ABSTRACT
Broad-range fungal PCR is a powerful tool for identifying pathogens directly from patient specimens; however, reported estimates of clinical utility vary and costs discourage universal testing. We investigated the diagnostic and clinical utility of broad-range fungal PCR by examining 9 years of results from sinonasal specimens, hypothesizing that this anatomic location would identify immunocompromised patients at high risk for invasive fungal disease. We retrospectively identified 644 PCRs and 1,446 fungal cultures from sinus sites. To determine the relative performance of each testing modality, we performed chart review on 52 patients having specimens submitted for culture and PCR on the same day. Positivity rates were significantly higher for PCR (37.1%) than culture (13.7%) but similar for formalin-fixed and fresh tissues (42.3% versus 34.6%). Relative to culture, PCR had significantly faster turnaround time to both preliminary (94.5 versus 108.8 h) and final positive (137.9 versus 278.5 h) results. Among chart-reviewed patients, 88% were immunocompromised, 65% had proven or probable fungal disease, and testing sensitivities for culture and PCR (67.5% and 85.0%) were not statistically different. Nevertheless, PCR identified pathogens not recovered by culture in 14.9% of cases and informed clinical decision-making in 16.7% of all reviewed cases, and sensitivity of PCR combined with culture (90.0%) was higher than that of culture alone. We conclude that broad-range fungal PCR is frequently informative for patients at risk of serious fungal disease and is complementary to and has faster turnaround time than culture. Formalin-fixed tissue does not adversely affect diagnostic yield, but anatomic site may impact assay positivity rates.
KEYWORDS: fungal PCR, molecular microbiology, sinusitis, invasive fungal infection, invasive fungal disease, clinical yield, frozen section, fungal stain
INTRODUCTION
Prompt, species-level identification of fungal pathogens helps providers tailor antifungal medication, with the potential for a significant patient survival benefit (1–3). Broad-range molecular assays for the detection and identification of fungal pathogens now constitute an important diagnostic modality that can be applied to both cultured isolates and direct patient specimens, including formalin-fixed paraffin-embedded (FFPE) tissue (4–6). Broad-range PCR-based detection of fungal pathogens may be particularly important when initial microscopy or cultures are negative (7, 8) or when organisms are unexpectedly identified in histopathology, since morphologic identification of fungi in direct specimens is highly error prone (9, 10). Nevertheless, the reported diagnostic yields of molecular assays have varied widely across studies (11), with estimated clinical sensitivity ranging from 57.1% for microscopy-negative specimens to 96.6% for patients with suspected culture-negative invasive fungal infections (IFI) and attendant specificity estimates ranging from 91.0% to 98.2% (6, 8, 12, 13). The clinical utility of broad-range fungal PCR is similarly debated, with molecular results reported to have changed management in from 0% to 58.2% of cases reviewed (6, 13).
Preanalytical variables likely account for some of these inconsistencies. Factors associated with increased diagnostic yield include larger specimen volume (6), microscopically observed inflammation and/or organisms (6, 7, 12), anatomic site selected (7), and specimen collection method, with better yields obtained from tissue than fine-needle aspirates (6) or body fluids (7). These uncontrolled, specimen-specific variables leave in question the optimal application of broad-range fungal PCR despite its high concordance with orthogonal fungal assays (6, 12, 13).
Acknowledging that specimen-specific factors may influence testing outcomes, in this study, we characterized the comparative clinical performance of molecular testing and conventional, culture-based methods by leveraging paired patient specimens that had been concordantly collected and submitted for laboratory diagnosis by each modality in parallel. Although we are unable to assess the contributions of these factors to test results, this study design enables their effects to be largely controlled for. We hypothesized that targeted application of testing in a patient population at high risk for invasive fungal sinusitis (IFS), a disease predominantly seen in immunocompromised patients with mortality on the order of 50% (1, 14, 15), would best demonstrate diagnostic yield and clinical value of testing and, therefore, specifically examined sinonasal samples. Testing results were integrated with retrospective chart review in order to evaluate diagnostic yield, clinical impact, turnaround time (TAT), likelihood of invasive fungal disease by European Organisation for Research and Treatment of Cancer (EORTC) criteria, and impact of formalin fixation on organism detection.
MATERIALS AND METHODS
Ethical approval.
This study was approved by the Institutional Review Board of the University of Washington (STUDY00002130).
Case identification.
All fungal cultures and broad-range PCRs performed from 1 January 2011 through December 2019 were identified in the Laboratory Information System (LIS). Analysis was performed using R version 4.0.4. Sinonasal specimens were identified by a natural language search of the “specimen description” field using asterisks (*) as wild cards (i) by using the keywords sinus, maxilla/maxillary, maxilla, frontal, septum, ethmoid, sphenoid, nasal, and turbinate, (ii) by excluding specimens with keywords indicating a nonsinus site of origin (brain, cortex, cerebellum, tumor, sulcus, petrosal, dura, lobe, trac*, gyrus, heart, atri*, catheter, pubic, CSF [cerebrospinal fluid], cavern, sputum, heel, and pyriform), and (iii) by excluding specimens collected by neurosurgeons at our institution since early versions of the script included intracranial specimens despite filtering by specimen description. Autopsy testing was excluded. Specimens were further segmented with the R script into either reference laboratory—i.e., specimens submitted by outside facilities—or local specimens based on associated ordering location metadata.
Microbiology assays.
All tests were performed using standard, validated protocols in the clinical laboratory certified under the Clinical Laboratory Improvement Amendments (CLIA) regulations, including culture, direct stain, and molecular assays. Specimens received for fungal culture were examined by direct fluorescent KOH-calcofluor stain upon receipt and inoculated onto inhibitory mold agar containing chloramphenicol (Hardy Diagnostics), brain heart infusion agar with blood (Hardy Diagnostics), and Sabouraud’s agar (Hardy Diagnostics). Usage of Sabouraud’s agar was discontinued as of August 2019 after internal studies demonstrated that its use did not change the rate of mold recovery (data not shown). Cultures were examined daily for the first 4 days postinoculation and twice weekly thereafter. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed for yeasts but not for molds, for which sequence-based identification could be performed in-house.
