatial working memory but not incremental spatial reference memory. J.Neuroscience, 2008. 28(14): p. 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hillman BG, et al. , Behavioral analysis of NR2C knockout mouse reveals deficit in acquisition of conditioned fear and working memory. Neurobiol Learn Mem, 2011. 95(4): p. 404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shelkar GP, et al. , Differential effect of NMDA receptor GluN2C and GluN2D subunit ablation on behavior and channel blocker-induced schizophrenia phenotypes. Sci Rep, 2019. 9(1): p. 7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto H, et al. , Loss of GluN2D subunit results in social recognition deficit, social stress, 5-HT2C receptor dysfunction, and anhedonia in mice. Neuropharmacology, 2017. 112(Pt A): p. 188–197. [DOI] [PubMed] [Google Scholar]
- 73.Salimando GJ, et al. , BNST GluN2D-Containing NMDA Receptors Influence Anxiety- and Depressive-like Behaviors and ModulateCell-Specific Excitatory/Inhibitory Synaptic Balance. J Neurosci, 2020. 40(20): p. 3949–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salmi M, et al. , Transient microstructural brain anomalies and epileptiform discharges in mice defective for epilepsy and language-related NMDA receptor subunit gene Grin2a. Epilepsia, 2018. 59(10): p. 1919–1930. [DOI] [PubMed] [Google Scholar]
- 75.Salmi M, et al. , Impaired vocal communication, sleep-related discharges, and transient alteration of slow-wave sleep in developing mice lacking the GluN2A subunit of N-methyl-d-aspartate receptors. Epilepsia, 2019. 60(7): p. 1424–1437. [DOI] [PubMed] [Google Scholar]
- 76.Single FN, et al. , Dysfunctions in mice by NMDA receptor point mutations NR1(N598Q) and NR1(N598R). J Neurosci, 2000. 20(7): p. 2558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Intson K, et al. , Progressive neuroanatomical changes caused by Grin1 loss-of-function mutation. Neurobiol Dis, 2019. 132: p. 104527. [DOI] [PubMed] [Google Scholar]
- 78.Kew JN, et al. , Functional consequences of reduction in NMDA receptor glycine affinity in mice carrying targeted point mutations in the glycine binding site. J Neurosci, 2000. 20(11): p. 4037–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furuse T, et al. , Phenotypic characterization of a new Grin1 mutant mouse generated by ENU mutagenesis. Eur J Neurosci, 2010. 31(7): p. 1281–91. [DOI] [PubMed] [Google Scholar]
- 80.Labrie V, Lipina T, and Roder JC, Mice with reduced NMDA receptor glycine affinity model some of the negative and cognitive symptoms of schizophrenia. Psychopharmacology (Berl), 2008. 200(2): p. 217–30. [DOI] [PubMed] [Google Scholar]
- 81.Labrie V, Clapcote SJ, and Roder JC, Mutant mice with reduced NMDA-NR1 glycine affinity or lack of D-amino acid oxidase function exhibit altered anxiety-like behaviors. Pharmacol Biochem Behav, 2009. 91(4): p. 610–20. [DOI] [PubMed] [Google Scholar]
- 82.Mielnik CA, et al. , Consequences of NMDA receptor deficiency can be rescued in the adult brain. Mol Psychiatry, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bertocchi I, et al. , Voltage-independent GluN2A-type NMDA receptor Ca(2+) signaling promotes audiogenic seizures, attentional and cognitive deficits in mice. Commun Biol, 2021. 4(1): p. 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pierson TM, et al. , GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol, 2014. 1(3): p. 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen W, et al. , GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J Hum Genet, 2017. 62(6): p. 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mullier B, et al. , GRIN2B gain of function mutations are sensitive to radiprodil, a negative allosteric modulator of GluN2B-containing NMDA receptors. Neuropharmacology, 2017. 123: p. 322–331. [DOI] [PubMed] [Google Scholar]
- 87.Xu Y, et al. , Recurrent seizure-related GRIN1 variant: molecular mechanism and targeted therapy Annals of Clinical and Translational Neurology, 2021. doi: 10.1002/acn3.51406 PMID: 34227748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang W, et al. , Positive allosteric modulators that target NMDA receptors rectify loss-of-function GRIN variants associated with neurological and neuropsychiatric disorders. Neuropharmacology, 2020. 177: p. 108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soto D, et al. , l-Serine dietary supplementation is associated with clinical improvement of loss-of-function GRIN2B-related pediatric encephalopathy. Sci Signal, 2019. 12(586). [DOI] [PubMed] [Google Scholar]
- 90.Elberger AJ and Deng J, Corpus callosum and visual cortex of mice with deletion of the NMDA-NR1 receptor: I. Accelerated development of callosal projection neurons. Brain Res Dev Brain Res, 2003. 144(2): p. 121–33. [DOI] [PubMed] [Google Scholar]
- 91.Gupta SC, et al. , The NMDA receptor GluN2C subunit controls cortical excitatory-inhibitory balance, neuronal oscillations and cognitive function. Sci Rep, 2016. 6: p. 38321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ikeda K, et al. , Reduced spontaneous activity of mice defective in the î4 subunit of the NMDA receptor channel. Mol.Brain Res, 1995. 33: p. 61–71. [DOI] [PubMed] [Google Scholar]
- 93.Das S, et al. , Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature, 1998. 393(6683): p. 377–81. [DOI] [PubMed] [Google Scholar]
- 94.Mohamad O, et al. , Regulatory roles of the NMDA receptor GluN3A subunit in locomotion, pain perception and cognitive functions in adult mice. J Physiol, 2013. 591(1): p. 149–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Otsu Y, et al. , Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science, 2019. 366(6462): p. 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Niemann S, et al. , Genetic ablation of NMDA receptor subunit NR3B in mouse reveals motoneuronal and nonmotoneuronal phenotypes. Eur J Neurosci, 2007. 26(6): p. 1407–20. [DOI] [PubMed] [Google Scholar]