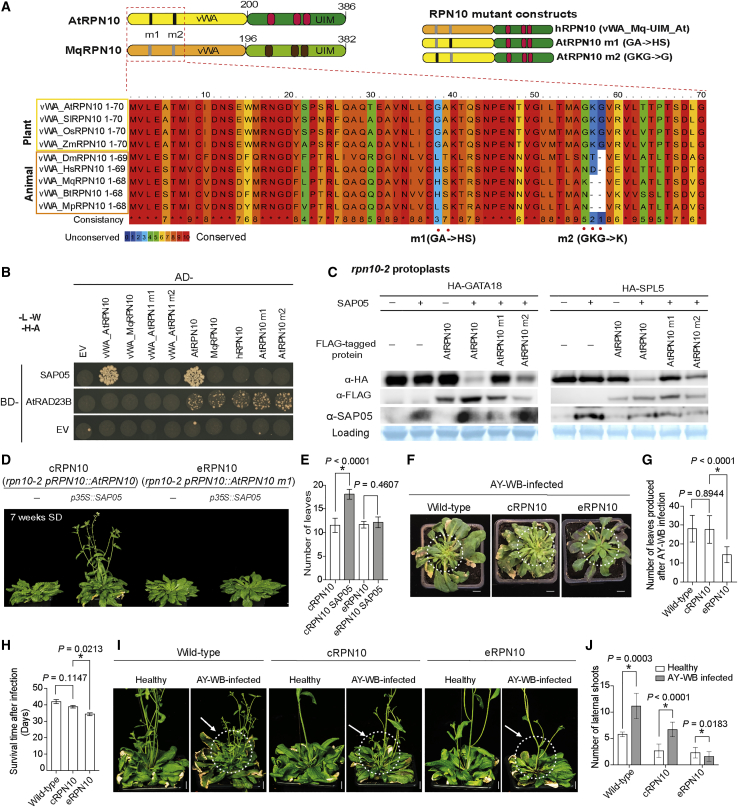
Figure 5.
Insect-directed engineering of At RPN10 confers resistance to SAP05 action
(A) Schematic of domain organizations of At and M. quadrilineatus RPN10 proteins and alignment of the first 70 residues of the vWA domains. Highly divergent residues are highlighted below the alignment. Alignments of full-length RPN10 homologs are shown in Figure S5D.
(B) Specific residues within the At RPN10 vWA domain are required for SAP05 interaction in Y2H assays. See the legends of Figures 2 and 5A for abbreviations. Yeast growth on -L-W medium is shown in Figure S6C.
(C) Specific residues within the At RPN10 vWA domain are required for SAP05 degradation of plant GATA and SPL in At protoplasts. See the legend of Figure 2E.
(D–J) Specific residues within the At RPN10 vWA domain are required for leaf and stem proliferation of At plants in the presence of constitutively expressed SAP05 (D and E) and during AY-WB phytoplasma infection (F–J). At plants included in these experiments were rpn10-2 null mutants complemented with wild-type AtRPN10 (cRPN10) or AtRPN10 m1 (eRPN10). Scale bars, 1 cm. Symptomatic leaves in (F) and lateral shoots in (I) are circled. Phenotypes were analyzed statistically for number of leaves of 4-week-old plants (E), number of newly produced rosette leaves after AY-WB infection (G), plant survival time after AY-WB inoculation (H), and numbers of lateral shoots in control and infected plants (J). Data are mean ± SD from 2 independent experiments. ∗p < 0.05, two-tailed Student’s t test.