Broad-range endpoint PCR with sequence-based identification was performed essentially as previously described (4, 5) using primers targeting 28S D1/D2, ITS1, and ITS2 loci. All analytical steps were batched, per laboratory protocol, when possible. Briefly, DNA was extracted from fresh tissue with standard QIAcube DNA extraction protocols (Qiagen) or from 50 to 100 μm of FFPE tissue with deparaffinization in xylene followed by alcohol rehydration prior to QIAcube extraction. Pre-PCR steps were performed in a dedicated clean room with decontamination of FFPE blocks prior to sectioning and extraction. Fixed volumes of extracted DNA (5 μl) were added to PCRs for all loci in technical duplicate with a parallel inhibition control reaction. If inhibition was observed, amplification was repeated using 1:5 and 1:10 template dilutions. Resulting ribosomal DNA (rDNA) amplicons were prepared for Sanger sequencing with the BigDye Xterminator reaction kit (Thermo Fisher Scientific). Taxonomic identification was performed following CLIA laboratory protocols by comparing DNA sequences to a curated, in-house database developed using publicly available sequences in GenBank, with emphasis on sequences from type strains. The most specific taxonomic identification possible was recorded. If different species were detected by amplification and sequencing of different loci, each analytically valid result was reported. Preliminary result entry was performed following two independent reviews by American Society for Clinical Pathology-certified medical laboratory scientists. Final result entry occurred after review by a pathologist.
Determination of TAT.
Turnaround time (TAT) was calculated from sample receipt to final result. FFPE tissue blocks were previously fixed, grossed, and processed in anatomic pathology laboratories and thus are not reflected in these TATs. Preliminary results were reported for both assays, typically to the genus level for culture and species level for PCR. Preliminary TAT was calculated in a convenience subset of sinus specimens for which every LIS update for each specimen was available by manually reviewing the daily result updates in the LIS line by line for the first presumptive organism identification by broad-range fungal PCRs (n = 237, including 58 FFPE) or positive fungal cultures (n = 109). Negative fungal cultures were excluded from this analysis, as these are all held for 28 days.
Chart review.
Medical records were reviewed for 54 patient encounters representing all 52 unique patients for whom specimens were submitted for both fungal PCR and culture and were collected on the same day. Multiple encounters for the same patient were included if occurring at least 3 weeks apart. Chart review evaluated pretest diagnosis, probability of IFI by EORTC criteria (16), pretest exposure to antifungal agents, fungal test results, clinical interpretation of test results, changes in management, and case outcomes.
In accordance with EORTC criteria for classification of IFI current as of 2020 (16), patients were characterized as having “proven” IFI if fungal organisms were known from a prior, recent sampling, were detected by direct stain in the microbiology laboratory (KOH-calcofluor) at the time of specimen collection, or were detected during intraoperative frozen section on the day of specimen collection. Otherwise, encounters were classified as “possible” or “probable” IFI based on established guidelines. For patients who did not meet criteria for determination of possible or probable IFI, such as nonimmunosuppressed hosts or those with well-controlled HIV, pretest probability of IFI was classified as “not applicable” (NA) unless IFI was proven (16). Clinical diagnosis of the case as true fungal infection or not a fungal infection (including alternative diagnoses such as presence of bacterial infection) was determined from the clinical notes, including relevant pathology reports, and used to establish concordance between test method and clinical interpretation.
Statistical analysis.
Statistical tests were performed in GraphPad Prism version 7.03 and R version 4.0.4 using stats package version 4.0.4. Nonparametric statistical tests were employed: Welch two-sample t test for unequal variances, Fisher’s exact test, or Mann-Whitney test for nonparametric ordinal data. Kruskal-Wallis test was used to detect differences in medians across groups. Pearson’s exact method for calculating confidence intervals (CIs) was performed in GraphPad.
Data availability.
A Health Insurance Portability and Accountability Act (HIPAA)-compliant version of the R script used for analysis may be obtained by contacting the corresponding author.
RESULTS
Aggregate positivity rates of fungal PCR and culture.
Over the 9-year study period, we identified a total of 644 fungal PCRs from 569 patients and 1,446 cultures from 949 patients originating from sinonasal specimen sites (Fig. 1). Testing performed for patients seen in our hospital system (i.e., local patients) comprised 101 PCRs from 71 separate patient encounters and 1,396 cultures from 918 patient encounters (Fig. 1) resulting in 220 unique isolates, 192 of which could be identified by phenotypic assays (see Fig. S1 in the supplemental material). Sequence-based identification was required for taxonomic identification for 25 of the 220 local isolates (11.4%); sequence-based identification was not performed on 3 isolates which could not be identified by traditional methods. The overall fungal PCR positivity rate for sinonasal specimens was 37.1%, significantly higher than the 13.7% fungal positivity observed for culture (P = 2.2 × 10−16 [Table 1]). The positivity rate for sinonasal FFPE specimens was 42.3% (n = 208) and was statistically equivalent (P = 0.0634) to the 34.6% positivity observed for fresh specimens (n = 436). Fungal PCRs performed on sinonasal specimens from local patients had a positivity rate (43.6%) similar to that of fungal PCRs performed for sinonasal specimens submitted from outside institutions (35.9%; P = 0.1440 [Table 1]). Among all local patient specimen collections, the PCR positivity rate was significantly higher than for culture (Table S1), particularly for local patients (43.6% versus 13.8%; P = 3.73 × 10−12).
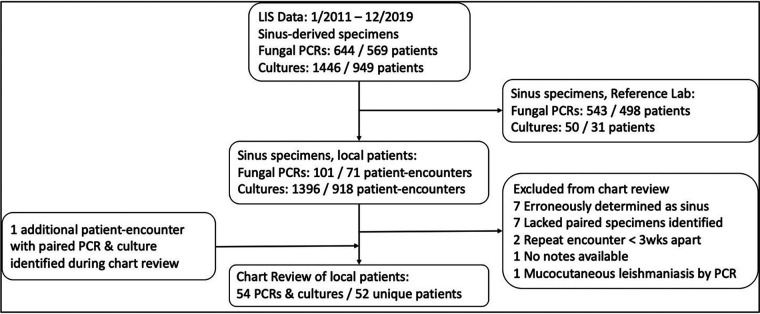
FIG 1.
Consort diagram for included patients and test results. Sinus specimens were identified from the LIS by a custom script to filter based primarily on specimen description. Sinus specimens were stratified into those submitted for reference laboratory testing or those from local patients. Local patient specimens, representing 71 patient encounters, were filtered for inclusion in chart review. During chart review, one patient had an additional (third) set of paired cultures and PCRs collected >3 weeks from either other set of specimens that was included for analysis.
TABLE 1.
Summary of chart review patients
| Pretest EORTC category | No. of: |
No. (%) positive by: |
Culture vs PCR P valueb | |||
|---|---|---|---|---|---|---|
| Unique patientsa | Patients with pretest antifungal exposure | Paired specimens | Culture | PCR | ||
| All | 52 | 43 | 54 | 29 (53.7) | 36 (66.7) | 0.238 |
| Proven IFI | 28 | 23 | 28 | 22 (78.6) | 25 (89.3) | 0.844 |
| Probable IFI | 9 | 7 | 10 | 5 (50.0) | 7 (70.0) | 0.726 |
| Possible IFI | 11 | 11 | 11 | 0 (0.0) | 1 (9.1) | NDd |
| Not applicablec | 5 | 1 | 5 | 2 (40.0) | 3 (60.0) | ND |
One patient had parallel PCR and culture performed three times with >3 weeks between each sampling, and each encounter was included as a single unique patient for overall counts, but testing was counted twice as “probable” and once as “proven” IFI based on chart review.
Fisher’s exact test.
One patient to whom EORTC criteria were not applicable had HIV and a CD4 count of 14 cells/μl, and pretest amphotericin B had a true positive detection of A. fumigatus by PCR only; all other results for patients in this category were clinically thought to reflect bystanders/colonizers.
ND, not determined.
Turnaround times of fungal PCR and culture.
To best compare turnaround times of culture and PCR, we focused on the subset of “paired” specimens from local patients, defined as those submitted on the same day for parallel testing by both of those assays. Fungal PCR yielded final results faster than culture (Fig. 2), with a median positive PCR TAT of 137.9 h (interquartile range [IQR], 113.3 to 158.6 h) and a positive culture TAT of 278.5 h (IQR, 209.5 to 494.0 h; P < 0.0001). Similarly, for negative PCRs and cultures, the median TATs were 100.5 h (IQR, 83.2 to 130.0 h) and 698.6 h (IQR 689.1 to 715.1 h), respectively (P < 0.0001). Fifteen cultures with outlier TATs of >300 h were identified, six of which required sequence-based identification (see the Supplemental Results).
FIG 2.
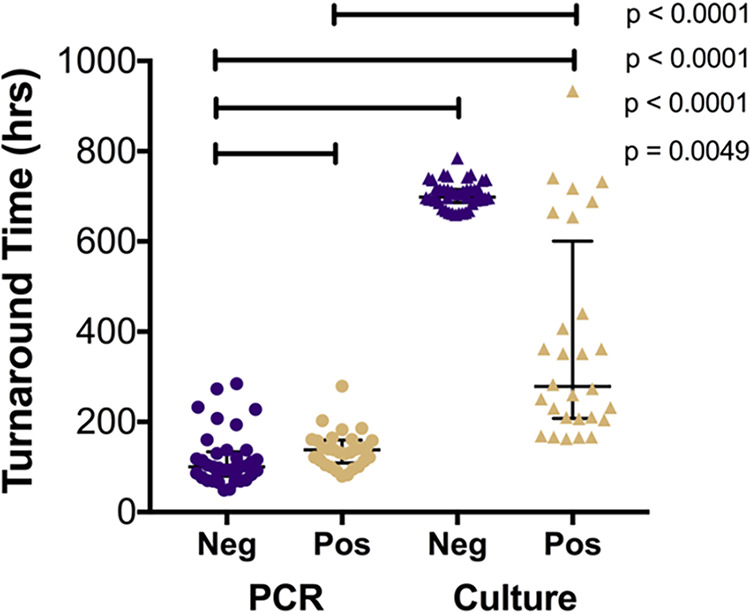
Turnaround times to final results for local patients. Turnaround times were determined for local patients receiving PCR (n = 66) and culture (n = 71) and were stratified by positive and negative results. Error bars represent median and IQR. The positive PCR median TAT, 137.9 h (n = 29), was faster than the median for final positive culture, 278.5 h (n = 28; P < 0.0001). Negative PCR median TAT, 100.5 h, was faster than the positive PCR median TAT (P = 0.0049), negative culture median TAT (698.6 h [P < 0.0001]), and positive culture median TAT (P < 0.0001). Tests for significance included Mann-Whitney rank sum test and Kruskal-Wallis test to detect different medians across groups (P < 0.0001).
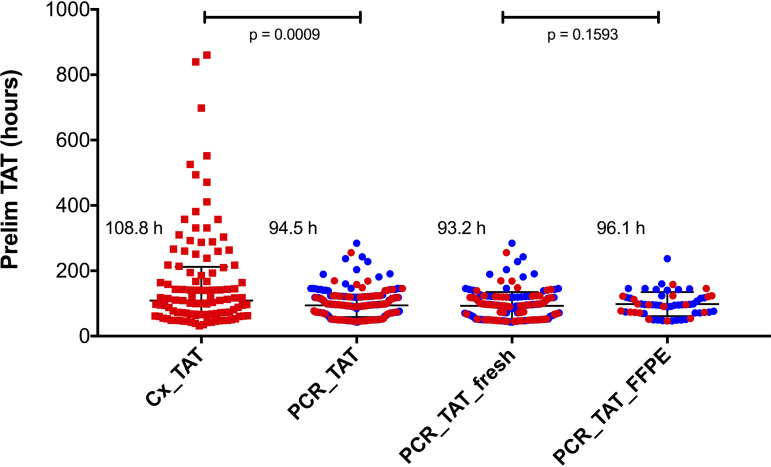
Recognizing that both assays can generate clinically informative preliminary results, we manually reviewed the LIS result history for a subset of broad-range PCRs (n = 237) and positive cultures (n = 109) to identify TAT to the first clinically actionable, preliminary result (Fig. 3). Preliminary results from culture were reported, at least to the genus level, for 99 of 109 positive cultures, with the majority reported as “mold identified” or “yeast identified” with further identification pending additional incubation and biochemical testing (yeasts) or sequence-based identification (molds). In contrast, positive fungal PCRs identified organism taxonomy, usually to the species or species complex level, in all 99 positive tests. The median TAT for preliminary result by PCR was 94.5 h (IQR, 60.0 to 117.9 h). Positive and negative PCRs had similar preliminary TATs, 95.0 h (IQR, 62.5 to 117.9 h) and 93.1 h (IQR, 52.4 to 116.1 h), respectively. Median preliminary TATs for PCR performed on FFPE (96.1 h; IQR, 72.5 to 120.3 h) and fresh tissue (93.2 h; IQR 52.4 to 115.6 h) were not statistically different from each other (P = 0.1593 [Fig. 3]). The median preliminary TAT for positive cultures was 108.8 h (IQR, 66.5 to 209.8 h) and was slower than PCR (P = 0.0009 [Fig. 3]).
FIG 3.
Turnaround time to first actionable preliminary result. Shown are the number of hours (TAT) from specimen receipt to first reporting of a positive cultures (n = 109) and preliminary result for PCR (n = 237). Red dots represent positive results; blue dots represent negative results. Median preliminary TATs were 108.8 h for culture and 94.5 h for PCR (P = 0.0009, Mann-Whitney Test). Median preliminary TATs for positive and negative PCR preliminary results were similar at 95.0 h (IQR, 62.5 to 117.9 h) and 93.1 (52.4 to 116.1 h). The median preliminary TAT for PCR performed on fresh tissue (n = 179) was not significantly different from that of PCR on FFPE (n = 96.1; P = 0.1593). Error bars show median and interquartile range.
Diagnostic yield of fungal testing.
To evaluate the success of fungal PCR and conventional fungal culture in diagnosing clinically assessed disease, we next conducted chart review of the 54 local patient encounters (52 patients; all specimen collections/encounters >3 weeks apart) which resulted in paired patient specimens being submitted for fungal PCR and culture-based testing. Forty-six patients (88.4%) were markedly immunocompromised: 41 had hematopoietic neoplasms, 1 presented with AIDS (CD4 count 14 cells/μl), and 2 were solid-organ transplant patients (Table S2). Three additional patients had diabetes mellitus type 2 without other immunocompromise, one had a solid neoplasm, one had well-controlled HIV, and one presented with a nasal lesion due to oxymetazoline hydrochloride (Afrin) use (Table S2). For chart review, patients were stratified by pretest probability of fungal detection by EORTC criteria for fungal infections (Table 1; full results in Table S1): 21 were classified as having proven IFI, 26 were classified as having possible or probable IFI, and there were 5 for whom EORTC criteria did not apply (Table 1 and Table S2).
The first result for all specimens was either a direct fungal stain, performed by protocol as part of the fungal culture assay in all cases (n = 54), or an intraoperative pathology consult (“frozen section”) performed in a subset of cases when requested by surgeons (n = 36). Direct stain was positive in 18 cases (33.3%), while intraoperative pathology consult (frozen section) was positive in 19 (52.7%) (Table S2). Either direct stain or frozen section was positive in 26 cases (48.1%), and 1 false-positive case, a frozen section, was identified (Table S2). Stain results were reported as fungal elements, fungal hyphae, or yeast given the known risks of major diagnostic errors identifying fungi in situ by morphology (9, 10). For one frozen section, further classification was attempted and erroneously described Fusarium as suggestive of a mucormycete based on morphology (Table S2, case 40). Positive percent agreement (PPA) for both staining methods with PCR and culture was high, usually >90% (Fig. 4), but between the two staining methods it was 57.9% (n = 36 [Fig. 4]). Notably, negative percent agreement (NPA) for either staining method with culture or PCR was low (<70%).
FIG 4.
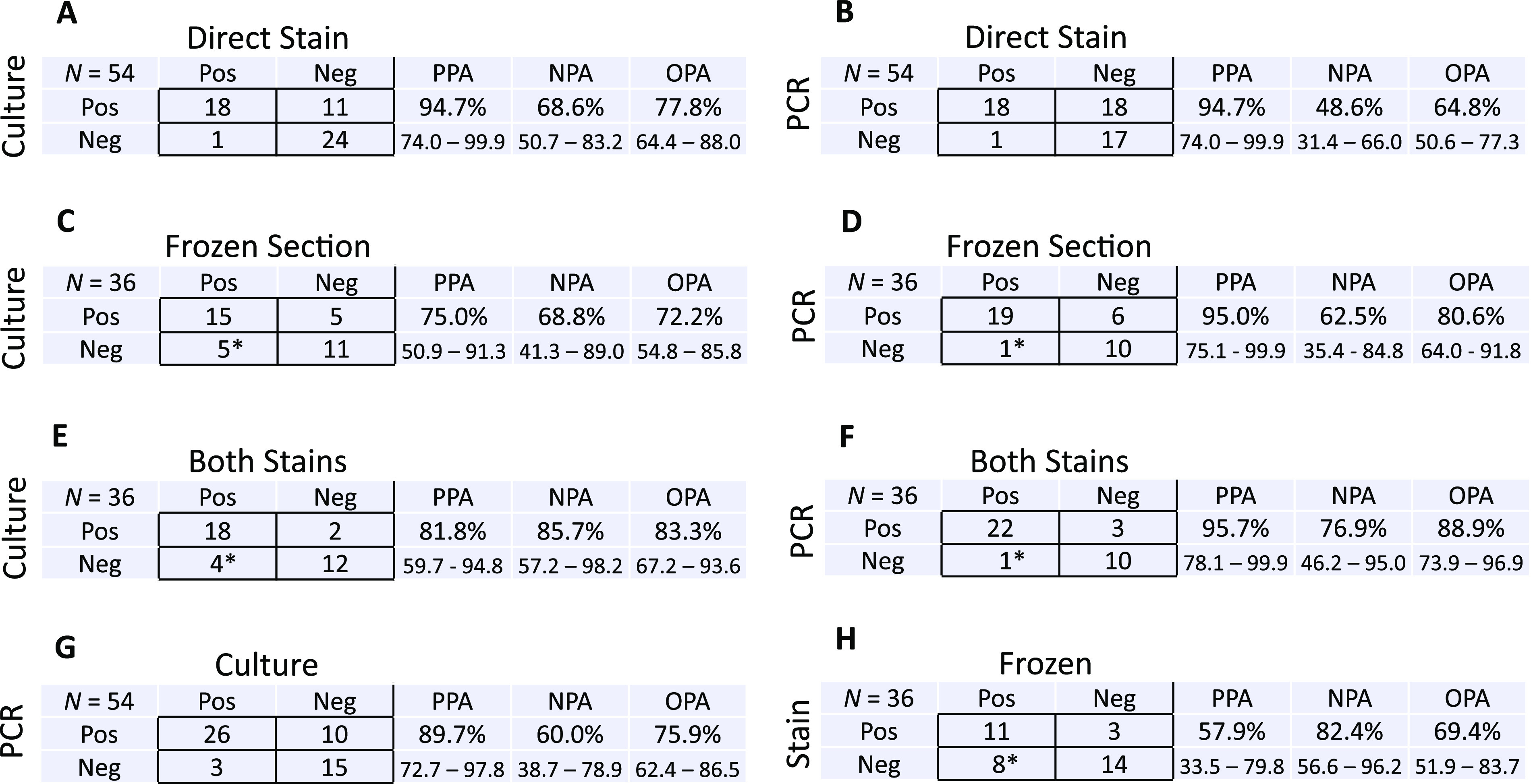
Test concordance across stain, frozen section, culture, and PCR. Contingency tables compare categorical results (pos/neg) for culture and direct stain (A), broad-range fungal PCR and direct stain (B), fungal culture with frozen section (C), broad-range fungal PCR and frozen section (D), fungal culture and both direct stain and frozen section performed in tandem (E), broad-range fungal PCR and both direct stain and frozen section performed in tandem (F), broad-range fungal PCR and fungal culture (G), and direct stain and frozen section (H). *, one false-positive frozen section result was reported.
PCR detected an organism in 36 cases (66.7%), and culture was positive in 29 cases (53.7%) (Table 1). Although positivity rates for PCR and culture were not different in this cohort (P = 0.2381), both rates were significantly higher than aggregate positivity rates for culture (13.7%; P < 0.0001) or PCR (37.1%; P < 0.0001) alone. PCR detected an organism in 11 cases for which both direct stain and frozen section were negative (20.4%) and did not detect an organism in 1 case for which yeast was seen on direct stain, although the organism was not thought to contribute to the patient’s pathology (Table S2, case 39). Culture detected an organism in 7 cases where both staining methods were negative (13.0%) and could not recover an organism in 4 stain-positive cases: 3 positive by frozen section and 1 positive by direct stain, all four of which were also positive by PCR (Fig. 4 and Table S2). Three polyfungal cases were detected by culture, one of which was also determined to be polyfungal by broad-range PCR (DNA of distinct organisms detected in different amplified loci).
To estimate each assay’s sensitivity and specificity for detection of clinical disease, we extracted from chart review the final posttest clinical assessment of whether the patient’s pathology was driven by a fungal infection (true clinical case), nonfungal infection, or noninfectious process (Table S2). The consensus diagnosis documented by the clinical team(s) integrated assessment of all laboratory, histopathology, and radiographic findings with the patient’s disease course. By this metric, broad-range PCR had a sensitivity of 85.0% (95% CI, 70.1 to 94.3%) and culture had a sensitivity of 67.5% (95% CI, 50.9 to 81.4%; P = 0.1136), while the specificity of either assay alone was 85.7% (95% CI, 57.2 to 98.2% [Fig. 5 and Fig. S2]). The sensitivity of culture performed jointly in combination with PCR was 90.0% (95% CI, 76.3 to 97.2%) if a positive result from either test was accepted. The specificity of such biphasic testing was 78.5% (95% CI, 49.2 to 95.3%). Notably, the sensitivity of culture combined with PCR was significantly higher than that of culture performed alone (P = 0.0269) but not different from that of PCR alone (P = 0.7370). The specificity of biphasic testing did not differ significantly from that of using either assay alone (P = 1.0 [Fig. 5 and Fig. S2]). Overall percent agreement between PCR and culture was 75.9%, with PPA of 89.7% and NPA of 60.0% (Fig. 5 and Fig. S2). In comparison to that of PCR, culture, or biphasic testing with PCR and culture performed jointly, the sensitivity of direct stain was much lower, 45.2% (95% CI, 29.9 to 61.3% [Fig. 5 and Fig. S2]). The sensitivity of frozen section, 67.9% (95% CI, 47.7 to 84.1%), was also less than that of the combination of PCR and culture (Fig. 5 and Fig. S2). Although the specificities were 100% (95% CI, 73.5 to 100%) for stain and 87.5% (95% CI, 47.4 to 99.7%) for frozen section (Fig. 5), both methods lacked precision in organism identification. The sensitivity and specificity for performing both stains in tandem and accepting a positive result from either test were 78.6% (95% CI, 59.1 to 91.7%) and 87.5% (95% CI, 47.4 to 99.7%), respectively.
FIG 5.
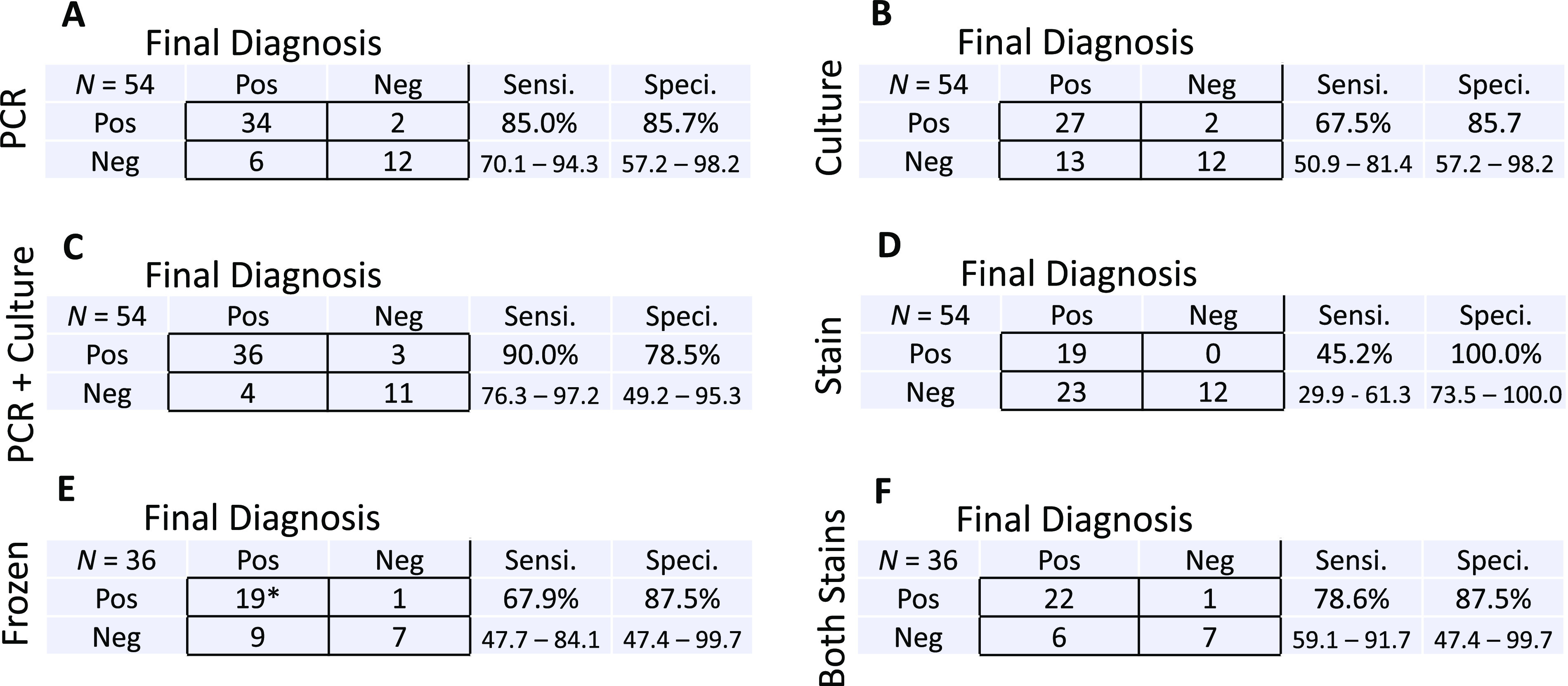
Estimated sensitivity and specificity of PCR, culture, and stains. Contingency tables compare categorical results (pos/neg) to final clinical diagnosis for broad-range fungal PCR (A), fungal culture (B), tandem submission of both broad-range fungal PCR and fungal culture (C), direct stain of specimens submitted for culture (D), frozen section results (E), and tandem submission of both frozen section and direct stain (F). Shown is the number of cases, N, with assay performed in the upper left of each table. Estimated sensitivity (Sensi.) and specificity (Speci.) are reported in each table, with corresponding 95% confidence intervals (percent) reported below. *, one frozen section erroneously suggested mucormycete morphology in a case of Fusarium infection.
Clinical impact of fungal PCR results.
Based on chart review, PCR was the primary driver of a change in the clinical management of nine cases. Broad-range PCR was the sole assay to identify a pathogen in seven cases, and in two additional cases it identified the pathogen faster than culture (Table S3). Conversely, culture detected two true positives missed by PCR. Out of 10 cases with discordant PCR and culture results, nine patients were immunocompromised and one had end-stage renal disease, ethanol abuse, and anemia on admission (patient 51 [Table S2]). Of the 13 negative cultures which were adjudicated as false negatives by the clinical assessment, 9 were for patients who had prior antifungal exposure. Of the six false-negative broad-range fungal PCRs, four were for patients who had prior antifungal exposure and two originated from the same patient with persistent sinus aspergillosis (patient 6 [Table S2]). In three cases, culture, PCR, or both assays detected organisms in nonimmunocompromised patients (patients 13, 39, and 50) that were deemed by the clinical team to represent bystanders/colonizers or superinfection and which led to de-escalation or nonescalation of therapy. Cumulative mortality in the chart reviewed cohort was 51.9%, including a 30-day (from testing) mortality rate of 15.4% and cumulative 1-year mortality rate of 36.5% (Fig. S3). Fungal infection was a significant factor in the deaths of 8 of 11 patients who died within 60 days from testing (21.2% [Table S3]).
DISCUSSION
Laboratory stewardship for molecular microbiology testing has emerged as an important consideration in clinical practice, with the goal of maximizing diagnostic yield while reducing both unnecessary testing and costs of patient care. Past studies have reported heterogeneous estimates of the clinical yield and positivity rates of broad-range fungal PCR. Comparing prior literature can be challenging, as some studies have included specimens from diverse anatomic sites and patient populations (7, 12) and consequently may be confounded by variations in pretest probability of infection and other preanalytical factors. Similarly, analytical differences may be due to variation in target loci used by performing laboratories, such as 18S (13) or 28S (6) ribosomal genes. This study sought to elucidate optimal application of broad-range fungal PCR in a single anatomic site at risk for fungal infection in immunocompromised patients. These findings were corroborated by chart review of a subset of patients from a single institution, thereby minimizing variation in both analytical and preanalytical parameters, like specimen handling and formalin fixation.
Our results indicated that broad-range fungal PCR in sinus sites had a relatively high positivity rate, ranging from 35.9% for sinus specimens submitted for reference testing to 43.6% for local cases, which was significantly higher than culture positivity (13.8% in local cases). We also observed that broad-range fungal PCR typically yielded both final and preliminary results faster than culture and that preliminary results were more specific from PCR than from culture. In comparison, negative fungal PCR results were available several weeks before negative cultures were finalized, adding value in ruling out fungal infection. Negative fungal PCR results were reported substantially faster than positive results, indicating the additional time required to sequence and analyze positive cases. Thus, an important diagnostic consideration for using PCR is added value in ruling out fungal infection. While direct stain and frozen section are important rapid diagnostic tools with TATs of <1 h (17), we found that both staining methods had limited sensitivity (Fig. 5 and Fig. S2) and could not provide meaningful taxonomic identifications (9, 10). PCR and culture consequently provide important clinical information that is not otherwise obtainable.
In addition, the PCR positivity rate for FFPE sinus tissue (42.3%) was statistically equivalent to that of fresh sinus tissue (34.6%). Several studies have demonstrated equivalent diagnostic yield of broad-range PCR performed in both FFPE and fresh tissue (6, 7, 12), which is surprising given that formalin induces DNA damage (18) and can decrease diagnostic yield (19, 20). Our observation of equivalent positivity between fresh and fixed specimens submitted for fungal PCR is consistent with these prior reports, suggesting that a pathologist’s ability to select optimal tissue for PCR-based pathogen detection can compensate for formalin damage to DNA.
In order to evaluate for clinical yield and real-world assay performance, and to control for underlying differences in the patient populations assessed by culture or PCR, we reviewed the charts of all patients from our hospital system with sinus specimens tested by both methods in parallel. To minimize selection bias created by PCR orders placed on FFPE after histopathological evaluation, we focused on assays initiated on the same day. Almost all (88.5%) of those patients were immunocompromised and significant overall mortality was observed, consistent with the hypothesis that sinus tissue submitted for fungal PCR represents a patient population with a higher-than-average pretest probability of invasive fungal infection.
In the focused chart review, fungal PCR was the only test to detect a pathogen in 14.9% of cases (8 of 54) and drove a change in treatment in 16.7% of cases. Similarly, direct stain was negative in 38 and 50% of cases found positive by culture or PCR, respectively, and frozen section was negative in ∼25% of positive cultures or PCRs. In aggregate, median TATs to final and preliminary results were faster for PCR than culture; however, chart review demonstrated that culture was also a major contributor to management decisions. The discrepancy between aggregate and chart review TAT data may be attributable to multiple factors. It could reflect a high frequency of infection with faster-growing molds like Aspergillus fumigatus, Fusarium spp., and mucormycetes, which accounted for 11.8%, 6.9%, and 5.5% of cultured isolates identified by chart review, respectively. Alternatively, the increasing role of isolate identification by sequence analysis may have prolonged final culture result TAT, while adding specificity and without delaying preliminary findings. Lastly, an increased number of slower growing fungi from lower-risk patients may not have been captured by the focused chart review (Supplemental Results). Discordant results for PCR, culture, direct stain, and frozen section were likely due to preanalytical factors, since the detected pathogens in such cases were common organisms expected to be detected by each method.
In our cohort, two PCRs and two cultures derived from three patient cases recovered fungal organisms deemed to be nonpathogens by the clinical infectious diseases team. We conclude that culture and PCR are similar in detecting organisms that are not primary drivers of pathology (e.g., colonizers/bystanders). While detection of nonpathogens in both PCR and culture is rare, their presence can be readily interpreted by specialist clinicians, particularly in consultation with laboratorians.
Collectively, these data demonstrate that broad-range fungal PCR is an important assay in the evaluation of patients at risk for fungal disease, that it is complementary to culture-based methods, and that the addition of PCR to culture provides superior sensitivity to culture alone. PCR assays have limitations and are unlikely to ever fully replace culture. For example, loci routinely sequenced from direct patient specimens cannot resolve closely related species within taxonomic complexes or subgenera without obtaining sequences from additional loci, as is the case with Fusarium spp. and Aspergillus spp. (Fig. S2), and cultured isolates are required to phenotypically determine antifungal susceptibilities. Nevertheless, our results show that broad-range fungal PCR has demonstrated ability to detect pathogens that are not recovered by culture, to reduce TAT, drive clinical decision-making, corroborate culture findings, and have high yield from FFPE specimens. These findings argue that clinicians caring for high-risk patients should submit tissue for PCR simultaneously with culture and/or coordinate with anatomic pathologists to ensure that FFPE with visible fungi is promptly submitted for molecular testing when cultures are negative or not ordered.
ACKNOWLEDGMENTS
J.A.L. and A.B. received funding from a University of Washington Department of Pathology Accountability Care Organization grant.
The funders had no role in study design, data collection, or interpretation.
We thank Noah Hoffman, Nathan Breit, and Tuan Q. Nguyen for clinical informatics support, Ashley Eckel and Benjamin Bradley for help with collection of anatomic pathology cases, Ian Humphreys for helpful discussions of ordering practices by head and neck surgeons, and the clinical staff of the UW microbiology and molecular microbiology and pathology laboratories for performing clinical testing for the included patients.
J.A.L. conceptualized the project, wrote the manuscript, curated and analyzed data, including chart review, assisted with the R script, acquired funding, and administered and supervised the project. A.B. conceptualized the project, curated and analyzed the data, including chart review, and acquired funding. J.A.M. prepared the R script, curated and analyzed the data, and helped craft the manuscript. P.C.M. contributed to the R script and data curation. K.K. and D.S. contributed to the methodology and analysis. K.S., L.B., S.J.S., and B.T.C. contributed to the methodology, analysis, and writing of the manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Joshua A. Lieberman, Email: joshuaal@uw.edu.
Kimberly E. Hanson, University of Utah
REFERENCES
- 1.Foshee J, Luminais C, Casey J, Farag A, Prestipino A, Iloreta AM, Nyquist G, Rosen M. 2016. An evaluation of invasive fungal sinusitis outcomes with subsite analysis and use of frozen section analysis. Int Forum Allergy Rhinol 6:807–811. 10.1002/alr.21714. [DOI] [PubMed] [Google Scholar]
- 2.Herbrecht R, Denning DW, Patterson TF, Bennett JE, Greene RE, Oestmann J-W, Kern WV, Marr KA, Ribaud P, Lortholary O, Sylvester R, Rubin RH, Wingard JR, Stark P, Durand C, Caillot D, Thiel E, Chandrasekar PH, Hodges MR, Schlamm HT, Troke PF, de Pauw B, Invasive Fungal Infections Group of the European Organisation for Research and Treatment of Cancer and the Global Aspergillus Study Group. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N Engl J Med 347:408–415. 10.1056/NEJMoa020191. [DOI] [PubMed] [Google Scholar]
- 3.Chamilos G, Lewis RE, Kontoyiannis DP. 2008. Delaying amphotericin B-based frontline therapy significantly increases mortality among patients with hematologic malignancy who have zygomycosis. Clin Infect Dis 47:503–509. 10.1086/590004. [DOI] [PubMed] [Google Scholar]
- 4.Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, Bui U, Limaye AP, Cookson BT. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J Clin Microbiol 39:4042–4051. 10.1128/JCM.39.11.4042-4051.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakeman JL, Bui U, Lafe K, Chen Y-C, Honeycutt RJ, Cookson BT. 2005. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol 43:3324–3333. 10.1128/JCM.43.7.3324-3333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez CA, Budvytiene I, Zemek AJ, Banaei N. 2017. Performance of targeted fungal sequencing for culture-independent diagnosis of invasive fungal disease. Clin Infect Dis 65:2035–2041. 10.1093/cid/cix728. [DOI] [PubMed] [Google Scholar]
- 7.Kerkhoff AD, Rutishauser RL, Miller S, Babik JM. 2020. Clinical utility of universal broad-range PCR amplicon sequencing for pathogen identification: a retrospective cohort study. Clin Infect Dis 71:1554–1557. 10.1093/cid/ciz1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lass-Flörl C, Mutschlechner W, Aigner M, Grif K, Marth C, Girschikofsky M, Grander W, Greil R, Russ G, Cerkl P, Eller M, Kropshofer G, Eschertzhuber S, Kathrein H, Schmid S, Beer R, Lorenz I, Theurl I, Nachbaur D. 2013. Utility of PCR in diagnosis of invasive fungal infections: real-life data from a multicenter study. J Clin Microbiol 51:863–868. 10.1128/JCM.02965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sangoi AR, Rogers WM, Longacre TA, Montoya JG, Baron EJ, Banaei N. 2009. Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am J Clin Pathol 131:364–375. 10.1309/AJCP99OOOZSNISCZ. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Yun NR, Kim K-H, Jeon JH, Kim E-C, Chung DH, Park WB, Oh M-D. 2010. Discrepancy between histology and culture in filamentous fungal infections. Med Mycol 48:886–888. 10.3109/13693780903512835. [DOI] [PubMed] [Google Scholar]
- 11.Terrero-Salcedo D, Powers-Fletcher MV. 2020. Updates in laboratory diagnostics for invasive fungal infections. J Clin Microbiol 58:e01487-19. 10.1128/JCM.01487-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basein T, Gardiner BJ, Andujar Vazquez GM, Joel Chandranesan AS, Rabson AR, Doron S, Snydman DR. 2018. Microbial identification using DNA target amplification and sequencing: clinical utility and impact on patient management. Open Forum Infect Dis 5:ofy257. 10.1093/ofid/ofy257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stempak LM, Vogel SA, Richter SS, Wyllie R, Procop GW. 2019. Routine broad-range fungal polymerase chain reaction with DNA sequencing in patients with suspected mycoses does not add value and is not cost-effective. Arch Pathol Lab Med 143:634–638. 10.5858/arpa.2017-0299-OA. [DOI] [PubMed] [Google Scholar]
- 14.Zuniga MG, Turner JH. 2014. Treatment outcomes in acute invasive fungal rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg 22:242–248. 10.1097/MOO.0000000000000048. [DOI] [PubMed] [Google Scholar]
- 15.Cho H-J, Jang M-S, Hong SD, Chung S-K, Kim HY, Dhong H-J. 2015. Prognostic factors for survival in patients with acute invasive fungal rhinosinusitis. Am J Rhinol Allergy 29:48–53. 10.2500/ajra.2015.29.4115. [DOI] [PubMed] [Google Scholar]
- 16.Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes D, Azoulay E, Bialek R, Bradsher RW, Jr, Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher B, Harrison T, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg B-J, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrisey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, Roilides E, Ruhnke M, Prokop CS, Shoham S, Slavin MA, Stevens DA, Thompson GR, III, Vazquez JA, Viscoli C, Walsh TJ, Warris A, Wheat LJ, White PL, Zaoutis TE, Pappas PG. 2020. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 71:1367–1376. 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comacle P, Belaz S, Jegoux F, Ruaux C, Le Gall F, Gangneux J-P, Robert-Gangneux F. 2016. Contribution of molecular tools for the diagnosis and epidemiology of fungal chronic rhinosinusitis. Med Mycol 54:794–800. 10.1093/mmy/myw041. [DOI] [PubMed] [Google Scholar]
- 18.Khan J, Lieberman JA, Lockwood CM. 2017. Variability in, variability out: best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin Chem Lab Med 55:608–621. 10.1515/cclm-2016-0471. [DOI] [PubMed] [Google Scholar]
- 19.Lu XJD, Liu KYP, Zhu YS, Cui C, Poh CF. 2018. Using ddPCR to assess the DNA yield of FFPE samples. Biomol Detect Quantif 16:5–11. 10.1016/j.bdq.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amemiya K, Hirotsu Y, Oyama T, Omata M. 2019. Relationship between formalin reagent and success rate of targeted sequencing analysis using formalin fixed paraffin embedded tissues. Clin Chim Acta 488:129–134. 10.1016/j.cca.2018.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, supplemental results, Fig. S1 to S3, and Tables S1 to S3. Download JCM.00955-21-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Data Availability Statement
A Health Insurance Portability and Accountability Act (HIPAA)-compliant version of the R script used for analysis may be obtained by contacting the corresponding author.